Abstract
In addition to T cells’ roles in immune response and autoimmune diseases, certain types of T cells, called regulatory T cells (Tregs), play important roles in microenvironment modulation for resolution and tissue regeneration. However, there are currently few options available other than introducing more Tregs or immunosuppressive drugs to locally enrich Tregs. Herein, poly(L-lactic acid) (PLLA) nanofibrous spongy microspheres (NF-SMS), PLLA/polyethylene glycol (PEG) co-functionalized mesoporous silica nanoparticles (MSN), and poly(lactic acid-co-glycolic acid) microspheres (PLGA MS) are integrated into one multi-biologic delivery vehicle for in situ Treg manipulation, where the MSNs and PLGA MS were utilized to distinctly release IL-2/TGF-β and miR-10a to locally recruit T cells and stimulate their differentiation into Tregs, while PLLA NF-SMS serve as an injectable scaffold for the adhesion and proliferation of these Tregs. In a mouse model of periodontitis, the injectable and biomoleculedelivering PLLA NF-SMS lead to Treg enrichment, expansion and Treg-mediated immune therapy against bone loss. This system can potentially be utilized in a wide variety of other immune and regenerative therapies.
Keywords: nanofiber, microcarrier, miRNA and protein delivery, regulatory t cells, bone
While the immune system plays a pivotal role in protecting the host by detecting and eliminating invading foreign pathogens, exacerbated immune response can lead to unregulated inflammation and cytokine activity, resulting in tissue damage or even host death.1 Tissue engineering is a promising field that develops biomaterial, biomolecule, and cell based therapies to regenerate lost or diseased tissue.2 However, when there is a lack of homeostasis or biocompatibility in a tissue engineering system, the host immune response could lead to unwanted outcomes. Therefore, immune-regulation should be an important consideration in the design of a tissue engineering system. Advanced biomaterials could be utilized to improve immunotherapy3,4 and regulate immune response.5
Regulatory T cells (Tregs), a small subpopulation of T cells, are a key regulator of the immune system in maintaining tolerance to self-antigens and reducing the symptoms of autoimmune disease by suppressing the activities of other immune cells.6 Accordingly, expansion, infusion, and modulation of Tregs have been proposed as a therapeutic approach to treating destructive inflammation and autoimmune diseases.7 However, traditional Treg-mediated immune therapy is hampered by a series of disadvantages, including in vitro expansion, infusion, high cost, and the side effects of immunosuppressive drugs. Interleukin 2 (IL-2) and transforming growth factor beta (TGF-β) are cytokines known to enhance Treg recruitment, proliferation, and differentiation.8,9 The co-administration of rapamycin with IL-2 and TGF-β has been shown to increase Treg numbers in vitro.10 However, there are significant concerns over the use of immunosuppressive drugs including rapamycin. For example, long-term rapamycin administration has been associated with metabolic, haematologic, and kidney dysfunctions and the onset of diabetes.11–13 Currently, there is no effective treatment capable of expanding and maintaining a high local number of Tregs over a long period of time without side effects.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs (~22 nucleotides long) that bind to partially complementary sites on target messenger RNAs to function as post-transcriptional repressors.14 miRNAs are actively pursued as candidates to treat various diseases, including cancer, metabolic, fibrotic and inflammatory diseases.15–19 miRNAs have also been successfully utilized to promote tissue regeneration in an animal model in our laboratory.20 However, no miRNA has been reported to induce Treg proliferation or differentiation. miR-10a has recently been found to be expressed in Tregs but not in naïve T cells, and is considered to be a Treg marker.21 We hypothesize that miR-10a can facilitate naïve T cells to differentiate to Tregs. We further hypothesize that the combined application of miR-10a and IL-2/TGF-β may synergistically recruit and highly efficiently induce naïve T cells to differentiate into Tregs. The traditional vectors for miRNA delivery often have low transfection efficiency, high toxicity or viral immunogenicity.22–24 Recently, our lab established a hyperbranched polymer (HP) polyplex system as a non-viral vector that can be encapsulated in microspheres to achieve long-term, sustained miRNA delivery.20 This non-viral vector also has high miRNA binding affinity, high transfection efficiency, and negligible cytotoxicity.20
In this study, we developed PLLA nanofibrous spongy microspheres (NF-SMS) to serve as an injectable scaffold to accommodate Tregs, endogenous stem/progenitor cells for regeneration, integrating mesoporous silica nanoparticles (MSN) to achieve a faster release of IL-2/TGF-β combination, and carrying controlled-release poly(lactic acid-co-glycolic acid) microspheres (PLGA MS) to release miR-10a/HP polyplexes over a longer time. The ultimate objectives of this Treg-mediated immune therapy are to increase the number of functional Tregs in situ and establish a local immune regulatory microenvironment. The NF-SMS have been previously demonstrated to be an advanced injectable cell microcarrier for tissue engineering and utilized to localize the release of biomolecules to minimize potential off-target side effects.25,26 Periodontitis is caused by an exacerbated host immune response against pathogens.27 There are not sufficient natural Tregs in vivo to counterbalance the progression of alveolar bone loss accelerated by periodontitis.28 Using a mouse periodontitis model, we tested the integrated, injectable scaffold/multi-biologic delivery system for the hypothesized role of miR-10a in Treg differentiation, synergistic effects between miR-10a and IL-2/TGF-β in Treg expansion, and ultimately rescuing alveolar bone loss in this exacerbated host immune response disease model.
RESULTS
Induction of mouse regulatory T cells using miR-10a/IL-2/TGF-β combination
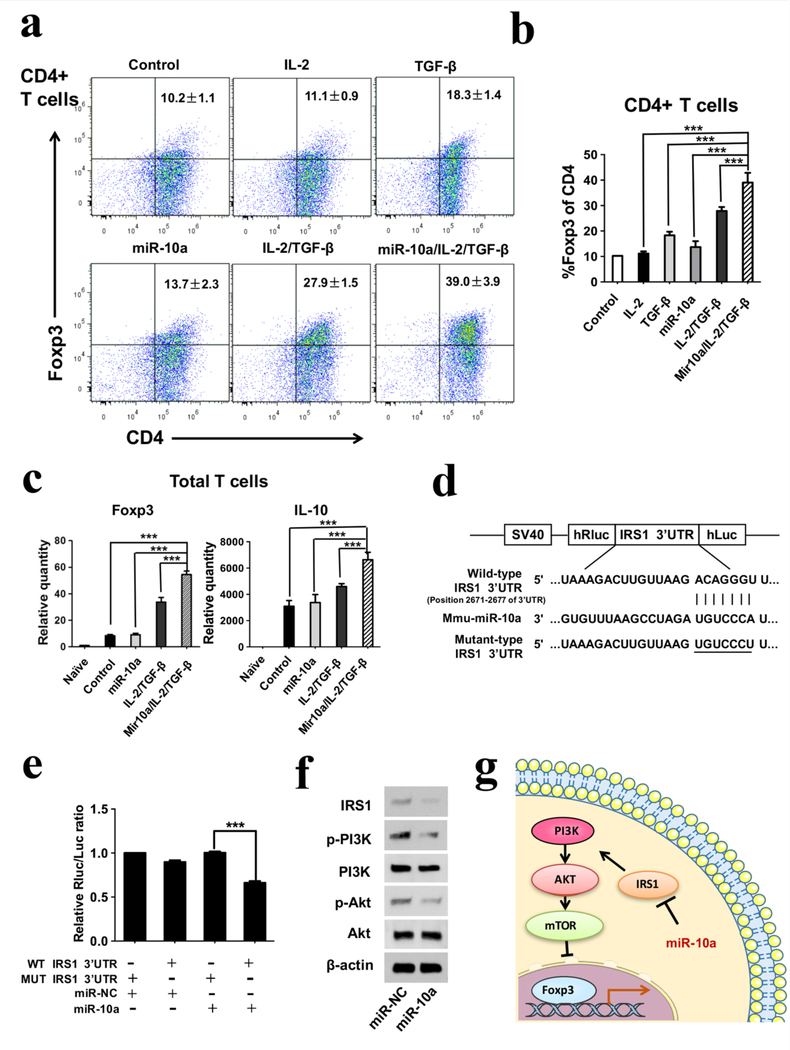
First, we tested whether the combination of miR-10a with IL-2/TGF-β could enhance the Treg differentiation over growth factors IL-2/TGF-β alone in vitro. Forkhead box p3 (Foxp3) is considered to be the master regulator in development and function of Tregs, and is recognized as a marker for Tregs.29 To examine Foxp3 expression, CD4+ T cells were isolated and cultured for 4 days, and then stained with CD4 and Foxp3 antibodies for flow cytometry analysis. The miR-10a and IL-2/TGF-β combination significantly enhanced Foxp3 expression compared to either miR10a or IL-2/TGF-β alone (Figure 1a & b), indicating a synergy of miR-10a and IL-2/TGF-β in enhancing Treg differentiation. In total, the percentage of Foxp3-positive cells increased from 10.2 ± 1.1% in the blank control to 39.0 ± 3.9% in the miR-10a/IL-2/TGF-β combination group. Next, we cultured total T cells with miR-10a/IL-2/TGF-β for 3 days, then measured the gene expression of the anti-inflammatory cytokine IL-10 and Foxp3 using real-time PCR. Both IL-10 and Foxp3 expressions were highly enhanced by the addition of miR-10a to IL-2/TGF-β (Figure 1c), indicating that the induced Tregs by miR-10a/IL-2/TGF-β combination were functional in immunomodulation to suppress inflammation.
Figure 1. Induction of regulatory T cells by miR-10a/IL-2/TGF-β and the mechanism of miR10a in Treg differentiation in vitro.
a, b) CD4+ T cells were isolated and cultured for 4 days, stained using CD4 and Foxp3 antibodies, and evaluated using flow cytometry analysis. The results showed that the miR-10a/IL-2/TGF-β group had the highest percentage of Foxp3 positive cells in CD4+ T cells. c) The gene expressions of anti-inflammatory cytokine IL-10 and Foxp3 in total T cells were highest in the miR-10a/IL-2/TGF-β group. d) The predicted miR-10a binding site within IRS1 3’UTR is shown. e) The suppression of luciferase activity by IRS1 3’UTR was dependent on miR-10a. (f) The expressions of IRS1, PI3K and Akt in total T cells were determined by Western blotting and were found to be inhibited by miR-10a. g) The schematic diagram describing the mechanism of miR-10a effect on Foxp3 expression and Treg differentiation.
