Abstract
Background
Genome-wide association studies (GWAS) have identified multiple loci associated with coronary artery disease (CAD) and myocardial infarction (MI), but only a few of these loci are current targets for on-market medications. To identify drugs suitable for repurposing and their targets, we created two unique pipelines integrating public data on 49 CAD/MI-GWAS loci, drug-gene interactions, side effects and chemical interactions.
Methods
We first used publicly available GWAS results on all phenotypes to predict relevant side effects, identified drug-gene interactions, and prioritized candidates for repurposing among existing drugs. Secondly, we prioritized gene product targets by calculating a druggability score to estimate how accessible pockets of CAD/MI associated gene products are, then used again the GWAS results to predict side effects, excluded loci with widespread cross-tissue expression to avoid housekeeping and genes involved in vital processes and accordingly ranked the remaining gene products.
Results
These pipelines ultimately led to three suggestions for drug repurposing: pentolinium, adenosine triphosphate and riociguat (to target CHRNB4, ACSS2 and GUCY1A3, respectively); and three proteins for drug development: LMOD1, HIP1 and PPP2R3A. Most current therapies for CAD/MI treatment were also “rediscovered”.
Conclusions
Integration of genomic and pharmacological data may prove beneficial for drug repurposing and development, as evidence from our pipelines suggests.
Keywords: coronary artery disease, myocardial infarction, drug interactions, pharmacogenetics, Genome Wide Association Study
Journal Subject Terms: Coronary Artery Disease, Functional Genomics, Genetic, Association Studies, Pharmacology, Myocardial Infarction
Introduction
Coronary artery disease (CAD) is a major cause of death worldwide, leading to a yearly estimated 8.5 million cases of myocardial infarction (MI) 1 and loss of an expected ~150 million disability-adjusted life years globally in 2020 2. Current therapeutics for prevention of CAD mainly comprise the control of risk factors, e.g. the prescription of HMG-CoA reductase inhibitors, known as statins, or PCSK9 inhibitors, to reduce low-density cholesterol (LDL-C) 3,4, 5. More recently, the CANTOS study has shown that also non-lipid pathways, such as inflammatory processes, also influence atherothrombotic development 6, 7. In addition, platelet inhibition may be used for prevention of coronary events in certain, high-risk patient groups.
The discrepancy between the overwhelming clinical need and the small number of agents used in the preventive treatment of CAD and MI is largely explained by a high attrition in drug development, which is mostly attributable to unacceptable side effects and/or lack of efficacy 8. Currently, it is estimated that only one in every 5000 new drug compounds makes it to market 9. Furthermore, this process may take 10–15 years and costs billions of dollars for conducting clinical trials to clear the stringent requirements set by health agencies around the world 10, 11. Drug development has therefore become an expensive and difficult process, hindering the clinical implementation of potentially beneficial new drugs. Novel approaches to support drug development have emerged in recent years based on genetic strategies. For example, one may now conduct in-silico druggability analyses on genetic data, using bioinformatics tools, in order to identify approved and already marketed drugs for treating a new phenotype other than the one the drug was originally developed for. This strategy is referred to as drug repositioning or repurposing, an approach proposed and improved in the past 15 years 12, 13, based on new discoveries including, more recently, genetic information 14, 15. In such case, where an existing drug targets a gene product or pathway of a disease different from the original indication, fewer clinical trials may need to be conducted to alter the label and indicate a treatment for another disease as safety has already been demonstrated. An example of repurposing is sildenafil, initially produced with the expectation of reducing angina, and later found to be effective to treat erectile dysfunction 16 and pulmonary hypertension 17, leading to the subsequent releases of Viagra® in 1998 and Revatio® in 2005 18. Other successful examples of repurposing include gemfibrozil, duloxetine, dapoxetine and thalidomide 19 (original indications, repurposed indications and evidence for repurposing available in Supplemental Table 1).
Genome wide association studies (GWAS) have identified multiple independent loci that contribute to the genetic susceptibility of CAD/MI 20–23. Many of these loci include genes involved in diverse and currently unexplored biological mechanisms. Thereby these loci represent novel drug targets for treatment and prevention of CAD/MI. A key challenge is to prioritize GWAS hits and their products for pharmacological intervention. In this process, bioinformatics methods may yield novel insights into the potential “druggability” 24 of each of these loci in order to translate genetic knowledge into clinical care.
When assessing the druggability of a GWAS hit, several factors need consideration. A particular gene may not be “druggable”, which means that developing a molecule to target this gene product is not feasible, due to the lack of a defined drug binding pocket (known as a pharmacophore), or the druggability cannot be assessed due to unavailable relevant protein structural information. Although a target may be druggable, it still may not be suitable for clinical exploration, as immediate toxicity issues, buffering effects, redundancy, robustness and possible undesired pleiotropic effects in downstream biological pathways need to be clarified. For example, inhibition of the cardiac expressed HERG gene causes severe QT-interval prolongation, which is now screened as a liability in all drug discovery programs 25. Other adverse events may be more subtle, for example genetic variability in HMGCR has recently been identified as a risk factor for type 2 diabetes (T2D), which partially explains the relationship between statin use and risk of developing T2D 26,27, 28. Today, GWAS have found associations between thousands of loci and hundreds of phenotypes, thereby enabling a robust exploration of possible pleiotropic effects for any given locus or SNP 29. Here, we present two unique pipelines integrating currently available public data on GWAS, drug-gene interactions, side effects and chemical interactions. The first pipeline aims to identify approved drugs that may be suitable for repurposing for treatment of CAD/MI, while the second pipeline ranks non-targeted genes for their suitability to be a target for development of new drugs. The pipelines make use of numerous sources of information made available publicly in the past few years.
