Abstract
Vanadium-dependent haloperoxidases (VHPOs) are fascinating enzymes that facilitate electrophilic halogen incorporation into electron-rich substrates, simply requiring vanadate, a halide source, and cosubstrate hydrogen peroxide for activity. Initially characterized in fungi and red algae, VHPOs were long believed to have limited regio-, chemo-, and enantioselectivity in the production of halogenated metabolites. However, the recent discovery of homologues in the biosynthetic gene clusters of the stereoselectively halogenated meroterpenoids from marine-derived Streptomyces bacteria has revised this paradigm. Their intriguing transformations have both enhanced and contributed to the fields of synthetic organic and natural product chemistry. We, herein, describe the expression, purification, and chemical assays of two characterized vanadium-dependent chloroperoxidase enzymes (NapH1 and Mcl24), and one homologue devoid of chlorination activity (NapH3), involved in the biosyntheses of halogenated meroterpenoid products.
1. INTRODUCTION
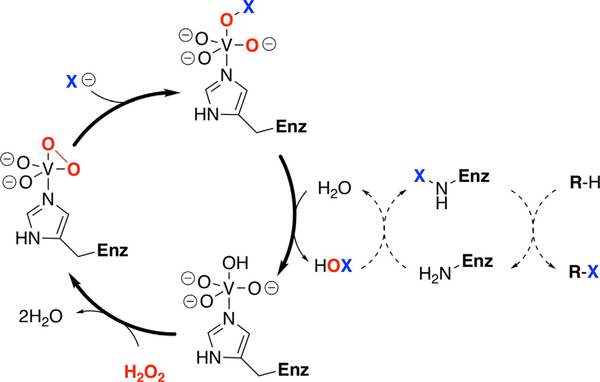
Halogenated natural products are particularly intriguing due to their incorporation of electronegative halides into hydrocarbon backbones (Gribble, 2010). These metabolites are particularly prevalent in marine environments due to high concentrations of halides in sea water. The discovery of novel halogenated metabolites is intriguing from a chemical diversity perspective, in addition to the intricate enzymology responsible for the addition of these group 17 elements. A variety of biosynthetic approaches to incorporate halogens into molecules have been elucidated (Agarwal et al., 2017), with one of the most intriguing mechanisms exhibited by the vanadiumdependent haloperoxidase (VHPO) class of enzymes (Winter & Moore, 2009). Utilizing a vanadate prosthetic group to coordinate cosubstrate hydrogen peroxide, VHPOs perform a two-electron oxidation of electrophilic halides to generate a vanadium-bound hypohalous acid. This electrophilic halenium source can then react with electron-rich scaffolds, facilitating halogenation, cyclization, and hydration reactions. Intriguingly, VHPOs are not redox active at the vanadium metal center, remaining in the V5+ oxidation state, and thus do not require additional regeneration systems nor suffer oxidative inactivation during catalysis (Fig. 1). VHPO enzymes are more specifically categorized based on the most electronegative element they can oxidize; the vanadium-dependent chloroperoxidases (VCPOs) discussed in this chapter can therefore oxidize chloride, bromide, and iodide.
Fig. 1.
Proposed catalytic mechanism for the two-electron oxidation of halide ions (X−) by VHPO enzymes. In the Streptomyces VCPO enzymes, the oxidized HOX species is suspected to be intercepted by a neighboring lysine, generating an enzyme-bound halogen species believed to be responsible for the specificity. In the fungal and algal VHPOs, which do not exhibit specificity, the oxidized HOX species is released to rather react directly with electron-rich substrates.
First identified in red algae and fungi, VHPO enzymes have been extensively studied for their roles in organohalogen production, and a thorough discussion of their history, characterization, biological potentials, and chemical applications are included within this series (Wever, Krenn, & Renirie, 2018). However, the discovery of VHPO genes within Streptomyces biosynthetic pathways of the regio- and stereospecifically halogenated meroterpenoid natural products napyradiomycin (Henkel & Zeeck, 1991; Shiomi et al., 1986) and merochlorin (Kaysser et al., 2012) was particularly intriguing. Heterologous expression of the nap (Winter et al., 2007) and mcl (Kaysser et al., 2012) gene clusters supported the hypothesis that VHPOs were responsible for halogenating the hybrid polyketide– terpenoid molecules. Subsequent in vitro characterization of recombinant VCPO enzymes NapH1 (Bernhardt, Okino, Winter, Miyanaga, & Moore, 2011), Mcl24 (Diethelm, Teufel, Kaysser, & Moore, 2014; Teufel et al., 2014), and homologue NapH3 (Miles et al., 2017) further validated this hypothesis through elucidation of the distinct catalytic transformations. Although few examples have been characterized thus far, Streptomyces VCPO enzymes are capable of remarkable chemistry, inspiring biomimetic organic total syntheses, while also benefiting significantly from the contributions of the fields of organic and natural product chemistry (Moore, 2018).
This chapter will cover both the characterized VCPOs, NapH1 and Mcl24, involved in halogenated meroterpenoid biosynthesis and draw comparisons to homologue NapH3, which is not known to facilitate halogenating chemistry despite high sequence identity. Details, explanations, and challenges regarding their cloning, expression, purification, and enzymatic activity will be further explored.