To investigate the mechanism of action of miR-10a on T cells, we investigated the pathways of Treg differentiation. Since PI3K-Akt-mTOR is a major pathway for the inhibition of Treg differentiation, miR-10a could promote the differentiation of T cells into Tregs by inhibiting this pathway. To examine the action of miR-10a in this pathway, TargetScan Release 7.0 (http://www.targetscan.org/) was used to predict potential target genes of miR-10a. Among the putative targets, we focused on insulin receptor substrate 1 (IRS-1), which plays a key role in transmitting signals to the PI3K-Akt-mTOR pathway. To determine whether miR-10a directly targets IRS-1, wild-type (WT) IRS-1 3’UTR and mutant (MUT) IRS-1 3’UTR luciferase reporters were co-transfected with miR-NC (negative control) and miR-10a (Figure 1d). The WT IRS-1 3’UTR luciferase reporter activity was significantly inhibited by miR-10a treatment, while the MUT IRS-1 3’UTR was not (Figure 1e). In the presence of IL-2/TGF-β, total T cells were transfected with a miR-10a mimic or miR-NC. After 3 days, Western blots showed that miR-10a reduced IRS-1 and the phosphorylation of both PI3K and Akt (Fig. 1f). Taken together, these results provide evidence that miR-10a downregulates the PI3K-Akt-mTOR pathway via directly targeting IRS-1 (Figure 1g). To investigate the mechanism of IRS-1 in regulating Treg differentiation, the IRS-1-specific siRNA (siR-IRS-1) was used to silence IRS-1 expression in CD4+ T cells. The intracellular IRS-1 level was substantially downregulated by siR-IRS-1 compared with its siR-NC (Supplementary Figure S9a). In contrast, Foxp3 protein level was significantly upregulated in response to the reduced IRS-1 expression. Moreover, the CD4+ T cells were transfected with IRS-1 plasmid to overexpress IRS-1, the cells transfected with negative control plasmid were used as the control group. The Western blot results showed that Foxp3 protein level was downregulated by the transfection of IRS-1 overexpressing plasmid compared with the control group (Supplementary Figure S9b). The results indicate that miR-10a functionally targets IRS-1 to regulate Foxp3 expression.
Development of multi-functional nanofibrous spongy microspheres (NF-SMS)
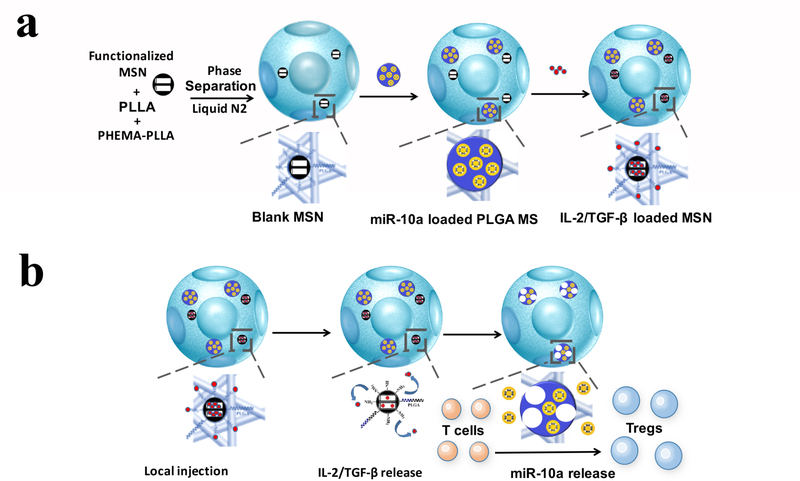
Since periodontal defects are irregular in shape and size, an injectable osteogenic scaffold would be ideal for the regeneration of alveolar bone loss in periodontitis. The high porosity and interpore connectivity in a scaffold are important for cell-seeding, migration, neovascularization and tissue regeneration,30,31 and NF morphology in a scaffold is osteogenic.32–37 We therefore selected the recently developed NF-SMS in our lab38 as the desired scaffold for alveolar bone regeneration in the periodontal disease model because of the NF morphology, interconnected pores and injectability. Due to the different physiological functions of miR-10a and IL-2/TGF-β, we rationalized that two distinct release rates for them would be beneficial for Treg expansion. We functionalized the NF-SMS with MSNs (Figure 2a) to relatively quickly relatively faster release the loaded two growth factors, IL-2 and TGF-β, to recruit naïve T cells and initiate the differentiation of the recruited cells into Tregs (Figure 2b). We also functionalized the NF-SMS with PLGA microspheres (Figure 2a) to release miR-10a/HP polyplexes over a longer period of time to further enhance Treg differentiation and the Treg phenotypic maturation during the entire transformation process from T cells to Tregs in the combined multi-functionalized NF-SMS.
Figure 2. Schematic illustration of the fabrication of multi-functionalized NF-SMS and their intended release and function.
a) Illustrative flow chart of fabricating multi-functionalized PLLA NF-SMS with MSN to incorporate growth factors and PLGA MS to incorporate microRNA/HP polyplexes. b) Schematic representation of the use of functionalized PLLA NF-SMS and their distinct releases of growth factors (IL-2/TGF-β) and miR-10a for T cells recruiting and transformation. The recruiting is achieved by release of grow factors loaded in MSN. The Treg transformation is achieved by both extracellular IL-2/TGF-β release and efficient intracellular miR-10a delivery.
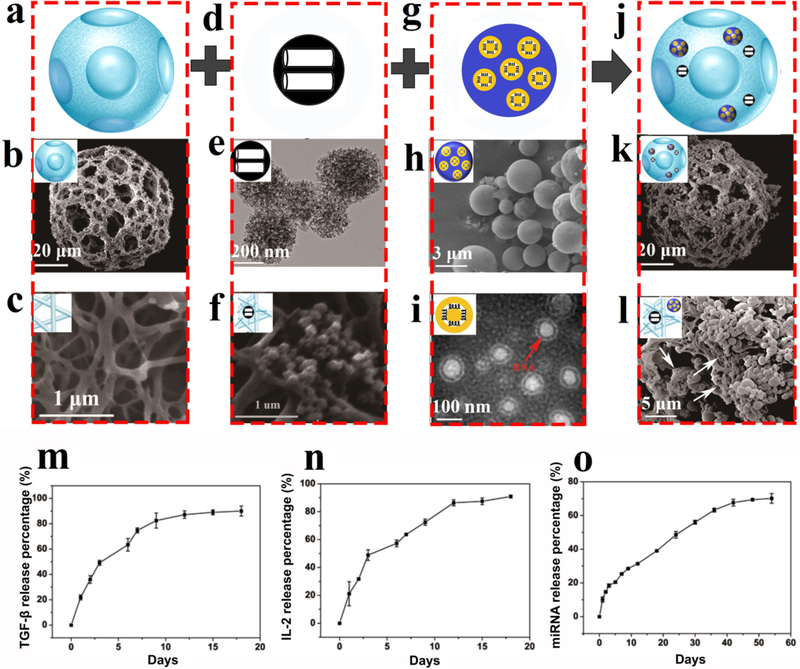
The injectable PLLA NF-SMS, which are approximately 60–90 μm in diameter with multiple pores at the single cell level in a nanofiber network, were fabricated according to our previously published protocol39 (Figure 3a-c). As nanocontainers for IL-2 and TGF-β, the MSN were intended to achieve a high loading capacity as well as a faster release rate to establish a broader distribution of these growth factors early in the process. MSNs were synthesized and extended to a larger size using 1, 3,5-trimethylbenzene (a pore expander) to incorporate IL-2 and TGF-β. The MSN were thoroughly characterized using dynamic light scattering (DLS), transmission electron microscopy (TEM), and N2 adsorption/desorption isotherms, demonstrating the spherical and mesoporous morphology of the MSN with a diameter of about 300 nm and a pore size of about 15 nm, which were loaded in the NF-SMS (Figure 3d-f, Supplementary Figure S1).
Figure 3. Characterization of multi-functionalized NF-SMS and the distinct release profiles of IL2/TGF-β and miR-10a.
a) The schematic image of a PLLA NF-SMS; b) An SEM image of a representative NF-SMS; c) A high-magnification image of the NF-SMS in b, showing the nanofibers with an average diameter of about 160 nm; d) the schematic and e) TEM image of MSN, showing the size of the MSN to be around 300 nm with small pores of about 15 nm; f) A highmagnification image of the MSN in e, showing the MSN attached on the PLLA nanofibers; g) An illustration of microRNA/HP polyplexes loaded in a PLGA microsphere. h) An SEM image of miR-10a/HP polyplexes loaded in PLGA microspheres; i) TEM image of microRNA encapsulated amino functionalized multi-armed cationic polymer polyplexes with a red arrow pointing to the miRNA (dark circle), showing the polyplexes to be about 100 nm in diameter sandwiched between inner and outer PEG layers; j) the schematic and k) The schematic and SEM image of MSN and PLGA particles immobilized on PLLA NF-SMS; l) A high-magnification image of MSN clusters (arrows) and PLGA MS on nanofibers; m-o) Time-dependent release kinetics of TGF-β (m), IL-2 (n) and miR-10a (o) for in vitro verification of their distinct release profiles. Each data is presented as the mean ± standard deviation (n = 3).
In order to achieve the controlled release of miR-10a, we synthesized an amino-functionalized multi-armed cationic polymer that self-assembles with miRNA to form advanced polyplexes for highly efficiently cell transfection.20 The chemical structure of this multi-armed cationic polymer was characterized using nuclear magnetic resonance (NMR) and Fourier transform infrared spectroscopy (FTIR) (Supplementary Figure S2), revealing successful synthesis of the polymer with short polyethylenimine (PEI) and defined polyethylene glycol (PEG) chains. The PLGA microspheres with a diameter of about 2 μm were prepared (Figure 3g&h) to encapsulate the miR10a-loaded polyplexes of about 100 nm in diameter (Figure 3i). The effective N/P (nitrogen in polymer/phosphate in nucleic acid) ratio for a cationic polymer to capture its target miRNA was examined using hydrodynamic diameter and zeta (ζ) potential measurements (Supplementary Figure S3), and a standard gel retardation assay (Supplementary Figure S4), demonstrating that substantial binding happens when the mass ratio between the cationic polymer and miRNA is larger than 1. Although the cationic surface facilitates the binding of a nanocarrier to the cell membrane via electrostatic interactions, a high positive charge may also impose a high cytotoxicity.40 Thus, we chose polymer/miRNA = 10:1 (ζ potential 12 mV) to compromise between binding efficiency and cytotoxicity for subsequent experiments. We transfected cationic polymer/Cy™3 dye-labeled miRNA-mimic polyplexes into total T cells. The red fluorescence shows that the cationic polymer can bind and transfer the miRNA into T cells (Supplementary Figure S5). Moreover, after the transfection of polymer/miR-10a polyplexes into total T cells for 1 day and 3 days, miR-10a gene expression in the culture was increased (Supplementary Figure S6).
The above two delivery systems were then immobilized on the injectable PLLA NF-SMS (Figure 3j&k) to form the multi-functional NF-SMS. In order to stably immobilize MSNs on the PLLA NF-SMS, the surface of the MSNs was modified using a PEG-graft-PLLA (PEG-PLLA) and an amino group taking advantage of the affinity of the PLLA block in the copolymer to PLLA in the NF-SMS as well as the affinity of the PEG block in the copolymer to the hydrophilic domains on MSN, which was confirmed using FTIR (Supplementary Figure S7). PLGA MSs loaded with HP/miRNA-polyplexes were immobilized on PLLA NF-SMS using a modified method previously developed in our laboratory.20 After immobilization, abundant PLGA microspheres and MSNs were found on the PLLA nanofiber pore surfaces (Figure 3l). To simplify the description of these multifunctional NF-SMS, we call them control NF-SMS (when no biomolecules are incorporated), miR10a NF-SMS, IL-2/TGF-β NF-SMS, or miR-10a/IL-2/TGF-β NF-SMS, respectively, without mentioning MSN and PLGA MS throughout the rest of this paper. The biocompatibility of NFSMS for T cells was tested by using Cell Counting Kit-8 (CCK-8) kit, after co-culturing NF-SMS and T cells for 3 days. The results show that the microspheres had no inhibitory effect on T cells, suggesting that the NF-SMS had no detectable level of cytotoxicity (Supplementary Figure S10). The IL-2/TGF-β loading (0.45 ug/mg for IL-2 and 0.87 ug/mg for TGF-β) and release profile of growth factors was quantified using an enzyme-linked immunosorbent assay (ELISA). There was a typical burst release during the first 4 days (50%), followed by a sustained release of 90% of the total loaded IL-2 and TGF-β by day 12 (Figure 3M & N). The miR-10a was loaded 0.0162 nmol/mg) and confirmed to be released relatively slowly, with about 90% released by day 50 by the measurement of absorbance at 260 nm (Figure 3O).