Methods
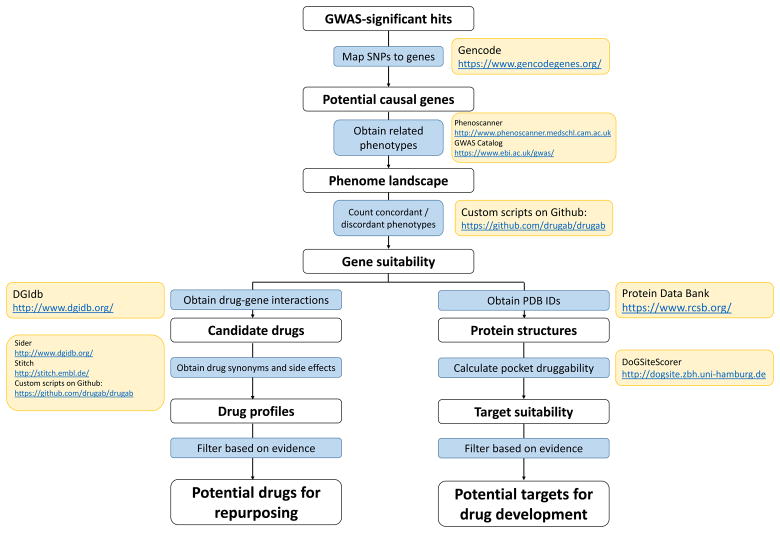
A fluxogram of the pipeline developed in this work is presented in Figure 1. Custom algorithms created in this project are available on GitHub (https://github.com/drugab/drugab). Methods are available in the Supplemental Material. This study does not make use of identifiable data, only public, summary-level data.
Figure 1.
Complete pipeline presented in the paper, with resources used for building up the pipeline. Codes that are not Unix commands were uploaded to Github and can be accessed at https://github.com/drugab/drugab.
Results
Candidates for repurposing
All 153 SNP regions, selected for their association with CAD/MI, queried on PhenoScanner had a nominally significant association with another phenotype, 68 of which with a positive score of associations in the same direction as for CAD/MI, i.e., the risk of developing these phenotypes can be decreased together with CAD/MI in case of targeting given gene products. We filtered the list further by using DGIdb 30 to identify existing medications targeting the gene products of these regions, and found drugs targeting 15 of these gene products (ABO, ACSS2, ARVCF, CDKN1A, CHRNB4, CKM, GUCY1A3, HDAC9, IL6R, LPL, MAP4, MTAP, PCSK9, SCARB1 and SLC22A4). Due to gene products affected by multiple drugs, the targeted genes showed interactions with 48 drugs. Further analysis of the results showed that 22 of them are not clinically available drugs, either due to halted development (N=15) or still under evaluation in clinical trials (N=7). After excluding these, 26 medicinal products mapping to 15 genes of interest remain.
Out of the 26 products on our list, three are not classified as drugs (L-carnitine, phosphatidylserine and adenine), leaving 23 marketed drugs that have 26 assigned Anatomical Therapeutic Chemical (ATC) codes in total. The most common ATC groups involved were cardiovascular (group C - 8 drugs), cancer (group L - 7 drugs) and nervous system (group N - 6 drugs). Table 1 presents drug-gene interactions and ATC codes for these drugs.
Table 1.
