Abstract
Chromosome segregation relies on forces generated by spindle microtubules that are translated into chromosome movement through interactions with kinetochores, highly conserved macromolecular machines that assemble on a specialized centromeric chromatin structure. Kinetochores not only have to stably attach to growing and shrinking microtubules, but they also need to recruit spindle assembly checkpoint proteins to halt cell cycle progression when there are attachment defects. Even the simplest kinetochore in budding yeast contains more than 50 unique components that are present in multiple copies, totaling more than 250 proteins in a single kinetochore. The complex nature of kinetochores makes it challenging to elucidate the contributions of individual components to its various functions. In addition, it is difficult to manipulate forces in vivo to understand how they regulate kinetochore–microtubule attachments and the checkpoint. To address these issues, we developed a technique to purify kinetochores from budding yeast that can be used to analyze kinetochore functions and composition as well as to reconstitute kinetochore–microtubule attachments in vitro.
1. INTRODUCTION
Accurate cell division requires equal distribution of the chromosomes between mother and daughter cells. Defects in kinetochore–microtubule attachments are costly and generate aneuploidy, which is the most common chromosomal abnormality in cancer cells (Orr & Compton, 2013). Chromosome segregation is mediated by interactions between a DNA-bound protein structure called the kinetochore and spindle microtubules (Cheeseman, 2014; Holland & Cleveland, 2009). The spindle is a complex and dynamic structure comprised of microtubules that grow and shrink due to the addition and removal of tubulin subunits (Petry, 2016; Prosser & Pelletier, 2017). The dynamic nature of the microtubule tips makes it especially challenging for the kinetochore to stay attached (Asbury, Tien, & Davis, 2011).
To ensure that each daughter cell inherits one copy of the genome, sister kinetochores on duplicated chromosomes must biorient and interact with microtubules from opposite spindle poles (Tanaka, 2010). However, microtubules initially capture kinetochores in a random manner, which would lead to segregation defects if left uncorrected (Nicklas, 1997). Cells therefore have mechanisms to detect and correct inappropriate attachments prior to anaphase. When kinetochores achieve biorientation, they come under tension due to the pulling forces from microtubules on linked sister kinetochores (Tanaka, 2010). Tension appears to be the signal that kinetochore–microtubule attachments are correct because attachments under tension are much more stable in vivo than those lacking tension (Li & Nicklas, 1995; Nicklas & Koch, 1969). This effect is partly due to an error correction system mediated by the Aurora B protein kinase that phosphorylates kinetochores lacking tension in order to destabilize incorrect attachments and give the cell a chance to make proper attachments (Akiyoshi, Nelson, & Biggins, 2013; Biggins & Murray, 2001; Kalantzaki et al., 2015; Tanaka, 2010; Tanaka et al., 2002). In addition, kinetochore–microtubule attachments under tension are directly stabilized via an intrinsic mechanical mechanism (Akiyoshi et al., 2010; Miller, Asbury, & Biggins, 2016). Accurate segregation is also ensured by a surveillance mechanism called the spindle assembly checkpoint (SAC) that monitors kinetochore–microtubule attachments (Hoyt, Totis, & Roberts, 1991; Li & Murray, 1991; Musacchio & Salmon, 2007). Unattached kinetochores recruit SAC proteins that generate a soluble wait signal that prevents cell cycle progression until all kinetochores are attached to spindle microtubules (Musacchio & Salmon, 2007). Kinetochores therefore carry out essential mechanical and signaling mechanisms to ensure faithful inheritance of chromosomes.
It has been challenging to elucidate the mechanisms that underlie the complex functions of kinetochores for a number of reasons. First, the kinetochore is an extremely large protein machine. Even the simplest kinetochore documented in budding yeast has greater than 50 components that are present in multiple copies (Biggins, 2013; Musacchio & Desai, 2017). It is therefore difficult to identify the precise proteins that attach to microtubules and to understand how they maintain attachments to dynamic microtubule tips. This complexity has also made it hard to identify the receptors for checkpoint proteins. Second, tension is difficult to manipulate in vivo, making it challenging to understand how mechanical forces regulate the interaction between kinetochores and microtubules. It has also been controversial how the SAC is triggered and silenced to control cell cycle progression (Etemad & Kops, 2016). To overcome these issues, we developed a method to purify native kinetochores that can be used for reconstitution studies. We have used the isolated kinetochores to directly probe how forces affect kinetochore–microtubule attachments as well as to identify SAC receptors and biochemical events that signal the checkpoint (Akiyoshi et al., 2010; London & Biggins, 2014; London, Ceto, Ranish, & Biggins, 2012; Miller et al., 2016; Sarangapani, Akiyoshi, Duggan, Biggins, & Asbury, 2013; Sarangapani et al., 2014). Additionally, using electron microscopy, we have begun to study the ultrastructure of the kinetochore (Gonen et al., 2012). We used budding yeast as a model system given that it is the simplest kinetochore, yet its functions and composition are generally conserved across diverse taxa.
2. OVERVIEW OF YEAST KINETOCHORES
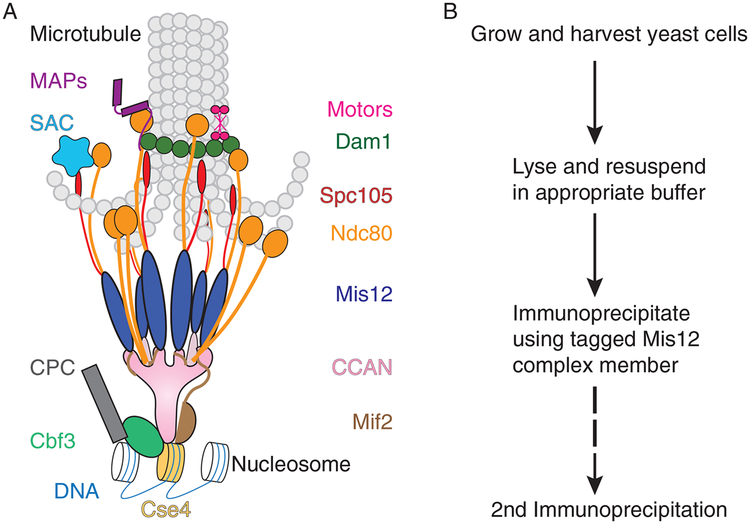
Kinetochores are conserved megadalton complexes that are constructed from multiple protein subcomplexes on a specific region of the DNA known as the centromere (CEN) (Biggins, 2013; Musacchio & Desai, 2017). In multicellular eukaryotes, CENs can be up to megabases of repetitive DNA (Grady et al., 1992; McKinley & Cheeseman, 2016). In contrast, the simplest CEN is a 125-bp defined sequence called a point centromere in the budding yeast Saccharomyces cerevisiae (Clarke & Carbon, 1983; McKinley & Cheeseman, 2016). Despite this extreme difference in the underlying centromeric DNA, the overall architecture is conserved across eukaryotes (van Hooff, Tromer, van Wijk, Snel, & Kops, 2017) (Table 1). There is a specialized centromeric nucleosome containing a histone H3 variant called CENP-ACse4 that directs kinetochore assembly (Biggins, 2013; McKinley & Cheeseman, 2016; Musacchio & Desai, 2017; van Hooff et al., 2017) (Fig. 1A). The CCAN (constitutive centromere-associated network) directly assembles onto the specialized chromatin to form the inner kinetochore (Foltz et al., 2006). This is the foundation for assembly of a highly conserved outer kinetochore network of the KNL1, MIND/Mis12, and Ndc80 (KMN) subcomplexes that mediate micro-tubule binding (Biggins, 2013; Cheeseman, Chappie, Wilson-Kubalek, & Desai, 2006; Musacchio & Desai, 2017; van Hooff et al., 2017). These conserved subunits constitute the “core” kinetochore components that make up most of the structural elements across all organisms (van Hooff et al., 2017) (Table 1).