Induction of mouse Tregs by biomolecules released from NF-SMS in vitro
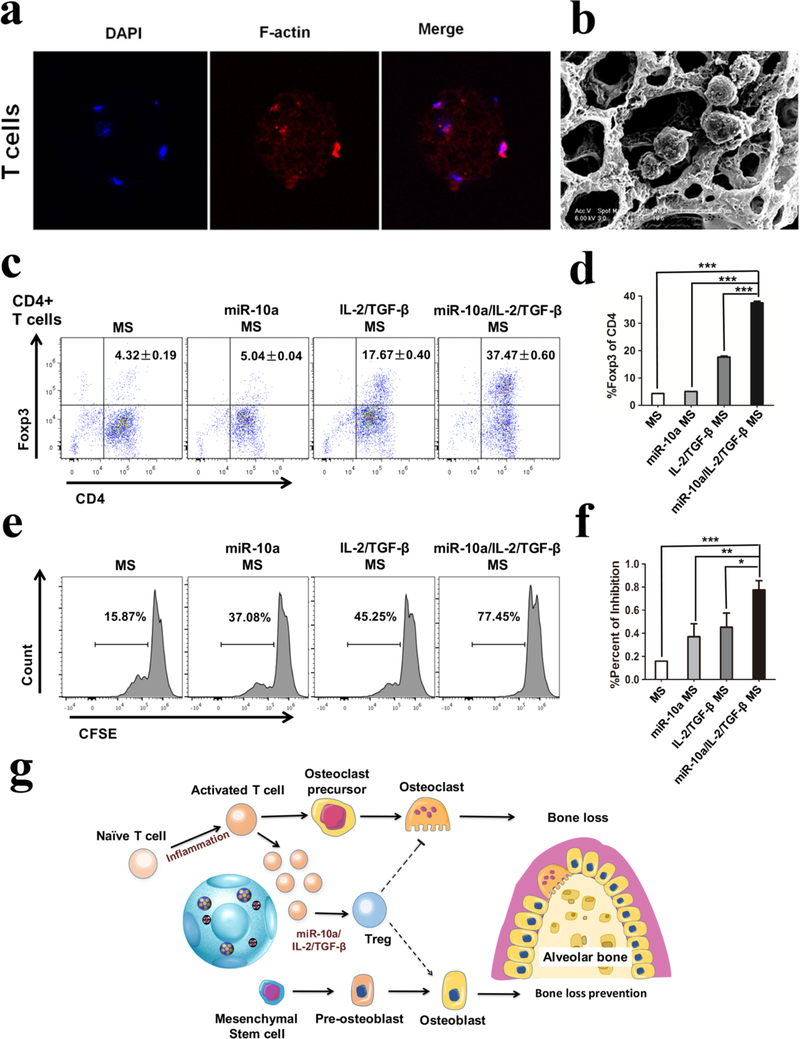
We tested the biocompatibility and carrying capacity of the NF-SMS for T cells. Total T cells were mixed with NF-SMS and cultured for 24h. The morphology of T cells in NF-SMS were observed using fluorescence confocal microscopy and SEM (Figure 4a & b). In order to validate whether our functionalized NF-SMS that sustain the release of the selected biomolecules could induce Treg differentiation, the naïve CD4+ T cells were cultured with miR-10a/IL-2/TGF-β NF-SMS at 37°C for 4 days, where the functionalized NF-SMS were separated from the CD4+ T cells by a permeable transwell insert. Flow cytometry analysis showed that the T cells could be induced to differentiate into Tregs by the released biomolecules in vitro, as measured with Foxp3 expression (Figure 4c and 4d). To further evaluate the function of the induced Tregs, they were cultured with naïve T cells. After 48 hours, the T cell suppression assay showed that these induced Tregs significantly inhibited the proliferation of the naïve T cells over control groups, demonstrating that the induced Tregs were functional (Figure 4e and 4f). These data support our hypothesis, showing that such IL-2/TGF-β release and miR-10a release synergistically facilitated the Treg differentiation, resulting in the generation of functional Treg, which is further hypothesized to suppress osteoclastogenesis and enhance osteoblast differentiation to rescue periodontitis-associated bone loss (Figure 4g), which will subsequently be tested in vivo.
Figure 4. Induction of Tregs by multi-functionalized PLLA NF-SMS in vitro.
a) T cells in multi-functionalized PLLA NF-SMS observed under confocal microscopy and b) SEM; c) and d) Induction of Tregs with different growth factor and miRNA combinations released from the functionalized NF-SMS in vitro; e) and f) Analysis of the induced Tregs showed that the cells could inhibit the proliferation of total T cells, where carboxyfluorescein diacetate succinimidyl ester (CFSE) is used to label generations of proliferating cells; g) Schematic illustration of our hypothesis that the release of IL-2/TGF-β and miR-10a from the multi-functionalized NF-SMS induces T cell transformation into Tregs and thereby rescues periodontal bone loss through Treg enhanced osteoblast activity and suppressed osteoclast activity.
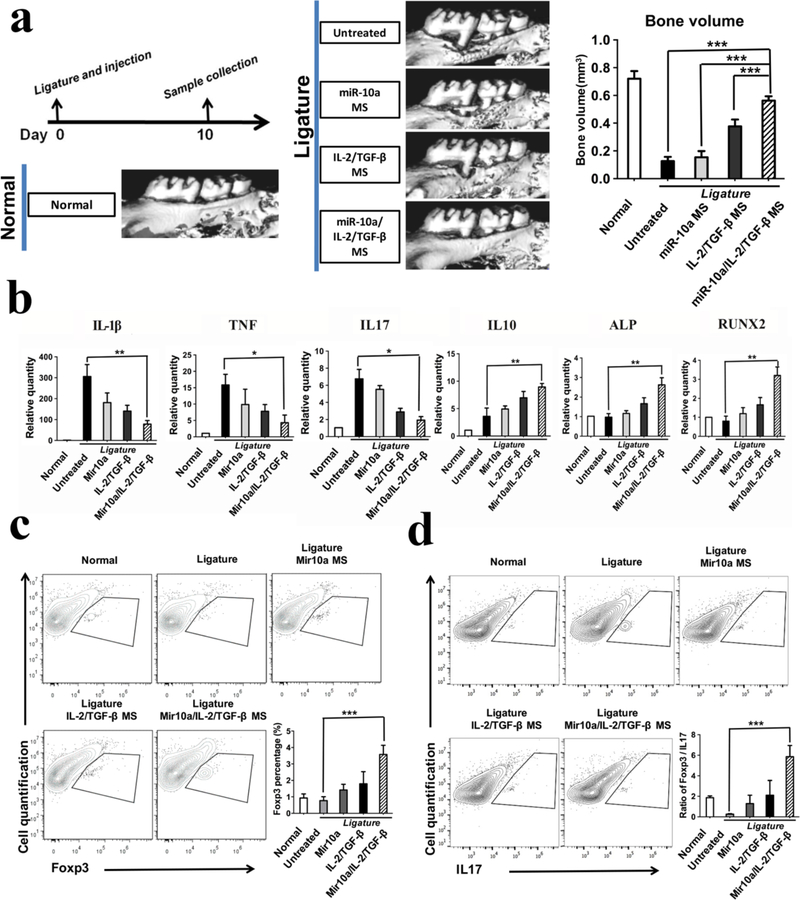
miR-10a/IL-2/TGF-β releasing NF-SMS rescuing bone resorption in a mouse periodontal disease model
To test our hypothesis and evaluate the effect of multi-functionalized NF-SMS on prevention of periodontitis-associated alveolar bone loss, we used a mouse periodontal disease model.41 Briefly, Silk sutures (5–0, Roboz Surgical Instrument Co. USA) were inserted between the first and second maxillary molars of mice with ends knotted for 10 days. Previous studies have shown that placement of the ligature between the molars resulted in a distinct increase in the number of total bacteria at day 10 41. Moreover, periodontal injury caused by silk ligature placement can mimic the complications induced by colonization of the oral cavity by harmful bacteria. The accumulation of dental plaque caused by ligature placement and invasion of pathogens into periodontal tissue can result in serious inflammation and exuberant host immune response that leads to bone loss 42. After ligature between the first and second maxillary molars, we injected 3μl functionalized NF-SMS into the gingival margin between these molars (Supplementary Figure S8) of 8-week-old C57BL/6 mice. Ten days later, microCT examination showed that the alveolar bone between the first and second molars was severely lost in the ligature without treatment group, indicating the establishment of a periodontal disease model (Figure 5a). The combined miR-10a/IL-2/TGF-β release from NF-SMS was significantly more effective than miR-10a release alone or IL-2/TGFβ release alone (also from NF-SMS) in rescuing bone resorption, indicating a synergistic effect between miR-10a and IL-2/TGF-β (Figure 5a). The results of the other four control groups, miR10a injection group without NF-SMS, IL-2/TGF-β injection group without NF-SMS, miR-10a/IL2/TGF-β injection group without NF-SMS, and NF-SMS injection group without biologics, show that there were no statistical differences in bone loss volume between these control groups and the untreated group (Supplementary Figure S12).
Figure 5. Multi-functionalized PLLA NF-SMS rescued bone resorption in a mouse periodontal disease model.
a) MicroCT results show bone loss between the first and second molars in the periodontitis model and the bone volume changes in various treatment groups; b) The gene expression of gingival tissues was quantified using real-time PCR, showing the effector T cell cytokines IL-1β, TNF, IL17, anti-inflammatory cytokine IL-10, and osteogenic markers ALP and RUNX2; c) The flow cytometry analysis of the isolated gingival tissues show the percentage of Foxp3+ cells (gated on CD4-expressing cells) in different treatment groups. d) The IL17+ cells (gated on CD4-expressing cells) and the ratios of Treg/Th17 cells of the isolated gingival tissues were shown.
The gene expression analysis of the isolated gingival tissues showed that the primary cytokines of effector T cells, IL-1β, IL6, IL8, IL17, TNF, IFN-γ and MCP-1 were upregulated in ligature without treatment group, and significantly decreased by the injection of the combined miR-10a/IL2/TGF-β releasing NF-SMS (Figure 5b and Supplementary Figure S11). In contrast, the expression of anti-inflammatory cytokine IL-10 and osteogenic markers, ALP and RUNX2 were significantly enhanced in the miR-10a/IL-2/TGF-β group in comparison to the miR-10a or IL-2/TGF-β alone groups (Figure 5b), suggesting that the effector T cell differentiation/activity was likely inhibited by the induced Tregs.