Predicted drug-gene interactions by DGIdb, ATC codes and nominally significant loci in the same LD block of the SNP presented
| Reported SNP | Gene | Interacting drug | ATC code 1 | ATC code 2 | ATC code 3 | GWAS predicted concordant phenotypes | GWAS predicted discordant phenotypes |
|---|---|---|---|---|---|---|---|
| rs11206510 | PCSK9 | EVOLOCUMAB | C10AX13 | LDL cholesterol, Total cholesterol, Triglycerides, Obesity class 2 | - | ||
| rs11206510 | PCSK9 | ALIROCUMAB | C10AX14 | LDL cholesterol, Total cholesterol, Triglycerides, Obesity class 2 | - | ||
| rs6088638 | ACSS2 | ADENOSINE TRIPHOSPHATE | C01EB10 | Triglycerides, Obesity class 2, Obesity class 1, BMI | - | ||
| rs264 | LPL | ORLISTAT | A08AB01 | Triglycerides, Type II diabetes, Obesity class 2, BMI | - | ||
| rs264 | LPL | CLOFIBRATE | C10AB01 | Triglycerides, Type II diabetes, Obesity class 2, BMI | - | ||
| rs264 | LPL | GEMFIBROZIL | C10AB04 | Triglycerides, Type II diabetes, Obesity class 2, BMI | - | ||
| rs4845625 | IL6R | TOCILIZUMAB | L04AC07 | LDL | - | ||
| rs1034565 | ARVCF | RISPERIDONE | N05AX08 | Type II diabetes, BMI | - | ||
| rs1034565 | ARVCF | BUPROPION | N06AX12 | Type II diabetes, BMI | - | ||
| rs11072794 | CHRNB4 | VARENICLINE | N07BA03 | Type II diabetes | - | ||
| rs11072794 | CHRNB4 | PENTOLINIUM | C02xxxx | Type II diabetes | - | ||
| rs11072794 | CHRNB4 | DEXTROMETHORPHAN | N07XX59 | R05DA09 | Type II diabetes | - | |
| rs11072794 | CHRNB4 | ETHANOL | V03AB16 | V03AZ01 | D08AX08 | Type II diabetes | - |
| rs11072794 | CHRNB4 | NICOTINE | N07BA01 | Type II diabetes | - | ||
| rs8111989 | CKM | CREATINE | C01EB06 | BMI | - | ||
| rs2023938 | HDAC9 | VORINOSTAT | L01XX38 | - | - | ||
| rs2023938 | HDAC9 | BELINOSTAT | L01XX49 | - | - | ||
| rs2023938 | HDAC9 | VALPROIC ACID | N03AG01 | - | - | ||
| rs2023938 | HDAC9 | PANOBINOSTAT | L01XX42 | - | - | ||
| rs2023938 | HDAC9 | ROMIDEPSIN | L01XX39 | - | - | ||
| rs7692387 | GUCY1A3 | RIOCIGUAT | C02KX05 | - | - | ||
| rs7642590 | MAP4 | PACLITAXEL | L01CD01 | Triglycerides | BMI | ||
| rs7642590 | MAP4 | DOCETAXEL | L01CD02 | Triglycerides | BMI | ||
| rs273909 | SLC22A4 | L-CARNITINE | A16AA01 | LDL, Total cholesterol, Triglycerides |
We determined side effects of the 23 drugs using different databases, in particular SIDER; accordingly, we suggested possible candidates for repurposing. We excluded chemotherapy compounds (N=6), based on their serious side effect profile and unsuitability for continuous use in cardiovascular indications. Further exclusions involved medications which are already marketed for CAD prevention (N=2), expected to cause tachycardia (N=4), MI (N=1), liver injury (N=3), kidney damage (N=1) and stroke (N=1). Full counts of indications and side effects are available in Supplemental Tables 2 and 3, respectively. Out of the remaining medications (N=5), we suggest three possible candidates for repurposing based on their positive impact on cardiovascular system seen in previous clinical trials. These include pentolinium (predicted as antagonist of the gene product of CHRNB4), adenosine triphosphate (targeting the gene product of ACSS2) and riociguat (antianginal agent works as a stimulator for the product of GUCY1A3).
“Re-discovery” of existing CAD drugs
After all steps of the pipeline, we identified drugs that are already prescribed for the treatment of CAD: simvastatin (representing statins), evolocumbad and alirocumab (representing PCSK9 inhibitors), irbesartan (representing angiotensin II receptor blockers) and gemfibrozil (a cholesterol-lowering agent more recently adjuvant in CAD treatment). They serve as positive control for the pipeline.
Druggability of docking pockets
We also investigated druggability of CAD/MI loci, by analyzing the chemical structures of the respective gene products in search of pockets suitable for docking with novel molecules. We obtained PDB structures for 60 out of the 153 proteins produced by the estimated genes these CAD/MI loci belong to. Six of those were not human, but from a homologous animal model, and we decided to keep them for the analyses. Fifty-two structures had good druggability scores (>=0.5), and the eight remaining structures with low druggability scores were excluded from further analyses. Thirty-seven structures of the 52 remaining are not targeted by drugs available in the market, according to DGIdb. The statement of the original DoGSiteScorer manuscript about threshold for druggability were confirmed (scores above 0.5 would be in theory druggable 31) by the 15 structures currently targeted by drugs, with scores ranging from 0.5–0.89. We also used the PhenoScanner ranking explained above for ranking the most promising candidates for drug development, and excluded a further 21 loci due to its predicted negative effect on related phenotypes. We then evaluated the remaining 16 loci for their function and tissue expression using Protein Atlas 32 to observe tissue expression of the gene products in different tissues, and excluded those with high levels of expression (more than 1SD from the average expression cross-tissue) across multiple tissues (e.g., brain, kidney, pancreas, muscle), since those are most likely housekeeping genes or necessary for cell cycle, and therefore not suitable for intervention 33. We concluded with this approach that the most suitable targets to be considered are leiomodin 1 (LMOD1), huntingtin-interacting protein 1 (HIP1) and protein phosphatase 2, regulatory subunit b-double prime, alpha (PPP2R3A) (druggability scores were 0.73, 0.79 and 0.85, respectively). Score of effect directions were 4, 3, and 7 for CAD/MI related phenotypes (max. possible score 8) and 24, 24, and 14 for all other phenotypes, respectively. Full description of all molecules that passed the filters are shown in Table 2. Druggability scores for all proteins with a PDB entry are presented on Supplemental Table 4.
Table 2.
