Abstract
Non-Structural Protein 1 (NS1) is a multifunctional protein involved in preventing host-interferon response in Influenza A Virus (IAV). Previous studies have indicated that NS1 also stimulates the translation of viral mRNA by binding to conserved sequences in the viral 5’-UTR. Additionally, NS1 binds to Poly (A) Binding Protein (PABP1) and eukaryotic Initiation Factor 4G (eIF4G). The interaction of NS1 with the viral 5’-UTR, PABP1, and eIF4G has been suggested to specifically enhance the translation of viral mRNAs. In contrast, we report that NS1 does not directly bind to sequences in the viral 5’-UTR indicating that NS1 is not responsible for providing the specificity to stimulate viral mRNA translation. We also monitored the interaction of NS1 with PABP1 using a new, quantitative FRET assay. Our data shows that NS1 binds to PABP1 with high affinity; however, the binding of double-stranded RNA (dsRNA) to NS1 weakens the binding of NS1 to PABP1. Correspondingly, the binding of PABP1 to NS1 weakens the binding of NS1 to double-stranded RNA (dsRNA). In contrast, the affinity of PABP1 for binding to Poly (A) RNA is not significantly changed by NS1. We propose that the modulation of NS1•PABP1 interaction by dsRNA may be important for the viral cycle.
Keywords: Non-structural Protein 1, Poly A Binding Protein 1, translation, mRNA, FRET
Introduction
Influenza A virus is a small RNA virus whose seasonal epidemics are responsible yearly for 250,000 to 500,000 deaths worldwide and millions of dollars of lost productivity, including medical expenses 1, 2. The IAV genome consists of eight negative-strand RNA segments, which codes for at least thirteen viral proteins 3. Previous studies have shown that IAV is capable of efficiently translating viral mRNAs while attenuating hosts’ protein production 4, 5. This switch from cellular to virus-specific protein synthesis in the infected cell is mainly brought about by the decreased production and increased degradation of cellular mRNAs 6–10. Nevertheless, there are still significant amounts of cellular mRNAs in the cytoplasm of infected cells, especially during the early phase of infection, suggesting that the viral mRNAs are selectively translated by the ribosomes 8, 11.
One of the proteins thought to stimulate the translation of viral mRNAs is the Non-Structural Protein 1 (NS1) of IAV 4, 12, 13. NS1 is composed of an RNA Binding Domain (RBD) (residues 1-74), a linker region (residues 75-109), and an effector domain (ED) (residues 110-230) with a variable C-terminal tail domain 13. Studies using temperature-sensitive mutant alleles of NS1 showed that most viral protein levels do not change at the permissive and restrictive temperatures; however, the levels of viral matrix protein (M1) and nucleocapsid protein (NP) are reduced at the restrictive temperature even though their mRNA levels do not change 14. This result suggests that NS1 is required for stimulating the translation of M1 and NP mRNAs. Cell-based reporter assays also showed that NS1 stimulates the translation of the M1 mRNA in virus-infected cells 15. The GGUAGAUA sequence in the 5’-untranslated region (UTR) of M1 mRNA was required for stimulating translation 15. Consistent with this result, UV crosslinking experiments and electrophoretic mobility shift assays (EMSA) showed that NS1 binds to the 5’-UTR of M1 and NP mRNAs 16. Finally, co-expression of NS1 protein in cells leads to a large enhancement in the translation of M1 and NP mRNAs but not of control mRNAs 17. Taken together, these data indicate that NS1 binds to a conserved sequence in the 5’-UTR of M1 and NP mRNAs to stimulate their translation.
In contrast to the above data, Krug and co-workers showed that mRNAs that are directly transfected or synthesized endogenously in the cytoplasm of IAV-infected cells are efficiently translated even if they don’t possess the 5’-UTR of IAV 18. Furthermore, co-transfection of plasmids expressing NS1 and luciferase showed that NS1 enhances luciferase activity by about 20-fold 19. Thus, transfection studies and cell-based reporter assays have yielded conflicting data, and it is unclear whether the 5’-UTR of IAV is required for NS1 to stimulate protein synthesis.
More recent studies using rabbit reticulocyte lysate (RRL)-based in vitro translation assays have shown that NS1 at high concentrations (4 μM) may act as a general enhancer of translation 20, 21. Interestingly, the RBD of NS1 is sufficient for enhancing translation; however, the RNA-binding residues Arg38 and Lys41 are not required. Since the RBD of NS1 binds to Poly A Binding Protein 1 (PABP1) 22, the authors speculated that NS1 stimulates translation by interacting with Poly A Binding Protein 1 (PABP1) 20. Another study using RRL also showed that NS1 is a general enhancer of translation 23. In contrast to the previous study with RRL, the RNA-binding residues Arg38 and Lys41 of NS1 were required to stimulate translation. NS1 was observed to be associated with the ribosomes and to bind directly to 28S and 18S rRNAs. These results led the authors to propose that NS1 stimulates translation by recruiting ribosomes to target mRNAs 23. Thus, there are different models to explain the mechanism of translation stimulation by NS1.
During protein synthesis, PABP1 binds to the 3’ poly A tail of the mRNA and it also interacts with eIF4G at the 5’-end of the mRNA (the 5’-m7G cap complex is composed of eIF4E, eIF4G, and eIF4A) to form a mRNA “closed-loop” structure 24. The “closed-loop” structure of the mRNA enhances translation by protecting the mRNA from degradation by nucleases, and by increasing the concentration of terminating ribosomes in the vicinity of the 5’-cap 24, 25. Interestingly, previous studies have shown that the RBD of NS1 binds to the homodimerization region of PABP1 22, and the linker-ED (residues 82-113) of NS1 binds to a region close to the N-terminus of eIF4G 26. Since NS1 can interact with both PABP1 and eIF4G, it has been proposed that NS1 stimulates translation by binding specifically to viral mRNAs and stabilizing the “closed-loop” structure 12, 22. To test this proposed model for NS1-dependent stimulation of translation, we performed quantitative RNA-binding studies to determine whether NS1 has any specificity for binding to the 5’UTR of IAV. Additionally, we developed a FRET-based quantitative assay to monitor the binding of NS1 to PABP1. Our studies show that NS1 does not preferentially bind to sequences in the 5’-UTR of IAV. We also show that NS1 binds with high affinity to PABP1, and this interaction is weakened by dsRNA that binds to NS1. Our results suggest that PABP1 competes with dsRNAs to bind to NS1, and the PABP1•NS1 complex may stabilize the “closed-loop” mRNA structure to enhance translation.
Materials and Methods
Purification of NS1 Protein
The pGEX-3X-NS1 (H3N2 strain) plasmid was generously provided by Prof. Robert Krug at UT Southwestern. The H3N2 NS1 protein was tagged with Glutathione-S-Transferase protein for purification and solubility 27. The GST-NS1C13S and GST-NS1C116S mutants were constructed by site-directed mutagenesis using the Quickchange protocol. For expression of an untagged NS1 protein, the genes were subcloned into the pMCSG10 vector resulting in an N-terminal 6X His-tagged GST protein followed by a Tobacco Etch Virus (TEV) protease site and the NS1 protein 28. Plasmid pGEX-3X-NS1 was used to overexpress and purify the GST-NS1 fusion protein from E. coli BL21 cells as described previously 27. The protein was concentrated and flash frozen in NS1 Buffer (25 mM Tris pH=7.5, 25 mM NaCl, 10% Glycerol, 0.25 mM TCEP or 0.5 mM DTT). Plasmid pMCSG10-GST-TEV-NS1, described above, was used to overexpress and purify the untagged NS1 protein from E. coli BL21 (DE3) cells. Briefly, the GST-TEV-NS1 protein was purified using Glutathione-Sepharose 4B beads (GE Healthcare) as described previously 27. The purified GST-TEV-NS1 protein was incubated with 1/10th molar amount of TEV-His protease at 30 ºC with gentle inverting for 2 hours. The TEV-His protease, GST-His, and uncleaved GST-TEV-NS1 proteins were removed using a Ni-NTA column. Fractions from the Ni-NTA column containing the untagged NS1 were identified by analyzing aliquots by 12% SDS-PAGE. The untagged NS1 protein was concentrated, flash-frozen and stored at −80 ºC.