2. MOLECULAR BIOLOGY OF STREPTOMYCES VCPO GENES
The identification of VCPO encoding genes in Streptomyces bacteria was facilitated by their coclustering within biosynthetic gene clusters associated with meroterpenoid natural products, thereby allowing for the context of their biosynthetic reactivity. All cloning of these VCPO enzymes were done using relatively GC-rich actinomycete gDNA; proper precautions should be taken including the proper choice of DNA polymerase and potential use of DMSO in the PCR reaction. The napH1 and napH3 genes were obtained by PCR amplification from Streptomyces sp. CNQ-525 gDNA. The napH1 gene was ligated into a pHis8 vector, for expression of protein with an N-terminal octahistidine tag (Jez, Ferrer, Bowman, Dixon, & Noel, 2000) using the NcoI and NotI restriction sites using standard methodology (Bernhardt et al., 2011). In this same publication, the authors analogously ligated napH3 into pHis8 that ultimately yielded soluble heterologous protein expression; however, no haloperoxidase activity was observed. Future examination of the annotated napH3 gene using GeneMark (Besemer & Borodovsky, 2005) identified additional putative translational start sites. A larger open reading frame was subsequently amplified using PCR, and ligated into a pET28a vector (Novagen) again for expression of protein with an N-terminal hexahistidine tag, using the NdeI and HindIII restriction sites (Miles et al., 2017). This construct also resulted in soluble protein expression and is believed to be the full, functional gene. The mcl24 gene was obtained by PCR amplification from Streptomyces sp. CNH-189 gDNA and was inserted into a pHis8 vector (Jez et al., 2000) using the BamHI and HindIII restriction sites, generating an N-terminal octahistidine-tagged protein (Teufel et al., 2014).
To probe the roles of key active site residues in Streptomyces VCPO enzymes based on sequence alignment to red algal VHPO enzymes, sitedirected mutagenesis was performed according to standard procedures. Owing to the high GC percentage of Streptomyces sp., the same precautions as detailed earlier should be taken, and with the expectation of a lower rate of success for mutations.
3. HETEROLOGOUS PROTEIN EXPRESSION AND PURIFICATION
To our satisfaction, these Streptomyces VCPO enzymes have thus far been readily expressed following transformation into Escherichia coli BL21-Gold (DE3) cells. However, this observation is not applicable to all VCPO enzymes, as recombinant protein expression of NapH4 from the napyradiomycin biosynthetic pathway (Bernhardt et al., 2011) and Mcl40 from the merochlorin biosynthetic pathway (unpublished results) have failed to yield soluble protein. Additional approaches to yield soluble and stable protein are currently under investigation within our lab. Although slight deviations exist in the literature procedures for the published VCPO enzymes, a general unified procedure for their recombinant overexpression and purification is outlined.
3.1. Recombinant VCPO Expression
3.1.1. Equipments
Erlenmeyer culture flasks (50 mL and 2.8 L)
Centrifuge tubes (500 mL, Thermo Scientific Nalgene)
Temperature-controlled centrifuge
Temperature-controlled shaker
3.1.2. Materials
E. coli BL21-Gold (DE3) competent cells
Lysogeny Broth (LB)
Terrific Broth (TB)
LB agar plates
50 mg/mL aqueous kanamycin solution
1 M isopropyl-β-D-thiogalactopyranoside (IPTG)
Buffer A (50 mM Tris pH 8.0, 500 mM NaCl, 25 mM imidazole)
3.1.3. Procedure
Transform E. coli BL21-Gold (DE3) competent cells with ~100 ng of vector according to the manufacturer’s protocol. Plate each transformation on LB agar plates with 50 μg/mL kanamycin and incubate overnight at 37°C.
Select one positive colony from the plate and inoculate 25 mL of LB media with 50 μg/mL kanamycin. Incubate overnight at 37°C with shaking at ~200 rpm.
To 1.5 L of TB media supplemented with 50 μg/mL kanamycin add 15 mL of the overnight LB culture. Incubate at 37°C with shaking at ~200 rpm until the OD600 is approximately 0.6 (typically 3–4 h).
Cool the incubator to 18°C and maintain shaking at ~200 rpm for 1 h.
Add IPTG to a final concentration of 100 μM to induce protein expression. Shake overnight (~16 h) at ~200 rpm.
Pellet cells by centrifugation (8000×g, 5 min, 4°C), decant the supernatant media, and resuspend the cell pellets in 40 mL buffer A (50 mM Tris pH 8.0, 500 mM NaCl, 25 mM imidazole).
Store the resuspended cell pellets at −80°C or proceed directly to protein purification.
3.1.4. Notes
Overnight LB starter cultures can instead be initiated from a glycerol stock; however, we consistently observed better results from selecting a freshly transformed colony.
A final concentration of 10% glycerol was added to buffer A for NapH1 to aid in protein stability, or to all overexpressed VCPO cell pellets if they were to be stored at −80°C.
Minimal difference in yields or activities was observed between stored and freshly purified protein preparations.