Table 1.
Yeast Kinetochore Components With Known Human Orthologs
| Subcomplex | Yeast protein | Human Name | Subcomplex | Yeast protein | Human Name | Subcomplex | Yeast protein | Human Name |
|---|---|---|---|---|---|---|---|---|
| Cbf1 | Cbf1 | RBPJ | CCAN | Ctf19 | CENP-P | Dam1 | Dam1 | |
| Cbf3 | Cbf2 | Okp1 | CENP-Q | Dad1 | ||||
| Cep3 | Mcm21 | CENP-0 | Dad3 | |||||
| Ctf13 | Ame1 | CENP-U | Ask1 | |||||
| Skp1 | Iml3 | CENP-L | Duo1 | |||||
| Nucleosome and associated | Cse4 | CENP-A | Chl4 | CENP-N | Hsk3 | |||
| Hta2 | Histone H2A | Nkp1 | Spc19 | |||||
| Htb2 | Histone H2B | Nkp2 | Spc34 | |||||
| Hht1 | Histone H3 | Mem 16 | CENP-H | Dad2 | ||||
| Hhf1 | Histone H4 | Ctf3 | CENP-I | Dad4 | ||||
| Psh1 | Mcm22 | CENP-K | Motors | Kip3 | Kinesin-8 | |||
| Scm3 | Cnn1 | CENP-T | Cikl | Kinesin-14 | ||||
| Mif2 | Mif2 | CENP-C | Wip1 | CENP-W | Kar3 | Kinesin-14 | ||
| CPC | Ipl1 | Aurora B | Mhf1 | CENP-S | Cin8 | Kinesin-5 | ||
| Sli15 | INCENP | Mhf2 | CENP-X | Kip1 | BimC | |||
| Nbl1 | Borealin | Mtw1 | Mtw1 | Mis12 | MAPs | Stu2 | ChTOG | |
| Birl | Survivin | Nnf1 | Nnf1 | Stu1 | CLASP | |||
| Ns11 | Nsl1 | Slk19 | ||||||
| Dsn1 | Dsn1 | Bik1 | CLIP-170 | |||||
| Ndc80 | Ndc80 | HEC1 | SAC | Mps1 | Mps1 | |||
| Nuf2 | Nuf2 | Bub1 | Bub1 | |||||
| Spc24 | Spc24 | Bub3 | Bub3 | |||||
| Spc25 | Spc25 | Mad1 | Mad1 | |||||
| Spc105 | Spc105 | KNL1 | Mad2 | Mad2 | ||||
| Kre28 | Zwint | Mad3 | BubR1 |
Listed are the yeast proteins known to localize to the kinetochore and their corresponding human homologs. Shaded in gray are “core” kinetochore components that are structural elements that are conserved across most eukaryotes (van Hooff et al., 2017).
Adapted from Biggins, S. (2013). The composition, functions, and regulation of the budding yeast kinetochore. Genetics, 194(4), 817–846. https://doi.org/10.1534/genetics.112.145276 with subcomplex designation adapted from Pekgoz Altunkaya, G., Malvezzi, F., Demianova, Z., Zimniak, T., Litos, G., Weissmann, F., et al. (2016). CCAN assembly configures composite binding interfaces to promote cross-linking of Ndc80 complexes at the kinetochore. Current Biology, 26(17), 2370–2378. https://doi.org/10.1016/j.cub.2016.07.005.
FIG. 1.
Composition and purification of yeast kinetochores. (A) Kinetochores are multiprotein structures that assemble specifically on a specialized nucleosome containing Cse4 (dark yellow). The deposition of Cse4 is facilitated by the Cbf3 complex (aqua) in conjunction with Cbf1 (not shown) and the yeast-specific Cse4 chaperone, Scm3 (not shown). The Cbf3 complex also recruits the chromosomal passenger complex (CPC) (dark gray), which destabilizes kinetochore attachments lacking tension. The Cse4 nucleosome and the adjacent DNA are bound by the larger CCAN (constitutive centromere-associated network) complex (pink) and the Mif2 protein (brown). A conserved interaction between these two facilitates recruitment of the Mis12 complex (dark blue), which brings in the Spc105 (red) and Ndc80 (orange) complexes. Under some conditions, Ndc80 can also be bound by the CCAN, but the bulk of the yeast Ndc80 localizes to the kinetochore via its interaction with the Mis12 complex. The Ndc80 and Spc105 complexes recruit the diffusible wait signal in the event of an unattached kinetochore, the SAC (spindle assembly checkpoint) (teal). Finally, to secure the attachment of the kinetochore to the microtubule, the Ndc80 complex recruits the Dam1 complex (dark green), which oligomerizes to form a ring around the microtubule. The sustained attachment of the kinetochore to the microtubule is mediated by many subcomplexes, including Ndc80, Dam1, and additional microtubule-associated proteins (MAPs) (purple) and molecular motors (hot pink) that transiently localize to the kinetochore as necessary. (B) In order to study the multitude of the kinetochore functions, we have devised a method to purify kinetochores. The workflow involves growing and harvesting cells, lysing them in the appropriate buffer, immunoprecipitating the kinetochores using a tagged component of the Mis12 complex (Dsn1 is preferred), eluting the kinetochores, and, if necessary, performing a second immunoprecipitation. Slight adjustments at every one of these steps of the protocol can alter the composition of the resulting kinetochores.
Budding yeast are an excellent model system to study kinetochores because the kinetochore protein composition is generally similar to that observed in more complex eukaryotes, but they only bind to a single microtubule (Winey et al., 1995) (Fig. 1A, for full comparison of yeast and human kinetochore proteins, see Table 1). Yeast kinetochore assembly is initiated by the direct binding of Cbf1 and the CBF3 complex to the CEN DNA (Biggins, 2013; Lechner & Carbon, 1991). CBF3 recruits the Cse4 chaperone, Scm3, to deposit the specialized nucleosome at the CEN (Camahort et al., 2007; Stoler et al., 2007) (Fig. 1A). Once the centromeric nucleosome is present, Mif2 and the rest of the CCAN assemble. This inner kineto-chore is the platform for the outer kinetochore components including KMN, Stu2, and the Dam1 subcomplex. Although Dam1 is not found outside of fungi, the vertebrate Ska complex has been proposed to be a functional ortholog (Auckland, Clarke, Royle, & McAinsh, 2017; van Hooff et al., 2017). Additionally, there are numerous other molecular motors and microtubule-associated proteins (MAPs) that cycle on and off the kinetochore at different times (He, Rines, Espelin, & Sorger, 2001; Tytell & Sorger, 2006) (Fig. 1A and Table 1).