Purification of Human PABP1
The human PABP1 (Accession: BC015958) in pANT7_cGST vector was purchased from DNASU. The PABP1 gene was subcloned into the pMCSG26, which contains a C-terminal 6X-His tag 29. PABP1 has four cysteines: C42, C128, C132, and C339. Three of the cysteines (C42, C128, and C132) were changed to serines by site-directed mutagenesis to create the single-cysteine PABP1 (scPABP1). Plasmid pMCSG26-PABP1 was transformed into E. coli Rosetta 2 (DE3) pLysS cells (Millipore). The cells were grown at 37 ºC in LB-Ampicillin-Chloramphenicol to an O.D.600 of 0.6-0.8, cooled to 18ºC and then induced with 0.3 mM IPTG for 12-16 hours. Cells were pelleted, flash-frozen and stored at −80 ºC. Cells were resuspended in PABP1 Lysis Buffer (25 mM Tris pH=7.5, 250 mM NaCl, 10% Glycerol, 5 mM β-Mercaptoethanol, 0.5 mM EDTA, 1 mM PMSF, 0.1% Triton X-100, 5 mM Imidazole) and disrupted by sonication. The cell lysate was centrifuged at 20,000 × g for 45 minutes at 4 ºC. The supernatant was incubated with 2 mL of Ni-NTA beads for 5 minutes on ice. The slurry was poured over a column and washed with 50 mL of PABP1 Wash Buffer (Lysis Buffer + 20mM Imidazole). Protein was eluted with PABP1 Elution Buffer (25 mM Tris pH=7.5, 250 mM NaCl, 10% Glycerol, 5 mM β-Mercaptoethanol, 0.5 mM EDTA, 250 mM imidazole). Fractions were collected and concentrated using a 10K MWCO concentrator until the volume was less than 1 mL. The protein sample was filtered and further purified using a Superdex 16/60 200 pg column at a flow rate of 1 mL/min using PABP1 Storage Buffer (25 mM Tris pH=7.5, 250 mM NaCl, 5% Glycerol, 0.25 mM TCEP). Sample peaks were collected and analyzed by a 10% SDS-PAGE. Pure fractions were pooled and concentrated using a 10K MWCO concentrator, aliquoted and flash-frozen. Concentrations of purified proteins were determined by Bradford assay (BioRad).
RNAs for Fluorescence Anisotropy
The following RNAs with a fluorescein dye attached to the 3’-end were purchased from GE Dharmacon: (1) Poly (G)18, (2) Poly (C)18, (3) Poly (A)18, (4) ssIAV: 5’-AGCAAAAGCAGG-3’, (5) ssCR1: 5’- GCUAUCCAGAUUCUGAUU-3’, and (6) M1: 5’-GGUAGAUA-3’. The dsRK1 with a fluorescein dye attached to the 5’-end was synthesized as two complementary RNAs: 5’-FL-CCAUCCUCUACAGGCG-3’ and 5’-FL-CGCCUGUAGAGGAUGG-3’. All RNAs were deprotected and purified by denaturing Urea-PAGE. The complementary RNAs for making dsIAV and dsCR1were synthesized by in vitro transcription using T7 RNA polymerase and the appropriate double-stranded DNA templates 30. The RNA transcripts were purified by denaturing Urea-PAGE. All RNAs were resuspended in water, and their concentrations were determined by measuring the absorbance at 260 nm. All RNAs were stored at −80 ºC in small aliquots. To make double-stranded RNA, equimolar amounts of sense and anti-sense RNAs were heated to 85 ºC in 50 mM Tris pH 8, 50 mM KCl and 1 mM DTT for 2 minutes then allowed to cool to room temperature slowly. The dsRNA was then aliquoted and stored frozen at −80 ºC until needed.
In vitro transcription of long RNAs
Influenza A Virus Matrix 1 and Matrix 2 cDNA was kindly provided by Prof. Beatriz Fontoura at UT Southwestern 31. The pGEM-T-M plasmid contains both the M1 and M2 proteins and the 5’-UTR with a 3’ 25-nt Poly (A) tail. In order to closely mimic the H3N2 version of M mRNA, a 3’-UTR (5’-AAAACTACCTTGTTTCTACT-3’) was added to the vector by PCR. For making the full-length M mRNA, the plasmid was digested with SphI-HF (New England Biolabs) and used for in vitro transcription reaction with SP6 RNA Polymerase (New England Biolabs) as per the manufacturer’s protocol. For making the full-length Renilla luciferase mRNA, the pRL-null (Promega) vector was digested with XbaI (New England Biolabs) and used as the template for in vitro transcription reaction with T7 RNA polymerase 30. The RNA was further purified using silicon-carbide columns (Norgen Biotek) and stored at −80 ºC. The integrity of the RNAs was verified by separating an aliquot on a 6% denaturing Urea-PAGE.
To make the M-Del and R-Del deletion mutants, the above-described plasmids for full-length M and Renilla luciferase mRNAs were used. Most of the protein coding region of M and Renilla luciferase genes were deleted by PCR. M-Del and R-Del retain only the sequence for the first 10 and the last 10 amino acids of the M1 protein and the Renilla luciferase genes, respectively. RNAs were purified as described previously and analyzed for integrity by a 6% denaturing Urea-PAGE.
Fluorescence Anisotropy
Fluorescence anisotropy studies were performed using a fixed concentration of fluorescein-labeled RNA and an increasing concentration of protein (NS1 or PABP1) in Anisotropy Buffer (50 mM Tris pH=8, 50 mM KCl, 50 ng/µL E. coli total tRNA, 1 mM DTT, 0.01% Tween-20) 32. Additionally, we repeated the NS1 binding experiments in the absence of the non-specific E.coli total tRNA to determine the effects of non-specific RNA. For the NS1 anisotropy experiments, the concentration of RNA was fixed at 10 nM, and the concentration of NS1 was titrated from 0 to 5 µM. For the PABP1 anisotropy experiments, the concentration of RNA was fixed at 5 nM, and the concentration of PABP1 was titrated from 0 to 500 nM. Some of the initial anisotropy experiments were performed using a Fluoromax-P fluorimeter equipped with automatic polarizers (JY Horiba). In this case, 700 µl of sample was placed in a quartz cuvette, excited at 494 nm and the polarized emission at 520 nm was measured with 5 nm band slits for both excitation and emission. Most of the anisotropy studies were performed using a Tecan Safire2 plate reader in a 96 well plate. The sample (final volume 200 µl) was excited at 470 nm, and the polarized emission at 520 nm was measured with 10 nm band slits for both excitation and emission. The G-factor was determined using a control sample with fluorescein-labeled RNA. The anisotropy values were subtracted from their initial value, plotted and fit the following quadratic equation to determine KD as described previously 32, 33:
Where [P+RNA]/[RNA] is the anisotropy value and [P] is the protein concentration. GraphPad Prism software (Graphpad Software Inc.) was used to perform the curve fits. All experiments were performed a minimum of 3 times with different protein batches to ensure reproducibility.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed by incubating either 2.5 μM or 5 μM of NS1 with fluorescein-labeled RNA (final conc. 300 nM Poly(G)18, 100 nM ssCR1, 100 nM dsCR1, 100 nM dsIAV, and 100 nM for M1 5’UTR) in Anisotropy Buffer to a final volume of 12.3 µL at room temperature for 1 hour. After incubation, 1.3 μL of ice-cold 50% glycerol was added to the mix. The protein-RNA complexes were separated from unbound RNA by electrophoresis on 4% non-denaturing gel using 79:1 acrylamide:bisacrylamide solution (Bio-Rad) made with 1× Tris-HCl + KCl buffer (45 mM Tris, pH =7.4, 50 mM KCl). In the case of PABP1 binding to Poly (A)18, we used a non-denaturing gel composed of 0.5% agarose and 2% acrylamide:bisacrylamide (79:1) made with 1× Tris-HCl + KCl buffer. After a 45 minutes pre-run at 4°C, the gel was run at 150 V constant voltage at 4°C with buffer recirculation. Some samples were analyzed by separating on a 0.7% Agarose gel at 4°C in 1× TBE buffer for 2 hours at 66 V constant voltage. The gels were visualized by scanning on a FLA9500 Typhoon using the Cy2 excitation laser at 600 PMT voltage and 50 µm resolution.