The percentage of CD4+ Foxp3+ or CD4+ IL17+ cells in gingival tissues was investigated by flow cytometry. The cells isolated from gingival tissues were stained against Anti-Mouse CD4 FITC, Anti-Mouse IL17 PE and Anti-Mouse Foxp3 APC (eBiosciences, USA). The percentage of CD4+ cells that also express Foxp3 (CD4+Foxp3+ cells) was significantly higher in the combined miR10a/IL-2/TGF-β release group compared with that untreated group. (Figure 5c). In contrast, the percentage of CD4+ IL17+ cells were lower in the miR-10a/IL-2/TGF-β group. Furthermore, the ratio of Treg/Th17 was downregulated in the untreated group and significantly upregulated in the miR-10a/IL-2/TGF-β group (Figure 5d).
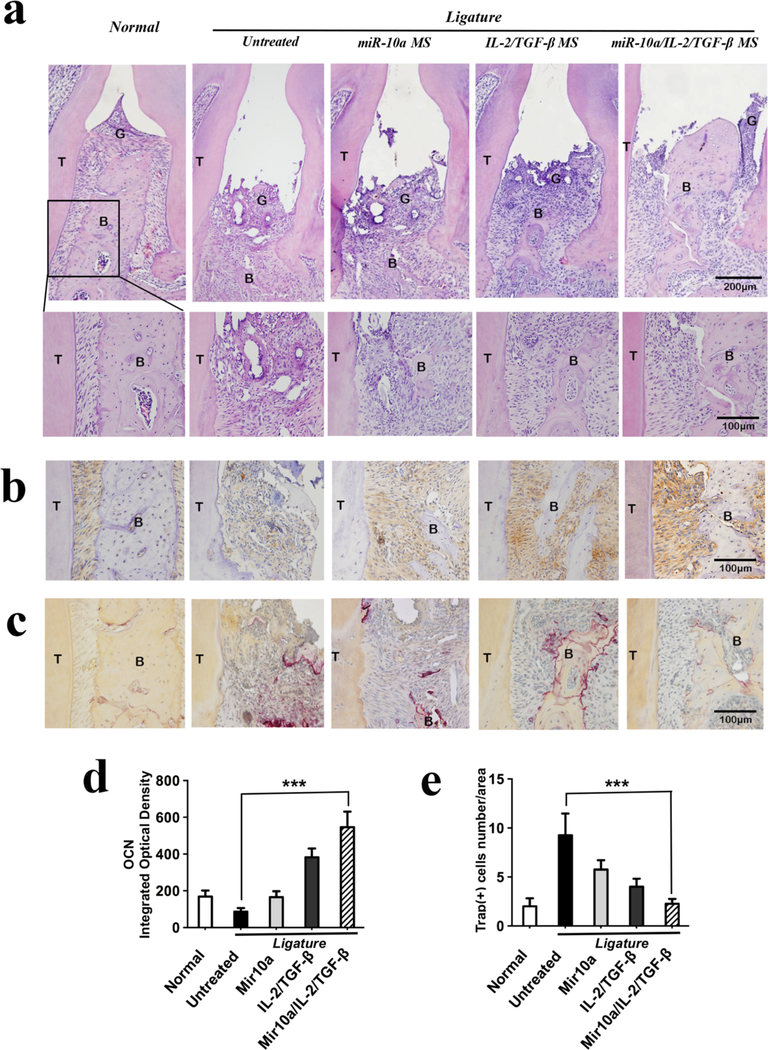
To further examine the bone loss and rescue outcome, the collected maxillae were stained using H&E (Figure 6a), OCN (Figure 6b&d) and TRAP (Figure 6c&e) methods. H&E staining showed that after ligation without treatment for 10 days, the alveolar bone between the two molars was severely lost. Only the combined miR-10a/IL-2/TGF-β release group could substantially rescue the bone loss, where the alveolar bone height was similar to that of the normal control group. The OCN immunostaining was clearly more positive in the combined miR-10a/IL-2/TGF-β release group than in untreated or other treated control groups. There were lower levels of OCN positive staining in miR-10a alone or IL-2/TGF-β alone groups. Moreover, the TRAP positive cell number was significantly decreased in miR-10a/IL-2/TGF-β release group than in the untreated group. Taken together, the higher number and possibly more mature Tregs in the combined miR-10a/IL-2/TGF-β treatment group substantially suppressed the destructive osteoclastogenesis and enhanced the osteoblastic activity, synergistically rescuing periodontal bone loss. There was gingival tissue loss in all ligation groups, which likely resulted from the ligation model but not the treatments.
Figure 6. Histological analysis of maxillae in mouse periodontal disease model.
a) H&E staining results show the alveolar bone areas between the first and second molars in different treatment groups after ligation for 10 days; b) OCN immunostaining was very positive in the combined miR-10a/IL-2/TGF-β release group compared to all the other groups. c) The TRAP staining indicated minimal osteoclasts in healthy maxillae and in the miR-10a/IL-2/TGF-β NF-SMS treated maxillae, while there were significant numbers of osteoclasts in the other treatment groups. d) Quantitative analysis of cells with positive OCN immunostaining using Image-Pro Plus 6.0 software. e) The quantified TRAP-positive cells. Labels: Bone area (B), Gingiva (G) and Tooth (T).
DISCUSSION
CD4+ T cells are crucial in regulating immune response to pathogens and maintaining immune balance. They are divided into conventional T helper cells and regulatory T cells (Tregs). T helper cells help the activity of other immune cells by releasing T cell cytokines and are associated with a variety of autoimmune diseases. Tregs play critical roles in preventing harmful immunopathological responses in multiple autoimmune diseases and in maintaining immunological homeostasis.6,43 Over the past a few years, a number of studies demonstrated that expansion of fully functional Tregs can prevent autoimmune diseases. Moreover, during the early stage of CD4+ T cell differentiation, the cell fate decision is heavily influenced by the local microenvironment, which ultimately pushes cells towards committing to different T cell lineages.44,45 However, there is limited technology to expand Tregs for controlling immune pathogenesis, especially in topical treatments of lesions.
This project aimed to develop a programmable delivery system that establishes a local microenvironment to enhance the differentiation of T cells into a sufficient quantity of induced Tregs. Growth factors, IL-2 and TGF-β, were loaded into MSNs and miR-10a/HP polyplexes were loaded into PLGA microspheres, and both were immobilized on an injectable NF-SMS. Foxp3 is reported to be an activation marker and a transcription factor for Tregs.46 IL-2 and TGF-β are two well-known cytokines in the immune system to recruit T cells and facilitate their proliferation and differentiation.8,9 The presence of TGF-β is essential for the expression of Foxp3 in naïve T cells.47 IL-2 is required for both function and proliferation of natural Tregs as well as the upregulation of Foxp3 expression in CD4+ T cells.48–50 The combined use of IL-2, TGF-β and rapamycin has been shown to induce Tregs in vitro.10 However, the use of the immunosuppressive rapamycin has been associated with metabolic, haematologic, and kidney dysfunctions and even the onset of diabetes.11–13 Therefore, novel and highly efficient Treg-expansion technologies without the side-effects of immunosuppressive drugs are highly desired.
miRNAs are endogenous small single-stranded non-coding RNAs, which function by directly binding to the untranslated 3’ region (3’UTR) of target mRNAs, leading to deactivation or degradation of the mRNAs.51 A variety of miRNAs have been investigated as novel therapeutics against human diseases because of their many advantages and few side effects.16 miRNAs have been found to be involved in the regulation of immune cell development and function, including fate determination, differentiation, lineage stability, plasticity, and immune homeostasis.52–54 However, application of miRNAs in immune disease treatment is still limited.16 miR-10a is highly expressed in natural Tregs but not in other T cells and attenuates the induction of Tregs into follicular helper T cells.21,55 We hypothesize that miR-10a is involved in Treg differentiation and the co-delivery of miR-10a and IL-2/TGF-β may synergistically induce Treg differentiation and maturation. In this study, the large pore size (15 nm) and availability of functional groups on MSNs allow our system to achieve high protein (IL-2 and TGF-β) loading capacity and the intended faster release to recruit T cells and initiate their early differentiation towards Tregs. Our non-viral vectors have high miRNA-binding affinity, low cytotoxicity, and high transfection efficiency.20 The use of PLGA microspheres allows us to achieve the desired longer-term delivery of miR-10a to synergistically enhance Treg differentiation and phenotypic maturation. Results show that miR10a can indeed promote Treg differentiation in vitro in the presence of IL-2 and TGF-β. This combined fast IL-2/TGF-β release and sustained miR-10a release system has also been validated in a mouse periodontitis model in this study, showing that miR-10a increases the number of Tregs in this inflammatory site. The target gene analysis reveals that miR-10a directly suppresses insulin receptor substrate 1 (IRS-1), which is involved in the PI3K pathway that inhibits Treg differentiation. Previous studies have shown that control of PI3K signaling is essential for maintaining Treg lineage homeostasis and stability.56 Moreover, periodontal disease has also been considered as a part of the insulin resistance syndrome and a complication of diabetes mellitus.57,58 Periodontitis can be associated with aggravation of insulin resistance.59 Insulin receptor substrate 1 (IRS-1) plays a critical role in insulin signaling, which transmits signals from the insulin to intracellular PI3K/Akt pathways.60 The results in this study showed that miR-10a directly suppresses IRS-1, which indicates that miR-10a can participate in the regulation of periodontal metabolic balance. Thus, this study has identified miR-10a to be an important regulator of T cell fate determination and Treg differentiation. Furthermore, we demonstrated the substantial therapeutic effect of miR-10a in the prevention of bone loss in a murine periodontal disease model by inducing Treg differentiation.
Periodontitis is a chronic inflammatory disease characterized by progressive destruction of gingival tissues, periodontal ligament, and alveolar bone. It is the most common bone pathology in humans. Furthermore, it is associated with cardiovascular disease, stroke, obesity, diabetes, and premature births.61 It has been demonstrated that periodontitis is caused by exuberant host immune response against periodontal pathogens.27 Inflammatory immune reactions in response to periodontal microorganisms are considered to be a defense mechanism of the host to protect against infections. However, the sustained release of inflammatory cytokines leads to gingival recession and alveolar bone loss. A series of studies has shown that accumulation of T cells in periodontal tissue is involved in the progression of periodontitis. Characteristic markers of Th1, Th2, Th17 and Treg subsets have been found in inflammatory periodontal tissues. Th1 and Th17 type cytokines are thought to up-regulate pro-inflammatory cytokines and receptor activator for nuclear factor kB ligand (RANKL) expression, which could induce osteoclastogenesis and alveolar bone loss in periodontitis.62 Tregs constitute a vital part of the process of immune regulation in periodontitis, ranging from infection to immunopathology and autoimmunity.