Here, we describe the method we use to purify native yeast kinetochores and modifications that can be made depending on the application (Fig. 1B). The purification technique allows the composition of both kinetochore populations and single particles to be analyzed. In addition, it is amenable to structural analysis by electron microscopy and facilitates biophysical and biochemical analyses to probe underlying mechanisms (Akiyoshi, Nelson, & Biggins, 2013; Akiyoshi et al., 2010; Gonen et al., 2012; London & Biggins, 2014; London et al., 2012; Miller et al., 2016; Sarangapani et al., 2013). Together, studying the structure and biophysical properties of native kinetochores in vitro deepens our understanding of the various roles of the kineto-chore within cells.
3. ISOLATION OF YEAST KINETOCHORES
Budding yeast kinetochores are isolated by immunoprecipitation of the Mis12 complex kinetochore component Dsn1 (Akiyoshi et al., 2010). The purification is specific to the Mis12 complex as tags on components of any other subcomplex do not yield complete kinetochore particles (Akiyoshi et al., 2010). Kinetochores can also be purified using epitope-tagged Mtw1, which is another component of the Mis12 complex (De Wulf, McAinsh, & Sorger, 2003). We make whole-cell extracts from cells containing epitope-tagged Dsn1 (see Table 2 for suggested epitope tags) and then immunoprecipitate Dsn1 using antibody-conjugated dynabeads (Fig. 1B). The immunoprecipitation results in the purification of native kinetochore particles that are capable of binding and tracking microtubules as well as holding force (Akiyoshi et al., 2010; Miller et al., 2016; Sarangapani et al., 2013, 2014). In addition, we find that complete kinetochore complexes with slightly different protein compositions can be isolated when subtle modifications are made to the overall procedure (Fig. 1B).
Table 2.
Tags Used for Kinetochore Purification
| Tag | Efficiently Eluted | Elution Peptide |
|---|---|---|
| 3-FLAG | Yes | MDYKDHDGDYKDHDIDYKDDDDK |
| 3-V5 | Yes | GKPIPNPLLGLDGKPIPNPLLGLDSTPIPN PLLGLDST |
| 3-HA | No | – |
Commonly used tags for kinetochore purification. All three tags have been used to purify kinetochores from budding yeast. 3-FLAG remains the most studied and robust tag used in kinetochore purifications and works better than the 3-V5 tag in our hands. Both these tags can be efficiently eluted using competitive peptides, which is crucial for all the biophysical assays. Although 3-HA can be used, it purifies more nonspecific components and is unable to be efficiently eluted.
3.1. MEDIA AND BUFFERS
The purification of kinetochores requires the growth of yeast cells followed by harvesting, lysis, and immunoprecipitation (Fig. 1B). The buffers used for the purification are either BH 0.15 or BL 0.175, which differ primarily in the concentration of the potassium salt and the presence or absence of chloride ions. In several steps in the protocol, the buffer is supplemented with dithiothreitol (DTT), various phosphatase inhibitors (20 × phosphatase inhibitor stock, microcystin-LR), and/or protease inhibitors (phenylmethylsulfonyl fluoride (PMSF), 1000 × protease stock) immediately prior to use.
3.1.1. Media
Yeast are grown using standard media and microbial techniques (Sherman, 1991). As required, cells can be arrested in mitosis due to incapacity to satisfy the SAC by the addition of benomyl (Cat# 381586, Millipore Sigma) for 2h at 23°C. 2 × Benomyl media are made by dissolving 60mg of benomyl in 2mL dimethyl sulfoxide (DMSO) and adding this directly to 1L of freshly autoclaved or boiling media. Because the media must be hot for the benomyl to dissolve, we make the media a day prior to the experiment. The benomyl will come out of solution if stored for too long so we use the media within a day or two.
3.1.2. Buffers
All buffers are made by mixing the component parts and filter-sterilizing prior to use. For short-term use, buffers can be stored at room temperature unless noted otherwise. Long-term storage should be at 4°C.
Buffer H 0.15—25mM HEPES (pH 8.0), 2mM MgCl2, 0.1mM EDTA (pH 8.0), 0.5mM EGTA-KOH (pH 8.0), 15% Glycerol, 0.1% IGEPAL CA-630 (Cat# I3021, Millipore Sigma), 150mM KCl.
Buffer L 0.175—25mM HEPES (pH 7.6), 6mM Mg(OAc)2, 0.1mM EDTA (pH 7.6), 0.5mM EGTA-KOH (pH 7.6), 15% Glycerol, 0.1% IGEPAL CA-630, 175mM l-glutamic acid potassium salt monohydrate (Cat# G1501, Millipore Sigma).
20 × Phosphatase inhibitor—20mM Sodium pyrophosphate (Cat# 221368, Millipore Sigma), 40mM Sodium-beta-glycerophosphate (Cat# G5422, Millipore Sigma) (make 1M stock in water), 2mM Na3VO4 (Cat# S6508, Millipore Sigma), 100mM NaF (Cat# 450022, Millipore Sigma).
1000 × Protease inhibitor stock—10mg Leupeptin (Cat# E18, EMD Millipore), 10mg pepstatin A (Cat# EI10, EMD Millipore), and 10mg chymostatin (Cat# EI6, EMD Millipore) are dissolved in 1mL DMSO and stored at −80°C.
500 × PMSF—100mM PMSF (Cat #P7626, Millipore Sigma) is dissolved in isopropanol and used immediately as a 500 × stock.
1000 × Microcystin-LR—Dissolve 0.5mg of microcystin-LR (Cat# 475815, Calbiochem) in 5mL of 100% ethanol to a final concentration of 100μM (1 ×). Store at −80°C and add immediately before use as this solution is extremely labile.
Cell wash buffer—BH 0.15 or BL 0.175+1 × phosphatase inhibitor+1 PMSF.
Cell lysis buffer—BH 0.15 or BL 0.175+1 ×phosphatase inhibitor+1 × PMSF+ 2× protease inhibitor+2× microcystin-LR.
IP wash buffer I—BH 0.15×or BL 0.175+1 × phosphatase inhibitor+1 PMSF+ 2mM DTT+1× protease inhibitor+1× microcystin-LR.
IP wash buffer II—BH 0.15 or BL 0.175+1× PMSF+1× protease inhibitor.
Elution peptide—3FLAG or 3V5 peptide (see Table 2 for×sequences) resuspended at 2.5mg/mL (5×) and 5mg/mL (10×), respectively, in BH 0.15 or BL 0.175. These can be stored at −20°C for short-term use and −80°C for long-term use.
3.1.3. Special equipment
To lyse cells, use one of the following:
-
a.
Oster classic series blender (Cat# 006684–000-N01, Oster) with mini-blend jar (Cat# 004937–000-NP0, Oster), ice crushing blade (Cat# 004961–011-NP0, Oster), and bottom cap (Cat# 148381-000-090, Oster).
-
b.
Freezer mill (Cat# 6875, SPEX).
-
c.
Bead beater (Cat# 607, BioSpec) with acid-washed glass beads (Cat# 11079105, BioSpec) soaked in 1/100 concentrated HCl overnight and then washed in water until pH 7. Beads are then baked overnight twice.
Additional special equipment
SW65Ti rotor with SW41 tubes, to clarify lysates ≥8mL
10-mL Syringe and 16.5-G needle to remove lysates from tubes
Protein G Dynabeads (Cat #10003D, Thermo Fisher), to conjugate to antibodies and use for immunoprecipitation
Magnetic Particle Concentrator (MPC-S, Cat #A13346, Thermo Fisher), to concentrate dynabeads for wash steps and elution
3.2. PREPARATION OF YEAST LYSATE
As discussed later, altering lysis methods can have an impact on the overall composition of the kinetochore. Therefore, select a lysis method based on the components being studied.