Filter Binding Assays
To perform filter binding studies, 50 pmoles of the RNA transcripts (M mRNA, Renilla luciferase mRNA, M-Del, and R-Del) were treated with rSAP (New England Biolabs) for 1 hour at 37 ºC and heat inactivated for 15 minutes at 65 ºC. The reaction was then supplemented with DTT and 10 µCi of [γ−32P]-ATP and incubated with T4 PNK (New England Biolabs) for 1 hour at 37 ºC. The RNAs were then purified using Norgen RNA Clean-up kit. A 1 µL sample was taken and read on a scintillation counter to determine the counts per minute (cpm). A fixed concentration of the unlabeled RNA was mixed with the radioactive RNA (≈2000 cpm, 5 nM), heated to 65 ºC and allowed to slow-cool to room temperature in Filter Binding Buffer (50 mM Tris pH = 8, 50 mM KCl, 50 ng/µL E. coli total tRNA, 50 ng/µL of BSA, 5 mM MgCl2, 1 mM DTT, 0.01% Tween-20). Twenty-seven microliters of the RNA in Filter Binding Buffer was added to 3 µL of increasing concentrations of untagged WT-NS1 or NS1-C13S and incubated for 1 hour at room temperature. Samples were then passed through a 96-well filter manifold (Schleicher & Schuell) having a nitrocellulose membrane on top (Spectrum Laboratories) and a Hybond-N+ nylon membrane on the bottom (Amersham) 34. The wells were washed three times with 100 µL of wash buffer (50 mM Tris pH=8, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.01% Tween-20). The membranes were dried and exposed to a phosphorimager screen and read using a Typhoon Scanner (GE). Data were fitted to the quadratic equation (see above) to calculate the KD (GraphPad Prism) 33.
Förster Resonance Energy Transfer
Untagged NS1-C13S was labeled with Cy3 maleimide (GE Bioscience), and scPABP1-His was labeled with Cy5 maleimide (GE Bioscience), according to manufacturer’s instructions and as described previously 35). The labeled proteins were aliquoted and flash-frozen until further use. FRET studies were performed by using 12.5 nM of NS1-C13S-Cy3 and titrating the concentration of scPABP1-His-Cy5 from 0 to 100 nM. The samples were incubated for 1 hour at room temperature in FRET Buffer (50 mM Tris pH=8, 50 mM KCl, 5 mM MgCl2, 1 mM DTT). Each sample (150 µl final volume) was transferred to a quartz cuvette and placed in a fluorometer (Jasco FP-8500). The samples were excited at 545 nm, and the fluorescence emission intensity from 560 to 720 nm was measured. The excitation and emission bandwidths were 5 nm. As control for background emission, we used in parallel samples containing only scPABP1-His-Cy5. The emission intensity at 564 nm (Cy3 maximal emission) and 667 nm (Cy5 maximal emission) were used to calculate the FRET efficiency (E-FRET) using the equation: EFRET= [IA/(IA+ID)] × 100 where IA is the emission intensity of acceptor (scPABP1-His-Cy5) and ID is the emission intensity of donor (NS1-C13S-Cy3). Values were then plotted and fitted to the quadratic equation (see above) to determine KD (GraphPad Prism) 33. For FRET studies in the presence of RNA, the RNAs were pre-incubated in Binding Buffer, heated to 65 ºC and slowly cooled to room temperature. The RNAs were then mixed with NS1-Cy3 and PABP1-Cy5, and the FRET experiments were performed as described above.
Results
Binding of NS1 and PABP1 to Model RNAs
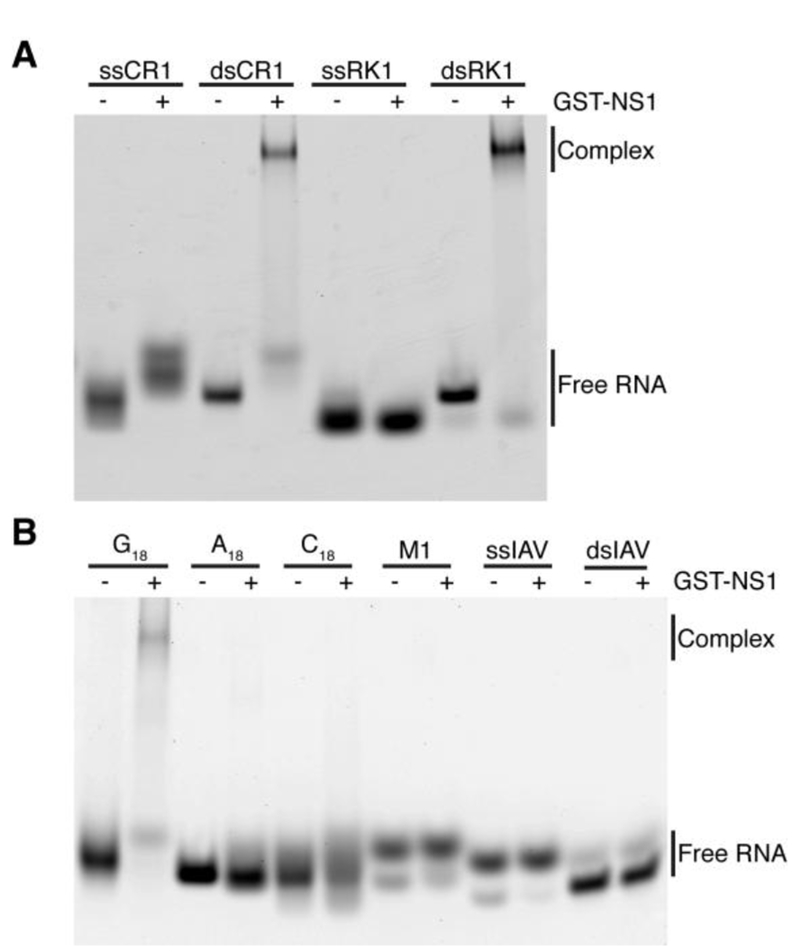
Previous biochemical and structural studies showed that NS1 binds non-specifically to double-stranded RNA 36–39. We initially used an electrophoretic mobility shift assay (EMSA) to monitor the binding of wild-type NS1 fused with GST (GST-NS1) to single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) 38. The GST-NS1 protein construct was chosen as it allows NS1 to remain soluble at high concentrations and has been used previously for binding studies 32. We used the following ssRNAs with a fluorescein dye attached to the 5’ or 3’ end (Table 1): (1) Poly (G)18; (2) Poly (A)18; (3) Poly (C)18; (4) M1 RNA, an 8 nucleotide RNA based on a unique sequence in the 5’-UTR of the positive-sense M mRNA; and (5) ssIAV, a 12 nucleotide RNA based on sequence alignment of the 5’-UTR of all 8 viral, positive-sense mRNA segments. Since NS1 binds to double-stranded RNAs, as a positive control, we tested the binding of NS1 to dsRK1 RNA, which is a 16 base pair dsRNA with 62% GC content that was used in a previous study 32; dsCR1 RNA, which is an 18 base pair dsRNA with 39% GC content; and dsIAV, which was made by annealing ssIAV with its reverse complement (50% GC content). As expected, NS1 binds to dsRK1, and to a lesser extent to dsCR1 suggesting that NS1 binds with higher affinity to GC-rich dsRNAs (Figure 1A). NS1 did not bind to dsIAV (Figure 1B). The dsIAV is unusual in that it has four consecutive AU base pairs in the middle of the sequence, which may not be the sequence preferred by NS1. Interestingly, NS1 binds to Poly (G)18 (Figure 1B), which is consistent with a previous study 27. NS1 did not bind to Poly (A)18, Poly (C)18, ssIAV, and M1 RNA (Figure 1B). It was previously reported that NS1 binds to Poly (A), but the binding was readily competed by Poly (G) indicating that the affinity of NS1 for Poly (A) is weak 27.