Most suitable drug targets according to predicted pocket interactions and nominally significant loci in the same LD block of the SNP presented
| Reported SNP | Genes | DoGSiteScorer pocket score | PDB code | GWAS predicted concordant phenotypes | GWAS predicted discordant phenotypes |
|---|---|---|---|---|---|
| rs1393786 | PPP2R3A | 0.85 | 4i5j | LDL cholesterol, Total cholesterol, Triglycerides, Obesity class 2, Obesity class 1, Obesity class 3, BMI | |
| rs2820315 | LMOD1 | 0.73 | 4z79 | Obesity class 2, Obesity class 1, Obesity class 3, BMI | |
| rs1167800 | HIP1 | 0.79 | 3i00 | Triglycerides, Obesity class 1, BMI | |
| rs15563 | UBE2Z | 0.73 | 5a4p | LDL cholesterol, Total cholesterol, Type II diabetes | |
| rs6544713 | ABCG8 | 0.89 | 5d07 | LDL cholesterol, Total cholesterol, Triglycerides | BMI |
| rs9326246 | BUD13, ZNF259, APO5A, APOA1 | 0.62 | 4uqt | LDL cholesterol, Total cholesterol, Triglycerides | LDL cholesterol |
| rs972158 | SNX10 | 0.73 | 4pzg | Triglycerides, Type II diabetes | |
| rs10797416 | SKI | 0.73 | 1sbx | LDL cholesterol, Total cholesterol | |
| rs12205331 | ANKS1A | 0.81 | 2lmr | Triglycerides, BMI | |
| rs7139492 | COL4A1 | 0.85 | 1li1 | Total cholesterol | |
| rs816889 | RND3 | 0.83 | 1m7b | Type II diabetes | |
| rs7173743 | MORF4L1 | 0.79 | 2f5j | Obesity class 2 | |
| rs10495907 | 7SK | Error | 2kx8 | LDL, Total cholesterol, Triglycerides, Type II diabetes, Obesity class 2, Obesity class 1, Obesity class 3, BMI | |
| rs2281727 | SMG6 | 0.89 | 4um2 | BMI | Total cholesterol |
| rs2294461 | LY86 | 0.82 | 3b2d | Type II diabetes | LDL |
| rs6984210 | BMP1 | 0.69 | 3edg |
Discussion
GWAS have identified multiple loci and genes that appear to play a causal role in CAD/MI. While new efforts may unveil other associated loci (and indeed have already, with the current loci count at 164 34, 35), it is essential to maximize the value of the current data to translate this knowledge into clinical care, and improve management of CAD/MI. One way to utilize genetic information is by identifying suitable targets for drugs and possible repurposing of already existing drugs. Here, we cross-referenced multiple bioinformatics databases to identify potentially druggable genes and related compounds that may be suitable for repurposing in order to treat CAD/MI.
Until very recently, drug development has not been guided by genetic profiles and risks, therefore expecting that GWAS hits perfectly correspond to current treatments is unrealistic. That said, we compiled a list of medications used in the treatment of CAD (N=79), obtained from different sources, searched for drug-gene interactions among those with any level of confidence (N=608), checked how many unique genes are represented in these interactions (N=251) and compared to the genes we obtained from the CAD loci, either original mapping or GENCODE annotation (N=144). Expectedly, our data indicate that the drugs from the ATC group “cardiovascular system” (C) are overrepresented among our results. Indeed, our pipeline was able to identify three of the main medication groups used for treatment of CAD: statins (e.g. simvastatin), PCSK9 inhibitors (e.g. evolocumab) and angiotensin II receptor blockers (e.g. irbesartan), which may serve as a validation of the method.
We found an overlap of 9 genes, namely APOA1, APOB APOC1, APOE, EDNRA, GUCY1A3, LIPA, LPL and PCSK9. Among those, our top results, in order, include PCSK9 (target of newest medications in the field, evolocumab and alirocumab), LPL (indirectly a target of gemfibrozil), APOC1 and APOE (indirectly representing statins, i.e. first in line drugs against CAD, and ritonavir) and GUCY1A3 (target of suggested for repurposing drug riociguat) (Supplemental Table 5). Starting with a set of 153 loci identified through GWAS experiments for association with CAD/MI, through a series of filtering steps, we add evidence of the value of our druggability approach and suggest specifically three hits to be targeted by three drug compounds that show promise for repurposing including adenosine triphosphate (ATP), pentolinium, and riociguat.
ATP is a promising novel candidate with a ranking score (+4) similar to those obtained for the main rediscovered agents. Here the route of administration (intravenous) is an obstacle that needs to be addressed in future studies. ATP, which canalizes the reactions involving ACCS2 gene product, is one of the top ranked repurposing candidates; this makes it the best promising agent suggested for CAD patients. It is has a role in regulating various biological cascades such as cardiac function, muscle contractility and blood circulation 36. Through the period of 80s and 90s, ATP was useful in managing several clinical conditions such as haemorrhagic shock, pulmonary hypertension and paroxysmal supraventricular tachycardias 37. ATP is not currently marketed in the United States but is available certain European countries; it is indicated as an adjunct therapy for low back pain in France 38 and was tested as therapeutic agent for patients with Alzheimer Disease in a recent clinical trial (https://clinicaltrials.gov/ct2/show/NCT02279511). According to DGIdb, ACSS2 gene is a suggested target for ATP; it synthesizes acetyl CoA from ATP and CoA through an acetyl-adenosine monophosphate (AMP). The Drug repurposing hub confirms the drug-gene interaction.