Blender
Grow yeast to mid-log phase prior to harvesting. Harvest no fewer than 5 × 1010 cells which will yield approximately 2.6×10−12 mol Dsn1 after purification. If scaling up, adjust volumes accordingly. Keep cells and extracts on ice throughout all the subsequent steps.
Wash cells first with 25mL ice-cold water containing 1 × PMSF and then with 25mL cell wash buffer. Freeze pellet in liquid nitrogen.
Fill capsule with dry ice to the 4oz. level and crush. Leave to prechill blender capsule and blades and confirm blades can still shred dry ice. Bring level back to about the 4oz. mark on capsule with more dry ice. Add frozen cell pellet and shred for 20min. Check dry ice levels every 2min, refilling dry ice as necessary to maintain temperature without overfilling. Scrape cell pellet-dry ice mixture from blades and base into the capsule. As soon as all the dry ice sublimates and the lysate resembles kneaded dough, resuspend in 8mL cell lysis buffer. Premature addition of the lysis buffer results in frozen bubbles that are detrimental to kinetochore integrity.
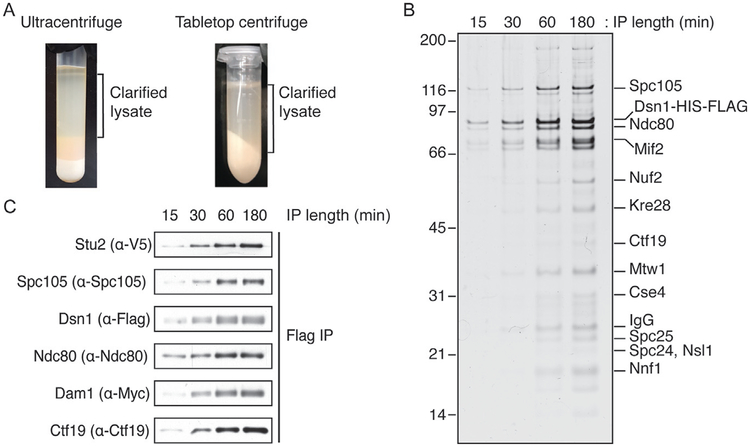
To clarify the lysate, the lysed cells need to be centrifuged to remove larger cell debris. If the resultant volume from the cell lysis is ≥8mL, the lysate should be loaded into the SW41 tubes and ultracentrifugated at 24,000rpm for 90min at 4°C. The clarified lysate is the largest layer above the cell debris and can be extracted using a syringe and needle (Fig. 2A).
FIG. 2.
The length of the immunoprecipitation affects kinetochore composition. (A) Following clarifying spins in either the ultracentrifuge or the tabletop centrifuge, the clarified yeast extract is present above a pellet but below a lipid layer as indicated. The yeast extract is removed through a side wall of the SW41 tube by using a needle and syringe. From the Eppendorf tube, the lysate can be pipetted out without disturbing the pellet. Kinetochores purified from yeast extracts by immunoprecipitation for the time indicated (minutes) at 4°C are visualized by silver-stained SDS-PAGE (B) or by immunoblot of the indicated proteins (C). In both cases, “core” kinetochore components (Table 1) are rapidly purified, but outer kinetochore components are enriched over time.
3.2.1. Freezer mill
Grow yeast to mid-log phase prior to harvesting. Harvest no fewer than 7.5 × 109 cells which will yield approximately 7.5 × 10−13 mol Dsn1 upon purification. If scaling up, adjust volumes accordingly. Keep cells and extracts on ice throughout all the subsequent steps.
Wash cells first with 3.75mL ice-cold water containing 1 × PMSF and then with 3.75mL cell wash buffer. Carefully resuspend pellet in an×equal volume of cell lysis buffer and pipette into liquid nitrogen dropwise to create “dots.”
Prechill freezer mill capsule(s) in liquid nitrogen, leaving the lid of the capsule(s) at room temperature. Remove capsule(s) one at a time from liquid nitrogen, pour out liquid nitrogen, and add “dots.” No liquid nitrogen should be left in the capsule as it will prevent the lid from being inserted. Secure lid and place capsule(s) into freezer mill. Work quickly to prevent melting of dots. Lyse cells for 2min and recover for 2min. Repeat 7–9 more times. Remove powder from capsule(s) and thaw on ice.
If the thawed cell lysate is ≥8mL, clarify by ultracentrifugation as with the blender method. If the lysed cells are ≤8mL, aliquot into Eppendorf tubes and centrifuge at maximum speed for 30min at 4°C in a tabletop centrifuge. The clarified lysate is the top layer and can be pipetted and collected into a single tube at this point (Fig. 2A).
3.2.2. Bead beat
Grow yeast to mid-log phase prior to harvesting. Harvest no fewer than 2.5 × 109 cells which will yield approximately 3.5 × 10−13 mol Dsn1 upon purification. If scaling up, adjust volumes accordingly. Keep cells and extracts on ice throughout all the subsequent steps.
Wash cells first with 1.25mL ice-cold water containing 1 × PMSF and then with 1.25mL cell wash buffer. Freeze pellet in liquid nitrogen.
Thaw pellet on ice and resuspend in 198μL cell lysis buffer per 2.5 × 109 cells. Aliquot 400μL into screw-top tubes (Cat# 02-681-339; Cat# 02-681-358, Fisher) and add equal volume of acid-washed glass beads. Lyse 35s in bead beater in cold room and recover on ice for 1.5min. Repeat three additional times. Poke holes in the bottoms of the tubes using a 16-G needle and place into labeled 1.5-mL Eppendorf tubes. Spin briefly (~3s) in tabletop centrifuge to collect lysate into the Eppendorf tubes.
Centrifuge the Eppendorf tubes at maximum speed for 30min at 4°C in a tabletop centrifuge. The clarified lysate is the top layer and can be pipetted and collected into a single tube at this point (Fig. 2A).
3.3. ANTIBODY–BEAD CONJUGATION
To purify the kinetochores, we use Protein G Dynabeads that are preconjugated with the antibodies required to purify epitope-tagged Dsn1. We find that purification of the 3FLAG epitope is the most efficient of the epitopes we have tested.
Wash 600μL Protein G Dynabeads 2× with 1mL 0.1M sodium phosphate buffer (pH 7.0). Resuspend beads in 90μL 0.1M sodium phosphate buffer (pH 7.0) and 30μL of 5mg/mL FLAG-M2 (Cat# F3165, Millipore Sigma) or V5 antibody (Cat# R960–25, Thermo Fisher Scientific). Gently agitate at room temperature for 15–40min. While agitation takes place, thaw dimethyl pimelimidate (DMP) (Cat #D8388, Millipore Sigma) from −80°C to room temperature for at least 15min.
Wash beads 2× in 1mL 0.1M sodium phosphate buffer (pH 7.0) containing 0.01% Tween 20 followed by 2× in 1mL 0.2M triethanolamine (pH 8.2) solution.