Table 1 –
Sequence of Model RNAs
| RNA | Sequence |
|---|---|
| Poly (A)18 | 5’-AAAAAAAAAAAAAAAAAU-FL-3’ |
| Poly (G)18 | 5’-GGGGGGGGGGGGGGGGGU-FL-3’ |
| Poly (C)18 | 5’-CCCCCCCCCCCCCCCCCU-FL-3’ |
| ssCR1 | 5’-GCUAUCCAGAUUCUGAUU-FL-3’ |
| dsCR1 | 5’-GCUAUCCAGAUUCUGAUU-FL-3’ 3’-CGAUAGGUCUAAGACUAAG-5’ |
| dsRK1 | 5’-FL-CCAUCCUCUACAGGCG-3’ 3’-GGUAGGAGAUGUCCGC-FL-5’ |
| M1 | 5’-GGUAGAUA-FL-3’ |
| ssIAV | 5’-AGCAAAAGCAGG-FL-3’ |
| dsIAV | 5’-AGCAAAAGCAGG-FL-3’ 3’-UCGUUUUCGUCC-5’ |
“FL” stands for fluorescein tag at either end.
Figure 1. Analysis of NS1 binding to RNA by EMSA.
(A) Binding of NS1 to single-stranded CR1 (ssCR1), double-stranded CR1 (dsCR1), single-stranded RK1 (ssRK1), and double-stranded RK1 (dsRK1). (B) Binding of NS1 to Poly (G)18, Poly (A)18, (3) Poly (C)18, M1 RNA, single-stranded IAV (ssIAV), and double-stranded IAV (dsIAV). “−” and “+” indicate the absence and presence of NS1, respectively. Free RNA and RNA-NS1 complex are indicated next to the gels.
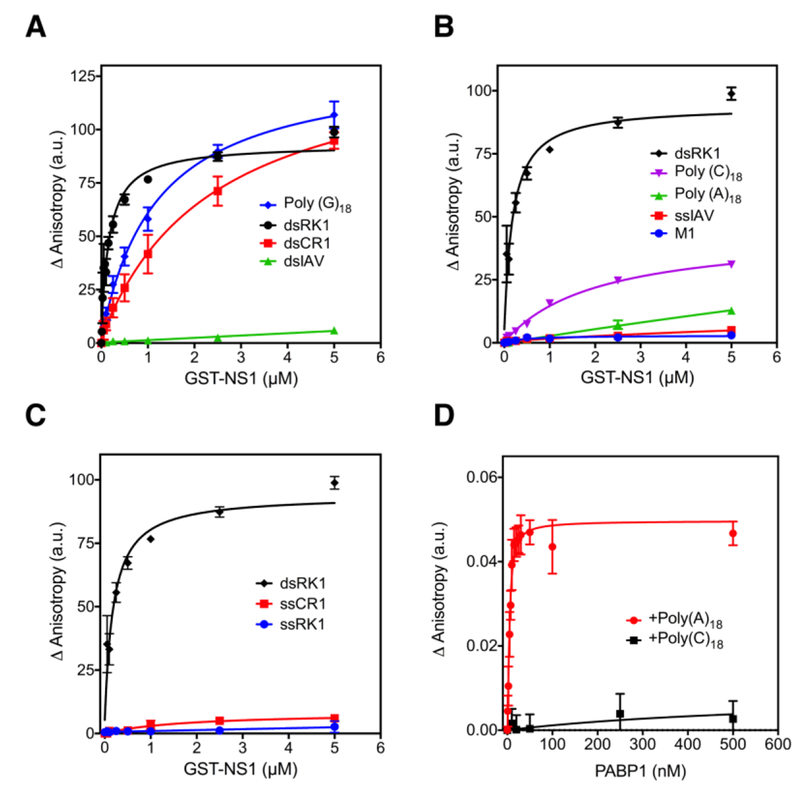
We next used a quantitative, fluorescence anisotropy based assay to determine the equilibrium dissociation constant (KD) for the binding of GST-NS1 to ssRNA and dsRNA 32. To verify that the double-stranded RNAs were, in fact, double-stranded, we analyzed the annealed dsRNAs using an 8% non-denaturing gel. All double-stranded RNAs ran higher than their single-stranded counterpart, confirming that our RNAs were double-stranded (Figure 1 and data not shown). Binding experiments were performed by incubating increasing amounts of GST-NS1 with 10 nM of fluorescein-tagged RNA, and the change in anisotropy was measured. NS1 bound to dsRK1 with KD = 170 ± 19 nM, which is identical to the value previously reported 32 (Figure 2A). However, NS1 bound weakly to dsCR1 (KD = 2.4 ± 0.3 µM) and to Poly (G)18 (KD = 1.2 ± 0.1 µM), and did not bind to viral dsIAV (Figure 2A). NS1 did not bind to any of the ssRNAs (Figure 2B and 2C) indicating that it only binds to dsRNAs. These results are consistent with our EMSA data (Figure 1). The above EMSA and anisotropy experiments were performed in the presence of total tRNA to avoid non-specific binding of NS1 to the model RNAs. Since NS1 is known to bind to RNA in a non-specific manner, we also performed binding studies in the absence of the total tRNA. We observed increased binding affinity by NS1 for the model RNAs without total tRNA (Figure S1); however, the overall trend was similar regardless of the presence or absence of tRNA (Table S1). We choose to perform our studies in the presence of tRNA because we wish to compare our results with previous data 32. Furthermore, for NS1 to bind to specific RNA sequences it would have to do so in the presence of other non-specific RNAs in vivo.
Figure 2. Binding of NS1 and PABP1 to RNA analyzed using a fluorescence anisotropy assay.
(A) Binding of NS1 to dsRK1, dsCR1, dsIAV, and Poly (G)18. (B) Binding of NS1 to dsRK1, Poly (C)18, Poly (A)18, ssIAV, and M1 RNA. (C) Binding of NS1 to dsRK1, ssCR1, and ssRK1. The final concentration of the RNAs was 10 nM, and the final concentration of NS1 was increased from 0 to 5 μM. (D) Binding of PABP1 to Poly (A)18, and Poly (C)18. The final concentration of the RNAs was 5 nM, and the final concentration of PABP1 was increased from 0 to 500 nM. The change in anisotropy is shown on the y-axis. The error bars represent the standard deviation from three independent experiments.
To monitor the interaction of NS1 with PABP1, we decided to develop a fluorescence resonance energy transfer (FRET)-based assay (described below). For the FRET assay, we made single-cysteine mutants of NS1. NS1 has two cysteines, C13 and C116; however, NS1-C116S mutation resulted in low solubility of the protein. Therefore, the C13S mutant was chosen for RNA binding studies. In general, GST-NS1-C13S binds to dsRNAs with the same specificity as GST-NS1, but with increased affinity. GST-NS1-C13S bound to dsRK1 (KD = 70 ± 7 nM), dsCR1 (KD = 350 ± 50 nM), and Poly (G)18 (KD = 600 ± 50 nM) (Figure S2 and S3). In agreement with our data with wild-type NS1, the NS1C13S mutant did not bind to dsIAV and ssRNAs. Additionally, arginine 38 and lysine 41 of NS1 are critical for binding to dsRNA 38. We constructed the GST-NS1-R38A-K41A mutant and, as expected, it did not bind to dsRNA (Figure S2A).