3.2. Purification of Recombinant Streptomyces VCPO Enzymes
3.2.1. Equipments
Beaker (variable size)
Magnetic stir bar and stir plate
Sonifier
Temperature-controlled centrifuge
Centrifuge tubes (50 mL, Oak Ridge)
HisTrap FF column (GE Healthcare) ӒKTA FPLC system (GE Healthcare)
Fraction collection tubes
Polyacrylamide gel apparatus
Amicon Ultra-15 10,000 MWCO spin concentrator (EMD Millipore) PD-10 desalting column (GE Healthcare)
3.2.2. Materials
Buffer A (50 mM Tris pH 8.0, 500 mM NaCl, 25 mM imidazole)
Buffer B (50 mM Tris pH 8.0, 500 mM NaCl, 500 mM imidazole)
12% SDS-polyacrylamide gels
NapH1 and NapH3 storage buffer (25 mM HEPES-KOH pH 8.0, 300 mM KCl, 10% glycerol)
Mcl24 storage buffer (25 mM HEPES-NaOH pH 7.6, 200 mM NaCl, 10% glycerol)
3.2.3. Procedure
The resuspended cell pellet in buffer A was transferred to a beaker equipped with a small magnetic stir bar and kept on ice. Cells were lysed by sonication using a Branson digital sonifier (40% intensity) with magnetic stirring using a 15 s on/45 s off cycle for 40 min.
Cell debris was pelleted by centrifugation (16,000×g, 30 min, 4°C).
The clarified supernatant was loaded onto a 5 mL HisTrap FF column (GE Healthcare), previously equilibrated with buffer A, at a flow rate of 2 mL/min using an ӒKTA FPLC system (GE Healthcare).
After loading the supernatant, the HisTrap column was washed with 5–10 column volumes (25–50 mL) of buffer A to remove unbound proteins. A gradient from 0% to 10% buffer B (50 mM Tris pH 8.0, 500 mM NaCl, 500 mM imidazole) was initiated over 10 mL, followed by five column volumes (25 mL) of 10% buffer B to elute weakly bound proteins.
Polyhistidine-tagged proteins were eluted using a gradient of 10%– 100% buffer B over 60 mL, collecting 5 mL fractions.
Fractions containing purified recombinant VCPO were identified using SDS-PAGE (12% acrylamide gel), pooled, and concentrated by centrifugation (3500×g, 4°C) using an Amicon Ultra-15 10,000 MWCO spin concentrator (EMD Millipore) to under 2.5 mL.
A PD-10 desalting column (GE Healthcare) was equilibrated following manufacturer instructions with >5 column volumes of cold storage buffer (NapH1 and NapH3: 25 mM HEPES-KOH pH 8.0, 300 mM KCl, 10% glycerol; Mcl24: 25 mM HEPES-NaOH pH 7.6, 200 mM NaCl, 10% glycerol).
The concentrated VCPO enzyme was applied to the PD-10 column, noting the volume added. Cold storage buffer was then added to bring the final volume applied to the column to precisely 2.5 mL. The buffer-exchanged enzyme was eluted using 3.5 mL of cold storage buffer, collecting a single fraction on ice. The PD-10 column can be regenerated through extensive washing (>10 column volumes) with storage buffer.
The recombinant VCPO enzyme was again concentrated (3500×g, 4°C) using an Amicon Ultra-15 10,000 MWCO spin concentrator (EMD Millipore) to approximately 1 mL.
Next, the protein concentration was determined via the Bradford method using bovine serum albumin as a standard.
Finally, the protein was aliquoted into smaller fractions (50–100 μL), frozen on dry ice, and stored at −80°C until needed.
3.2.4. Notes
This procedure is also amenable to manual batch protein purification using a stepwise imidazole elution gradient and Ni-NTA resin.
Removing the N-terminal polyhistidine tag, possible through the use of a thrombin cleavage site, has little impact on protein activity or stability and was forgone for all published meroterpenoid biochemical assays.
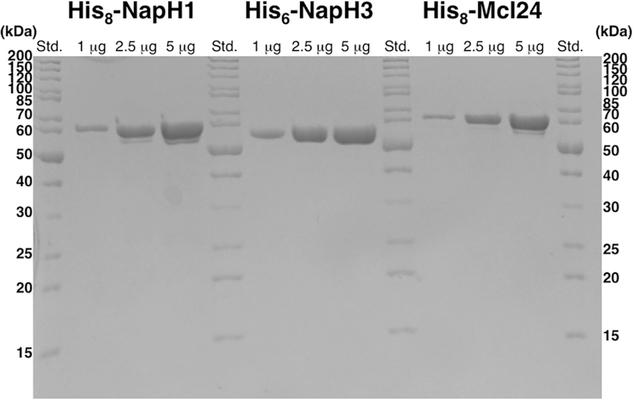
To investigate their unique chemistry, the PD-10 desalting procedure described earlier is suitable for the removal of imidazole and retaining active VCPO enzymes in >90% purity (Fig. 2). Sephadex G-200 size-exclusion chromatography can also be used to additionally purify VCPO enzymes to greater purity with negligible impact on protein activity.
When thawing VCPO aliquots for activity assays, quickly thaw them using your fingers, and then subsequently place in ice until needed. NapH3 shows stability to successive freeze–thaw cycles; however, both NapH1 and Mcl24 will precipitate over time and should be used shortly after thawing (<1 h).