Tregs were identified in human periodontitis tissues by the expression of Foxp3, IL-10 and TGFβ, and were considered to affect the maintenance of periodontal health.63 The production of certain cytokines, such as IL-10 and TGF-β, can inhibit the differentiation of osteoclasts.64 In addition, Tregs play a role in the inhibition of RANKL in periodontitis tissues, and have an immunosuppressive function against osteoclast differentiation and bone loss.65,66 Previous studies showed that the inhibition of Tregs resulted in more severe alveolar bone resorption in a periodontitis animal model.28 Thus, Tregs were considered to have the potential to minimize tissue damage through modulating the host immune response and bone reconstruction in periodontitis.28,67 However, the methods of expanding Tregs to control destructive immune pathogenesis remain very limited, especially in topical applications. This work has developed a method to distinctively release two different categories of biomolecules (a miRNA and two growth factors) to locally enrich Tregs to rescue bone loss from the exacerbated destructive immune response in periodontitis. As an alternative strategy to current anti-inflammation treatments for periodontitis, we developed a technology to enrich the local Tregs in the periodontal tissues. We found that an environment rich in IL-2, TGF-β and miR-10a enhanced the recruitment and differentiation of Tregs. We further developed a distinctly controllable release of IL-2/TGF-β combination and a non-viral vector carried miR-10a from the injectable NF-SMS. The localized MSN on PLLA NF-SMS achieved the fast release of IL-2 and TGF-β with retained bioactivity likely due to the known confinement effect of the stable nano-sized pores in MSN, which supports the microenvironment necessary to recruit nearby T cells and facilitate their differentiation to Tregs. The in-situ conversion of conventional T cells into Tregs can be enhanced by the relatively longer-term release of miR10a/HP polyplexes from biodegradable PLGA microspheres (MS) on the NF-SMS and subsequent intracellular-delivery of the miR-10a. The polyplexes protect miRNAs from extracellular degradation on their journey into cells, enhance their endocytosis, and finally release miR-10a in the cytosol of target cells. These polyplexes are very stable, enable their encapsulation into biodegradable PLGA MS under rigorous sonication to achieve the controllable release of bioactive miR-10a for desired long durations. The injectable 3D delivery system effectively gathers the surrounding T cells and biologic release induces the cells to differentiate into Tregs. As a result, the injectable NF-SMS with programmable multi-biologic deliveries enhanced Treg expansion and triggered a Treg-mediated immune therapy against the bone loss in the periodontitis model.
CONCLUSIONS
We have developed an injectable, micro-porous and nanofibrous scaffold capable of efficiently delivering miR-10a non-virally combined with a growth factor combination (IL-2/TGF-β) with distinctly controlled releasing kinetics. The experimental data show that such released miRNA and growth factors can create a synergistic microenvironment to direct naïve T cells to differentiate into Tregs, supporting our hypothesis. Using a mouse periodontal disease model, we have demonstrated that this biologic delivery NF-SMS system is able to enrich Tregs in vivo, mediate immune homeostasis, suppress exacerbated host immune response under periodontal disease conditions, and thereby rescue alveolar bone loss. This advanced injectable scaffold with rationally designed multiple biologic delivery capacities may also be utilized for in situ expansion of Tregs to modulate microenvironment of stem/progenitor cells for the regeneration of other tissues or to treat various autoimmune diseases.
MATERIALS AND METHODS
Materials.
(3s)-cis-3,6-dimethyl-1,4-dioxane-2,5-dione (L-lactide) was purchased from SigmaAldrich Inc. (St. Louis, MO) and purified by recrystallization from toluene. Stannous 2ethylhexanoate (Sn(Oct)2) and 2-hydroxyethyl methacrylate (HEMA) were purchased from Sigma-Aldrich Inc. and distilled under reduced pressure before use. 1,4-Dioxane (99%) was purchased from Aldrich Chemical (Milwaukee, WI) and dehydrated with calcium chloride. 2,2’-Azoisobutyronitrile (AIBN) was purchased from Sigma-Aldrich Inc. and recrystallized from ethanol. PLLA (inherent viscosity 1.6 dL/g) was purchased from Boehringer Ingelheim (Ingelheim, Germany). Poly(ethylene glycol) monomethyl ether methacrylate (PEGMA), FITC labeled BSA, 3-aminopropyltriethoxysilane (APTS), 3-(Trimethoxysilyl)propyl methacrylate (TMSPMA), Ncetyltrimethylammonium bromide (CTAB), tetraethyl orthosilicate (TEOS), 1,3,5trimethylbenzene (TMB) and ammonium hydroxide, polyethyleneimine (PEI, Mw 800 Da and 25 kDa), poly(ethylene glycol) (PEG, Mw 2000, 5000, 10000, and 20000 Da), and hyperbranched bis-MPA polyester (H20: 16 hydroxyls, H30: 32 hydroxyls, and H40: 64 hydroxyls) were purchased from Sigma–Aldrich Inc. and used without further treatment.
Mesoporous silica nanoparticles
(MSN) with template (MSN-T) were prepared using basecatalyzed sol-gel method with CTAB as a template.68–70 Adding 52.8 mL ammonium hydroxide (29 wt% NH3 in water) to 1000 mL deionized water to achieve a pH value of approximately 11. After the temperature was raised to 323 K, 1.12 g CTAB and subsequently 5.8 mL TEOS were added under rapid stirring for 2 hours. After being aged overnight, the mixture was centrifuged and washed with distilled water and ethanol thoroughly. The obtained silica nanoparticles were sonicated for 30 min to be dispersed in ethanol. Then 20 mL mixture of water and TMB (v/v=1:1) was added and placed in an autoclave at 140°C for 4 days without stirring. The collected white powder was finally washed with ethanol and water five times each.
Modification of MSN with functional groups.
Amino groups and C=C double bonds functionalized MSN (NH2-MSN–C=C) were obtained by refluxing 1 g of the resultant MSN-T with 2 mL of APTES and 0.1 ml MPS in 240 mL of ethanol for 12 h, filtered, washed with copious ethanol, and dried at 45 °C under vacuum overnight.71
Polymer functionalization of MSN (PLLA-PEG-MSN-NH2).
Hydroxyethyl methacrylate-graftpoly(L-lactic acid) macromonomer (HEMA-g-PLLA) was synthesized using previously published methods.72 Briefly, 100 mg of NH2-MSN–C=C, 0.3 mL of as-synthesized HEMA-g-PLLA, 0.3 ml of PEGMA, and 7.5 mg of AIBN were successively added into 40 mL of acetonitrile. After 30 min of bubbling, the mixture was refluxed at 80°C for 2 h. The product was separated from the mixture by filtration and washing with copious ethanol. To remove structure-template CTAB and TMB, the product was subsequently refluxed in ammonium nitrate/ethanol solution (NH4NO3/C2H5OH, 10 mol/L) for 6 h. The mixture was filtered and dried at 45°C under vacuum for 12 h, yielding the final product of PLLA-PEG-MSN-NH2 as a white powder.
Fabrication of MSN functionalized nanofibrous spongy microspheres (MSN/NF-SMS).38,73
Poly(hydroxyethyl methacrylate)-graft-poly(L-lactic acid) (PHEMA-g-PLLA) were synthesized using a previously published method.72 As-synthesized PHEMA-g-PLLA, PLLA and PLLA-PEGMSN-NH2 (1:1:0.85 in mass ratio) were dissolved in tetrahydrofuran at 50°C with a concentration of 2.5% (wt/v). After sonication for 30 min and incubation under 50 °C for 30 min, the mixed solution was rapidly poured into glycerol (50 °C) with three times the volume of the mixed solution under rigorous stirring (speed 7, MAXIMA, Fisher Scientific). After 1 min, the mixture was quickly poured into liquid nitrogen and was kept there for 10 min. Then a water-ice mixture (1,000 ml) was added to exchange with solvent for 24 h. The obtained microspheres were sieved and washed with excessive distilled water six times to remove glycerol residue. The collected microspheres (NF-SMS) were then lyophilized for three days.
Preparation of polymer/miR-10a polyplexes and agarose gel retardation.
The hyperbranched polymer (HP) vector was synthesized following our previous publication.20 Different amounts of designed HP solutions (1.0 mg/mL) were added slowly to miRNA-10a solutions containing 60 pmol of miRNA-10a, which was incubated at room temperature for 30 min to form HP/miRNA10a polyplexes. 10 μL polyplex solution was mixed with 4 μL of 1× loading buffer, loaded onto 0.7% agarose gel containing GelRed, and electrophoresed with a Tris-acetate (TAE) running buffer (pH 8) at 80 V for 45 min. RNA bands were visualized using an UV (254 nm) illuminator and photographed using a BioSpectrum Imaging System (USA).
Particle size and zeta potential measurements.
The size of the polyplexes and their zeta potential were measured on a Beckman Coulter DelsaNano C Submicron Particle Size Analyzer at room temperature. The polyplexes at different polymer/RNA ratios were prepared by adding different volumes of RNA solution and polymer solution (1.0 mg/mL), then incubated at room temperature for 30 min. The polyplexes were then diluted with RNase-free water to 1.0 mL volume for measurements.
Preparation of polymer/miR-10a polyplexes-containing PLGA microspheres (MS).
PLGA microspheres (MS) encapsulating HP/miRNA polyplexes were prepared using a water-in-oil-inwater (w/o/w) double emulsion technique.20 Briefly, 30 mg of PLGA (molecular weight = 64k; LA/GA ratio = 50/50) was dissolved in 1 mL dichloromethane. 100 μL aqueous solution of polymer/miRNA polyplexes with the weight ratio of HP/miRNA at 10:1 containing 1.8 nmol of miR-10a (Ambion, USA) was added into the above solution and emulsified with a probe sonicator at 50W (Virsonic 100, Gardiner, NY). The resulting primary w/o emulsion was then poured into 10 mL PVA solution (1% wt/vol) under sonication at 90W to produce the double (w/o/w) emulsion, was magnetically stirred at room temperature to evaporate the solvents for 12 h. The PLGA microspheres were collected by centrifugation, followed by washing three times with water, and freeze-drying.
Preparation of PLGA MS and MSN loaded PLLA NF-SMS.
The PLGA MS containing HP/miR-10a polyplexes were incorporated onto the as-synthesized MSN/NF-SMS using a post seeding method.74, 75 Briefly, PLGA MS and MSN/NF-SMS were dispersed in hexane with 0.1% span 80 separately. Then the two suspensions were mixed together. After 2 h incubation under vacuum, the dried power was subjected to vapor of a mixed solvent of hexane/THF (9:1 by volume) for 30 min. Then the PLGA MS (with miR-10a or without miR-10a) and MSN dually loaded PLLA NF-SMS were dried under vacuum for 3 days to remove solvents.
Characterizations.
The morphology and size of MSNs with and without functionalization or growth factor loading were characterized using Philips CM200 transmission electron microscope operated at 200 kV. Samples were prepared by dropping 5 μL of sample suspension onto carboncoated copper grids. The TEM images were taken without any staining. The surface functionalization of MSN was characterized using KBr pellets and an AVATAR 320 FT-IR spectrometer. The FT-IR spectra were recorded on a Perkin Elmer BX spectrometer. The synthesized polymers were characterized by 1H NMR analysis using a Varian INOVA-400 spectrometer at room temperature. The morphology and size of the PLGA MS and PLLA NF-SMS before and after PLGA MS and MSN dual incorporation were observed under scanning electron microscopy (Philips XL30 FEG SEM) at an accelerating voltage of 15 kV. Before observation, the samples were coated with gold for 120 s using a sputter coater (DeskII, Denton vacuum Inc), with gas pressure at 50 mtorr and current at 40 mA.
miR-10a/IL-2/TGF-β in vitro release kinetics.