To make sure that PABP1 is functional, we tested the binding of PABP1 to RNA. A fixed concentration of Poly (A)18 was titrated with increasing concentrations of the untagged wild-type PABP1 (WT-PABP1) and the change in anisotropy was measured (Figure 2D). The KD for WT-PABP1 binding to Poly (A)18 is 2.0 ± 0.5 nM, which is consistent with a previous study 40. We used Poly (C)18 as a negative control RNA, and as expected, WT-PABP1 did not bind to this RNA (Figure 2D). The FRET experiments (described below) require a single-cysteine mutant of PABP1 for the site-specific attachment of a fluorescent dye. We constructed a single-cysteine PABP1 mutant (scPABP1) that maintains a cysteine at position 339, but the cysteines at positions 42, 128, and 132 were changed to serines by site-directed mutagenesis. The protein was then purified and tested for binding to Poly (A)18. The KD for scPABP1 binding to Poly (A)18 is 5.1 ± 0.6 nM (Figure S3D), which is similar to the result obtained with wild-type PABP1 indicating that the cysteine to serine substitutions do not inhibit the function of scPABP1.
NS1 does not bind specifically to viral mRNA
Although anisotropy experiments demonstrated that NS1 does not bind to short, single-stranded regions of viral RNA, it is possible that it may bind to larger viral mRNAs that may have unique secondary structures. To test this idea, we constructed two model mRNAs, M-Del and R-Del that are each ≈150 nucleotides in length. M-Del has the 5’-UTR of M mRNA, followed by the first 10 and the last 10 amino acid sequence of the M1 protein, then the 3’-UTR, and a 25 nucleotide 3’-Poly (A) tail. R-Del is a control RNA derived from a Renilla luciferase reporter mRNA construct and has non-viral 5’ and 3’ UTRs, a 25 nucleotide 3’-Poly (A) tail and a truncated Renilla luciferase ORF.
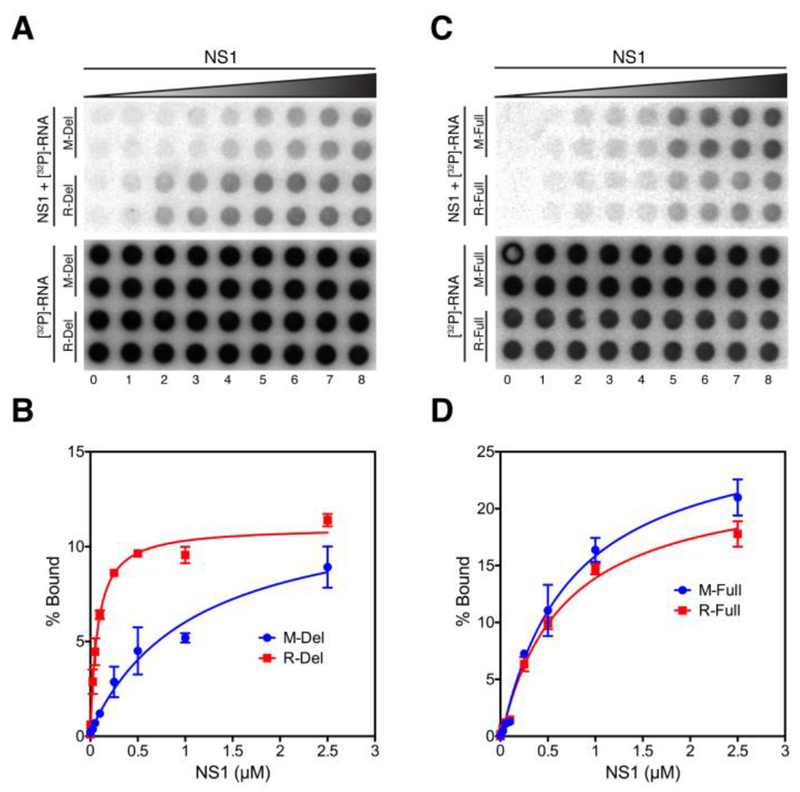
Because anisotropy is limited in its ability to detect large protein-RNA interactions, we used a filter binding assay to determine the KD for NS1 binding to M-Del and R-Del RNAs 34. The KD for untagged NS1 binding to M-Del is 1085 ± 230 nM and to R-Del is 72 ± 7 nM (Figure 3B). For NS1-C13S, the KD is 690 ± 103 nM for M-Del and 110 ± 20 nM for R-Del (Figure S4B). These results show that in general NS1 binds stronger to the non-viral, control R-Del RNA than to the viral M-Del RNA. Secondary structure analysis using mFold program 41 showed that R-Del forms a longer dsRNA region than M-Del, possibly explaining the higher affinity of NS1 for R-Del (Figures S5 and S6).
Figure 3. Binding of NS1 to viral and non-viral mRNAs analyzed using a filter binding assay.
(A) Binding of NS1 to M-Del and R-Del mRNAs. 5’-[32P]-labeled M-Del and R-Del mRNAs were incubated with increasing concentration of NS1, and the samples were filtered through a nitrocellulose membrane (top), and a nylon membrane (bottom). The two rows for each RNA represents experiments done in duplicate. (B) Binding curves for M-Del and R-Del. (C) Binding of NS1 to M-Full and R-Full mRNAs. The experiment was performed as described in (A). (D) Binding curves for M-Full and R-Full. The error bars represent the standard deviation from two independent experiments.
Since M-Del and R-Del may truncate critical binding sites for NS1, we tested the binding of NS1 to the full-length M and R mRNAs (M-Full and R-Full, respectively). Interestingly, NS1 binds to M-Full (KD = 720 ± 90 nM) and R-Full (KD = 642 ± 62 nM) with similar affinities (Figure 3D). NS1-C13S also binds to the full-length RNAs with similar affinities (Figure S4D). These results indicate that NS1 does not preferentially bind to viral mRNAs. We also determined the KD for PABP1 and scPABP1 binding to M-Del and R-Del. As expected, PABP1 and scPABP1 bind to both M-Del and R-Del with high affinity because they have a poly A tail at the 3’-end (KD ≈ 5 nM) (Figure S7).