To cross-link the antibody of choice to the Protein G Dynabeads, resuspend 5.18mg DMP in 1mL of 0.2M triethanolamine (pH 8.2) and use the resulting solution to resuspend beads. Incubate for precisely 30min with rotational mixing at room temperature. The amount of DMP and the time are both crucial as excess of either can result in overcrosslinking.
Quench reaction by removing solution and resuspending in 1mL Tris–HCl (pH 7.5) and incubate by rotational mixing for 15min.
Wash beads 3× in 1mL PBS 0.1% Tween 20 and resuspend them in 600μL PBS 0.1% Tween 20. At this point, beads are ready to be used and can be stored at 4°C for up to a week. Longer storage can result in lower yield and/or higher background. There is no need to remove PBS 0.1% Tween 20 prior to immunoprecipitations and the slurry can be resuspended and added directly to whole-cell extracts.
3.4. IMMUNOPRECIPITATION OF KINETOCHORES
To ensure equal levels of kinetochores at the end of the purification, we always measure the protein concentration of the whole-cell extract by a Bradford assay (Cat# 500–0006, Bio-Rad) and then normalize the amount of lysate to the amount of beads. The ratio is optimized for the binding capacity of the FLAG dynabeads to Dsn1.
Normalize the amount of protein lysate to the amount of dynabeads. We find that a ratio of 12.6mg of clarified lysate sufficiently saturates 15μL of preconjugated dynabeads for purification of kinetochores.
Rotate yeast lysate with dynabeads at 4°C for anywhere between 15min and 3h. As will be discussed later, time can affect the composition of the kinetochores purified (Fig. 2B and C).
Wash beads 4× with IP wash buffer I using the MPC-S to concentrate beads after each wash. Transfer beads to a new tube with the last wash.
Wash beads 3× with IP wash buffer II using the MPC-S to concentrate beads after each wash. Transfer beads to a new tube with the last wash. At this point, the kinetochores are purified and ready to use. If any further reactions need to be performed with the kinetochores (such as kinase assays, phosphatase treatments, or rebinding assays), they should be done prior to elution from the dynabeads (London & Biggins, 2014; London et al., 2012; Miller et al., 2016).
The method of eluting and analyzing the kinetochore particles varies depending on the intended use postelution. See Section 3.5 for full details.
If necessary, the FLAG-eluted kinetochores can be immunoprecipitated a second time to further enrich for complete kinetochore particles. Using dynabeads conjugated with a different antibody (V5 is preferred), the preparations can be enriched for whichever kinetochore component is tagged with the corresponding epitope. The eluted kinetochores are incubated with the dynabeads for between 15min and 1h. Repeat steps 3–5 on the immunoprecipitation protocol.
3.5. ELUTION AND ANALYSIS OF KINETOCHORES
The method of elution is dependent on what the kinetochores will be used for post-elution and what concentration is desired. Here we outline some of the possible elution methods and how they can be analyzed postelution.
Mass spectrometry. After the last wash, wash beads 1 × in 50mM HEPES (pH 8.0). Elute using an equal volume of 2mg/mL RapiGest SF (Cat# 186001861, Waters Corp, Milford, MA) in 50mM HEPES (pH 8.0). This surfactant is mass spectrometry compatible and can be removed from the sample at low pH (Yu, Gilar, Lee, Bouvier, & Gebler, 2003). While the surfactant is strong enough to elute the proteins coimmunoprecipitating with Dsn1, it cannot disrupt the antibody–antigen bond so there is no enrichment of Dsn1 in the eluate. These samples can be stored at −20°C.
-
Biophysics, silver-stained gels, or immunoblots. The kinetochores should be eluted using an equal volume of 1× elution peptide to the beads and mixing the beads for 30min at room temperature. Adjusting the amount of purified peptide up to 2.5mg/mL (5×) can increase the sample eluted. Additionally, samples can be concentrated by reducing the elution volume down to one-third of the bead volume. This is not scalable and results in a reduction in overall yield, despite the increase in concentration.
To visualize most of the core kinetochore components, running the samples on a 4%–12% gradient gel and silver staining the gel is sufficient (Cat# NP0329BOX, Life Technologies) (Fig. 2B). Add sample buffer to 1 × +5% beta-mercaptoethanol to purified kinetochores and boil for 5min. Make BSA standards by using a 100ng/μL BSA stock to make an 8ng/μL standard and then serially diluting twofold down to 0.5ng/μL. The 0.5ng/μL sample is then diluted fivefold to create a 0.1ng/μL standard. Load purified kinetochore samples along with 10μL of each of the BSA standards to provide a range from 1 to 80ng to estimate the kinetochore concentration once stained. We find the Silver Quest Silver Staining Kit from Life Technologies (Cat# 6070) provides the best visualization. Samples in sample buffer can be stored at −20°C, while purified kinetochores without buffer can be stored at −20°C for the short term and −80°C for the long term.
Immunoblots. If the kinetochores are to be analyzed by immunoblot (Fig. 2C), the beads can be boiled for 3min in 1× sample buffer+5% beta-mercaptoethanol. This analysis is necessary to assay for the levels of components of the Dam1 complex, which are not visible by silver staining. The concentration can be increased by reducing the volume used in the elution to one-third of the bead volume. Prior to loading in an SDS-PAGE gel, samples should be boiled for 5min. Gel percentage can be varied based on the molecular weight of components of interest. We typically use 7.5%, 10%, and 12.5% acrylamide gels. These samples can be stored at −20°C.
4. DIFFERENCES IN KINETOCHORE COMPOSITION BASED ON PURIFICATION
The protocol listed above is our standard protocol. However, altering some steps can lead to a different composition of the purified kinetochores. Depending on the intended use for the purified kinetochores, these changes could help determine how to best optimize the purification.
4.1. KINETOCHORE MUTATIONS
The Mis12 complex is the key to purification of kinetochores. Purification of any subcomponent of the Mis12 complex, but not any other complex, yields intact kinetochores (Akiyoshi et al., 2010). This may be due to the importance of the Mis12 complex in binding to the CCAN and providing a platform for the outer kinetochore to assemble (Biggins, 2013; Musacchio & Desai, 2017). Crucially, this interaction between the Mis12 complex and the CCAN is regulated by two conserved Aurora B kinase phosphorylation sites on Dsn1 at S240 and S250 (Akiyoshi, Nelson, & Biggins, 2013; Dimitrova, Jenni, Valverde, Khin, & Harrison, 2016). Mutation of these two sites to create a phosphomimetic Dsn1–2D mutant protein (S240D, S250D) leads to enrichment of inner kinetochore components in the kinetochore purification (Akiyoshi, Nelson, & Biggins, 2013).
4.2. TIME
Kinetochores can be purified by incubating clarified lysate with dynabeads for as little as 15min (Fig. 2B and C). These kinetochore particles still contain all the components, but they are comparatively less concentrated than kinetochores purified following longer incubations (Fig. 2B and C). Increasing incubation time to up to an hour results in complete purification of the KMN complex and outer components such as Dam1 and Stu2 (Fig. 2C). If the kinetochore purification is being performed to assess functionality of outer kinetochore components, performing the immunoprecipitation for 3h enriches for the most outer kinetochore components (Fig. 2C).