2 mg of NF-SMS containing 0.8 nmol miR-10a (preloaded with HP/miR-10a polyplex-containing PLGA MS) were incubated in 70% ethanol for 30 min and washed with RNase-free water three times. The NF-SMS were dispersed in 1 mL RNase-free PBS (pH 7.4, 0.1 M) and added with 1μg IL-2 (R&D Systems, USA) and 2μg TGF-β (PeproTech, USA). The mixture was incubated at room temperature under gentle shaking at 60 rpm. After 24h of incubation, the miR-10a/IL-2/TGF-β NF-SMS were washed with RNase-free PBS and centrifuged to discard the supernatant, and then immersed in 1 mL RNase-free PBS, kept at 37– on an orbital shaker with a speed of 60 rpm. At predetermined time intervals, 100 μL supernatant was collected and replaced with equivalent volumes of pre-warmed fresh RNase-free PBS for each sample. The released TGF-β, IL-2, and miR-10a were detected by using a human TGF-β enzyme-linked immunosorbent assay (ELISA) kit (Boster, China) human IL-2 ELISA kit (PeproTech, USA), and The QuantiFluor® RNA System (Promega, USA), respectively, following the manufacturer’s instructions. The concentration of the samples was determined using a Varioskan Flash multimode reader.
Isolation of total T cells and CD4+ T cells.
Spleens from 4–6-week-old female C57BL/6J mice (Charles River Laboratories, Wilmington, MA) were collected. Splenocytes were filtered through 70 μm nylon cell strainers. Following red blood cell lysis, total T cells and CD4+ T cells were purified from splenocytes by negative selection using the EasySep™ Mouse T Cell and CD4+ T Cell Isolation Kits (Stem Cell Technologies, Canada), following the manufacturer’s protocol. The purified total T cells and CD4+ T cells were cultured in RPMI medium supplemented with 10% FBS (Invitrogen, USA).
T cell culture.
The freshly purified CD4+ T cells and total T cells were activated with plate-bound anti-CD3 and soluble anti-CD28 antibodies (eBioscience, USA). Aqueous solution of a selected HP polymer (1.0 mg mL−1) was added slowly to miR-10a solution at the weight ratio of polymer/miRNA at 10:1, and the mixture was incubated at room temperature for 30 min before use. The cells were cultured in the presence of 20 ng/mL TGF-β1, 10 ng/mL IL-2 (BD Biosicences, USA), with or without 20 nmol miR-10a-5p (Ambion, USA). Four days later, the CD4+ T cells were analyzed using iCyt Synergy Flow sorter (Sony Biotechnology Inc, Japan). The total T cells were collected for gene expression analyses.
Transfection of polymer/miRNA into T cells.
Cy3™ Dye-Labeled Pre-miR Negative Control (Ambion, USA) and HP polymer solution (1.0 mg/mL) were mixed and incubated at room temperature for 30 min for HP/miRNA complex formation. The complexes were then added into the culture medium of total T cells. After 24h of incubation, the cells were fixed with 4% paraformaldehyde, treated with 0.1% Triton X-100 for 5 min, blocked with 2% bovine serum albumin (BSA) for 15 min, mounted with Vectashield Mounting medium containing DAPI (Vector Laboratories, USA), and imaged under a laser scanning confocal microscope (Nikon TS-100, Tokyo, Japan).
To further verify the transfection efficiency, total T cells were cultured with polymer/miR-10a complexes for 1 day and 3 days. After incubation, RNA of the cells was extracted, and miR-10a gene expression was measured using real-time PCR.
Co-culture T cells with IL-2/TGF-β, miR-10a and miR-10a/IL-2/TGF-β NF-SMS.
The control NF-SMS (with MSN and PLGA MS loaded but with neither IL-2/TGF-β nor miR-10a) were prewetted in the cell culture medium for 2 h. 1×106Naïve T cells were mixed with these NF-SMS in a ratio of 10:1 and cultured at 37 – on an orbital shaker for 24h. The morphology of T cells in NF-SMS were observed under fluorescence confocal microscope and SEM. Briefly, for F-actin staining, the cells were fixed with 4% paraformaldehyde, treated with 0.1% Triton X-100 for 5 min, blocked with 2% bovine serum albumin (BSA) for 15 min, incubated with Alexa Fluor 555 phalloidin (Life Technologies, USA) for 30 min, mounted with Vectashield Mounting medium containing DAPI (Vector Laboratories, USA), and observed under a laser scanning confocal microscope (Nikon TS-100, Tokyo, Japan). For SEM observation, the cells were fixed in 2.5% glutaraldehyde, treated in 1% osmium tetroxide for 1h, dehydrated in ethanol (50%, 75%, 90%, 95% and 100%) and hexamethyldisilizane (HMDS), coated with gold and observed under an SEM microscope (Philips XL30 FEG, Holland) at 10 kV.
CD4+ T cells were co-cultured with control NF-SMS, miR-10a NF-SMS, IL-2/TGF-β NF-SMS, or miR-10a/IL-2/TGF-β NF-SMS, respectively. The control or biomolecule-loaded NF-SMS were separated from the CD4+ T cells with a permeable transwell insert. The cells were activated with plate-bound anti-CD3 and soluble anti-CD28 antibodies (eBioscience, USA). Four days later, the cells were collected for flow cytometry analysis and T cell Suppression Assay.
Flow Cytometry.
Cell suspension was centrifuged at 4°C and the supernatant was discarded. The cells were resuspended in 500 μl of Fixation/Permeabilization Solution (Fixation/Permeabilization kit, eBiosciences, USA), kept at 4°C for 60 min, and washed with 1X Permeabilization Buffer. After being blocked with rat anti-mouse DC16/CD32 mouse Fc block (BD Biosciences, USA) at 4°C for 5 min, the cells were double-stained against Anti-Mouse CD4 FITC (eBiosciences, USA), Anti-Mouse/Rat Foxp3 PE (eBiosciences, USA), following the manufacturer’s instructions. To determine the percentage of Tregs and IL17 positive cells in gingival tissues, the isolated cells were stained against Anti-Mouse CD4 FITC (eBiosciences, USA), Anti-Mouse IL17 PE (eBiosciences, USA) and Anti-Mouse Foxp3 APC (eBiosciences, USA). Briefly, the cells were incubated with the antibodies in Flow Cytometry Staining Buffer in the dark at room temperature for 1 h. Then the cells were washed twice and the resuspended cells in Flow Cytometry Staining Buffer were analyzed using an iCyt Synergy Flow sorter (Sony Biotechnology Inc. Japan).
Luciferase reporter assay.
To determine whether miR-10a directly targets the 3’UTRs of IRS-1, IRS-1 mRNA 3’UTRs with the miR-10a-5p binding sequences were amplified using PCR with SacI and XhoI restriction endonucleases and ligated into the SacI and XhoI sites of the pmiR-RB-REPORT™ vector (Ribobio, China). miR-10a-5p mimics or corresponding No-target Control with the vector were co-transfected into 293T cells by using Lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions. After 48 h, the cells were assayed using DualLuciferase® Reporter Assay Kit (Promega, USA) following the manufacturer’s instructions. Luminescent signals were quantified by luminometer Veritas 9100–002 (Turner Biosystems instrument, USA), and each value from the hRluc luciferase construct was normalized with a hluc luciferase assay.
RNA extraction and real-time PCR.
Total RNA from cells or gingival tissues was extracted using RNeasy Mini Kit (Qiagen, USA). 2 μg total RNA was converted to cDNA using reverse transcription reagents (Taqman, Applied Biosystems, CA). Gene expression was analyzed using Real-time PCR, where a total volume of 30 μL Taqman Universal PCR Master Mix (Applied Biosystems), 2 μL cDNA and 1.5 μL pre-designed primers and probes (Applied Biosystems) were incubated for 10 min at 25–, 30 min reverse transcription at 48– and 5 min inactivation at 95–. Real-time PCR reactions were performed using an ABI Prism 7500 Real time PCR System (Applied Biosystems) using gene-specific primers–probe sets (Foxp3: Mm00475162_m1, IL-10: Mm00439614_m1, IL-1β: Mm00434228_m1, Tumor Necrosis Factor (TNF): Mm00443258_m1,IL-17: Mm00439618_m1, IL6: Mm00446190_m1, IL8: Mm00441263_m1, Interferon gamma (IFN-γ): Mm01168134_m1, Monocyte chemoattractant protein-1 (MCP-1): Mm00441242_m1, runt-related transcription factor 2 (Runx2): Mm00501584_m1, alkaline phosphatase (ALP): Mm00475834_m1 , miR-10a: Mm04238220_s1, and the housekeeping gene GAPDH: Mm00484668_m1 as an internal control).
Western blot analysis.
Total protein of cells was extracted using EpiQuik whole-cell extraction kit (Epigentek, USA). The protein was examined using an SDS-PAGE Electrophoresis System (Bio-Rad, USA). Briefly, 30-μg protein was separated on 4–15% Tris-HCl gels (Bio-Rad, USA) and transferred to polyvinylidene difluoride membranes (PVDF, Millipore, USA). The membranes were blocked in blocking buffer (5% non-fat dry milk in TBS-T buffer) for 1 hour at room temperature and then incubated with the following antibodies in 1:1000 dilutions at 4 °C overnight: IRS-1, PI3K, p-PI3K, Akt, p-Akt (Cell signaling, USA) and β-actin (Santa Cruz, USA) antibodies. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (1:10000, Santa Cruz, USA) was used for 1 h at room temperature based on the source of the corresponding primary antibody. The immunoblots were detected by a SuperSignal West Pico Chemiluminescent Substrate (Thermo, USA).
T cell Suppression Assay.
Purified T cells from mouse spleens were labelled with carboxyfluorescein diacetate succinimidyl ester (CFSE) using the CellTrace CFSE Cell Proliferation Kit (Molecular Probes, USA). The CFSE-labelled T cells were cultured with antiCD3/CD28-coated sulfate latex beads. After the addition of cells that were co-cultured with the 4 types of NF-SMS (control, miR-10a, IL-2/TGF-β, and miR-10a/IL-2/TGF-β NF-SMS) at a ratio of 10:1 for 3 days, the suppression of T cell proliferation was assayed using flow cytometry.
Mouse model of periodontitis.
All experimental procedures have been approved by the University of Michigan Committee on Use and Care of Laboratory Animals. Five to six-week-old female C57BL/6J mice (Charles River Laboratories International, Inc., USA) were divided into 9 groups of 6 mice each. One group was untreated as a normal control, and eight groups were ligatured as experimental periodontal disease model and treatment groups. Briefly, Silk sutures (5–0, Roboz Surgical Instrument Co. USA) were inserted between the first and second maxillary molars of mice, and selected NF-SMS were injected into the local area of periodontal tissues. The eight periodontitis groups were treated as following: periodontitis group (untreated, no NF-SMS injection); miR-10a injection group; IL-2/TGF-β injection group; miR-10a/IL-2/TGF-β injection group; NF-SMS injection group without biologics; miR-10a NF-SMS injection group; IL-2/TGFβ NF-SMS injection group; miR-10a/IL-2/TGF-β NF-SMS injection group. The sutures were placed between the molars for 10 days. Mice were anesthetized using CO2 inhalation, gingival tissues and maxillae were collected for flow cytometry, gene expression and histological analyses, respectively.
