Abstract
Background:
R132H mutation of isocitrate dehydrogenase 1 (IDH1) are found in ~75% of low-grade gliomas and secondary glioblastomas as well as in several other types of cancer. More chemotypes of inhibitors of IDH1(R132H) are therefore needed.
Objective:
To develop a new class of IDH1(R132H) inhibitors as potent antitumor agents.
Method:
A biochemical assay was developed to find inhibitors of IDH1(R132H) mutant enzyme. Chemical synthesis and structure activity relationship studies were used to find compounds with improved potency. Antitumor activities of selected compounds were evaluated.
Results:
A series of aromatic sulfonamide compounds were found to be novel, potent inhibitors of IDH1(R132H) with Ki values as low as 0.6 μM. Structure activity relationships of these compounds are discussed. Enzyme kinetics studies showed that one compound is a competitive inhibitor against the substrate α-KG and a non-competitive inhibitor against the cofactor NADPH. Several inhibitors were found to have no activity against wild-type IDH1, showing a high selectivity. Two potent inhibitors exhibited strong activity against proliferation of BT142 glioma cells with IDH1 R132H mutation, while these compounds did not significantly affect growth of glioma cells without IDH1 mutation.
Conclusion:
This novel series of IDH1(R132H) inhibitors have potential to be further developed for the treatment of glioma with IDH1 mutation.
Keywords: Isocitrate dehydrogenase, Oncogenic mutation, Enzyme inhibitor, Medicinal chemistry
INTRODUCTION
Point mutations of isocitrate dehydrogenase (IDH) 1 and 2, located in cytoplasm and mitochondria, respectively, have been frequently found in cancer including glioma, leukemia and sarcomas. [1–6] In particular, mutations of IDH1 have been identified in ~75% low-grade gliomas (grade II and III), [2,3] which grow slowly but eventually develop to become secondary glioblastoma multiforme (GBM), which is highly invasive grade IV glioma with a very low 5-year survival rate of <10%. R132H (Arg132His) is the predominant (~90%) form of mutation in gliomas. There is therefore a pressing need to find effective treatments for GBM.
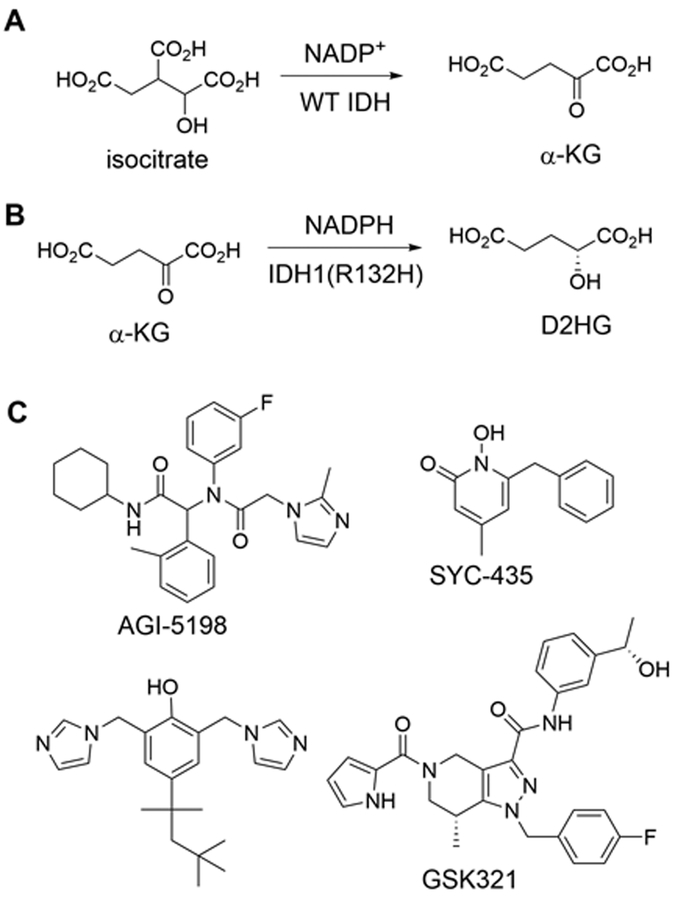
IDH is one of the key enzymes in tricarboxylic acid cycle for aerobic metabolism of carbohydrates and fats. Biochemically, IDH catalyzes oxidative decarboxylation of isocitric acid to produce α-ketoglutaric acid (α-KG) (Figure 1A). Mutant IDH enzymes including IDH1(R132H) almost lose the function of the wild-type (WT) enzyme. However, these mutant proteins can reduce α-KG to D-2-hydroxyglutaric acid (D2HG), using NADPH as the cofactor (Figure 1B). [4,7] This new function leads to a high cellular level of D2HG, which is an inhibitor of α-KG dependent histone demethylases as well as DNA methyl hydroxylases, [8] causing a genome-wide histone/DNA hypermethylation phenotype. Recent studies showed introducing R132H IDH1 mutation recapitulated the phenotype and blocked cell differentiation of the recipient cells. [9,10] These lines of evidence have indicated that IDH mutation is a key step towards oncogenesis and a potential drug target for intervention.
Figure 1.
Reactions catalyzed by (A) WT IDH1 and (B) IDH1(R132H); (C) Representative inhibitors of mutant IDH1.
There has been a significant amount of interest in discovering inhibitors of mutant IDH during the past few years. [11–16] Figure 1C shows structurally distinct inhibitors of mutant IDH. Several inhibitors have been in clinical trials against IDH mutated cancer. [17] Given the limited number of available inhibitors as well as the high potential to become potential therapeutics for these cancers, more chemotypes of inhibitors of mutant IDH are needed. Here, we report the discovery and structure activity relationships (SAR) of aromatic sulfonamides as a new class of inhibitors of IDH1(R132H). Antitumor activities of selected compounds are also reported against primary tumor cells from GBM patients bearing IDH1 R132H mutation.
MATERIALS AND METHODS
All reagents were purchased from Alfa Aesar (Ward Hill, MA) or Aldrich (Milwaukee, WI). All compounds were characterized by 1H spectrum on a Varian (Palo Alto, CA) 400-MR spectrometer. The purities were determined by a Shimadzu Prominence HPLC with a Zorbax C18 or C8 column (4.6 × 250 mm) or 1H (at 400 MHz) absolute spin-count quantitative NMR analysis with imidazole as an internal standard. The purities of all synthesized compounds were found to be >95%.