-
Typical production yields of recombinant VCPO proteins are as follows:
NapH1: 5–10 mg/L; NapH3: 70–100 mg/L; Mcl24: 100–140 mg/L.
Fig. 2.
SDS-PAGE gel (12%) of purified His8-NapH1 (left), His6-NapH3 (middle), and His8-Mcl24 (right). The bands are consistent with the predicted molecular weights of 58.2, 53.8, and 57.6 kDa, respectively.
4. ENZYMATIC ASSAYS OF HETEROLOGOUSLY EXPRESSED STREPTOMYCES VCPO ENZYMES
4.1. Monochlorodimedone Assay
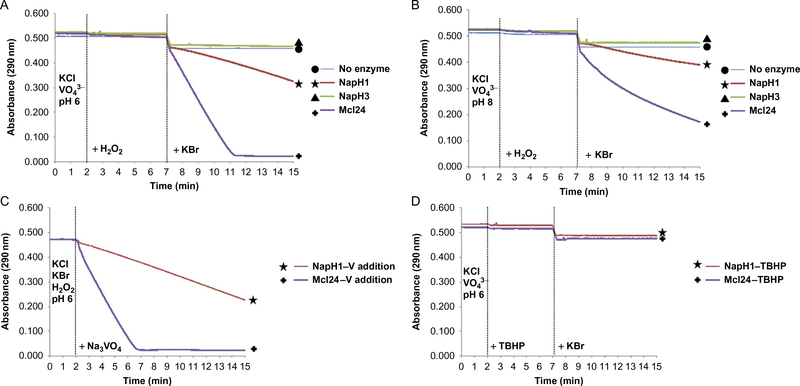
The classical way of investigating the halogenation potential of VHPO enzymes has involved the halogenation of monochlorodimedone (MCD). This assay is more thoroughly described in the algal VHPO article within the same series (Wever et al., 2018); however, a brief discussion of its application to marine Streptomyces VCPOs will be mentioned here. Algal and fungal VHPO enzymes generate a diffusible hypohalous acid (HOX species) that departs the active site. This can then halogenate the α-carbon between the two carbonyl moieties of MCD, disrupting the conjugation and diminishing the absorbance at 290 nm. However, a diffusible hypohalous acid would fail to provide the regio- and stereoselectivity synonymous with Streptomyces VCPO enzymatic transformations. Instead, the “Cl+” species generated is proposed to be transiently intercepted by a neighboring lysine residue, generating a chloramine intermediate capable of facilitating this specific chemistry (Diethelm et al., 2014). When applied to the Streptomyces VCPO enzymes capable of chlorination reactions, NapH1 and Mcl24, the MCD assay fails to show any activity in the presence of chloride anions; instead exhibiting a decrease in absorbance at 290 nm only in the presence of bromide. Although the literature sample size is limited to these two enzymes, this trend has held consistent for additional Streptomyces VCPO enzymes currently under investigation within our lab that catalyze meroterpenoid chlorination chemistry. Our best attempt to rationalize this result suggests the weak enzymatic capture of the oxidized bromine species, allowing a freely dissociable hypobromite ion to exit the enzyme and appear active via the MCD assay. This is not the only discrepancy in the application of the MCD assay to analyze haloperoxidase enzymes (Wagner, Molitor, & König, 2008). However, despite the contradictory halide specificity results of the MCD assays and their applications to specific bacterial VCPOs, these experiments are useful to validate the proper folding and general halogenation activity of these enzymes prior to thorough in vitro interrogation with substrates. They can also provide insight into the oxidation rates of these VCPOs at different pH, a significant factor when analyzing the chemistry facilitated by Mcl24 (Fig. 3A and B).
Fig. 3.
Monochlorodimedone (MCD) assays of VCPO enzymes and no enzyme controls in: (A) MES buffer, pH 6.0; (B) HEPES buffer, pH 8.0; (C) initial VCPO preincubation with hydrogen peroxide followed by addition of exogenous sodium vanadate at 2 min (both chloride and bromide ions present); and (D) substitution of hydrogen peroxide with tert-butyl hydrogen peroxide (TBHP) in MES buffer, pH 6.0 to assess hydrogen peroxide dependency.
4.1.1. Equipments
Quartz cuvette
Spectrophotometer capable of kinetic scanning
4.1.2. Materials
0.5 M buffer at desired pH
2 M potassium chloride
2 M potassium bromide
500 μM aqueous MCD solution
100 mM hydrogen peroxide
1 mM sodium orthovanadate (Na3VO4)
1 mg/mL enzyme solution
4.1.3. Procedure
Add all reaction components except MCD to a 1-mL quartz cuvette. Zero the UV spectrometer at 290 nm to this solution.
To a 1.5-mL microcentrifuge tube, add assay buffer (50 mM), MCD (50 μΜ), potassium chloride (200 mM), and sodium orthovanadate (10 μΜ) to the final concentrations shown in brackets. Add 10 μg of VCPO enzyme to this solution and transfer it to a quartz cuvette.
Record the absorbance at 290 nm of the solution every 1 s throughout the course of the assay using a kinetic scanning program. Monitor the initial 2 min to obtain a baseline MCD absorbance.