4.3. LYSIS METHOD
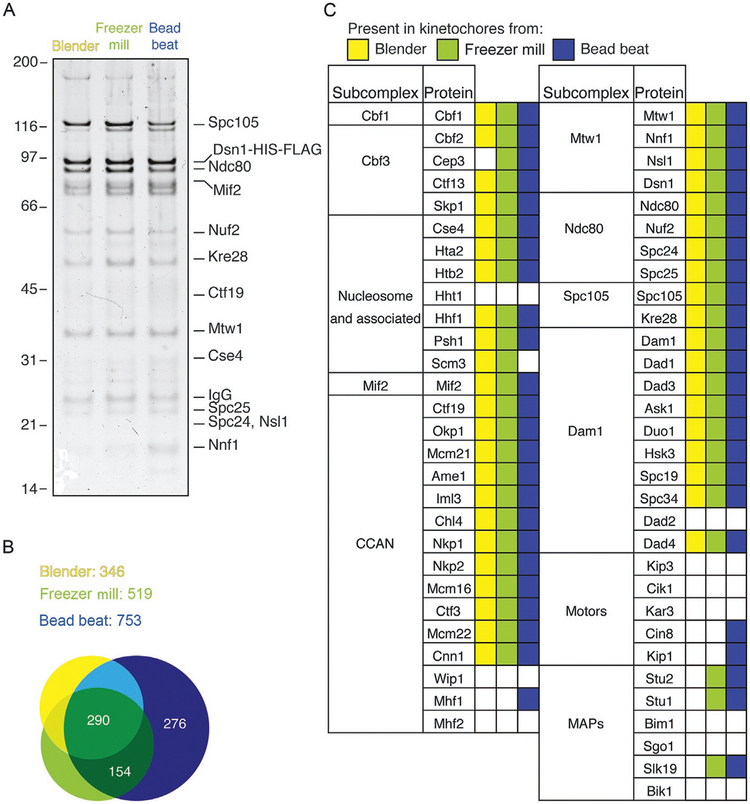
The choice for how to lyse the cells has a significant impact on the composition of the resulting kinetochores. Purification using the blender to lyse cells results in the fewest number of proteins coimmunoprecipitated with Dsn1 (Fig. 3). This set includes all of the “core” kinetochore components and provides an excellent tool to study kinetochore function (Akiyoshi et al., 2010) (Fig. 3A and B; Table 1). However, this purification results in kinetochores lacking some MAPs and motors, which have been shown to play critical roles at the kinetochore in addition to elsewhere in the cell (Fig. 3C) (Miller et al., 2016). Using the freezer mill or bead beater for cell lysis leads to more copurifying proteins, including MAPs and motors. Purifications from both lysis methods encompass almost all the proteins purified by immunoprecipitation from blender lysates (Fig. 3B). In terms of number of unique proteins purified, lysates from the bead beat method yield the highest number and include the motors Cin8 and Kip1 (Fig. 3C). While the additional proteins purified may have a strong effect on kinetochore function, they also contain numerous undesirable contaminants (unpublished data). Purified kinetochores from freezer mill lysates therefore maximize for additional crucial kinetochore components while minimizing additional contaminants (Fig. 3B and C).
FIG. 3.
Lysis method affects the number of unique proteins coimmunoprecipitated with Dsn1. (A) Kinetochores purified from yeast extract by immunoprecipitation of Dsn1-HIS-FLAG for 3h at 4°C are visualized by silver-stained SDS-PAGE. (B) Shotgun mass spectrometric analysis was performed on kinetochore purifications from the three lysis methods of purification and the number of unique proteins detected is indicated. Purification from blender lysates yields the fewest unique proteins, followed by freezer mill, with bead beat yielding the highest number of different proteins coimmunoprecipitated. There is a large overlap of the three sets indicating that all three lysis methods yield kinetochores with differences in the additional components purified. (C) Summary of kinetochore composition comparing the three lysis methods shows that the vast majority of the known kinetochore components are detected by all methods. However, the bead beat lysis, and to a smaller extent the freezer mill lysis, yields more of the transiently associated outer kinetochore components.
4.4. BUFFERS
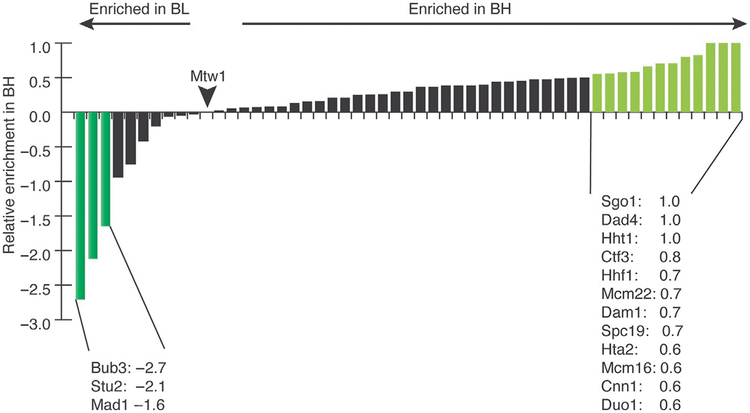
The two buffers used in the kinetochore purifications, BH 0.15 and BL 0.175, differ in overall salt concentration, pH, and the size of the anion in the salts used. By mass spectrometry analysis of freezer mill-purified kinetochores, it appears that neither buffer results in more or less components purified, but affects the apparent stoichiometry of the resulting kinetochores (Fig. 4). Purification using BH 0.15 results in more detectable peptides of the “core” kinetochore components relative to the number of Mtw1 peptides purified (Fig. 4). In contrast, purification using BL 0.175 enriches for kinetochore components that are more transiently associated with the kinetochore (Fig. 4).
FIG. 4.
Purification in buffer BL 0.175 leads to enrichment of some transient components. Nonquantitative shotgun mass spectrometry was performed on kinetochores purified using the freezer mill lysis method in either buffer BH 0.15 or BL 0.175. The resulting number of total peptides for every protein was normalized to the number of peptides of the Mis12 complex component, Mtw1. To compare across the two sets, we plotted a relative enrichment of a certain protein in BH 0.15 compared to BL 0.175 using the formula: (normalized BH 0.15 peptides−normalized BL 0.175 peptides)/normalized BH 0.15 peptides. By this analysis, most proteins were slightly more enriched in BH 0.15 vs BL 0.175. There were 12 proteins that were more than twofold enriched in BH 0.15 and 3 proteins that were more than twofold enriched in BL 0.175. The proteins enriched in BH 0.15 can be classified into three broad categories: the Dam1 complex, the Ctf3-Cnn1 arm of the kinetochore, and histones/chromatin. The major proteins enriched with the buffer BL 0.175 are the SAC proteins and the MAP, Stu2.
4.5. CELL CYCLE PHASE
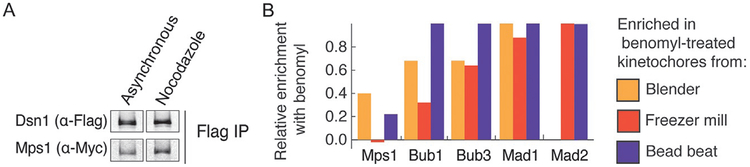
Kinetochores, in addition to promoting attachment to microtubules, also serve as hubs for checkpoint protein signaling when there are incorrect/incomplete attachments (Hoyt et al., 1991; Li & Murray, 1991). To gain a better understanding of the SAC, cells can be arrested by preventing SAC satisfaction. We use benomyl, a microtubule-depolymerizing drug, to create unattached kinetochores and enrich for kinetochores containing checkpoint proteins (Fig. 5A and B). Checkpoint proteins copurify with kinetochores purified from benomyl-treated cells, regardless of the lysis method used to purify kinetochores (Fig. 5B). The hierarchical recruitment and posttranslational modifications essential to checkpoint signaling and maintenance can thus be studied in vitro (London & Biggins, 2014; London et al., 2012).