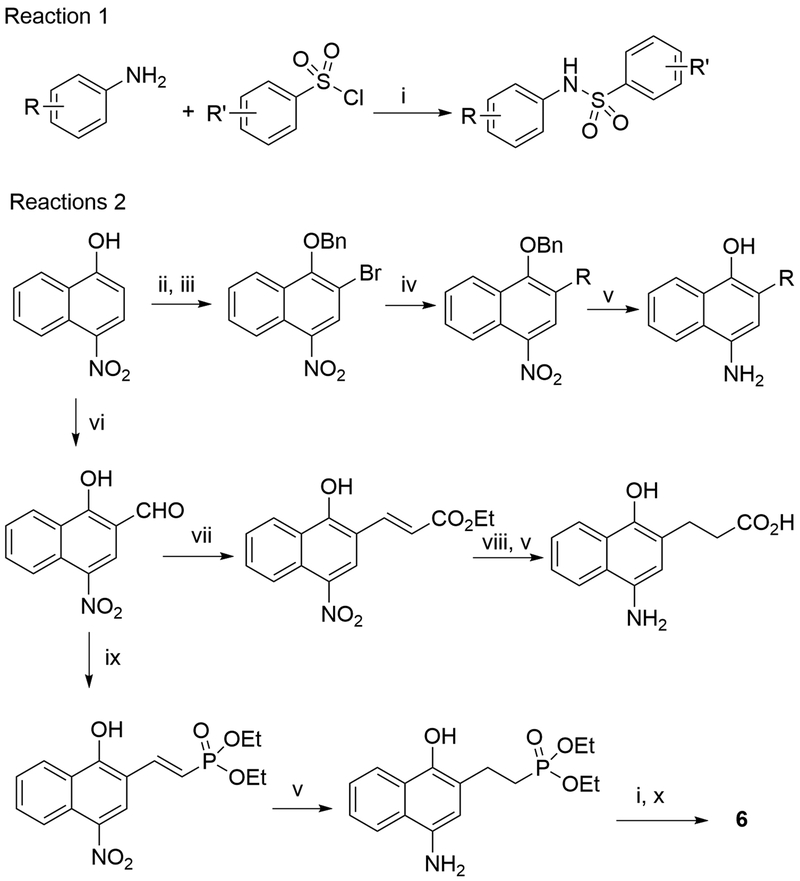
Chemical synthesis of compounds 1-20 is illustrated in Scheme 1 and described in detail below.
Scheme 1.
Synthetic methods.a
aReagents and conditions: (i) pyridine; (ii) Br2, AcOH; (iii) BnBr, K2CO3, acetone, reflux; (iv) for synthesizing 3 and 4, RB(OH)2, Pd(dppf)Cl2, K2CO3, dioxane, reflux; or for synthesizing 1, 14 and 15, phenol, CuI, N, N-dimethylglycine, Cs2CO3, dioxane, reflux; (v) 10% Pd/C, H2; (vi) paraformaldehyde, MgCl2, NEt3, CH3CN, reflux; (vii) triethyl phosphonoacetate, NaH, THF; (viii) NaOH, MeOH; (ix) tetraethyl methylenediphosphonate, NaH, THF; (x) bromotrimethylsilane, CH2Cl2.
General Method for Reaction 1.
(i) To a solution of an aromatic amine compound (1.38 mmol) in pyridine (3 mL), an aromatic sulfonyl chloride (1.38 mmol) solution in pyridine (3 mL) was added dropwise at 0 oC under N2. The mixture was stirred overnight. Pyridine was removed under reduced pressure. The oily residue was dissolved in ethyl acetate (20 mL), washed with water (2 × 10 mL) and brine, and concentrated. The residue was subjected to column chromatography (silica gel) to give a sulfonamide product in 66–89% yield.
General Methods for Reactions 2.
(ii) To a mixture of 4-nitro-1-naphthanol (10 mmol) in acetic acid (25 mL), Br2 (10 mmol, 1.60 g) in acetic acid (5 mL) was added dropwise at 5–10 oC. The mixture was allowed to stir at room temperature for 20min. The solvent was removed under reduced pressure to yield 3-bromo-4-nitro-1-naphthanol as a yellow solid in 99% yield. It can be use directly for next step without further purification.
(iii) A mixture of 3-bromo-4-nitro-1-naphthanol (1 mmol), BnBr (3 mmol, 0.35 mL), and K2CO3 (2 mmol) in acetone (3 mL) was heated to 70 oC for 20 h in a sealed tube. The mixture was diluted with ethyl acetate (20 mL), washed with water (2 × 10 mL), dried over anhydrous Na2SO4 and concentrated. Hexane (1 mL) was added into the resulting residue to give benzyl protected 3-bromo-4-nitro-1-naphthanol as a yellow solid 5 in ~90% yield.
(iv) For coupling with a boronic acid: A mixture of the above product (0.67 mmol), an aromatic boronic acid (1 mmol), K2CO3 (2 mmol), Pd(dppf)Cl2 (0.03 mmol) in dioxane (2 mL) was heated to 120 oC for 36 h in a sealed tube filled with N2. The Suzuki coupling product was obtained with column chromatography (silica gel) in 66–81% yield. For coupling with phenol: A mixture of the above product (0.28 mmol), Phenol (0.42 mmol), CuI (0.28 mmol), N, N-dimethylglycine (0.28 mmol) Cs2CO3 (0.56 mmol) in dioxane (2 mL) was heated to 105 oC for 20 h in a sealed tube filled with N2. The solvent was removed under reduced pressure, diluted with ethyl acetate and the pH was adjusted to 3–4 with HCl (aq). The product was washed with NH4Cl, and water, and purified with column chromatograph (silica gel) in 58–76% yield.
(v) A mixture of the product from (iv) (1 mmol), 2 N HCl (0.2 mL) and 10% Pd/C (20 mg) in 5 mL CH3OH was stirred overnight under H2 atmosphere. The catalyst was filtered off and the filtrate was concentrated and dried under reduced pressure to yield 3-substituted 4-amino-1-naphthanol in quantitative yield.
(vi) A mixture of 4-nitro-1-naphthanol (1 mmol), paraformaldehyde (5 mmol), MgCl2 (5 mmol), Et3N (5 mmol) in CH3CN was refluxed overnight under N2. The solvent was removed under reduced pressure. 1 N HCl (10 mL) and ethyl acetate (20 mL) was added. The mixture was stirred for 5 min until the solid was completely dissolved. The organic phase was collected, washed with water and brine, dried over Na2SO4, filtered and concentrated to obtain crude 2-formyl-4-nitro-1-naphthanol, which can be used directly for the next reaction.
(vii, viii) To a solution of 2-formyl-4-nitro-1-naphthanol (1 mmol), triethyl phosphonoacetate (1.5 mmol) in 4 mL THF, NaH (2 mmol 80mg 60% in mineral oil) was added portion wise at 0 oC. After addition, the mixture was allowed to stir overnight at room temperature. Then it was diluted with ethyl acetate, neturalized with HCl(aq.), washed with water and concentrated. The residue was purified through column chromatograph (silica gel, ethyl acetate). The product was readily hydrolyzed by NaOH (2N) in MeOH/THF to give 3-(4-nitro-1-hydroxy-2-naphthyl)propionic acid.