The proposed repurposing candidates also included riociguat (targeting GUCY1A3) and pentolinium (targeting CHRNB4), albeit with lower rankings (scores of 0 – +1 for all phenotypes), which may suggest additive roles for these medications in CAD. The findings of several GWAS suggest the gene GUCY1A3, coding for the alpha-3 subunit of soluble guanylate cyclase in chromosome 4 as a drug target to manage individuals with CAD/MI. The variant rs7692387 was strongly associated with CAD 23 and later found to modulate GUCY1A3 promoter activity 39 and rs13139571 was identified as a risk factor for hypertension 40. This gene codes for a protein that acts as a major receptor for nitric oxide and other nitro-derivative products (e.g. nitroglycerin) to induce vasodilatation and platelet inhibition 41. GUCY1A3 is highly expressed in the vascular smooth-muscle cells and has the potential to modulate vascular tone and to induce venous and arterial relaxation. Interaction of nitric oxide with different isoforms of soluble guanylate cyclase 1 (GUCY1) plays an important role in regulating platelet aggregation 42 as well as accelerating thrombus formation 43. Deletion of GUCY1A3 is known to cause asymptomatic moyamoya (intracranial stenosis) angiopathy 42. Its deletion is also known to cause myocardial infarction 43, and detailed molecular analyses identified how this variant modulates expression of the gene, soluble guanylyl protein levels, activity of the enzyme and platelet function 39. In addition, an exonic variant rs201558687 was reported as protective marker against pulmonary hypertension 44. This finding suggests a role for GUCY1A3 gene in pulmonary hypertension in addition to its association with CAD/MI.
The newly marketed drug riociguat was approved by the U.S. Food and Drug Administration (FDA) in 2013 45 and later by European Medicines Agency in 2014 to manage patients with pulmonary arterial hypertension (PAH) and chronic thromboembolic pulmonary hypertension (CTEPH) 46. Riociguat is a positive allosteric modulator for GUCY1A3. It is a unique drug that acts as guanylate cyclase stimulator 47; this mechanism makes a hope to repurpose the usage of riociguat as an antianginal agent 48. Hypotension, headache and dizziness are the common side effects of riociguat 49. A clinical trial was prepared to study the effects of riociguat for CAD (clinicaltrials.gov ID: NCT01165931) but the study was cancelled before recruitment for unknown reasons. Further evidence is provided by the Drug Repurposing Hub 50, which mentions that molsidomine, a drug not available in DGIdb and predicted to interact with GUCY1A3, is prescribed against CAD.
Pentolinium is an old antihypertensive agent indicated to control malignant hypertension and hypertensive crises, in particular, throughout surgery 51. Pentolinium is predicted to antagonize CHRNB4. It was marketed by WYETH AYERST under the trade name of Ansolysen then it was decided to stop its manufacturing in January 1982. The manufacturer did not indicate the reasons of discontinuation but possibly because of induction of severe postural and exertional hypotension 52. Moreover, the drug is rather non-specific (it targets different subunits of nicotinic acetylcholine receptors (nAChRs) at a time 53, in particular α3, α10, β2 and β4), and had to compete with newer medications with higher efficacy in lowering blood pressure. It has a potent peripheral ganglionic blocking action and acts as an antagonist to nicotinic receptors which inhibit the release of both adrenaline and noradrenaline 54. Although these receptors are abundant on somatic and central nervous system they are also expressed on aortic valves and atrial appendages 55. Nicotinic receptors are considered part of a superfamily of ligand-gated ion channels which mediate fast signal transmission at synapses 56. Blockage of the receptor results in relaxation as well as vasodilatation of smooth muscles. A single nucleotide polymorphism (SNP, rs11072794) in the gene cholinergic receptor, neuronal nicotinic, beta polypeptide 4 (CHRNB4) - in chromosome 15- which codes for nicotinic receptors was found to be a risk factor for CAD/MI 20 and nominally significantly associates to type 2 diabetes (T2D) 57 in a GWAS study.
The SNP rs11072794, located in an intronic region, is in complete LD (r2=1) with other two variants (rs899997 and rs12899940), that are located in regulatory regions that possibly affect gene function; rs12899940 is located in promoter flanking region, while rs899997 is located in a transcription factor binding site. The SNP rs899997 was recently identified as a risk factor to develop coronary artery disease and ischemic stroke according to recent analysis of three different genome-wide association studies; the METASTROKE, CARDIoGRAM, and C4D consortia 58. Further, another marker (rs8023822) in CHRNB4 gene was also detected as a susceptibility loci for CAD in T2D in a meta-analysis that involve several GWAS studies among Scottish population 59. These significant associations between different loci in CHRNB4 and CAD suggest the gene product as a good drug target; therefore, pentolinium can possibly repurposed in the management of CAD/MI conditions. In this case, however, we found no further evidence from the Drug Repurposing Hub.