NS1 binds to PABP1 with high affinity
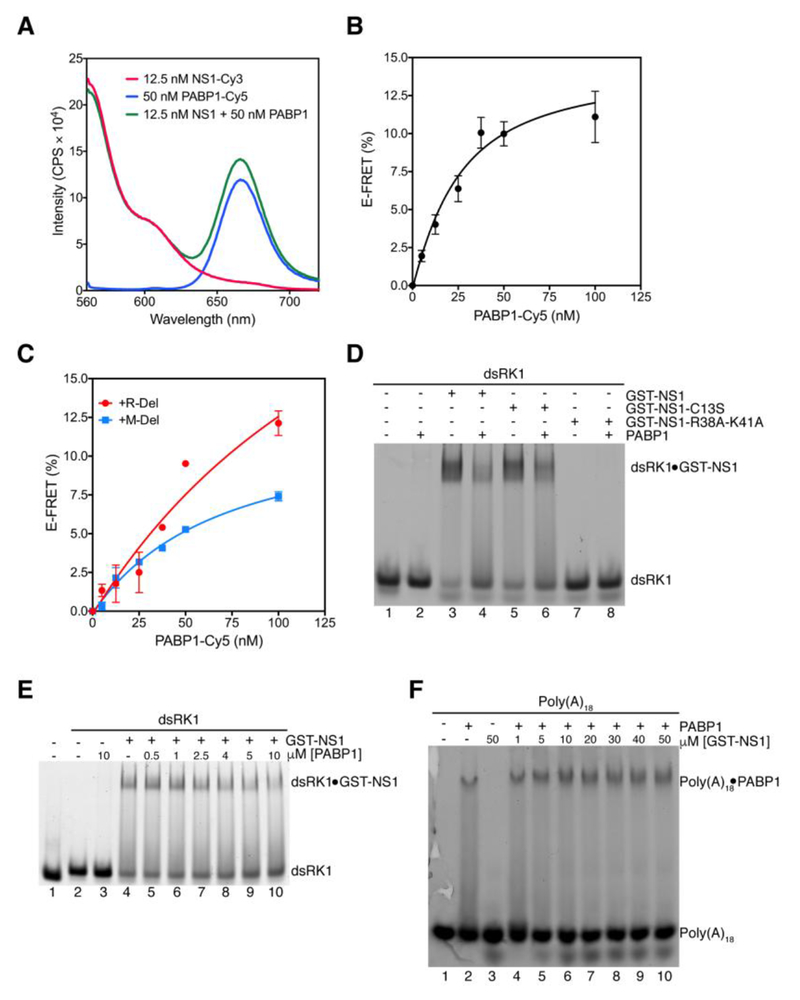
Previous qualitative studies showed that NS1 interacts with PABP1 22. To quantitatively determine the binding affinity of NS1 for PABP1, we developed a FRET-based assay. We labeled NS1-C13S with the fluorescent dye cyanine 3 (Cy3) and scPABP1 with the fluorescent dye cyanine 5 (Cy5). These dyes were chosen as they are efficient FRET pairs and can be used over a wide range of distances. NS1-Cy3 and PABP1-Cy5 were incubated either alone or together, and the samples were excited at 545 nm, and the fluorescence emission intensity was scanned from 560 to 720 nm (Figure 4A). When NS1-Cy3 and PABP1-Cy5 are present together, we observed a decrease in Cy3 emission intensity (≈560 nm) and an increase in Cy5 emission intensity (≈660 nm) compared to the control samples. This indicates that NS1-Cy3 binds to PABP1-Cy5 resulting in the FRET. To determine the KD for NS1 binding to PABP1, we used a low, fixed concentration of NS1-Cy3 and titrated the concentration of PABP1-Cy5 and measured the fluorescence emission intensity at 564 nm (Cy3) and 667 nm (Cy5). We then calculated the efficiency of FRET (E-FRET) at each concentration of PABP1-Cy5 to determine the KD. The KD for the NS1-PABP1 interaction in the absence of RNA is 19 ± 5 nM, indicating strong binding (Figure 4B).
Figure 4. Binding of NS1 to PABP1 monitored using a FRET assay.
(A) Emission spectrum showing the changes in fluorescence intensity because of NS1 binding to PABP1. NS1 was labeled with Cy3 and PABP1 was labeled with Cy5. Red trace, NS1-Cy3 only control reaction; blue trace, PABP1-Cy5 only control reaction; green trace, NS1-Cy3 mixed with PABP1-Cy5. The samples were excited at 545 nm, and the fluorescence emission intensity from 560 nm to 720 nm was monitored. The binding of NS1 to PABP1 results in increased emission at 665 nm because of the FRET. (B) Binding curve for NS1 binding to PABP1. The x-axis shows the concentration of PABP1 and the y-axis shows the FRET efficiency (E-FRET) in %. The error bars represent the standard deviation from four independent experiments. (C) Binding curve for NS1 binding to PABP1 in the presence of 250 nM M-Del, and 250 nM R-Del. The error bars represent the standard deviation from two independent experiments. (D) EMSA showing that the binding of dsRK1 RNA and PABP1 to NS1 is mutually exclusive. Lane 1, dsRK1; lane 2, dsRK1 + PABP1; lane 3, dsRK1 + GST-NS1; lane 4, dsRK1 + GST-NS1 + PABP1; lane 5, dsRK1 + GST-NS1-C13S; lane 6, dsRK1 + GST-NS1-C13S + PABP1; lane 7, dsRK1 + GST-NS1-R38A-K41A; lane 8, dsRK1 + GST-NS1-R38A-K41A + PABP1. “−” and “+” indicate the absence and presence of protein, respectively. The final concentrations in the reactions are: 100 nM dsRK1, 2.5 μM GST-NS1, 2.5 μM GST-NS1-C13S, 2.5 μM GST-NS1-R38A-K41A, and 4 μM PABP1. (E) EMSA showing PABP1 competes with dsRK1 RNA to bind to NS1. Lane 1, 100 nM ssRK1 forward strand; lane 2, 100 nM dsRK1; lane 3, 100 nM dsRK1 + 10 μM PABP1; lane 4, 100 nM dsRK1 + 2.5 μM GST-NS1; lanes 5 to 10 contain 100 nM dsRK1 + 2.5 μM GST-NS1 and the indicated concentration of PABP1. (F) EMSA showing that PABP1 binds to Poly (A) in the presence of NS1. Lane 1, 100 nM Poly (A)18 RNA; lane 2, 100 nM Poly (A)18 RNA + 500 nM PABP1; lane 3, 100 nM Poly (A)18 RNA + 50 μM GST-NS1; lanes 4 to 10 contain 100 nM Poly (A)18 RNA + 500 nM PABP1 and the indicated concentration of GST-NS1.
To test whether the NS1-PABP1 interaction is modulated by viral and non-viral RNA, we performed the binding reaction in the presence of 250 nM of purified M-Del and R-Del. The KD for NS1 binding to PABP1 increased by 3-fold with M-Del (KD = 60 ± 15 nM), whereas it increased by ≈9-fold with R-Del (KD > 170 nM) (Figure 4C). As reported above, NS1 binds to R-Del with a 15-fold higher affinity than to M-Del. The binding of NS1 to R-Del may sterically interfere with NS1 binding to PABP1 because the RBD of NS1 is used for both interactions 22. We verified the idea that the binding of NS1 to dsRNA and PABP1 are mutually exclusive using EMSA. As shown above (Figure 1A), NS1 binds to dsRK1, and the complex has a lower mobility in the native gel compared to the unbound dsRK1 (Figure 4D). More than 90% of dsRK1 is bound to NS1 and NS1-C13S; whereas, NS1-R38A-K41A does not bind to dsRK1. Interestingly, in the presence of PABP1 less than 10% of dsRK1 is bound to NS1 demonstrating that the NS1•PABP1 complex cannot bind to dsRNA. To confirm our results, we performed the dsRK1-NS1 binding reaction in the presence of increasing concentrations of PABP1. As shown in Figure 4E, the amount of dsRK1•NS1 complex formed progressively decreased as the concentration of PABP1 was increased indicating that NS1 cannot bind to dsRK1 and PABP1 simultaneously.
Finally, to determine whether the binding of PABP1 to Poly (A)18 is affected by NS1, we performed anisotropy and EMSA studies. The KD for PABP1 binding to Poly (A)18 was similar in the absence and in the presence of excess NS1 indicating that NS1 does not interfere with the binding of PABP1 to Poly (A)18 (Figure S8). Consistent with the anisotropy data, EMSA showed that PABP1 binds to Poly (A)18 even in the presence of NS1 (Figure 4F). Interestingly, in the presence of NS1 the band corresponding to Poly (A)18 •PABP1 shifts to a slightly slower mobility band, which may represent the formation of the Poly (A)18 •PABP1•NS1 complex (Figure 4F).
Discussion
Previous studies showed that only mRNAs containing the viral 5’-UTR are efficiently translated after infection by IAV or by co-expressing NS1 15, 42, 43. NS1 has been proposed to directly bind to the 5’-UTR of viral mRNAs to stimulate translation 15–17. The 5’-UTR of IAV mRNAs have a 12-nt sequence that is highly conserved and could serve as the NS1 binding site 44. However, our studies show that NS1 does not bind to the conserved 12-nt ssIAV or dsIAV sequence. Additionally, NS1 does not show any preference to bind to viral M mRNA in comparison to control mRNA. Taken together, our results indicate that NS1 does not interact directly with conserved sequences in the 5’-UTR of viral mRNAs to stimulate translation. The discrepancy of our results with earlier data, as pointed out by Krug and co-workers 18, is caused by the protection of reporter mRNAs that have the viral 5’-UTR from cleavage by the viral polymerase compared to reporter mRNAs with the non-viral 5’-UTR 45. Nevertheless, another study done in the absence of the viral polymerase showed that the expression of NS1 stimulates the translation of NP and M1 mRNAs but not of control mRNA lacking the viral 5’-UTR 17. The reason for the observed NS1-dependent increase in protein synthesis of mRNAs with the viral 5’-UTR is not clear.