(ix, x) To a solution of 2-formyl-4-nitro-1-naphthanol (1 mmol), tetraethyl methylenediphosphonate (1.5 mmol) in 4 mL THF, NaH (2 mmol 80mg 60% in mineral oil) was added portion wise at 0 oC. After addition, the mixture was allowed to stir overnight at room temperature. Then it was diluted with ethyl acetate, neturalized with HCl(aq.), washed with water and concentrated. The residue was purified through column chromatograph (silica gel, ethyl acetate) to give 4-nitro-2-diethylphosphonoethylenyl-1-naphthanol, which was hydrogenated and reacted with 4-bromo-phenylsulfonyl chloride using the above conditions to give diethyl ester of compound 6. To a solution of the ester (0.5 mmol) in CH2Cl2 (7 mL), bromotrimethylsilane (2.5 mmol) was added dropwise at 0 oC under N2. The mixture was allowed to stir for 36 h at room temperature under N2. The solvent was removed under reduced pressure. MeOH (5 mL) was added to the residue and stirred for 5 min. Upon removal of the solvent, the residue was stirred with CH2Cl2 to give compound 6 as an off-white powder in 40% overall yield.
Compound characterization
4-bromo-N-(4-hydroxy-3-phenoxynaphthalen-1-yl)benzenesulfonamide (compound 1).
1H NMR (400 MHz, CDCl3): δ 8.22 (d, J = 7.2 Hz, 1H), 7.80 (d, J = 7.2 Hz, 1H), 7.53–7.40 (m, 6H), 7.35 (t, J = 7.2 Hz, 2H), 7.19 (t, J = 7.2 Hz, 1H), 6.90 (m, 3H), 6.43 (s, 1H), and 6.06 (s, 1H); 13C NMR (100 MHz, CDCl3): 142.3, 138.4, 138.1, 136.4, 132.3, 132.2, 130.0, 128.9, 128.7, 126.5, 126.3, 123.9, 123.1, 122.2, 122.0, 120.8, 118.5, and 117.6; MS (ESI) [M+H]+ 471.3.
4-bromo-N-(3-bromo-4-hydroxynaphthalen-1-yl)benzenesulfonamide (2).
1H NMR (400 MHz, CDCl3): δ 8.20 (d, J = 7.5 Hz, 1H), 7.63 (d, J = 7.5 Hz, 1H), 7.58–7.39 (m, 7H), 6.88 (s, 1H), and 6.10 (s, 1H); 13C NMR (100 MHz, CDCl3): 148.5, 138.3, 132.4, 130.4, 129.0, 128.3, 128.0, 127.8, 126.9, 124.5, 124.2, 123.1, 122.0, and 121.9; MS (ESI) [M+H]+ 458.1.
N-(4-hydroxy-3-(p-tolyl)naphthalen-1-yl)-4-methylbenzenesulfonamide (3).
1H NMR (400 MHz, CDCl3): δ 8.45 (s, 1H), 8.20–8.14 (m, 2H), 7.98 (d, J = 7.2 Hz, 2H), 7.70 (t, J = 7.2 Hz, 1H), 7.62 (t, J = 7.2 Hz, 1H), 7.58 (d, J = 7.2 Hz, 2H), 7.40 (d, J = 7.2 Hz, 2H), 7.30–7.20 (m, 3H), 2.48 (s, 3H), and 2.40 (s, 3H); 13C NMR (100 MHz, CDCl3): 163.2, 146.3, 144.1, 140.9, 138.2, 133.5, 133.4, 133.1, 132.3, 131.8, 130.8, 129.9, 129.6, 129.3, 128.3, 127.4, 127.3, 126.1, 21.7, and 21.5; MS (ESI) [M+H]+ 404.5.
4-bromo-N-(1-hydroxy-[2,2’-binaphthalen]-4-yl)benzenesulfonamide (4).
1H NMR (400 MHz, CDCl3): δ 8.32 (d, J = 7.2 Hz, 1H), 8.00 (d, J = 7.2 Hz, 1H), 7.97–7.80 (m, 4H), 7.62–7.39 (m, 10H), 7.09 (s, 1H), 6.60 (s, 1H), and 6.10 (s, 1H); 13C NMR (100 MHz, CDCl3): 150.9, 146.2, 144.1, 140.7, 138.7, 135.2, 134.2, 133.5, 133.4, 133.1, 132.3, 131.9, 130.8, 129.9, 129.6, 129.3, 128.3, 127.4, 127.3, 126.3, 124.6, 126.1, 119.0, and 115.5; MS (ESI) [M+H]+ 505.4.
3-(4-((4-bromophenyl)sulfonamido)-1-hydroxynaphthalen-2-yl)propanoic acid (5).
1H NMR (400 MHz, DMSO-d6): δ 9.95 (s, 1H), 9.36 (br, 1H), 8.14 (d, J = 7.2 Hz, 1H), 7.80 (d, J = 7.2 Hz, 1H), 7.70 (t, J = 7.2 Hz, 2H), 7.50 (d, J = 7.2 Hz, 2H), 7.40 (d, J = 7.2 Hz, 2H), 7.38 (d, J = 7.2 Hz, 2H), 6.78 (s, 1H), 2.84 (t, J = 4.8 Hz, 2H), and 2.45 (t, J = 7.2 Hz, 2H); 13C NMR (100 MHz, CDCl3): 176.5, 150.3, 143.3, 138.7, 133.9, 131.0, 129.7, 129.5, 127.9, 126.7, 125.7, 124.0, 123.7, 121.8, 120.1, 34.1 and 24.6; MS (ESI) [M+H]+ 451.3.
(2-(4-((4-bromophenyl)sulfonamido)-1-hydroxynaphthalen-2-yl)ethyl)phosphonic acid (6).
1H NMR (400 MHz, CDCl3): δ 9.63 (s, 1H), 8.12 (d, J = 7.2 Hz, 1H), 7.90 (d, J = 7.2 Hz, 1H), 7.42 (t, J = 7.2 Hz, 2H), 7.40–7.30 (m, 2H), 7.22 (d, J = 7.2 Hz, 2H), 6.60 (br, 1H), 2.78–2.70 (m, 2H), and 1.63–1.58 (m, 2H); 13C NMR (100 MHz, CDCl3): 143.3, 137.9, 136.5, 132.2, 129.6, 128.9, 128.3, 127.7, 125.4, 123.7, 122.7, 121.8, 120.1, 107.5, 29.6 (d, JC-P = 98 Hz), and 27.9 (d, JC-P = 33 Hz); 31P NMR (162 MHz, DMSO-d6): 26.0; MS (ESI) [M-H]− 485.3.