FIG. 5.
Benomyl treatment leads to enhanced copurification of SAC proteins. (A) Immunoblot of Dsn1-HIS-FLAG and 10myc-Mps1 from kinetochores purified from bead beat lysis method. (B) Mass spectrometry analysis of kinetochores purified from untreated vs benomyl-treated cells using the blender, freezer mill, or bead beat lysis method. To measure SAC protein enrichment, peptides of each SAC protein were normalized to the number of peptides of Mtw1 from the respective sample. Enrichment of the SAC proteins following benomyl treatment for each type of lysis was calculated by: (normalized benomyl peptides−normalized untreated peptides)/normalized benomyl peptides.
4.6. TWO-STEP IMMUNOPRECIPITATION
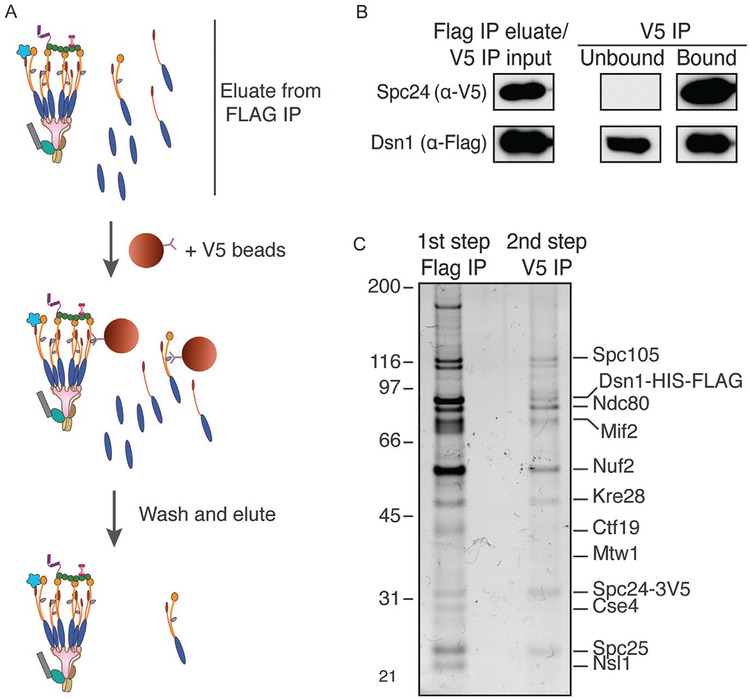
The immunoprecipitation of Dsn1 purifies free Dsn1 as well as Dsn1-containing sub-complexes in addition to more complete kinetochore particles. These complexes can potentially dilute the effect seen with complete kinetochores in follow-up experiments. To minimize these subcomplexes, a second immunoprecipitation can be performed using a different epitope tag on a separate kinetochore component such as Spc24 (Fig. 6A). This second selection step eliminates free Dsn1 complexes (Fig. 6B). As expected, the resulting kinetochore particles have closer to a 1:1:1 stoichiometry among Dsn1, Spc105, and Ndc80 (Fig. 6C).
FIG. 6.
Two-step immunoprecipitation of kinetochores. (A) Workflow for two-step immunoprecipitation using a second immunoprecipitation of Spc24–3V5 following purification of Dsn1-HIS-FLAG. (B) Immunoblot of two-step immunoprecipitation shows full depletion of Spc24–3V5 but not Dsn1-HIS-FLAG. (C) Silver staining of SDS-PAGE of one-step and two-step purified kinetochores.
5. CONCLUSIONS
The kinetochore purification protocol has proven to be a very powerful tool for studying the yeast kinetochore and uncovering specific regulatory events and components that shape its critical mitotic functions (Akiyoshi, Nelson, & Biggins, 2013; Akiyoshi, Nelson, Duggan, et al., 2013; Akiyoshi et al., 2010; London & Biggins, 2014; London et al., 2012; Miller et al., 2016; Sarangapani et al., 2013, 2014). Isolated kinetochores can be used to determine when and how different components are recruited to the kinetochore and what modifications are required. Purifying kineto-chores from strains containing loss-of-function mutants, as well as phosphodeficient or phosphomimetic mutants, allows the dependency relationships between the various kinetochore components to be analyzed (Akiyoshi, Nelson, & Biggins, 2013; Akiyoshi, Nelson, Duggan, et al., 2013; London & Biggins, 2014; London et al., 2012; Sarangapani et al., 2013). If these modifications are lethal in vivo, the purified kinetochores can be treated with kinases and phosphatases postpurification to determine the importance of phosphorylation in the recruitment or maintenance of different kinetochore components (London et al., 2012). Analysis of these kinetochores by mass spectrometry, immunoblotting, or silver staining of gels can provide powerful insight into the regulation of kinetochores (Akiyoshi, Nelson, & Biggins, 2013; Akiyoshi, Nelson, Duggan, et al., 2013; Akiyoshi et al., 2010; London & Biggins, 2014; London et al., 2012).
In addition to the compositional analysis that can be performed on the purified kinetochores, they are also functional for biophysical assays. Furthermore, compositional analyses highlight population differences, while the biophysical assays provide insight at a single-particle level. One such tool is total internal reflection fluorescence microscopy, which can be used to visualize single kinetochore particles (Kudalkar, Davis, & Asbury, 2016) and measure the ability of the kinetochores to bind and track microtubules (Akiyoshi et al., 2010). By tagging various kinetochore components with different fluorophores, we can also determine how frequently they are present together when the kinetochore is bound to either the microtubule lattice or the tip and whether they remain bound to disassembling microtubules (Sarangapani et al., 2014). Another tool is an optical trap, which can be used to measure the strength of the kinetochore–microtubule attachment. Kinetochores are bound to antibody-coated polystyrene beads that can be moved to the end of dynamic or taxol-stabilized microtubules and placed under a force using the optical trap (Akiyoshi et al., 2010; Miller et al., 2016; Sarangapani et al., 2013, 2014). By ramping up the force and recording when the linkage is ruptured, an average strength of the attachment can be determined in the presence and absence of various kinetochore components. Alternatively, by placing the beads under a constant force, we can determine how long the kinetochores can sustain the attachment. This assay also can be used to gain a greater understanding of how the kinetochores can alter microtubule dynamics (Akiyoshi et al., 2010; Miller et al., 2016; Sarangapani et al., 2013, 2014). Finally, the purified kinetochores have been visualized by negative-stain EM and provide a way to potentially obtain higher structural information in the future.
In summary, the yeast kinetochore purification has helped broaden our understanding of kinetochores and the mechanisms they use to carry out their complex functions. In the future, this technique may be adapted to purify kinetochores from other organisms to perform similar biochemical and biophysical assays. It should also be useful for developing new assays to further dissect the dynamic process of chromosome segregation in the future.