Add hydrogen peroxide to a final concentration of 1 mM with thorough pipette mixing. If no decrease in absorbance is observed 5 min following peroxide addition, add potassium bromide to a final concentration of 200 mM.
4.1.4. Notes
A quartz cuvette is needed to minimize UV absorbance/interference within the assay.
The two buffers used to assay these three VCPO enzymes at different pH were: 0.5 M 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.0 and 0.5 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 8.0.
Both hydrogen peroxide and vanadium salts must be exogenously supplied to initiate the oxidative activities of recombinant NapH1 and Mcl24. The assays as described earlier highlight the requirement of hydrogen peroxide for catalysis; to interrogate the necessity of exogenous vanadate, an analogous assay can be established, adjusting the order of addition of MCD assay components. The addition of potassium bromide (200 mM final concentration) and substitution of sodium vanadate with hydrogen peroxide in step 2, establishing an MCD baseline absorbance for 2 min as in step 3, and adding sodium vanadate in step 4 can interrogate the presence of bound vanadium in the recombinant protein (Fig. 3C). These results show there is minimal time required to form the histidine-coordinated vanadate prosthetic group in both halogenases.
Hydrogen peroxide has yet to be conclusively demonstrated to be the native oxidant for VCPO enzymes in vivo. Although this remains to be thoroughly examined in vitro or in vivo, the substitution of H2O2 with small alkyl hydrogen peroxides (tert-butyl hydrogen peroxide, TBHP, Fig. 3D) fails to diminish the absorbance at 290 nm in the MCD assay with either halide ion. This is in contrast to the results discussed within Section 4.2.3, where Mcl24 appears to be capable of using TBHP as an oxidizing source, albeit at a significantly reduced result.
4.2. Chemical Assays
Inarguably, the most intriguing feature of these Streptomyces VCPO enzymes involves their capability to specifically halogenate meroterpenoid substrates in a variety of ways. While genetic evidence initially highlighted these enzymes and their roles in meroterpenoid halogenation and biosynthesis, in vitro characterization has uncovered a diverse suite of chemical activities deserving of individual discussion.
4.2.1. NapH1 Cyclization of Naphthomevalin to Napyradiomycin A1
In the napyradiomycin suite of molecules, VCPO NapH1 has been shown to catalyze the chloronium-induced cyclization of the dimethylallyl (prenyl) sidechain of naphthomevalin to generate the tricyclic compound napyradiomycin A1 (Bernhardt et al., 2011) (Fig. 4). This activity was originally demonstrated on methylated naphthomevalin analogue SF2415B1 isolated from Streptomyces aculeolatus NRRL 18442 (Winter et al., 2007), generating 7-methyl-napyradiomycin A1 (SF2415B3). In vitro assay conditions indicate NapH1 halogenation is diastereoselective at the newly formed stereocenter at the chlorine atom, implying a specific facial chloronium formation and cyclization by the adjacent hydroxy group. Incubation of NapH1 with SF2415B1 in the presence of bromide facilitated a product consistent with bromonium-induced cyclization, which is a naturally produced napyradiomycin analogue (Farnaes et al., 2014). How-ever, this reaction failed to go to completion in vitro due to probable self-bromination and inactivation of key residues in the VCPO required for activity and/or stability.
Fig. 4.
NapH1 chlorination and cyclization of naphthomevalin (or methylated analogue SF2451B1) to tricyclic napyradiomycin A1 (or SF2451B3). (:B represents a general base in the reaction).
4.2.1.1. Procedure
For the in vitro preparative synthesis of SF2415B3 as reported (Bernhardt et al., 2011), starting material SF2415B1 (4.6 mg, 10 μmol) was added to an 100 mL aqueous solution of 50 mM MES buffer, pH 6.0, 50 mM potassium chloride, 10 μM sodium vanadate, and 1 mM hydrogen peroxide in a round-bottom flask equipped with a stir bar. The reaction was initiated through the addition of recombinant NapH1 (50 μg) and stirred at 30°C for 8 h.
The reaction was quenched by extraction with ethyl acetate (3×100 mL). Pooled organic layers were dried over magnesium sulfate, filtered, and concentrated in vacuo. RP–HPLC with a semipreparative Phenomenex Synergi Polar-RP 4 μm, 250×10 mm column purified enzymatic SF2415B3 using a gradient of 60%–100% acetonitrile in water over 30 min.
4.2.1.2. Notes
The structural differences between the starting material and product are diagnostic enough to facilitate the application of thin-layer chromatography (TLC) to monitor the course of the reaction instead of solely liquid chromatography-based methods. Naphthomevalin Rf 0.35, napyradiomycin A1 Rf 0.40 (4:1 hexane:EtOAc).
As indicated in the procedure, this reaction is amenable to excess hydrogen peroxide. However, the stoichiometry of this cosubstrate can be decreased to 1 molar equivalent relative to the initial starting material without significant loss of yield. Depending on the substrate used in the reaction, care must be taken to avoid oxidative degradation due to excess hydrogen peroxide.