Micro CT analysis.
Formalin-fixed maxillae were transferred to 70% ethanol, analyzed using SCANCO CT-100 Micro-CT system at the University of Michigan School of Dentistry Micro-CT Core. The alveolar bone volume was collected and measured from the disto-buccal root of the first molar to the mesio-buccal and mesio-palatal root of the second molar. The X-ray generator was operated as follows: voxel size 15 mm, 70 kVp, 114 mA, 0.5mm AL filter and integration time 500 ms. Bone volume was analyzed using the manufacturer’s software, and a fixed global threshold of 28% (280 on a greyscale of 0–1,000) was used to identify bone areas.
Bone histological analysis.
For histological analysis, the formalin-fixed maxillae were decalcified with 10% EDTA for 2 weeks, dehydrated through a graded series of ethanol, embedded in paraffin, and sectioned at a thickness of 5 μm. These sections were stained with hematoxylin and eosin (H&E), OCN antibody, or tartrate-resistant acid phosphatase (TRAP). TRAP staining was performed using an Acid Phosphatase, Leukocyte (TRAP) kit (Sigma-Aldrich, USA) following the protocol from the manufacturer. TRAP-positive multinucleated cells labeled with dark purple staining in the cytoplasm were identified as osteoclasts.
Statistical analyses.
All numerical data are expressed as means ± SD. Statistical differences between groups were compared using one-way ANOVA (SPSS 13.0). Differences were considered statistically significant if P < 0.05.
Supplementary Material
Acknowledgments
Funding Sources
The authors would like to acknowledge the financial support from the US National Institutes of Health (R01DE015384, R01DE022327, R01HL114038 and R01HL136231: PXM) and the US National Science Foundation (NSF DMR-1206575: PXM). ZL was partially supported by a fellowship from the Peking University School of Stomatology.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on ACS Publications website at DOI: 10.1021/acsnano.xxxxxx
The structural characterization of mesoporous silica nanoparticles (MSN) including their morphology, size, and pore volume distributions before and after functionalization; chemical structure of the HP polymer; zeta potential, diameter, agarose gel electrophoresis, and T cell transfection of HP/miRNA polyplexes; chemical structural characterization of MSN before and after amino group modification; histological images of injected microspheres in gingival tissue; Western blot images of IRS-1 and Foxp3; biocompatibility of NF-SMS; gene expression levels of additional cytokines (IL6, IL8, IFN-γ and MCP-1) in response to different treatments; microCT images and bone loss volumes of additional control groups; methods used in the supplemental data.
REFERENCES
- (1).Kim KD; Zhao J; Auh S; Yang X; Du P; Tang H; Fu YX, Adaptive Immune Cells Temper Initial Innate Responses. Nat Med 2007, 13, 1248–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ma PX, Biomimetic Materials for Tissue Engineering. Adv Drug Delivery Rev 2008, 60, 184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Stephan SB; Taber AM; Jileaeva I; Pegues EP; Sentman CL; Stephan MT, Biopolymer Implants Enhance the Efficacy of Adoptive T-Cell Therapy. Nat Biotechnol 2015, 33, 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Perica K; Bieler JG; Schutz C; Varela JC; Douglass J; Skora A; Chiu YL; Oelke M; Kinzler K; Zhou S; Vogelstein B; Schneck JP, Enrichment and Expansion with Nanoscale Artificial Antigen Presenting Cells for Adoptive Immunotherapy. ACS Nano 2015, 9, 6861–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Liu Q; Chen X; Jia J; Zhang W; Yang T; Wang L; Ma G, Ph-Responsive Poly(D,LLactic-Co-Glycolic Acid) Nanoparticles with Rapid Antigen Release Behavior Promote Immune Response. ACS Nano 2015, 9, 4925–4938. [DOI] [PubMed] [Google Scholar]
- (6).Sakaguchi S; Sakaguchi N; Shimizu J; Yamazaki S; Sakihama T; Itoh M; Kuniyasu Y; Nomura T; Toda M; Takahashi T, Immunologic Tolerance Maintained by Cd25+ Cd4+ Regulatory T Cells: Their Common Role in Controlling Autoimmunity, Tumor Immunity, and Transplantation Tolerance. Immunol Rev 2001, 182, 18–32. [DOI] [PubMed] [Google Scholar]
- (7).Sakaguchi S; Yamaguchi T; Nomura T; Ono M, Regulatory T Cells and Immune Tolerance. Cell 2008, 133, 775–787. [DOI] [PubMed] [Google Scholar]
- (8).Liao W; Lin JX; Leonard WJ, Il-2 Family Cytokines: New Insights into the Complex Roles of Il-2 as a Broad Regulator of T Helper Cell Differentiation. Curr Opin Immunol 2011, 23, 598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Li MO; Sanjabi S; Flavell RA, Transforming Growth Factor-Beta Controls Development, Homeostasis, and Tolerance of T Cells by Regulatory T Cell-Dependent and -Independent Mechanisms. Immunity 2006, 25, 455–471. [DOI] [PubMed] [Google Scholar]
- (10).Jhunjhunwala S; Balmert SC; Raimondi G; Dons E; Nichols EE; Thomson AW; Little SR, Controlled Release Formulations of Il-2, Tgf-Beta1 and Rapamycin for the Induction of Regulatory T Cells. J Controlled Release 2012, 159, 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Stallone G; Infante B; Grandaliano G; Gesualdo L, Management of Side Effects of Sirolimus Therapy. Transplantation 2009, 87, S23–S26. [DOI] [PubMed] [Google Scholar]
- (12).Teutonico A; Schena PF; Di Paolo S, Glucose Metabolism in Renal Transplant Recipients: Effect of Calcineurin Inhibitor Withdrawal and Conversion to Sirolimus. J Am Soc Nephrol 2005, 16, 3128–3135. [DOI] [PubMed] [Google Scholar]
- (13).Johnston O; Rose CL; Webster AC; Gill JS, Sirolimus Is Associated with New-Onset Diabetes in Kidney Transplant Recipients. J Am Soc Nephrol 2008, 19, 1411–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chen K; Rajewsky N, The Evolution of Gene Regulation by Transcription Factors and Micrornas. Nat Rev Genet 2007, 8, 93–103. [DOI] [PubMed] [Google Scholar]
- (15).Ibrahim AF; Weirauch U; Thomas M; Grunweller A; Hartmann RK; Aigner A, Microrna Replacement Therapy for Mir-145 and Mir-33a Is Efficacious in a Model of Colon Carcinoma. Cancer Res 2011, 71, 5214–5224. [DOI] [PubMed] [Google Scholar]
- (16).van Rooij E; Purcell AL; Levin AA, Developing Microrna Therapeutics. Circ Res 2012, 110, 496–507. [DOI] [PubMed] [Google Scholar]
- (17).Thum T; Gross C; Fiedler J; Fischer T; Kissler S; Bussen M; Galuppo P; Just S; Rottbauer W; Frantz S; Castoldi M; Soutschek J; Koteliansky V; Rosenwald A; Basson MA; Licht JD; Pena JTR; Rouhanifard SH; Muckenthaler MU; Tuschl T, et al. , Microrna-21 Contributes to Myocardial Disease by Stimulating Map Kinase Signalling in Fibroblasts. Nature 2008, 456, 980–984. [DOI] [PubMed] [Google Scholar]
- (18).Najafi-Shoushtari SH; Kristo F; Li YX; Shioda T; Cohen DE; Gerszten RE; Naar AM, Microrna-33 and the Srebp Host Genes Cooperate to Control Cholesterol Homeostasis. Science 2010, 328, 1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Marquart TJ; Allen RM; Ory DS; Baldan A, Mir-33 Links Srebp-2 Induction to Repression of Sterol Transporters. Proc Natl Acad Sci U S A 2010, 107, 12228–12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhang X; Li Y; Chen YE; Chen J; Ma PX, Cell-Free 3d Scaffold with Two-Stage Delivery of Mirna-26a to Regenerate Critical-Sized Bone Defects. Nat Commun 2016, 7, 10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Jeker LT; Zhou X; Gershberg K; de Kouchkovsky D; Morar MM; Stadthagen G; Lund AH; Bluestone JA, Microrna 10a Marks Regulatory T Cells. PLoS One 2012, 7, e36684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kirtane AR; Panyam J, Polymer Nanoparticles: Weighing up Gene Delivery. Nat Nanotechnol 2013, 8, 805–6. [DOI] [PubMed] [Google Scholar]
- (23).Nayak S; Herzog RW, Progress and Prospects: Immune Responses to Viral Vectors. Gene Ther 2010, 17, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Choi KY; Silvestre OF; Huang X; Min KH; Howard GP; Hida N; Jin AJ; Carvajal N; Lee SW; Hong JI; Chen X, Versatile Rna Interference Nanoplatform for Systemic Delivery of Rnas. ACS Nano 2014, 8, 4559–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kuang R; Zhang Z; Jin X; Hu J; Gupte MJ; Ni L; Ma PX, Nanofibrous Spongy Microspheres Enhance Odontogenic Differentiation of Human Dental Pulp Stem Cells. Adv Healthcare Mater 2015, 4, 1993–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Feng G; Zhang Z; Dang M; Zhang X; Doleyres Y; Song Y; Chen D; Ma PX, Injectable Nanofibrous Spongy Microspheres for Nr4a1 Plasmid DNA Transfection to Reverse Fibrotic Degeneration and Support Disc Regeneration. Biomaterials 2017, 131, 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Graves DT; Li J; Cochran DL, Inflammation and Uncoupling as Mechanisms of Periodontal Bone Loss. J Dent Res 2011, 90, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Garlet GP; Cardoso CR; Mariano FS; Claudino M; de Assis GF; Campanelli AP; Avila-Campos MJ; Silva JS, Regulatory T Cells Attenuate Experimental Periodontitis Progression in Mice. J Clin Periodontol 2010, 37, 591–600. [DOI] [PubMed] [Google Scholar]
- (29).Ramsdell F; Ziegler SF, Foxp3 and Scurfy: How It All Began. Nat Rev Immunol 2014, 14, 343–349. [DOI] [PubMed] [Google Scholar]
- (30).Chen VJ; Ma PX, Nano-Fibrous Poly(L-Lactic Acid) Scaffolds with Interconnected Spherical Macropores. Biomaterials 2004, 25, 2065–2073. [DOI] [PubMed] [Google Scholar]
- (31).Wei G; Ma PX, Macroporous and Nanofibrous Polymer Scaffolds and Polymer/Bone-Like Apatite Composite Scaffolds Generated by Sugar Spheres. J Biomed Mater Res A 2006, 78, 306–315. [DOI] [PubMed] [Google Scholar]
- (32).Woo KM; Chen VJ; Ma PX, Nano-Fibrous Scaffolding Architecture Selectively Enhances Protein Adsorption Contributing to Cell Attachment. J Biomed Mater Res A 2003, 67, 531–537. [DOI] [PubMed] [Google Scholar]