3-(1-hydroxy-4-((4-methoxyphenyl)sulfonamido)naphthalen-2-yl)propanoic acid (7).
1H NMR (400 MHz, CDCl3): δ 11.0 (br, 1H), 8.42 (br, 1H), 8.27 (d, J = 8.3 Hz, 1H), 7.69 (d, J = 8.3 Hz, 1H), 7.59 (d, J = 8.9 Hz, 2H), 7.41 (t, J = 7.3 Hz, 1H), 7.34 (t, J = 7.3 Hz, 1H), 6.98 (s, 1H), 6.79 (d, J = 8.9 Hz, 2H), 6.71 (s, 1H), 3.78 (s, 3H), 2.91 (t, J = 5.5 Hz, 2H), and 2.69 (t, J = 5.5 Hz, 2H); 13C NMR (100 MHz, CDCl3): 177.2, 163.1, 149.8, 131.0, 130.3, 129.7, 129.5, 127.9, 126.7, 125.7, 124.0, 123.2, 121.8, 120.2, 114.0, 55.7, 35.1 and 24.3; MS (ESI) [M+H]+ 402.4.
N-(3-bromo-4-hydroxynaphthalen-1-yl)-4-fluorobenzenesulfonamide (8).
1H NMR (400 MHz, CDCl3): δ 10.5 (br, 1H), 8.15 (d, J = 7.0 Hz, 1H), 7.80 (d, J = 7.0 Hz, 1H), 7.64 (d, J = 7.0 Hz, 2H), 7.48 (t, J = 8.0 Hz, 1H), 7.40 (t, J = 8.0 Hz, 1H), 7.30 (t, J = 8.0 Hz, 2H), and 7.03 (d, J = 8.0 Hz, 1H); 13C NMR (100 MHz, CDCl3): 165.4 (d, 1JF-C = 254 Hz), 148.4, 135.2, 130.4, 130.2 (d, 3JF-C = 9.7 Hz), 128.0, 127.8, 126.8, 124.4 (d, 2JF-C = 17.6 Hz), 123.0, 122.0, 116.5, 116.3, and 102.6; MS (ESI) [M+H]+ 397.2.
N-(3-bromo-4-hydroxynaphthalen-1-yl)-4-methylbenzenesulfonamide (9).
1H NMR (400 MHz, CDCl3): δ 10.0 (s, 1H), 9.95 (s, 1H), 8.15 (d, J = 7.5 Hz, 1H), 7.85 (d, J = 7.5 Hz, 1H), 7.50–7.39 (m, 4H), 7.25 (d, J = 7.0 Hz, 2H), 7.01 (s, 1H), and 2.30 (s, 3H); 13C NMR (100 MHz, CDCl3): 148.1, 144.1, 136.2, 130.5, 129.7, 127.63, 127.60, 127.54, 126.7, 124.8, 124.5, 122.9, 122.3, 102.6, and 21.7; MS (ESI) [M+H]+ 393.3.
N-(3-bromo-4-hydroxynaphthalen-1-yl)-[1,1’-biphenyl]-4-sulfonamide (10).
1H NMR (400 MHz, CDCl3): δ 8.20 (d, J = 7.5 Hz, 1H), 7.76 (d, J = 7.5 Hz, 2H), 7.67 (d, J = 7.5 Hz, 1H), 7.60–7.39 (m, 10H), 6.47 (s, 1H), and 5.59 (s, 1H); 13C NMR (100 MHz, CDCl3): 148.2, 142.1, 139.1, 134.4, 129.0, 128.5, 127.86, 127.84, 127.57, 127.53, 127.39, 127.27, 126.6, 122.8, 122.0, 118.6, 113.9, and 110.0; MS (ESI) [M+H]+ 455.3.
N-(3-bromo-4-hydroxynaphthalen-1-yl)naphthalene-2-sulfonamide (11).
1H NMR (400 MHz, CDCl3): δ 10.15 (s, 1H), 10.0 (s, 1H), 8.20 (s, 1H), 8.13–7.97 (m, 4H), 7.90 (d, J = 7.5 Hz, 1H), 7.75 (d, J = 7.5 Hz, 1H), 7.63 (t, J = 7.5 Hz, 1H), 7.58 (t, J = 7.5 Hz, 1H), 7.42 (t, J = 7.5 Hz, 1H), 7.38 (t, J = 7.5 Hz, 1H), and 7.01 (s, 1H); 13C NMR (100 MHz, CDCl3): 148.4, 136.2, 135.0, 132.1, 130.6, 129.5, 129.4, 129.08, 129.06, 128.0, 127.8, 127.7, 126.8, 125.3, 124.6, 122.9, 122.5, 122.2, 122.1, and 102.6; MS (ESI) [M+H]+ 429.3.
N-(4-hydroxynaphthalen-1-yl)-4-methylbenzenesulfonamide (12).
1H NMR (400 MHz, DMSO-d6) δ 10.28 (s, 1H), 9.82 (s, 1H), 8.08 (d, J = 7.6 Hz, 1H), 7.81 (d, J = 7.6, 3H), 7.61–7.52 (m, 2H), 7.45 (d, J = 7.6 Hz, 2H), 7.14 (d, J = 7.6 Hz, 1H), 7.10 (d, J = 8.1 Hz, 1H), and 2.41 (s, 3H); 13C NMR (100 MHz, DMSO-d6) 146.4, 142.3, 136.2, 132.0, 130.7, 128.7, 128.1, 127.6, 127.5, 126.9, 124.6, 123.1, 122.1, 118.9, and 21.6; MS (ESI) [M+H]+ 314.4.
N-(4-hydroxynaphthalen-1-yl)benzenesulfonamide (13).
1H NMR (400 MHz, DMSO-d6) δ 10.28 (s, 1H), 9.82 (s, 1H), 8.07 (d, J = 7.8 Hz, 1H), 7.85 (d, J = 7.8, 1H), 7.63 (d, J = 7.2 Hz, 2H), 7.56 (d, J = 7.6 Hz, 1H), 7.50–7.46 (m, 2H), 7.39–7.37 (m, 2H), 6.86 (d, J = 7.6 Hz, 1H), and 6.71 (d, J = 8.1 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) 151.2, 139.1, 131.3, 130.4, 127.8, 125.6, 124.9, 124.4, 123.7, 123.6, 122.05, 122.01, 120.9, and 106.1; MS (ESI) [M+H]+ 300.3.
4-bromo-N-(4-hydroxy-3-phenoxyphenyl)benzenesulfonamide (14).