NapH1 also catalyzes analogous chlorination and cyclization reactions on the des-methylated SF2415B1 substrate naphthomevalin, as highlighted in Fig. 4 (Miles et al., 2017). However, NapH1 failed to show any catalytic activity on the linear terpene alcohol precursor ()-nerolidol or the related meroterpenoid-like compound lapachol (Bernhardt et al., 2011), highlighting the extreme selectivity NapH1 has for its requisite substrate. Reactions catalyzed by NapH1 are also stereoselective, evidenced by the single enantiomer of napyradiomycin A1 generated from the synthetic, racemic naphthomevalin substrate. The concomitant, unreacted naphthomevalin recovered from a NapH1 assay, shows an opposite circular dichroism spectrum to naphthomevalin naturally isolated from Streptomyces bacteria or that produced in vitro by NapH3 reaction (Miles et al., 2017).
Ethyl acetate was an optimal choice for the extraction of meroterpenoid products from the NapH1 reaction due to their high solubility in this solvent, facile phase separation of ethyl acetate and the aqueous reaction, and high volatility when concentrating in vacuo.
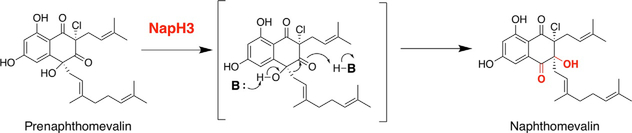
4.2.2. NapH3 α-Hydroxyketone Rearrangement to Generate Naphthomevalin
Although this reaction falls outside the scope of halogenation, the significance of NapH3 catalysis to the napyradiomycin biosynthetic pathway cannot be understated. In both the MCD assay and in vitro incubation with meroterpenoid substrates, NapH3 is devoid of halogenating activity with any halide ion. Rather, this VHPO homolog, which shares 57% sequence identity with the VCPO NapH1, facilitates an α-hydroxyketone rearrangement of the geranyl moiety of prenaphthomevalin to generate naphthomevalin, the substrate for NapH1 (Miles et al., 2017) (Fig. 5). Intriguingly, NapH3 does not require exogenous vanadium or any other cofactor/cosubstrate to facilitate its prenyl migration reaction. Computational calculations have shown that this reaction is energetically favorable, and incubation of synthetic prenaphthomevalin in polar protic solvents can also facilitate this suprafacial 1,2-shift; however, the rate of this shift is dramatically accelerated in the presence of NapH3. This shift was also shown to be enantioselective when incubated with racemic prenaphthomevalin based on circular dichroism analyses.
Fig. 5.
NapH3 α-hydroxyketone rearrangement of synthetic “prenaphthomevalin” to generate naphthomevalin (B: represents a general base in the reaction).
4.2.2.1. Procedure
For the in vitro preparative scale enzymatic synthesis of naphthomevalin (Miles et al., 2017), prenaphthomevalin (7 mg, 18 μmol) dissolved in DMSO (200 μL) was added to a 10 mL solution of 50 mM HEPESNaOH pH 8.0, 150 mM potassium chloride, and 10% glycerol. The reaction was initiated through the addition of recombinant NapH3 (20 mg) and stirred at room temperature for 2 h.
The reaction was quenched by extraction with ethyl acetate (3×10 mL). The solvent was removed in vacuo, and the crude mixture resuspended in methanol for reversed-phase HPLC purification on a semipreparative Phenomenex Luna C18(2), 5 μm, 250×10 mm column. Enzymatic naphthomevalin was purified at a flow rate of 2.5 mL/min using a gradient of 5%–100% B (A=0.1% aqueous trifluoroacetic acid, B=0.1% trifluoroacetic acid in acetonitrile) over 20 min.
4.2.2.2. Notes
To monitor the completion of this reaction, RP–HPLC is the most efficient approach and allows for clean separation of substrate and product. Both starting material and product have comparable TLC retention times and thorough NMR analyses can differentiate the conversion of the shift.
4.2.3. Multitasking Catalytic Activities of Mcl24 to Generate Merochlorins A, B, and X
VCPO Mcl24 from the merochlorin biosynthetic pathway shows an incredible ability to multitask, displaying all aforementioned VCPO function-alities (oxidative halogenation, terpene cyclization, and α-hydroxyketone rearrangement) within one enzyme (Diethelm et al., 2014; Miles et al., 2017; Teufel et al., 2014) (Fig. 6). At pH 6, Mcl24 catalyzes the halogenation, oxidative dearomatization, and terpene cyclization of premerochlorin to generate two dominant monochlorinated merochlorins. These structurally divergent molecules arise due to the final terpene cyclization cascade ending with a carbon–carbon bond, or oxygen–carbon bond in merochlorins A and B, respectively (Fig. 6). Biomimetic total syntheses have followed and supported this biosynthetic hypothesis for both merochlorin A (Pepper & George, 2013) and B (Meier, Strych, & Trauner, 2014) metabolites. However, under basic conditions (pH 8) the product distribution of Mcl24 is altered to predominantly catalyze the formation of merochlorin X, an α-hydroxyketone rearranged intermediate that may be further tailored by additional biosynthetic enzymes to generate merochlorins C and D (Fig. 6). The major mechanistic difference during the biosyntheses of these molecules involves the addition of water to the oxidatively dearomatized molecule, which Mcl24 catalyzes more favorably at a basic pH. Hydration of the benzylic cation facilitates dichlorination and the corresponding rearrangement.
Fig. 6.
