Abstract
The cause of posterior urethral valves (PUV) is unknown, but genetic factors are suspected given their familial occurrence. We examined cases of isolated PUV to identify novel copy number variants (CNVs). We identified 56 cases of isolated PUV from all live-births in New York State (1998–2005). Samples were genotyped using Illumina HumanOmni2.5 microarrays. Autosomal and sex-linked CNVs were identified using PennCNV and cnvPartition software. CNVs were prioritized for follow-up if they were absent from in-house controls, contained ≥10 consecutive probes, were ≥20 Kb in size, had ≤20% overlap with variants detected in other birth defect phenotypes screened in our lab, and were rare in population reference controls. We identified 47 rare candidate PUV-associated CNVs in 32 cases; one case had a 3.9 Mb deletion encompassing BMP7. Mutationsin BMP7 have been associated with severe anomalies in the mouse urethra. Other interesting CNVs, each detected in a single PUV case included: a deletion of PIK3R3 and TSPAN1, duplication/triplication in FGF12, duplication of FAT1—a gene essential for normal growth and development, a large deletion (>2 Mb) on chromosome 17q that involves TBX2 and TBX4, and large duplications (>1 Mb) on chromosomes 3q and 6q. Our finding of previously unreported novel CNVs in PUV suggests that genetic factors may play a larger role than previously understood. Our data show a potential role of CNVs in up to 57% of cases examined. Investigation of genes in these CNVs may provide further insights into genetic variants that contribute to PUV.
Keywords: posterior urethral valve, congenital urinary tract obstruction, copy number variant, urethra, urinary tract malformation
INTRODUCTION
Posterior urethral valves (PUV) are the most common cause of bilateral renal obstruction. They occur exclusively in male births with an estimated incidence of one in 5,000–8,000 [Krishnan et al., 2006]. Although PUV are relatively uncommon, they remain an important contributor to pediatric morbidity. Approximately 24–45% of patients with PUV will develop renal insufficiency [Yohannes and Hanna, 2002] and 13–28% will develop end-stage renal disease during childhood [Indudhara et al., 1998].
The majority of PUV cases reported in the literature are sporadic, but there have been several reports showing that PUV run in families, occurring in both twin and non-twin siblings as well as in successive generations [Weber et al., 2005; Schreuder et al., 2008]. As some cases are familial, it is recommended that brothers of affected males be screened using antenatal ultrasound to provide timely and optimal care [Thomalla et al., 1989; Schreuder et al., 2008]. Suggested modes of PUV inheritance have included autosomal recessive, autosomal dominant with reduced penetrance, X-linked recessive, and polygenic or multifactorial inheritance [Trembath and Rijhsinghani, 2002; Weber et al., 2005].
Very few studies have investigated gene variants and PUV [Laksmi et al., 2010; Caruana et al., 2015]. To our knowledge only one study has examined copy number variants (CNVs) in the etiology of birth defects of the kidney and urinary tract, including 29 patients with PUV with or without other associated defects [Caruana et al., 2015]. Our study was conducted to identify recurring, potentially causal CNVs.
MATERIALS AND METHODS
Cases
New York State (NYS) has mandatory reporting of major structural birth defects identified within the first 2 years of life to the NYS Congenital Malformations Registry (CMR). Each birth defect is coded using the expanded British Pediatric Association (BPA) coding system based on hospital provided descriptions entered as a text field and reviewed by a clinician as needed. To identify isolated PUV cases, we searched for a BPA code corresponding to congenital PUV or posterior urethral obstruction (BPA code 753.600). To ensure cases were not missed because of an error in BPA coding, we also searched the text field for “posterior urethral valves.” In total we identified 104 PUV cases from all male live-births occurring in NYS from 1998 through 2005 (n = 1,036,842). We excluded those with a known genetic syndrome (n = 6) and those with other major birth defects, such as heart defects (n = 8) or other genitourinary defects (n = 31) that were not specifically a result of PUV. Cases with PUV plus other genitourinary defects that could have occurred as a consequence of PUV such as hydronephrosis, ureteral dilation, ureteral anomaly not otherwise specified, and renal pelvic dilation/obstruction were not excluded. This resulted in a total of 59 cases with isolated PUV to examine. We excluded two additional cases to meet the platform sample number requirements from the genotyping lab resulting in a total of 57 isolated PUV cases for the current study.