Regarding novel targets, we were able to elaborate a ranking of the most suitable candidates. The most promising candidates for targeting are LMOD1, HIP1 and PPP2R3A. Leiomodin 1 (LMOD1) is related to smooth muscle contraction and cardiac conduction 60 and has been identified to have smooth muscle cell-specific eQTLs in SNP rs34091558 61. SNP rs2820315 at LMOD1 has reached genome-wide significance in the latest GWAS in CAD/MI 62. Huntingtin-interacting protein 1 (HIP1) codes for a protein significantly expressed in coronary artery endothelium cell and play a major role in cell endocytosis 63, having one of the top significant cis-eQTL expression patterns among CAD/MI loci 64. Finally, protein phosphatase 2, regulatory subunit b-double prime, alpha (PPP2R3A) is abundantly expressed in heart and skeletal muscles and responsible for intracellular signal regulation 65, being associated to several regulatory networks of CAD 66.
The pipelines are modular and can be easily generalized for other diseases. Once determined an appropriate set of bona fide associated SNPs for a given trait, the pipelines can provide candidates for repurposing and most suitable targets for drug development. However, a number of limitations need to be considered in translating our results to clinical studies. First, our approach relies on the well-informed yet unproven relationship between CAD/MI loci and a nearby gene and the druggability of its gene product. It is worth mentioning that the genes investigated in both pipelines were mapped to variants by positional mapping in their original study. We extended this analysis using GENCODE annotations to identify additional genes, a comprehensive resource that also integrates regulatory data in the process of annotation. However, variants can act on elements regulating genes that are not physically located in their immediate vicinity, but often even outside the locus 67. Experiments on chromosomal conformation can capture these dynamics by generating a map of tissue-specific genomic regions that physically interact 68–71. The ongoing generation and integration of such maps on disease-relevant cell-types will enable identification of target genes that might not have been unraveled by using current approaches, which in turn might improve results of our pipelines. Second, it needs to be investigated as to whether the drugs discussed modulate the gene product in a beneficial way. Third, we illustrate in our examples that some of the drugs are pleiotropic or not suitable for chronic application, and that they may be only a starting point for further developments. Fourth, the databases used were complete versions at the moment of usage, but those are ongoing efforts that will improve coverage and reliability, so iterative development may yield more reliable results. Finally, we focused this investigation on repurposing of established drugs. We expect that multiple GWAS loci, pointing to currently unexplored mechanisms, can be addressed by new drug development on antibodies, like what has been successfully achieved in case of PCSK9 72–74. In conclusion, we have found evidence for repurposing of drugs and candidates for drug development in the context of CAD/MI, suggesting that in-silico analysis using existing databases and genetic findings may be useful to accelerate translation into clinical practice. Clinical trials are now needed to explore the potential value of these agents.
Supplementary Material
Acknowledgments
DH is supported by the National Council for the Improvement of Higher Education (CAPES) and Science without Borders Project, process no 13259/13-0. FA is supported by a Dekker scholarship (Junior Staff Member 2014T001, Dutch Heart Foundation) and UCL Hospitals NIHR Biomedical Research Centre. JHM is supported by NIH process R01 LM010098.
Sources of Funding: The research leading to these results has received funding from the European Union Seventh Framework Programme FP7/2007–2013 under grant agreement n° HEALTH-F2-2013-601456 (CVgenes-at-target).
Footnotes
Disclosures: None
References
- 1.Vos T, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perk J, et al. European Guidelines on cardiovascular disease prevention in clinical practice (version 2012) Eur Heart J. 2012;33:1635–1701. doi: 10.1093/eurheartj/ehs092. [DOI] [PubMed] [Google Scholar]
- 3.Pursnani A, et al. Guideline-Based Statin Eligibility, Coronary Artery Calcification, and Cardiovascular Events. JAMA. 2015;314:134–41. doi: 10.1001/jama.2015.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz GG, et al. Effect of alirocumab, a monoclonal antibody to PCSK9, on long-term cardiovascular outcomes following acute coronary syndromes: rationale and design of the ODYSSEY outcomes trial. Am Heart J. 2014;168:682–9. doi: 10.1016/j.ahj.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 5.Raal FJ, et al. Inhibition of PCSK9 with evolocumab in homozygous familial hypercholesterolaemia (TESLA Part B): a randomised, double-blind, placebo-controlled trial. Lancet. 2015;385:341–50. doi: 10.1016/S0140-6736(14)61374-X. [DOI] [PubMed] [Google Scholar]
- 6.Ridker PM, et al. Interleukin-1beta inhibition and the prevention of recurrent cardiovascular events: rationale and design of the Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS) Am Heart J. 2011;162:597–605. doi: 10.1016/j.ahj.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Weber C, et al. CANTOS Trial Validates the Inflammatory Pathogenesis of Atherosclerosis: Setting the Stage for a New Chapter in Therapeutic Targeting. Circ Res. 2017;121:1119–1121. doi: 10.1161/CIRCRESAHA.117.311984. [DOI] [PubMed] [Google Scholar]