Mcl24 reaction on “premerochlorin” substrate at different pH to synthesize merochlorins A, B, and X. Following dichlorination and oxidative dearomatization, Mcl24 branches in how the proposed benzylic cation (bottom middle) is quenched, either through terpene cyclizations to generate merochlorins A and B, or through hydration and an α-hydroxyketone rearrangement to generate merochlorin X. (B: represents a general base in the reaction.)
When applied in vitro, Mcl24 generates a large number of by-products because of the substantial oxidative instability of the premerochlorin substrate. Minimizing the amount of hydrogen peroxide within the reaction to the stoichiometric equivalents required to facilitate the chemistry has improved yields substantially and should be explored when investigating the oxidation of sensitive naphthol-derived meroterpenoids with VCPO enzymes. Surprisingly, hydrogen peroxide can be substituted with the organic peroxide TBHP, generating small quantities of the major three merochlorins, although the rate of catalysis is dramatically reduced (Fig. 7). Investigation into appropriate hydrogen peroxide replacements and their substitution onVCPOactivity is currently ongoing.
Fig. 7.
RP-HPLC analyses of in vitro Mcl24 assays with synthetic “premerochlorin” under various conditions at both pH 6 and 8, and comparison to isolated merochlorin A, B, and X standards (UV absorbance at 340 nm).
4.2.3.1. Procedure
For the in vitro analytical scale production of merochlorin molecules, 1 mM premerochlorin (in a 100 mM DMSO stock solution) was added to a 0.5 mL solution of 50 mM buffer (pH 6.0—MES, pH 8.0— HEPES), 150 mM potassium chloride, 10% glycerol, 100 μM sodium vanadate, and 1.4 mg Mcl24 (50 μM) in a 1.5-mL microfuge tube and maintained under an inert atmosphere. To this solution, 3 mM hydrogen peroxide was added, mixed by pipetting, and left at room temperature (25°C) for 3 h.
A 50 μL aliquot of the reaction mixture was quenched into 50 μL of acetonitrile, mixed thoroughly, and centrifuged at 13,000g for 2 min to pellet precipitated protein. A 10 μL aliquot of the supernatant of the quenched reaction was analyzed by analytical HPLC (Phenomenex Luna C18(2) 5 μm, 1004.6 mm column) using the following separation gradient: 5% B for 5 min, 5%–85% B over 13 min, 85%–100% B over 2 min, and 100% B over 3 min (A¼0.1% aqueous formic acid, B¼0.1% formic acid in acetonitrile).
4.2.3.2. Notes
The reported large-scale enzymatic preparations of merochlorins A, B (Diethelm et al., 2014), and X (Miles et al., 2017) differ slightly from the aforementioned procedure, using superstoichiometric quantities of hydrogen peroxide. Because of the oxidative instability of premerochlorin to these conditions, we have been able to decrease the amount of hydrogen peroxide to the minimum required for catalysis while retaining comparable merochlorin production. This approach is recommended for future large-scale enzymatic preparations of meroterpenoid products to minimize decomposition.
When preparing assays using oxygen-sensitive premerochlorin, maintain solutions under an inert atmosphere to minimize atmospheric oxidation of the unstable substrate prior to hydrogen peroxide addition.
As seen in Fig. 7, a substantial amount of other products are formed from an in vitro Mcl24 reaction with the premerochlorin substrate. Additional iso-, di-, and des-chlorinated merochlorin analogues have been isolated from in vitro reactions, prepared using biomimetic chemical chlorinating conditions, and produced in vivo by Streptomyces bacteria (Diethelm et al., 2014).
5. CONCLUSIONS AND FUTURE OUTLOOK
Streptomyces VCPO enzymes have shown remarkable abilities to facilitate specific and complex halogenation reactions in the biosyntheses of merochlorin and napyradiomycin molecules. Despite the novel and interesting chemistry facilitated by these enzymes, we have only begun to scratch the surface of understanding their potential as halogenating biocatalysts. Even within these two biosynthetic gene clusters, additional VHPOs remain to be functionally characterized. There is considerable genetic evidence to suggest the involvement of Mcl40 in the chloronium-mediated macrocyclization of merochlorin D to merochlorin C, generating an unprecedented chlorinated 15-membered ether. The activity of the last uncharacterized VCPO in the napyradiomycin biosynthetic gene cluster, NapH4, also remains to be explored. Similarly halogenated actinomycete meroterpenoids such as napyradiomycin B1 and azamerone (Cho, Kwon, Williams, Jensen,&Fenical, 2006) are anticipated to haveVCPOenzymology implicated in their biosyntheses.Analogous α-hydroxyketone rearrangements may also be catalyzed by NapH3 homologues in the biosynthetic gene clusters of nonhalogenated meroterpenoids naphterpin (Shin-ya et al., 1990) and marinone (Pathirana, Jensen, & Fenical, 1992). Although the study of these enzymes in bacteria is still in its infancy, we hope that future work can continue to uncover additional applications of these specific VHPOenzymes and their unique roles in the enantioselective construction of complex natural product molecules.
ACKNOWLEDGMENTS
Research on meroterpenoid natural products has been generously supported by NIH Grant R01-AI047818 (B.S.M.) and funding from the Natural Sciences and Research Council of Canada (NSERC-PDF to S.M.K.M.).