1H NMR (400 MHz, CDCl3): δ 7.56–7.48 (m, 4H), 7.44–7.30 (m, 2H) , 7.19–7.14 (m, 1H), 6.92–6.84 (m, 2H), 6.76–6.68 (m, 3H), 6.53–6.50 (m, 1H), and 5.71 (br, 1H); 13C NMR (100 MHz, CDCl3): 155.9, 146.1, 144.0, 137.7, 133.1, 132.4, 130.2, 129.0, 124.4, 120.8, 120.0, 118.4, 116.9, and 114.6; MS (ESI) [M+H]+ 421.3.
4-bromo-N-(4-methoxy-3-phenoxyphenyl)benzenesulfonamide (15).
1H NMR (400 MHz, CDCl3): δ 7.55–7.50 (m, 4H), 7.31–7.26 (m, 2H), 7.08 (t, J = 7.6 Hz, 1H), 6.87 (d, J = 1.2 Hz, 2H), 6.80 (dd, J = 7.6, 1.2 Hz, 2H), 6.60 (s, 1H), 6.54 (s, 1H), and 3.81 (s, 3H); 13C NMR (100 MHz, CDCl3): 157.4, 146.7, 141.0, 138.9, 138.7, 133.1, 132.4, 130.2, 129.1, 124.6, 120.8, 120.0, 116.4, 111.7, and 56.8; MS (ESI) [M+H]+ 435.3.
4-bromo-N-(4-methoxyphenyl)benzenesulfonamide (16).
1H NMR (400 MHz, CDCl3): δ 9.97 (s, 1H), 8.16 (d, J = 7.2 Hz, 2H), 7.63 (d, J = 7.2 Hz, 2H), 6.94 (d, J = 1.2 Hz, 2H), 6.81 (dd, J = 7.6, 1.2 Hz, 2H), and 3.83 (s, 3H); 13C NMR (100 MHz, CDCl3): 153.4, 138.7, 132.9, 130.7, 129.5, 126.3, 124.5, 115.4, and 55.8; MS (ESI) [M+H]+ 343.2.
4-bromo-N-(4-hydroxyphenyl)benzenesulfonamide (17).
1H NMR (400 MHz, CDCl3): δ 9.97 (s, 1H), 9.90 (s, 1H), 8.18 (d, J = 7.2 Hz, 2H), 7.69 (d, J = 7.6 Hz, 2H), 6.81 (d, J = 7.2 Hz, 2H), and 6.60 (d, J = 7.6, Hz, 2H); 13C NMR (100 MHz, CDCl3): 148.4, 138.7, 131.3, 130.3, 129.1, 126.3, 124.5, and 117.4; MS (ESI) [M+H]+ 329.2.
4-bromo-N-(3-bromophenyl)benzenesulfonamide (18).
1H NMR (400 MHz, CDCl3): δ 10.02 (s, 1H), 9.96 (s, 1H), 8.18 (d, J = 7.2 Hz, 2H), 7.61 (d, J = 7.2 Hz, 2H), 7.01–6.70 (m, 4H); 13C NMR (100 MHz, CDCl3): 140.4, 138.7, 131.7, 130.3, 129.1, 126.3, 124.5, 121.6, 118.4, and 115.3; MS (ESI) [M+H]+ 392.1.
4-bromo-N-phenylbenzenesulfonamide (19).
1H NMR (400 MHz, CDCl3): δ 10.01 (s, 1H), 8.07 (d, J = 7.2 Hz, 2H), 7.62 (d, J = 7.2 Hz, 2H), 7.20–7.15 (m, 2H), and 6.85–6.80 (m, 3H); 13C NMR (100 MHz, CDCl3): 137.7, 136.0, 132.3, 129.4, 128.7, 128.1, 125.6, and 121.7; MS (ESI) [M+H]+ 313.2.
4-bromo-N-(3-chloro-4-fluorophenyl)benzenesulfonamide (20).
1H NMR (400 MHz, CDCl3): δ 9.95 (s, 1H), 8.06 (d, J = 7.2 Hz, 2H), 7.66 (d, J = 7.2 Hz, 2H), and 7.30–6.60 (m, 3H); 13C NMR (100 MHz, CDCl3): 151.6 (d, 1JF-C = 237 Hz), 143.2, 141.2, 132.0, 127.8, 125.7, 121.0, 116.8, 116.4, and 114.3; MS (ESI) [M+H]+ 365.6.
Enzyme Inhibition.
Inhibitory activities of compounds were tested against mutant and wild-type IDH1 using our previous methods. [13]
Steady-state kinetic study.
Steady-state kinetic inhibition experiment was performed by determining initial velocities of reactions catalyzed by IDH1(R132H) with varying the concentrations of compound 9, α-KG and NADPH. Data were imported into SigmaPlot (version 13, Systat Software, Inc.) and fitted to competitive, noncompetitive and uncompetitive inhibition models. The best kinetic models were determined by the highest R2 and lowest AICc values. Lineweaver-Burk or Michaelis-Menten plots were generated by Sigmaplot.
Inhibition of proliferation of glioma stem-like cells.
Two glioma cell lines BT142 and BXD-3752 were cultured as neurospheres in serum-free cell growth media at 37 °C in a 5% CO2 atmosphere with 100% humidity as described previously in Ref. 13. In brief, 2000 cells/well were added into 96-well plates and treated with increasing concentrations of a compound for 14 days. Cell viability was determined by Cell Counting Kit-8 (Dojindo Molecular Technologies, Rockville, MD) according to the manufacturer’s instructions.
RESULTS and DISCUSSION
Biochemical assay and inhibitor discovery.
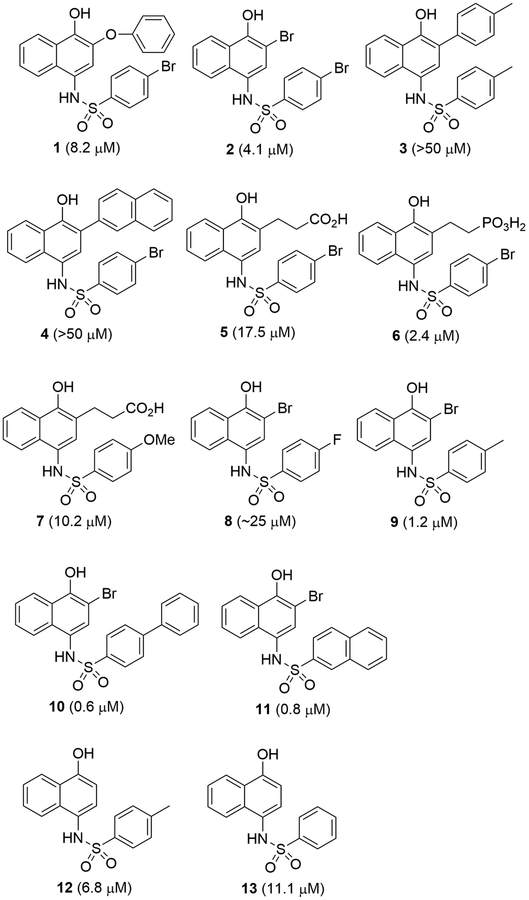
A biochemical assay was used to screen for new inhibitors of IDH1(R132H), which was expressed in E. coli and purified using our previous methods. [12–14] The assay is based on an initial linear consumption of NADPH, which has a maximal UV absorbance at 340 nm. Thus, an initial velocity of the enzyme reaction can be determined to be the decreasing rate of [NADPH] monitored by UV. We performed compound screening at 50 μM followed by IC50 (concentration at which a compound can inhibit the enzyme activity by 50%) and Ki (inhibition constant) determination for active hits, compound 1 (Figure 2) was found to be a novel inhibitor of IDH1(R132H) with a Ki value of 8.2 μM.