- 8.Arrowsmith J, et al. Trial watch: phase II and phase III attrition rates 2011–2012. Nat Rev Drug Discov. 2013;12:569–569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 9.Walmsley RM, et al. How accurate is in vitro prediction of carcinogenicity? Br J Pharmacol. 2011;162:1250–1258. doi: 10.1111/j.1476-5381.2010.01131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipsky MS, et al. From idea to market: the drug approval process. J Am Board Fam Pract. 2001;14:362–367. [PubMed] [Google Scholar]
- 11.Mullard A. Industry R&D returns slip. Nat Rev Drug Discov. 2016;15:7–7. [Google Scholar]
- 12.Rockey WM, et al. Progress toward virtual screening for drug side effects. Proteins. 2002;48:664–71. doi: 10.1002/prot.10186. [DOI] [PubMed] [Google Scholar]
- 13.Li YY, et al. A Large-Scale Computational Approach to Drug Repositioning. Genome Inform. 2006;17:239–247. [PubMed] [Google Scholar]
- 14.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol. 2012;30:317–20. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 15.Oprea T, et al. Drug repurposing: far beyond new targets for old drugs. AAPS J. 2012;14:759–763. doi: 10.1208/s12248-012-9390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arrowsmith J, et al. Drug Repositioning: The Business Case and Current Strategies to Repurpose Shelved Candidates and Marketed Drugs. Drug Repositioning: Bringing New Life to Shelved Assets and Existing Drugs. 2012:9. [Google Scholar]
- 17.Melnikova I. Rare diseases and orphan drugs. Nat Rev Drug Discov. 2012;11:267–268. doi: 10.1038/nrd3654. [DOI] [PubMed] [Google Scholar]
- 18.Galiè N, et al. Sildenafil citrate therapy for pulmonary arterial hypertension. N Engl J Med. 2005;353:2148–2157. doi: 10.1056/NEJMoa050010. [DOI] [PubMed] [Google Scholar]
- 19.Ashburn TT, et al. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 20.Deloukas P, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikpay M, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajduk PJ, et al. Predicting protein druggability. Drug Discov Today. 2005;10:1675–1682. doi: 10.1016/S1359-6446(05)03624-X. [DOI] [PubMed] [Google Scholar]
- 25.Guth BD, et al. Dealing with hERG liabilities early: diverse approaches to an important goal in drug development. Br J Pharmacol. 2010;159:22–4. doi: 10.1111/j.1476-5381.2009.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preiss D, et al. Risk of incident diabetes with intensive-dose compared with moderate-dose statin therapy: a meta-analysis. JAMA. 2011;305:2556–2564. doi: 10.1001/jama.2011.860. [DOI] [PubMed] [Google Scholar]
- 27.Medina MW, et al. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118:355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai R, et al. Statins worsen glycemic control of T2DM in target LDL-c level and LDL-c reduction dependent manners: a meta-analysis. Expert Opin Pharmacother. 2016;17:1839–49. doi: 10.1080/14656566.2016.1220539. [DOI] [PubMed] [Google Scholar]
- 29.Tragante V, et al. Harnessing publicly available genetic data to prioritize lipid modifying therapeutic targets for prevention of coronary heart disease based on dysglycemic risk. Hum Genet. 2016;135:1453–467. doi: 10.1007/s00439-016-1647-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffith M, et al. DGIdb: mining the druggable genome. Nat Methods. 2013;10:1209–10. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volkamer A, et al. DoGSiteScorer: a web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics. 2012;28:2074–2075. doi: 10.1093/bioinformatics/bts310. [DOI] [PubMed] [Google Scholar]
- 32.Uhlen M, et al. Towards a knowledge-based human protein atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 33.Uhlén M, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 34.Nelson CP, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 35.van der Harst P, et al. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agteresch HJ, et al. Adenosine triphosphate: established and potential clinical applications. Drugs. 1999;58:211–32. doi: 10.2165/00003495-199958020-00002. [DOI] [PubMed] [Google Scholar]
- 37.Belhassen B, et al. Adenosine triphosphate and adenosine: perspectives in the acute management of paroxysmal supraventricular tachycardia. Clin Cardiol. 1985;8:460–4. doi: 10.1002/clc.4960080903. [DOI] [PubMed] [Google Scholar]
- 38.Jager R, et al. Oral adenosine-5′-triphosphate (ATP) administration increases blood flow following exercise in animals and humans. J Int Soc Sports Nutr. 2014;11:28. doi: 10.1186/1550-2783-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessler T, et al. Functional Characterization of the GUCY1A3 Coronary Artery Disease Risk Locus. Circulation. 2017;136:476–489. doi: 10.1161/CIRCULATIONAHA.116.024152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.International Consortium for Blood Pressure Genome-Wide Association Studies, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zabel U, et al. Human soluble guanylate cyclase: functional expression and revised isoenzyme family. Biochem J. 1998;335(Pt 1):51–7. doi: 10.1042/bj3350051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herve D, et al. Loss of alpha1beta1 soluble guanylate cyclase, the major nitric oxide receptor, leads to moyamoya and achalasia. Am J Hum Genet. 2014;94:385–94. doi: 10.1016/j.ajhg.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erdmann J, et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature. 2013;504:432. doi: 10.1038/nature12722. [DOI] [PubMed] [Google Scholar]