REFERENCES
- Agarwal V, Miles ZD, Winter JM, Eustáquio AS, El Gamal AA, & Moore BS (2017). Enzymatic halogenation and dehalogenation reactions: Pervasive and mechanistically diverse. Chemical Reviews, 117, 5619–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt P, Okino T, Winter JM, Miyanaga A, & Moore BS (2011). Stereoselective vanadium-dependent chloroperoxidase in bacterial antibiotic biosynthesis. Journal of the American Chemical Society, 133, 4268–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, & Borodovsky M (2005). GeneMark: Web software for gene finding in prokaryotes, eukaryotes and virus. Nucleic Acids Research, 33, W451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JY, Kwon HC, Williams PG, Jensen PR, & Fenical W (2006). Azamerone, a terpenoid phthalazinone from a marine-derived bacterium related to the genus Streptomyces (Actinomycetales). Organic Letters, 8, 2471–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethelm S, Teufel R, Kaysser L, & Moore BS (2014). A multitasking vanadiumdependent chloroperoxidase as an inspiration for the chemical synthesis of the merochlorins. Angewandte Chemie International Edition, 53, 11203–11206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnaes L, Coufal NG, Kauffman CA, Rheingold AL, DiPasquale AG, Jensen PR, et al. (2014). Napyradiomycin derivatives, produced by a marine-derived actinomycete, illustrate cytotoxicity by induction of apoptosis. Journal of Natural Products, 77, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble GW (2010). Naturally occurring organohalogen compounds: A comprehensive update. Vienna: Springer. [Google Scholar]
- Henkel T, & Zeeck A (1991). Secondary metabolites by chemical screening, 15. Structure and absolute configuration of naphthomevalin, a new dihydro-naphthoquinone antibiotic from Streptomyces sp. The Journal of Antibiotics, 44, 665–669. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer JL, Bowman ME, Dixon RA, & Noel JP (2000). Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry, 39, 890–902. [DOI] [PubMed] [Google Scholar]
- Kaysser L, Bernhardt P, Nam SJ, Loesgen S, Ruby JG, Skewes-Cox P, et al. (2012). Merochlorins A–D, cyclic meroterpenoid antibiotics biosynthesized in divergent pathways with vanadium-dependent chloroperoxidases. Journal of the American Chemical Society, 134, 11988–11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R, Strych S, & Trauner D (2014). Biomimetic synthesis of (±)-merochlorin B. Organic Letters, 16, 2634–2637. [DOI] [PubMed] [Google Scholar]
- Miles ZD, Diethelm S, Pepper HP, Huang DM, George JH, & Moore BS (2017). A unifying paradigm for naphthoquinone-based meroterpenoid (bio)synthesis. Nature Chemistry, 9, 1235–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BS (2018). Asymmetric alkene and arene halofunctionalization reactions in meroterpenoid biosynthesis. Synlett, 29, 401–409. 10.1055/s-0036-1590919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathirana C, Jensen PR, & Fenical W (1992). Marinone and debromomarinone: Antibiotic sesquiterpenoid naphthoquinones of a new structure class from a marine bacterium. Tetrahedron Letters, 33, 7663–7666. [Google Scholar]
- Pepper HP, & George JH (2013). Biomimetic total synthesis of (±)-merochlorin A. Angewandte Chemie International Edition, 52, 12170–12173. [DOI] [PubMed] [Google Scholar]
- Shin-ya K, Imai S, Furihata K, Hayakawa Y, Kato Y, Vanduyne GD, et al. (1990). Isolation and structural elucidation of an antioxidative agent, naphterpin. The Journal of Antibiotics, 43, 444–447. [DOI] [PubMed] [Google Scholar]
- Shiomi K, Iinuma H, Hamada M, Naganawa H, Manabe M, Matsuki C, et al. (1986). Novel antibiotics napyradiomycins. Production, isolation, physico-chemical properties and biological activity. The Journal of Antibiotics, 39, 487–493. [DOI] [PubMed] [Google Scholar]
- Teufel R, Kaysser L, Villaume MT, Diethelm S, Carbullido MK, Baran PS, et al. (2014). One-pot enzymatic synthesis of merochlorin A and B. Angewandte Chemie International Edition, 53, 11019–11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Molitor IM, & König GM (2008). Critical view on the monochlorodimedone assay utilized to detect haloperoxidase activity. Phytochemistry, 69, 323–332. [DOI] [PubMed] [Google Scholar]
- Wever R, Krenn BE, & Renirie R (2018). Marine vanadium-dependent haloperoxidases, their isolation, characterization, and application. Methods in Enzymology, 605, in press. [DOI] [PubMed] [Google Scholar]
- Winter JM, Moffitt MC, Zazopoulos E, McAlpine JB, Dorrestein PC, & Moore BS (2007). Molecular basis for chloronium-mediated meroterpene cyclization—Cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. Journal of Biological Chemistry, 282, 16362–16368. [DOI] [PubMed] [Google Scholar]
- Winter JM, & Moore BS (2009). Exploring the chemistry and biology of vanadiumdependent haloperoxidases. Journal of Biological Chemistry, 284, 18577–18581. [DOI] [PMC free article] [PubMed] [Google Scholar]