ACKNOWLEDGMENTS
We are grateful to all members of the Biggins’ lab for ideas about the kinetochore purification procedure. This work was supported by NIH R01GM064386 to S.B., an American Cancer Society fellowship (PF-14-152-01-CCG) and National Institutes of Health Inter-Disciplinary Training Grant award (T32CA080416) through the National Cancer Institute to A.G., and the National Science Foundation Graduate Research Fellowship Program to L.B.K. S.B. is also an investigator of the Howard Hughes Medical Institute.
REFERENCES
- Akiyoshi B, Nelson CR, & Biggins S (2013). The Aurora B kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics, 194(3), 785–789. 10.1534/genetics.113.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B, Nelson CR, Duggan N, Ceto S, Ranish JA, & Biggins S (2013). The Mub1/Ubr2 ubiquitin ligase complex regulates the conserved Dsn1 kinetochore protein. PLoS Genetics, 9(2), e1003216 10.1371/journal.pgen.1003216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, et al. (2010). Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature, 468(7323), 576–579. 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury CL, Tien JF, & Davis TN (2011). Kinetochores’ gripping feat: Conformational wave or biased diffusion? Trends in Cell Biology, 21(1), 38–46. 10.1016/j.tcb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auckland P, Clarke NI, Royle SJ, & McAinsh AD (2017). Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment. The Journal of Cell Biology, 216(6), 1623–1639. 10.1083/jcb.201607096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S (2013). The composition, functions, and regulation of the budding yeast kineto-chore. Genetics, 194(4), 817–846. 10.1534/genetics.112.145276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, & Murray AW (2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes & Development, 15(23), 3118–3129. 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, & Gerton JL (2007). Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Molecular Cell, 26(6), 853–865. 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM (2014). The kinetochore. Cold Spring Harbor Perspectives in Biology, 6(7), a015826 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, & Desai A (2006). The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell, 127(5), 983–997. 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Clarke L, & Carbon J (1983). Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature, 305(5929), 23–28. [DOI] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, & Sorger PK (2003). Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes & Development, 17(23), 2902–2921. 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova YN, Jenni S, Valverde R, Khin Y, & Harrison SC (2016). Structure of the MIND complex defines a regulatory focus for yeast kinetochore assembly. Cell, 167(4), 1014–1027. e1012. 10.1016/j.cell.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad B, & Kops GJ (2016). Attachment issues: Kinetochore transformations and spindle checkpoint silencing. Current Opinion in Cell Biology, 39, 101–108. 10.1016/j.ceb.2016.02.016. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR 3rd, & Cleveland DW (2006). The human CENP-A centromeric nucleosome-associated complex. Nature Cell Biology, 8(5), 458–469. 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Gonen S, Akiyoshi B, Iadanza MG, Shi D, Duggan N, Biggins S, et al. (2012). The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nature Structural & Molecular Biology, 19(9), 925–929. 10.1038/nsmb.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady DL, Ratliff RL, Robinson DL, McCanlies EC, Meyne J, & Moyzis RK (1992). Highly conserved repetitive DNA sequences are present at human centromeres. Proceedings of the National Academy of Sciences of the United States of America, 89(5), 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, & Sorger PK (2001). Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell, 106(2), 195–206. [DOI] [PubMed] [Google Scholar]
- Holland AJ, & Cleveland DW (2009). Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nature Reviews. Molecular Cell Biology, 10(7), 478–487. 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, & Roberts BT (1991). S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell, 66(3), 507–517. 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Kalantzaki M, Kitamura E, Zhang T, Mino A, Novak B, & Tanaka TU (2015). Kinetochore-microtubule error correction is driven by differentially regulated interaction modes. Nature Cell Biology, 17(4), 421–433. 10.1038/ncb3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudalkar EM, Davis TN, & Asbury CL (2016). Single-molecule total internal reflection fluorescence microscopy. Cold Spring Harbor Protocols, 2016(5), pdb top077800. 10.1101/pdb.top077800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, & Carbon J (1991). A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell, 64(4), 717–725. [DOI] [PubMed] [Google Scholar]
- Li R, & Murray AW (1991). Feedback control of mitosis in budding yeast. Cell, 66(3), 519–531. 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X, & Nicklas RB (1995). Mitotic forces control a cell-cycle checkpoint. Nature, 373(6515), 630–632. 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- London N, & Biggins S (2014). Mad1 kinetochore recruitment by Mps1-mediated phosphor-ylation of Bub1 signals the spindle checkpoint. Genes & Development, 28(2), 140–152. 10.1101/gad.233700.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Ceto S, Ranish JA, & Biggins S (2012). Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Current Biology, 22(10), 900–906. 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, & Cheeseman IM (2016). The molecular basis for centromere identity and function. Nature Reviews. Molecular Cell Biology, 17(1), 16–29. 10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MP, Asbury CL, & Biggins S (2016). A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell, 165(6), 1428–1439. 10.1016/j.cell.2016.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, & Desai A (2017). A molecular view of kinetochore assembly and function. Biology (Basel), 6(1), 1–47. 10.3390/biology6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, & Salmon ED (2007). The spindle-assembly checkpoint in space and time. Nature Reviews. Molecular Cell Biology, 8(5), 379–393. 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nicklas RB (1997). How cells get the right chromosomes. Science, 275(5300), 632–637. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, & Koch CA (1969). Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of Mal-oriented chromosomes. The Journal of Cell Biology, 43(1), 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr B, & Compton DA (2013). A double-edged sword: How oncogenes and tumor suppressor genes can contribute to chromosomal instability. Frontiers in Oncology, 3, 164 10.3389/fonc.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S (2016). Mechanisms of mitotic spindle assembly. Annual Review of Biochemistry, 85, 659–683. 10.1146/annurev-biochem-060815-014528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, & Pelletier L (2017). Mitotic spindle assembly in animal cells: A fine balancing act. Nature Reviews. Molecular Cell Biology, 18(3), 187–201. 10.1038/nrm.2016.162. [DOI] [PubMed] [Google Scholar]
- Sarangapani KK, Akiyoshi B, Duggan NM, Biggins S, & Asbury CL (2013). Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proceedings of the National Academy of Sciences of the United States of America, 110(18), 7282–7287. 10.1073/pnas.1220700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani KK, Duro E, Deng Y, Alves Fde L, Ye Q, Opoku KN, et al. (2014). Sister kinetochores are mechanically fused during meiosis I in yeast. Science, 346(6206), 248–251. 10.1126/science.1256729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F (1991). Getting started with yeast. Methods in Enzymology, 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, & Baker RE (2007). Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proceedings of the National Academy of Sciences of the United States of America, 104(25), 10571–10576. 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU (2010). Kinetochore-microtubule interactions: Steps towards bi-orientation. The EMBO Journal, 29(24), 4070–4082. 10.1038/emboj.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, et al. (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell, 108(3), 317–329. [DOI] [PubMed] [Google Scholar]
- Tytell JD, & Sorger PK (2006). Analysis of kinesin motor function at budding yeast kinetochores. The Journal of Cell Biology, 172(6), 861–874. 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooff JJ, Tromer E, van Wijk LM, Snel B, & Kops GJ (2017). Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Reports, 18(9), 1559–1571. https://doi.org/10.15252/embr.201744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH Jr., McDonald KL, et al. (1995). Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. The Journal of Cell Biology, 129(6), 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YQ, Gilar M, Lee PJ, Bouvier ES, & Gebler JC (2003). Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Analytical Chemistry, 75(21), 6023–6028. 10.1021/ac0346196. [DOI] [PubMed] [Google Scholar]