Interestingly, NS1 is also a general enhancer of translation 20, 21, 23. IAV mRNAs have m7G cap at the 5’-end and a poly (A) tail at the 3’-end, similar to cellular mRNAs 46–48. eIF4F consisting of eIF4E, eIF4G, and eIF4A binds to 5’-m7G cap, and PABP1 binds to the 3’-poly(A) tail of IAV mRNAs 49. The general enhancement of translation may be the result of NS1 binding directly to PABP1 and eIF4G 22, 26. We show that NS1 binds to PABP1 with a high binding affinity (KD = 19 ± 5 nM). Although we have not determined the binding affinity of NS1 for eIF4G, it is likely that the interaction of NS1 with both PABP1 and eIF4G strengthens the circularization of the mRNA (Figure 5) 22. Surprisingly, the binding affinity of NS1 for PABP1 is reduced significantly by dsRNA that binds to NS1. The RBD of NS1 binds to both dsRNA and PABP1, and our results suggest that the binding is mutually exclusive possibly because the binding of NS1 to dsRNA may sterically block binding to PABP1 (Figure 5). This interpretation is consistent with our observation that the binding of NS1 to PABP1 lowers the affinity of NS1 for dsRNA. In contrast, PABP1 binds with a similar affinity to Poly (A)18 in the presence of NS1 suggesting that PABP1 will be recruited to the poly A tails of mRNAs even during IAV infection. This is consistent with the fact that the RRM1 and 2 of PABP1 is mainly responsible for binding to the Poly (A) tail, and they are located at a distance from the homodimerization region to which NS1 binds 50. Therefore, PABP1 should be capable of binding both the Poly (A) tail and NS1 simultaneously (Figure 4F).
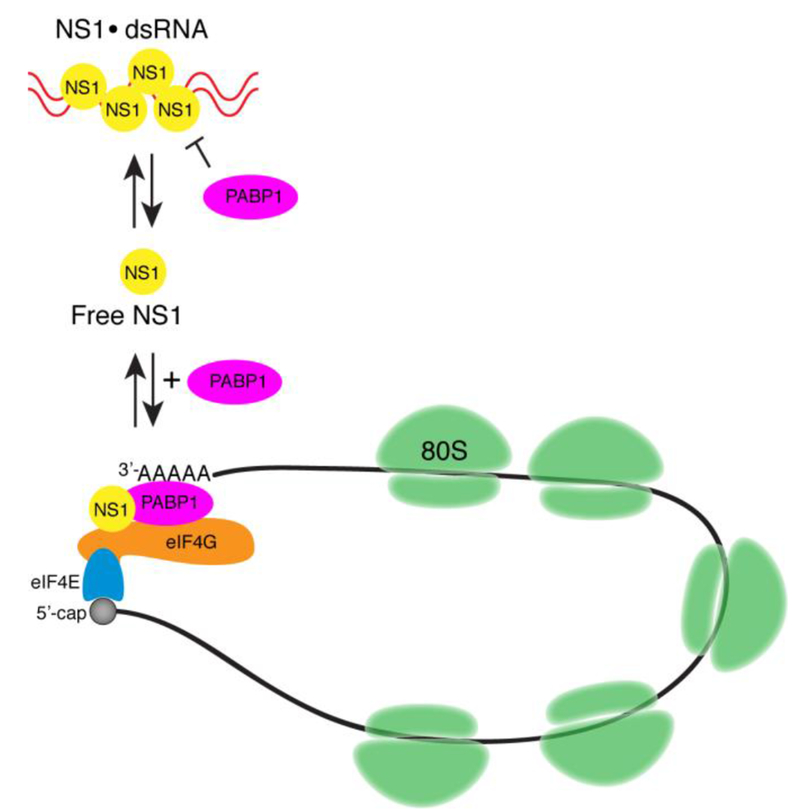
Figure 5. Model for the stimulation of translation initiation by NS1.
NS1 (yellow) bound to dsRNA (red) cannot bind to PABP1 (pink). NS1 that is not bound to dsRNA synergistically stimulates mRNA translation by binding to eIF4G (orange) and PABP1 to promote mRNA circularization. Also indicated are the m7G cap at the 5’-end of the mRNA (black circle), eIF4E (blue), and 80S ribosomes (green).
One of the main functions of NS1 is to bind to viral dsRNAs to prevent the establishment of an antiviral response by the cell 13. Studies have shown that NS1 binds to dsRNA to form long, tube-like oligomers with the dsRNA in the center 51, 52. Since these NS1•dsRNA structures are not translated, it is consistent with our result that dsRNA-bound NS1 has reduced affinity for PABP1. The dsRNA sequesters NS1 and the NS1•dsRNA cannot efficiently interact with PABP1. In contrast, PABP1 has a high affinity for NS1 that is not bound to dsRNA. This pool of free NS1 may be responsible for stimulating protein synthesis by directly binding to PABP1 and eIF4G to stabilize the “closed-loop” mRNA structure 22. Alternatively, NS1 bound to the ribosome 23 may interact with PABP1 bound to the 3’ poly A tail of the mRNA to circularize the mRNA and stimulate translation.
Supplementary Material
Acknowledgments
We thank Robert Krug for providing us the GST-NS1 plasmid and Beatriz Fontoura for providing us the viral M mRNA plasmid. We thank Xinying Shi for technical help, and Norman Zhu for helping to optimize the purification of PABP1. We are grateful for the financial support from the San Diego Fellows, Molecular Biophysics Training Grant (NIH Grant T32 GM008326), and NIH R03 (AI123873).
Footnotes
Associated Content
Supporting Information
The Supporting Information consists of:
A table with experimentally derived KD values, additional NS1 and PABP1 binding data, and RNA secondary structures predicted by mFold.
References
- [1].(2016) Influenza Fact Sheet, World Health Organization http://www.who.int/mediacentre/factsheets/fs211/en/.