- (33).Woo KM; Jun JH; Chen VJ; Seo J; Baek JH; Ryoo HM; Kim GS; Somerman MJ; Ma PX, Nano-Fibrous Scaffolding Promotes Osteoblast Differentiation and Biomineralization. Biomaterials 2007, 28, 335–343. [DOI] [PubMed] [Google Scholar]
- (34).Woo KM; Chen VJ; Jung HM; Kim TI; Shin HI; Baek JH; Ryoo HM; Ma PX, Comparative Evaluation of Nanofibrous Scaffolding for Bone Regeneration in Critical-Size Calvarial Defects. Tissue Eng Part A 2009, 15, 2155–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Smith LA; Liu X; Hu J; Ma PX, The Enhancement of Human Embryonic Stem Cell Osteogenic Differentiation with Nano-Fibrous Scaffolding. Biomaterials 2010, 31, 5526–5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Smith LA; Liu X; Hu J; Ma PX, The Influence of Three-Dimensional Nanofibrous Scaffolds on the Osteogenic Differentiation of Embryonic Stem Cells. Biomaterials 2009, 30, 2516–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chen VJ; Smith LA; Ma PX, Bone Regeneration on Computer-Designed Nano-Fibrous Scaffolds. Biomaterials 2006, 27, 3973–3979. [DOI] [PubMed] [Google Scholar]
- (38).Zhang Z; Marson RL; Ge Z; Glotzer SC; Ma PX, Simultaneous Nano- and Microscale Control of Nanofibrous Microspheres Self-Assembled from Star-Shaped Polymers. Adv Mater 2015, 27, 3947–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Zhang Z; Gupte MJ; Jin X; Ma PX, Injectable Peptide Decorated Functional Nanofibrous Hollow Microspheres to Direct Stem Cell Differentiation and Tissue Regeneration. Adv Funct Mater 2015, 25, 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).O’ Neill M; Guo J; Byrne C; Darcy R; O’ Driscoll C, Mechanistic Studies on the Uptake and Intracellular Trafficking of Novel Cyclodextrin Transfection Complexes by Intestinal Epithelial Cells. Int J Pharm 2011, 413, 174–183. [DOI] [PubMed] [Google Scholar]
- (41).Jiao Y; Darzi Y; Tawaratsumida K; Marchesan JT; Hasegawa M; Moon H; Chen GY; Nunez G; Giannobile WV; Raes J; Inohara N, Induction of Bone Loss by PathobiontMediated Nod1 Signaling in the Oral Cavity. Cell Host Microbe 2013, 13, 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Graves DT; Kang J; Andriankaja O; Wada K; Rossa C Jr., Animal Models to Study Host-Bacteria Interactions Involved in Periodontitis. Front Oral Biol 2012, 15, 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Maloy KJ; Powrie F, Regulatory T Cells in the Control of Immune Pathology. Nat Immunol 2001, 2, 816–822. [DOI] [PubMed] [Google Scholar]
- (44).Maldonado RA; Irvine DJ; Schreiber R; Glimcher LH, A Role for the Immunological Synapse in Lineage Commitment of Cd4 Lymphocytes. Nature 2004, 431, 527–532. [DOI] [PubMed] [Google Scholar]
- (45).O’Shea JJ; Paul WE, Mechanisms Underlying Lineage Commitment and Plasticity of Helper Cd4+ T Cells. Science 2010, 327, 1098–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Fontenot JD; Gavin MA; Rudensky AY, Foxp3 Programs the Development and Function of Cd4+Cd25+ Regulatory T Cells. Nat Immunol 2003, 4, 330–336. [DOI] [PubMed] [Google Scholar]
- (47).Selvaraj RK; Geiger TL, A Kinetic and Dynamic Aanalysis of Foxp3 Induced in T Cells by Tgf-Beta. J Immunol 2007, 178, 7667–7677. [DOI] [PubMed] [Google Scholar]
- (48).Aoki CA; Roifman CM; Lian ZX; Bowlus CL; Norman GL; Shoenfeld Y; Mackay IR; Gershwin ME, Il-2 Receptor Alpha Deficiency and Features of Primary Biliary Cirrhosis. J Autoimmun 2006, 27, 50–53. [DOI] [PubMed] [Google Scholar]
- (49).Frick SU; Domogalla MP; Baier G; Wurm FR; Mailaender V; Landfester K; Steinbrink K, Interleukin-2 Functionalized Nanocapsules for T Cell-Based Immunotherapy. ACS Nano 2016, 10, 9216–9226. [DOI] [PubMed] [Google Scholar]
- (50).Kanamori M; Nakatsukasa H; Okada M; Lu Q; Yoshimura A, Induced Regulatory T Cells: Their Development, Stability, and Applications. Trends Immunol 2016, 37, 803–811. [DOI] [PubMed] [Google Scholar]
- (51).Bartel DP, Micrornas: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- (52).Baltimore D; Boldin MP; O’Connell RM; Rao DS; Taganov KD, Micrornas: New Regulators of Immune Cell Development and Function. Nat Immunol 2008, 9, 839–845. [DOI] [PubMed] [Google Scholar]
- (53).Liston A; Lu LF; O’Carroll D; Tarakhovsky A; Rudensky AY, Dicer-Dependent Microrna Pathway Safeguards Regulatory T Cell Function. J Exp Med 2008, 205, 1993–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Zhou X; Jeker LT; Fife BT; Zhu S; Anderson MS; McManus MT; Bluestone JA, Selective Mirna Disruption in T Reg Cells Leads to Uncontrolled Autoimmunity. J Exp Med 2008, 205, 1983–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Takahashi H; Kanno T; Nakayamada S; Hirahara K; Sciume G; Muljo SA; Kuchen S; Casellas R; Wei L; Kanno Y; O’Shea JJ, Tgf-Beta and Retinoic Acid Induce the Microrna Mir-10a, Which Targets Bcl-6 and Constrains the Plasticity of Helper T Cells. Nat Immunol 2012, 13, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Huynh A; DuPage M; Priyadharshini B; Sage PT; Quiros J; Borges CM; Townamchai N; Gerriets VA; Rathmell JC; Sharpe AH; Bluestone JA; Turka LA, Control of Pi(3) Kinase in Treg Cells Maintains Homeostasis and Lineage Stability. Nat Immunol 2015, 16, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Demmer RT; Jacobs DR Jr.; Singh R; Zuk A; Rosenbaum M; Papapanou PN; Desvarieux M, Periodontal Bacteria and Prediabetes Prevalence in Origins: The Oral Infections, Glucose Intolerance, and Insulin Resistance Study. J Dent Res 2015, 94, 201S–211S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Li Y; Lu Z; Zhang X; Yu H; Kirkwood KL; Lopes-Virella MF; Huang Y, Metabolic Syndrome Exacerbates Inflammation and Bone Loss in Periodontitis. J Dent Res 2015, 94, 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Gurav AN, Periodontitis and Insulin Resistance: Casual or Causal Relationship? Diabetes Metab J 2012, 36, 404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Bhattacharya S; Dey D; Roy SS, Molecular Mechanism of Insulin Resistance. J Biosci 2007, 32, 405–413. [DOI] [PubMed] [Google Scholar]
- (61).Iacopino AM; Cutler CW, Pathophysiological Relationships between Periodontitis and Systemic Disease: Recent Concepts Involving Serum Lipids. J Periodontol 2000, 71, 1375–1384. [DOI] [PubMed] [Google Scholar]
- (62).Beklen A; Ainola M; Hukkanen M; Gurgan C; Sorsa T; Konttinen YT, Mmps, Il-1, and Tnf Are Regulated by Il-17 in Periodontitis. J Dent Res 2007, 86, 347–351. [DOI] [PubMed] [Google Scholar]
- (63).Nakajima T; Ueki-Maruyama K; Oda T; Ohsawa Y; Ito H; Seymour GJ; Yamazaki K, Regulatory T-Cells Infiltrate Periodontal Disease Tissues. J Dent Res 2005, 84, 639–643. [DOI] [PubMed] [Google Scholar]
- (64).Bozec A; Zaiss MM; Kagwiria R; Voll R; Rauh M; Chen Z; Mueller-Schmucker S; Kroczek RA; Heinzerling L; Moser M; Mellor AL; David JP; Schett G, T Cell Costimulation Molecules Cd80/86 Inhibit Osteoclast Differentiation by Inducing the Ido/Tryptophan Pathway. Sci Transl Med 2014, 6, 235ra60. [DOI] [PubMed] [Google Scholar]
- (65).Ernst CW; Lee JE; Nakanishi T; Karimbux NY; Rezende TM; Stashenko P; Seki M; Taubman MA; Kawai T, Diminished Forkhead Box P3/Cd25 Double-Positive T Regulatory Cells Are Associated with the Increased Nuclear Factor-Kappab Ligand (Rankl+) T Cells in Bone Resorption Lesion of Periodontal Disease. Clin Exp Immunol 2007, 148, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Bozec A; Zaiss MM, T Regulatory Cells in Bone Remodelling. Curr Osteoporos Rep 2017, 15, 121–125. [DOI] [PubMed] [Google Scholar]
- (67).Glowacki AJ; Yoshizawa S; Jhunjhunwala S; Vieira AE; Garlet GP; Sfeir C; Little SR, Prevention of Inflammation-Mediated Bone Loss in Murine and Canine Periodontal Disease Via Recruitment of Regulatory Lymphocytes. Proc Natl Acad Sci U S A 2013, 110, 18525–18530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Vallet-Regi M; Ramila A; del Real RP; Perez-Pariente J, A New Property of Mcm-41: Drug Delivery System. Chemistry of Materials 2001, 13, 308–311. [Google Scholar]
- (69).Kim MH; Na HK; Kim YK; Ryoo SR; Cho HS; Lee KE; Jeon H; Ryoo R; Min DH, Facile Synthesis of Monodispersed Mesoporous Silica Nanoparticles with Ultralarge Pores and Their Application in Gene Delivery. Acs Nano 2011, 5, 3568–3576. [DOI] [PubMed] [Google Scholar]
- (70).Chen X; Soeriyadi AH; Lu X; Sagnella SM; Kavallaris M; Gooding JJ, Dual Bioresponsive Mesoporous Silica Nanocarrier as an “and” Logic Gate for Targeted Drug Delivery Cancer Cells. Adv Funct Mater 2014, 24, 6999–7006. [Google Scholar]
- (71).Bernardos A; Mondragon L; Aznar E; Marcos MD; Martinez-Manez R; Sancenon F; Soto J; Barat JM; Perez-Paya E; Guillem C; Amoros P, Enzyme-Responsive Intracellular Controlled Release Using Nanometric Silica Mesoporous Supports Capped with “Saccharides”. ACS Nano 2010, 4, 6353–6368. [DOI] [PubMed] [Google Scholar]
- (72).Liu XH; Ma PX, The Nanofibrous Architecture of Poly(L-Lactic Acid)-Based Functional Copolymers. Biomaterials 2010, 31, 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Zhang ZP; Gupte MJ; Jin XB; Ma PX, Injectable Peptide Decorated Functional Nanofibrous Hollow Microspheres to Direct Stem Cell Differentiation and Tissue Regeneration. Adv Funct Mater 2015, 25, 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Wei G; Jin Q; Giannobile WV; Ma PX, The Enhancement of Osteogenesis by NanoFibrous Scaffolds Incorporating rhBMP-7 Nanospheres. Biomaterials 2007, 28, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Wei G; Jin Q; Giannobile WV; Ma PX, Nano-Fibrous Scaffold for Controlled Delivery of Recombinant Human PDGF-BB. J Controlled Release 2006, 112, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.