Birth certificates were used to extract demographic information. Maternal and demographic characteristics were compared between PUV cases and a random sample of NYS male live-births (n = 3,916) using Fisher’s exact test or t-test, where applicable. NYS Department of Health Institutional Review Board approved this study (IRB 07-007).
Genotyping
A random ID number was assigned to each case and all personally identifying information was removed prior to genotyping and analysis. A total of 57 PUV cases, three unaffected controls and one quality control sample were batched and genotyped along with 114 cases with other unrelated phenotypes. Using an established method [Saavedra-Matiz et al., 2013], DNA was extracted from two 3-mm newborn dried blood spot (DBS) punches yielding approximately 200–1200 ng of DNA per subject for array genotyping. The Biomedical Genomics Center Core Facility at the University of Minnesota genotyped all samples using Illumina Human-Omni2.5-8_v1 bead arrays and the Infinium HD assay protocol. Illumina GenomeStudio v2011.1 was used for data analysis. Genotypes were set to missing if the GenCall score was <0.15. Genotypes were clustered based on the data generated in this project, which included a total of 174 DBS samples. Genotypes and clusters were manually reviewed, re-clustered, edited, and excluded where appropriate according to parameters and quality control metrics as described in Illumina’s Infinium Genotyping Data Analysis Technical Note [Illumina].
CNV Calling and Annotation
Autosomal CNVs were called using PennCNV v2011/05/03 [Wang et al., 2007] and Illumina’s cnvPartition algorithm v3.1.6. For both algorithms, data were GC-wave adjusted, and the minimum number of probes required for a CNV call was three. The confidence threshold for CNV calling was set to the default value of 10 for PennCNV and 35 for cnvPartition. Sex chromosome CNVs were called using PennCNV after recomputing Log R ratio (LRR) and B allele frequency (BAF) values using sex-specific centroids. Median values for R and theta were computed for each marker on the X and Y chromosomes in males and females separately and then applied using in-house software that implemented the standard formulas (ref = PMID 16899659) to generate new LRR and BAF values. These new values were fed into PennCNV as “autosomal” probes using custom sex-specific population frequency of B allele (.pfb) and GC content (.gcmodel) files. The PennCNV function clean_cnv.pl was run with default parameters to merge adjACEnt CNV calls. Autosomal CNV call files were annotated using custom C++ programs as previously described [Rigler et al., 2015] to compare concordance between CNV calling algorithms, count the number of cases and controls carrying overlapping CNVs in the current study, determine overlap with an in-house database of CNVs generated from cases and controls of other unrelated defects, determine overlap with the Database of Genomic Variants archive (DGV2), and identify intersecting transcripts and genes [Iafrate et al., 2004]. Transcripts included full-length coding transcripts and full-length non-coding transcripts with a well characterized biotype downloaded from GENCODE (version 19, accessed via UCSC genome browser May 2014) [Harrow et al., 2012]. Genes were defined as those included in the Consensus Coding Sequence project (CCDS; release 15, accessed via UCSC genome browser June 2014) [Pruitt et al., 2009]. Each sex chromosome CNV call was manually reviewed and annotated.
CNV Selection and Prioritization
We examined CNVs if they were absent from our in-house controls (i.e., PUV CNVs of the same type and with the same predicted breakpoints as in-house controls), if they contained at least 10 consecutive single nucleotide polymorphism (SNP) probes, were at least 20 Kb in size, had ≤20% overlap with common variants in HapMap [Altshuler et al., 2010] and Children’s Hospital of Philadelphia (CHOP) [Shaikh et al., 2009] CNV datasets, and had 20% overlap with any variant previously detected in other birth defect phenotypes screened in our lab. The remaining CNVs were then uploaded to the DGV2 genome browser, using build37/hg19 coordinates, and were examined for overlap with known CNVs. For follow-up, we prioritized CNVs that did not have significant overlap (>50%) with any catalogued CNV of the same type in DGV2 (date accessed July 23, 2013). Additionally, CNVs with >50% overlap with a DGV2 entry were prioritized if the non-overlapped portion intersected a gene with no overlap with any DGV2 entry. We also looked for significant overlap (>50%) with CNVs identified in an internal set of 1928 population controls that were genotyped on the same Omni 2.5 platform (624 individuals ascertained by the Social Development Research Groupand1,304 individuals participating in studies of Youth Drug Abuse, ADHD, and Related Disorders) and processed using the same CNV calling pipeline.