Figure 2.
Structures and inhibitory activities of compounds 1-13.
Chemical synthesis.
Methods for synthesizing these sulfonamide compounds are illustrated in Scheme 1. All sulfonamide compounds were synthesized by the reaction of a commercially available sulfonyl chloride and a substituted naphthyl or phenyl amine (Reaction 1). Except for a few that are commercially available, these amines were synthesized according to Reactions 2. 4-Nitro-1-naphthol or -phenol was treated with Br2 to add a 2-bromo group and 1-OH of the product was protected with a benzyl group. An ensuing metal-catalyzed reaction can substitute the 2-Br with the aromatic group (found in compounds 1, 3, 4, 14 and 15). The product was hydrogenated to give the desired 2-substituted naphthyl or phenyl amine for Reaction 1. 4-Nitro-1-naphthol was formylated at 2-position by treatment with paraformaldehyde in the presence of MgCl2 and triethylamine. The 2-formyl group can be converted to side chains in compounds 5 - 7, using methods shown in Reactions 2.
Structure activity relationship (SAR) studies.
We next performed structure activity relationship (SAR) studies based on the structure of 1, in an effort to find more potent inhibitors. Compounds 2 - 7 (Figure 2) were synthesized to see the effects of 2-substituents. Compound 2 with a 2-Br exhibited ~2-fold increased inhibitory activity (Ki = 4.1 μM). However, compounds 3 and 4 having a 4-tolyl and 2-naphthyl group at this position, respectively, were inactive, showing a direct aromatic substituent is disfavored. Compound 5 bearing a carboxylethyl group showed reduced activity with Ki of 17.5 μM, as compared with compounds 1 and 2. However, compound 6 with a 2-phosphono-ethyl substituent was found to have a considerably improved activity of 2.4 μM. Compound 7 with a 4-methoxybenzenesulfonyl moiety showed increased activity (Ki = 10.2 μM), as compared to 5. Because highly polar and negatively charged phosphonic compounds are generally not cell permeable, 2-Br was used for optimization of other positions.
2-Bromo compounds 8 - 11 with different aromatic sulfonyl groups were synthesized and tested for their activities inhibiting IDH1(R132H). As shown in Figure 2, compound 8 with a small, more electron-withdrawing 4-F substituted benzenesulfonyl was found to be significantly less active (Ki ~25 μM) than compound 2 with a 4-Br. On the other hand, compound 9 with a 4-methylbenzenesulfonyl is a strong inhibitor with a Ki value of 1.2 μM, ~4× more active than 2. Compounds 10 and 11 bearing a 4-biphenyl and 2-naphthyl sulfonyl group showed the most potent inhibitory activity with Ki values of 0.6 and 0.8 μM. These results suggest that electron-donating and/or bulky aromatic groups are more favored at this position.
Compound 12 (Ki = 6.8 μM, Figure 2) is ~6-fold less active than compound 9, showing the 2-Br group in 9 is significantly more favorable than the 2-H. In addition, as compared to 12, reduced activity of compound 13 (Ki = 11.1 μM) indicates that the 4-methyl group in the benzenesulfonyl moiety is a favorable substituent.
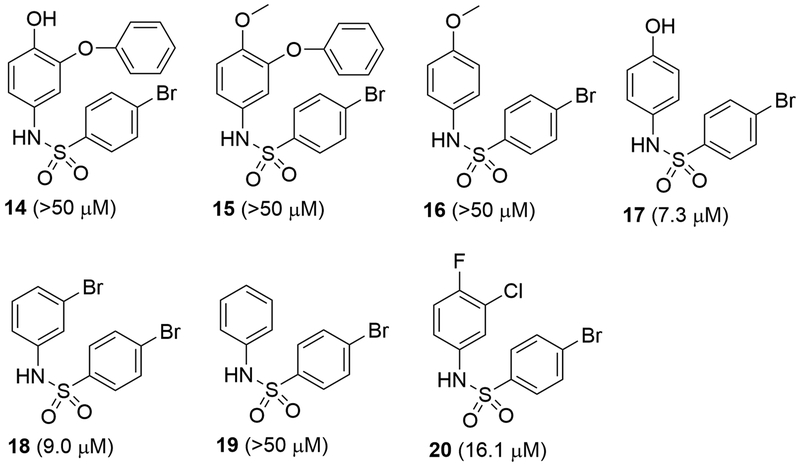
A series of phenylamine sulfonamide compounds 14 – 20 shown in Figure 3 were synthesized to further probe the SARs of this class of compounds. Devoid of activity for compound 14 clearly indicates the importance of the corresponding naphthyl ring in compound 1, in which the additional aromatic ring possibly provides favorable hydrophobic interactions with the protein. 1-Methoxy group in compound 15 as well as 16 does not seem to render an improved potency. However, compound 17 having a 1-OH showed enhanced activity (Ki = 7.3 μM), which is considerably more active than 16 with a 1-OMe. This shows the hydroxy group contributes significantly to the inhibition. Nevertheless, compound 18, which does not carry a -OH but has a 2-Br group, exhibited a comparable activity (Ki = 9.0 μM) to that of compound 17. Therefore, it can be inferred from these results that the 2-Br group also plays an important role in enzyme activity inhibition. This point can be further supported by the activities of compounds 19 and 20. Without the corresponding -Br group, compound 19 is inactive. Compound 20 having a smaller -Cl at this position is a weaker inhibitor of IDH1(R132H) (Ki = 16.1 μM), although it is possible that the -F group also contributes to the inhibition.
Figure 3.
Structures and activities of compounds 14-20.