- [2].Li L, Wong JY, Wu P, Bond HS, Lau EHY, Sullivan SG, and Cowling BJ (2017) Heterogeneity in Estimates of the Impact of Influenza on Population Mortality: A Systematic Review, Am J Epidemiol. 187, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Eisfeld AJ, Neumann G, and Kawaoka Y (2015) At the centre: influenza A virus ribonucleoproteins, Nat Rev Microbiol 13, 28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yanguez E, and Nieto A (2011) So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell, Virus Res 156, 1–12. [DOI] [PubMed] [Google Scholar]
- [5].Katze MG, and Krug RM (1990) Translational control in influenza virus-infected cells, Enzyme 44, 265–277. [DOI] [PubMed] [Google Scholar]
- [6].Rodriguez A, Perez-Gonzalez A, and Nieto A (2007) Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II, J Virol 81, 5315–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Krug RM, Broni BA, and Bouloy M (1979) Are the 5’ ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs?, Cell 18, 329–334. [DOI] [PubMed] [Google Scholar]
- [8].Katze MG, and Krug RM (1984) Metabolism and expression of RNA polymerase II transcripts in influenza virus-infected cells, Mol Cell Biol 4, 2198–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen Z, Li Y, and Krug RM (1999) Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3’-end processing machinery, EMBO J 18, 2273–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nemeroff ME, Barabino SM, Li Y, Keller W, and Krug RM (1998) Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3’end formation of cellular pre-mRNAs, Mol Cell 1, 991–1000. [DOI] [PubMed] [Google Scholar]
- [11].Katze MG, DeCorato D, and Krug RM (1986) Cellular mRNA translation is blocked at both initiation and elongation after infection by influenza virus or adenovirus, J Virol 60, 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Smith RW, and Gray NK (2010) Poly(A)-binding protein (PABP): a common viral target, Biochem J 426, 1–12. [DOI] [PubMed] [Google Scholar]
- [13].Hale BG, Randall RE, Ortin J, and Jackson D (2008) The multifunctional NS1 protein of influenza A viruses, J Gen Virol 89, 2359–2376. [DOI] [PubMed] [Google Scholar]
- [14].Wolstenholme AJ, Barrett T, Nichol ST, and Mahy BW (1980) Influenza virus-specific RNA and protein syntheses in cells infected with temperature-sensitive mutants defective in the genome segment encoding nonstructural proteins, J Virol 35, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Enami K, Sato TA, Nakada S, and Enami M (1994) Influenza virus NS1 protein stimulates translation of the M1 protein, J Virol 68, 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Park YW, and Katze MG (1995) Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation, J Biol Chem 270, 28433–28439. [DOI] [PubMed] [Google Scholar]
- [17].de la Luna S, Fortes P, Beloso A, and Ortin J (1995) Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs, J Virol 69, 2427–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cassetti MC, Noah DL, Montelione GT, and Krug RM (2001) Efficient translation of mRNAs in influenza A virus-infected cells is independent of the viral 5’ untranslated region, Virology 289, 180–185. [DOI] [PubMed] [Google Scholar]
- [19].Salvatore M, Basler CF, Parisien JP, Horvath CM, Bourmakina S, Zheng H, Muster T, Palese P, and Garcia-Sastre A (2002) Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis, J Virol 76, 1206–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kainov DE, Muller KH, Theisen LL, Anastasina M, Kaloinen M, and Muller CP (2011) Differential effects of NS1 proteins of human pandemic H1N1/2009, avian highly pathogenic H5N1, and low pathogenic H5N2 influenza A viruses on cellular pre-mRNA polyadenylation and mRNA translation, J Biol Chem 286, 7239–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Anastasina M, Terenin I, Butcher SJ, and Kainov DE (2014) A technique to increase protein yield in a rabbit reticulocyte lysate translation system, Biotechniques 56, 36–39. [DOI] [PubMed] [Google Scholar]
- [22].Burgui I, Aragon T, Ortin J, and Nieto A (2003) PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes, J Gen Virol 84, 3263–3274. [DOI] [PubMed] [Google Scholar]
- [23].Panthu B, Terrier O, Carron C, Traversier A, Corbin A, Balvay L, Lina B, Rosa-Calatrava M, and Ohlmann T (2017) The NS1 Protein from Influenza Virus Stimulates Translation Initiation by Enhancing Ribosome Recruitment to mRNAs, J Mol Biol 429, 3334–3352. [DOI] [PubMed] [Google Scholar]
- [24].Hinnebusch AG, and Lorsch JR (2012) The mechanism of eukaryotic translation initiation: new insights and challenges, Cold Spring Harb Perspect Biol 4 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Uchida N, Hoshino S, Imataka H, Sonenberg N, and Katada T (2002) A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation, J Biol Chem 277, 50286–50292. [DOI] [PubMed] [Google Scholar]
- [26].Aragon T, de la Luna S, Novoa I, Carrasco L, Ortin J, and Nieto A (2000) Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus, Mol Cell Biol 20, 6259–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Qiu Y, and Krug RM (1994) The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A), J Virol 68, 2425–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, and Mark ID (2009) A family of LIC vectors for high-throughput cloning and purification of proteins, Methods Mol Biol 498, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Eschenfeldt WH, Maltseva N, Stols L, Donnelly MI, Gu M, Nocek B, Tan K, Kim Y, and Joachimiak A (2010) Cleavable C-terminal His-tag vectors for structure determination, J Struct Funct Genomics 11, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Milligan JF, and Uhlenbeck OC (1989) Synthesis of small RNAs using T7 RNA polymerase, Methods Enzymol 180, 51–62. [DOI] [PubMed] [Google Scholar]
- [31].Tsai PL, Chiou NT, Kuss S, Garcia-Sastre A, Lynch KW, and Fontoura BM (2013) Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing, PLoS Pathog 9, e1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cho EJ, Xia S, Ma LC, Robertus J, Krug RM, Anslyn EV, Montelione GT, and Ellington AD (2012) Identification of influenza virus inhibitors targeting NS1A utilizing fluorescence polarization-based high-throughput assay, J Biomol Screen 17, 448–459. [DOI] [PubMed] [Google Scholar]
- [33].Pollard TD (2010) A guide to simple and informative binding assays, Mol Biol Cell 21, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wong I, and Lohman TM (1993) A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions, Proc Natl Acad Sci U S A 90, 5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen E, Sharma MR, Shi X, Agrawal RK, and Joseph S (2014) Fragile X mental retardation protein regulates translation by binding directly to the ribosome, Mol Cell 54, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hatada E, and Fukuda R (1992) Binding of influenza A virus NS1 protein to dsRNA in vitro, J Gen Virol 73 (Pt 12), 3325–3329. [DOI] [PubMed] [Google Scholar]
- [37].Lu Y, Wambach M, Katze MG, and Krug RM (1995) Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor, Virology 214, 222–228. [DOI] [PubMed] [Google Scholar]
- [38].Wang W, Riedel K, Lynch P, Chien CY, Montelione GT, and Krug RM (1999) RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids, RNA 5, 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cheng A, Wong SM, and Yuan YA (2009) Structural basis for dsRNA recognition by NS1 protein of influenza A virus, Cell Res 19, 187–195. [DOI] [PubMed] [Google Scholar]
- [40].Gorlach M, Burd CG, and Dreyfuss G (1994) The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity, Exp Cell Res 211, 400–407. [DOI] [PubMed] [Google Scholar]
- [41].Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction, Nucleic Acids Res 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Garfinkel MS, and Katze MG (1992) Translational control by influenza virus. Selective and cap-dependent translation of viral mRNAs in infected cells, J Biol Chem 267, 9383–9390. [PubMed] [Google Scholar]
- [43].Garfinkel MS, and Katze MG (1993) Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5’-untranslated region, J Biol Chem 268, 22223–22226. [PubMed] [Google Scholar]
- [44].Desselberger U, Racaniello VR, Zazra JJ, and Palese P (1980) The 3’ and 5’-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity, Gene 8, 315–328. [DOI] [PubMed] [Google Scholar]
- [45].Shih SR, and Krug RM (1996) Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins, Virology 226, 430–435. [DOI] [PubMed] [Google Scholar]
- [46].Neumann G, Brownlee GG, Fodor E, and Kawaoka Y (2004) Orthomyxovirus replication, transcription, and polyadenylation, Curr Top Microbiol Immunol 283, 121–143. [DOI] [PubMed] [Google Scholar]
- [47].Poon LL, Pritlove DC, Fodor E, and Brownlee GG (1999) Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template, J Virol 73, 3473–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Krug RM, Morgan MA, and Shatkin AJ (1976) Influenza viral mRNA contains internal N6-methyladenosine and 5’-terminal 7-methylguanosine in cap structures, J Virol 20, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bier K, York A, and Fodor E (2011) Cellular cap-binding proteins associate with influenza virus mRNAs, J Gen Virol 92, 1627–1634. [DOI] [PubMed] [Google Scholar]
- [50].Deo RC, Bonanno JB, Sonenberg N, and Burley SK (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein, Cell 98, 835–845. [DOI] [PubMed] [Google Scholar]
- [51].Bornholdt ZA, and Prasad BV (2008) X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus, Nature 456, 985–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Carrillo B, Choi JM, Bornholdt ZA, Sankaran B, Rice AP, and Prasad BV (2014) The influenza A virus protein NS1 displays structural polymorphism, J Virol 88, 4113–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.