- 44.Wilkins MR, et al. alpha1-A680T variant in GUCY1A3 as a candidate conferring protection from pulmonary hypertension among Kyrgyz highlanders. Circ Cardiovasc Genet. 2014;7:920–9. doi: 10.1161/CIRCGENETICS.114.000763. [DOI] [PubMed] [Google Scholar]
- 45.Traynor K. Riociguat approved for pulmonary hypertension. Am J Health Syst Pharm. 2013;70:1960. doi: 10.2146/news130073. [DOI] [PubMed] [Google Scholar]
- 46.Bishop BM. Riociguat for pulmonary arterial hypertension and chronic thromboembolic pulmonary hypertension. Am J Health Syst Pharm. 2014;71:1839–44. doi: 10.2146/ajhp130777. [DOI] [PubMed] [Google Scholar]
- 47.Guha M. First-in-class guanylate cyclase stimulator approved for PAH. Nat Biotechnol. 2013;31:1064. doi: 10.1038/nbt1213-1064b. [DOI] [PubMed] [Google Scholar]
- 48.Rask-Andersen M, et al. The druggable genome: Evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu Rev Pharmacol Toxicol. 2014;54:9–26. doi: 10.1146/annurev-pharmtox-011613-135943. [DOI] [PubMed] [Google Scholar]
- 49.Galie N, et al. PATENT PLUS: a blinded, randomised and extension study of riociguat plus sildenafil in pulmonary arterial hypertension. Eur Respir J. 2015;45:1314–22. doi: 10.1183/09031936.00105914. [DOI] [PubMed] [Google Scholar]
- 50.Corsello SM, et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med. 2017;23:405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haas E, et al. Effects of an antihypertensive drug, pentolinium. Am J Physiol. 1959;196:763–8. doi: 10.1152/ajplegacy.1959.196.4.763. [DOI] [PubMed] [Google Scholar]
- 52.Freis ED, et al. Results of Prolonged Treatment with Pentolinium Tartrate. Circulation. 1956;13:856–65. doi: 10.1161/01.cir.13.6.856. [DOI] [PubMed] [Google Scholar]
- 53.Cachelin AB, et al. Beta-subunits co-determine the sensitivity of rat neuronal nicotinic receptors to antagonists. Pflugers Arch. 1995;429:449–51. doi: 10.1007/BF00374164. [DOI] [PubMed] [Google Scholar]
- 54.Sethi OP, et al. Analysis of mode of action of some nicotinic blocking drugs. Jpn J Pharmacol. 1973;23:437–51. doi: 10.1254/jjp.23.437. [DOI] [PubMed] [Google Scholar]
- 55.Nebion AG. Top 10 tissues expressing genes. 2016 [Google Scholar]
- 56.Khakh BS, et al. State-dependent cross-inhibition between transmitter-gated cation channels. Nature. 2000;406:405–10. doi: 10.1038/35019066. [DOI] [PubMed] [Google Scholar]
- 57.Mahajan A, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–44. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dichgans M, et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke. 2014;45:24–36. doi: 10.1161/STROKEAHA.113.002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Natalie VZ. PhD. 2013. The genetic determinants of cardiovascular disease in patients with type 2 diabetes; p. 309. [Google Scholar]
- 60.Matic LP, et al. Phenotypic Modulation of Smooth Muscle Cells in Atherosclerosis Is Associated With Downregulation of LMOD1, SYNPO2, PDLIM7, PLN, and SYNM. Arterioscler Thromb Vasc Biol. 2016;36:1947–1961. doi: 10.1161/ATVBAHA.116.307893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller CL, et al. Integrative functional genomics identifies regulatory mechanisms at coronary artery disease loci. Nat Commun. 2016;7:12092. doi: 10.1038/ncomms12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howson JM, et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet. 2017;49:1113–1119. doi: 10.1038/ng.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Metzler M, et al. Disruption of the endocytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J. 2003;22:3254–66. doi: 10.1093/emboj/cdg334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joehanes R, et al. Integrated genome-wide analysis of expression quantitative trait loci aids interpretation of genomic association studies. Genome Biol. 2017;18:16. doi: 10.1186/s13059-016-1142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hendrix P, et al. Structure and expression of a 72-kDa regulatory subunit of protein phosphatase 2A. Evidence for different size forms produced by alternative splicing. J Biol Chem. 1993;268:15267–76. [PubMed] [Google Scholar]
- 66.Zhao Y, et al. Network-based identification and prioritization of key regulators of coronary artery disease loci. Arterioscler Thromb Vasc Biol. 2016;36:928–41. doi: 10.1161/ATVBAHA.115.306725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watanabe K, et al. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dekker J, et al. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 69.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 70.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 71.Rivera CM, et al. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Everett BM, et al. Reducing LDL with PCSK9 inhibitors—the clinical benefit of lipid drugs. N Engl J Med. 2015;373:1588–1591. doi: 10.1056/NEJMp1508120. [DOI] [PubMed] [Google Scholar]
- 73.Schwartz GG, et al. Effect of alirocumab, a monoclonal antibody to PCSK9, on long-term cardiovascular outcomes following acute coronary syndromes: rationale and design of the ODYSSEY outcomes trial. Am Heart J. 2014;168:682–689 e1. doi: 10.1016/j.ahj.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 74.Raal FJ, et al. PCSK9 inhibition with evolocumab (AMG 145) in heterozygous familial hypercholesterolaemia (RUTHERFORD-2): a randomised, double-blind, placebo-controlled trial. Lancet. 2015;385:331–340. doi: 10.1016/S0140-6736(14)61399-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.