Enzyme kinetics studies
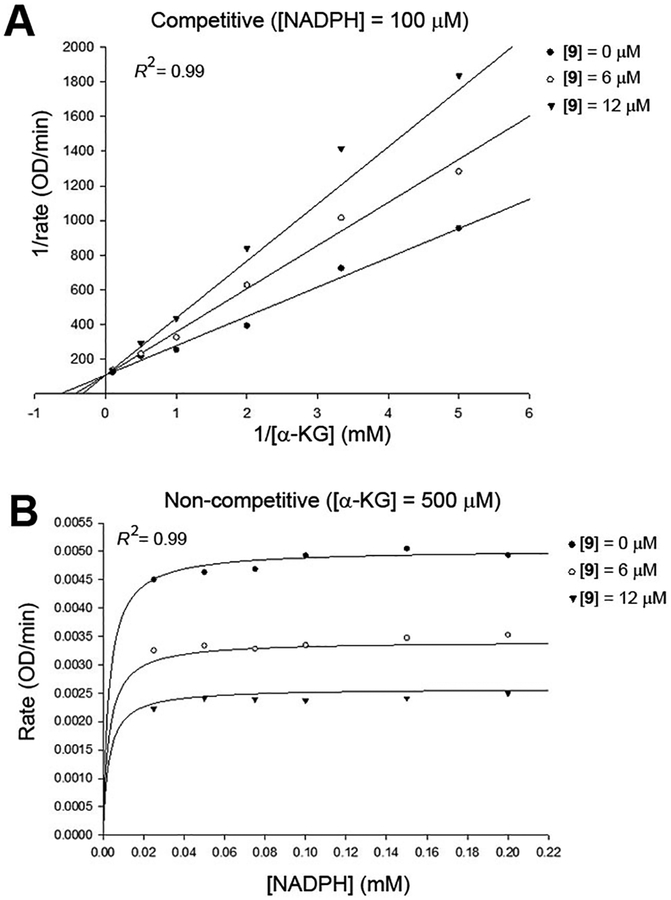
To find mode of action of this series of compounds, we did steady-state enzyme kinetic studies for compound 9. Initial velocities of IDH1(R132H) were determined with increasing concentrations of 9, the substrate α-KG and cofactor NADPH. The data were fitted and analyzed to competitive, uncompetitive and noncompetitive kinetics models by the program SigmaPlot. The mode of action for compound 9 to inhibit the enzyme can be determined by the best fitting, as judged by the R2 and AICc values, using a Lineweaver-Burk or Michaelis-Menten plot. With this method, compound 9 was found to be a competitive inhibitor against the substrate α-KG, as shown in Figure 4A. It was also found to exhibit a noncompetitive mode of action against the cofactor NADPH (Figure 4B).
Figure 4.
The best fitting models for enzyme kinetic studies of compound 9. (A) Competitive inhibition model with variable concentrations of α-KG using a Lineweaver-Burk plot; and (B) Noncompetitive inhibition model with variable concentrations of NADPH using a Michaelis-Menten plot.
Evaluation of anti-glioma activity
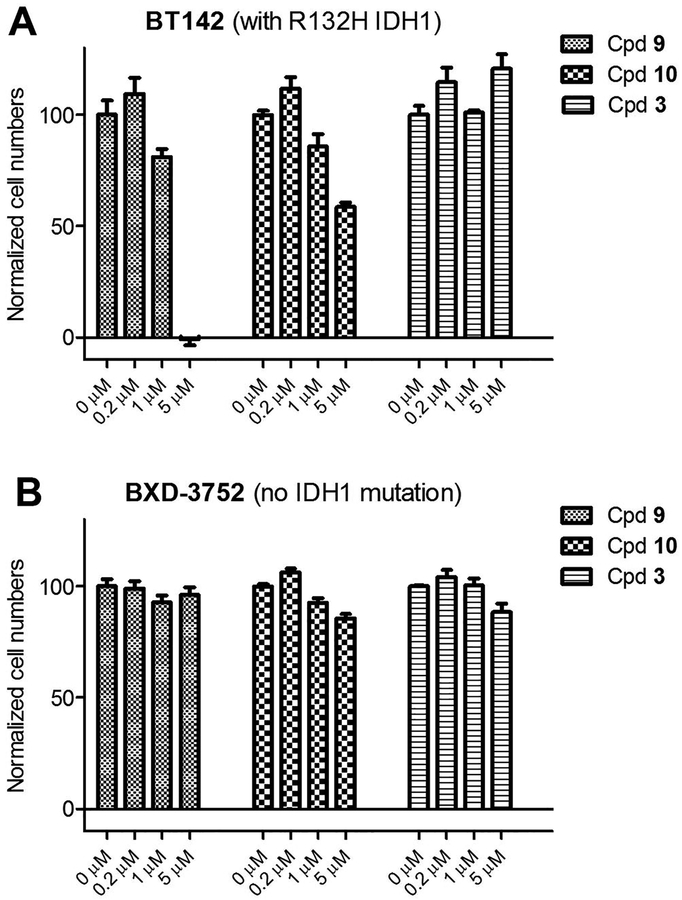
Finally, we tested antitumor activities of potent inhibitors 9 and 10 against proliferation of BT142 glioma cells harboring R132H IDH1 mutation (Figure 5). In the meantime, activity of these compounds against BXD-3752 glioma cells without an IDH1 mutation was also evaluated. Inactive compound 3 was included in these assays as a control to find possible off-target toxicity of these sulfonamide compounds. These glioma cells were derived from patient samples and cultured in serum-free media to form colony-like “neurospheres”. [13,14] Stem-like cancer cells were found to be enriched in these neurospheres, which have the ability to self-renew and differentiate. Inhibition of these stem-like tumor cells is of importance, given roles of these cells in drug resistance, relapse and metastasis. As shown in Figure 5A, compounds 9 and 10 at 5 μM strongly inhibited the growth of BT142 glioma cells with R132H IDH1 mutation, while these two compounds did not show activity against BXD-3752 cells. In addition, inactive compound 3 did not exhibit activity against proliferation of both glioma cells. These results show that potent inhibitors of IDH1(R132H) have selective activity against glioma cells containing the IDH1 mutation.
Figure 5.
Activity of compounds 9, 10 and 3 against BT142 and BXD-3752 glioma cells.
CONCLUSION
In summary, a series of aromatic sulfonamide compounds have been found to be novel, potent inhibitors of R132H mutant IDH1 with Ki values as low as 0.6 μM. This work is of interest because this specific mutation has been found to be a drug target in ~75% low-grade gliomas as well as secondary GBM. Several structure activity relationships have been concluded, which might be useful to guide further inhibitor optimization. For the aromatic amine moiety, 1-hydroxy and 2-bromo groups have been found to be critical to the inhibition of IDH1(R132H). 1-hydroxy-naphthylamine is superior to the corresponding phenylamine, presumably due to enhance hydrophobic interactions. For the benzenesulfonyl moiety, a bulkier (such as phenyl in 10) and/or an electron-releasing group (such as methyl in 9) at the 4-position are favorable. Enzyme kinetics studies showed that compound 9 is a competitive inhibitor of IDH1(R132H) against the substrate α-KG and a non-competitive inhibitor against the cofactor NADPH. In addition, potent sulfonamide inhibitors of IDH1(R132H) are highly selective, showing no or negligible inhibitory activity against WT or R132C IDH1 (Table 1). Potent inhibitors 9 and 10 exhibited strong activity against proliferation of BT142 glioma cells with IDH1 R132H mutation, while these compounds did not significantly affect growth of BXD-3752 cells without IDH1 mutation. These results suggest further characterization and optimization of these compounds are warranted with a goal to find a clinically useful drug targeting IDH1 mutated gliomas.
Table 1.
Activity (Ki in μM) against WT and mutant IDH1.
| Cpd | R132H | R132C | WT |
|---|---|---|---|
| 2 | 4.1 | >50 | >50 |
| 9 | 1.2 | >50 | >50 |
| 10 | 0.6 | 36 | >50 |
| 11 | 0.8 | >50 | >50 |
Acknowledgements
This work was supported by grants RP150129 and RP140469 from Cancer Prevention and Research Institute of Texas (CPRIT) and a grant (R01NS080963) from National Institute of Neurological Disorders and Stroke (NINDS/NIH) to Y.S.
Footnotes
Conflict of Interest
The author(s) confirm that this article content has no conflicts of interest.
References
- 1.Garber K Oncometabolite? IDH1 discoveries raise possibility of new metabolism targets in brain cancers and leukemia. J. Natl. Cancer Inst 2010, 102, 926–928. [DOI] [PubMed] [Google Scholar]
- 2.Yan H; Parsons DW; Jin G; McLendon R; Rasheed BA; Yuan W; Kos I; Batinic-Haberle I; Jones S; Riggins GJ; Friedman H; Friedman A; Reardon D; Herndon J; Kinzler KW; Velculescu VE; Vogelstein B; Bigner DD IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med 2009, 360, 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartmann C; Meyer J; Balss J; Capper D; Mueller W; Christians A; Felsberg J; Wolter M; Mawrin C; Wick W; Weller M; Herold-Mende C; Unterberg A; Jeuken JW; Wesseling P; Reifenberger G; von Deimling A Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009, 118, 469–474. [DOI] [PubMed] [Google Scholar]
- 4.Gross S; Cairns RA; Minden MD; Driggers EM; Bittinger MA; Jang HG; Sasaki M; Jin S; Schenkein DP; Su SM; Dang L; Fantin VR; Mak TW Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med 2010, 207, 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujii T; Khawaja MR; DiNardo C.; Atkins JT; Janku F Targeting isocitrate dehydrogenase (IDH) in cancer. Discov. Med 2016, 12, 373–80. [PubMed] [Google Scholar]
- 6.Clark O; Yen K; Mellinghoff IK Molecular Pathways: Isocitrate Dehydrogenase Mutations in Cancer. Clin. Cancer Res 2016, 22, 1837–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang L; White DW; Gross S; Bennett BD; Bittinger MA; Driggers EM; Fantin VR; Jang HG; Jin S; Keenan MC; Marks KM; Prins RM; Ward PS; Yen KE; Liau LM; Rabinowitz JD; Cantley LC; Thompson CB; van der Heiden MG; Su SM Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM & Xiong Y Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011, 19, 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu C; Ward PS; Kapoor GS; Rohle D; Turcan S; Abdel-Wahab O; Edwards CR; Khanin R; Figueroa ME; Melnick A; Wellen KE; O’Rourke DM; Berger SL; Chan TA; Levine RL; Mellinghoff IK; Thompson CB IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turcan S; Rohle D; Goenka A; Walsh LA; Fang F; Yilmaz E; Campos C; Fabius AW; Lu C; Ward PS; Thompson CB; Kaufman A; Guryanova O; Levine R; Heguy A; Viale A; Morris LG; Huse JT; Mellinghoff IK; Chan TA IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popovici-Muller J, Saunders JO, Salituro FG, Travins JM, Yan S, Zhao F, Gross S, Dang L, Yen KE, Yang H, Straley KS, Jin S, Kunii K, Fantin VR, Zhang S, Pan Q, Shi D, Biller SA & Su SM Discovery of the First Potent Inhibitors of Mutant IDH1 That Lower Tumor 2-HG in Vivo. ACS Med. Chem. Lett 2012, 3, 850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng B; Yao Y; Liu Z; Deng L; Anglin JL; Jiang H; Prasad BV; Song Y Crystallographic Investigation and Selective Inhibition of Mutant Isocitrate Dehydrogenase. ACS Med. Chem. Lett 2013, 4, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z; Yao Y; Kogiso M; Zheng B; Deng L; Qiu JJ; Dong S; Lv H; Gallo JM; Li X-N; Song Y Inhibition of cancer-associated mutant isocitrate dehydrogenases: synthesis, structure-activity relationship, and selective antitumor activity. J. Med. Chem 2014, 57, 8307–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu F; Zheng B; Jiang H; Kogiso M; Yao Y; Zhou C; Song Y Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds. J. Med. Chem 2015, 57, 6899–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng G; Shen J; Yin M; McManus J; Mathieu M; Gee P; He T; Shi C; Bedel O; McLean LR; Le-Strat F; Zhang Y; Marquette JP;Gao Q; Zhang B; Rak A; Hoffmann D; Rooney E; Vassort A; Englaro W; Li Y; Patel V; Adrian F; Gross S; Wiederschain D; Cheng H; Licht S Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by an allosteric small molecule. J. Biol. Chem 2015, 290, 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okoye-Okafor UC; Bartholdy B; Cartier J; Gao EN; Pietrak B; Rendina AR; Rominger C; Quinn C; Smallwood A; Wiggall KJ; Reif AJ; Schmidt SJ; Qi H; Zhao H; Joberty G; Faelth-Savitski M; Bantscheff M; Drewes G; Duraiswami C; Brady P; Groy A; Narayanagari SR; Antony-Debre I; Mitchell K; Wang HR; Kao YR; Christopeit M; Carvajal L; Barreyro L; Paietta E; Makishima H; Will B; Concha N; Adams ND; Schwartz B; McCabe MT; Maciejewski J; Verma A; Steidl U New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat. Chem. Biol 2015, 11, 878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Y; Wu F; Wu J Targeting histone methylation for cancer therapy: enzymes, inhibitors, biological activity and perspectives. J. Hematol. Oncol 2016, 9, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]