CNV Validation
A subset of the autosomal CNVs that met the above criteria were validated in the laboratory using two to three quantitative real-time PCR (qPCR) TaqManassays (Applied Biosystems, Carlsbad, CA) per region. Genomic DNA was extracted from one 3-mm DBS,[10] diluted 1:10 in water, and amplified using TaqMan Environmental MasterMix (ABI) in5 μl reaction volumes. A fragment of the RNaseP H1 RNA gene was co-amplified and used as an internal control (TaqMan Copy Number Reference Assay, ABI). Assays were run in quadruplicate on either an ABI 7900HT or an ABI QuantStudio. Seventeen autosomal CNVs were chosen for validation based on their size and/or overlap with genes deemed potentially biologically relevant to PUV. CopyCaller software v2.0 (ABI) was used to analyze the real-time data using relative quantitation (2-ΔΔCt method). The manual Ct threshold was set to 0.2 with the automatic baseline on. CopyCaller software parameters were as follows: the median ΔCt for each experiment was used as the calibrator, wells with an RNaseP Ct >38 were excluded and the zero copy ΔCt threshold was set to 6. The average copy number and a software-generated confidence value were calculated for each subject. Samples with confidence values ≥0.95 were considered valid; samples with confidence values <0.95 were rerun in quadruplicate. Since multiple assays targeted each CNV, in all cases, no single sample contained all low confidence calls throughout a CNV region. One probe (Hs03380682_cn) was excluded due to discordant results obtained when retesting multiple samples with low confidence calls (Supplemental Table SI). All assays were tested in each of the 57 cases and 32 unaffected NYS control subjects. We subsequently screened all validated CNVs against an additional 149 control samples from unaffected NYS births using at least one assay targeting the area of interest. Therefore, 181 unaffected controls were screened for at least one candidate CNV region.
RESULTS
We identified a total of 110 patients with PUV in the NYS CMR from the population of 1,036,842 male live-births resulting in a birth prevalence of 1 in 9,500 male live-births. After genotyping the 57 PUV cases, we discovered a trisomy 21 case that was not reported as such in the CMR, and we subsequently excluded this case from the results to meet our initial selection criteria of isolated PUV.
Isolated PUV cases (n = 56) did not differ significantly from the random sample of male control infants from NYS (n = 3,916) in maternal age, rACE/ethnicity, education, body mass index, or smoking status (Table I). PUV cases were significantly more likely than the control subjects to be born early (37.6 vs. 39.1 weeks, respectively) and to be of low birth-weight (<2,500 g; 17.9 vs. 6.2%, respectively). While cases were more likely than controls to be small for gestational age (SGA, defined as less than the 10th percentile of birth weight for gestational age) [Kramer et al., 2001] (18.2 vs. 11.6%, respectively), this difference did not reach statistical significance (P-value = 0.10).
TABLE I.
Demographic Characteristics of Patients With Isolated Posterior Urethral Valves As Compared to a Random Sample of New York State Male Live-Births
| PUV patients (n = 56) | NY state male births (n = 3,916) | P-value | |
|---|---|---|---|
| Maternal age years, n (%) | 0.5 | ||
| <20 | 7 (12.5) | 333 (8.5) | |
| 20–34 | 39 (69.6) | 2868 (73.2) | |
| ≥35 | 10 (17.9) | 715 (18.3) | |
| Maternal race/ethnicity, n (%) | 0.4 | ||
| Non-Hispanic White | 28 (50.0) | 2259 (57.8) | |
| African American | 12 (21.4) | 706 (18.1) | |
| Hispanic | 13 (23.2) | 646 (16.5) | |
| Asian/other | 3 (5.4) | 298 (7.6) | |
| Maternal education years, n (%) | 0.3 | ||
| <12 | 14 (25.5) | 660 (17.0) | |
| 12 | 14 (25.5) | 1138 (29.4) | |
| >12 | 27 (49.1) | 2080 (53.6) | |
| Nulliparous, n (%) | 22 (39.3) | 1613 (41.2) | 0.9 |
| Maternal smoking, n (%) | 3 (5.4) | 375 (9.6) | 0.4 |
| Maternal prepregnancy BMI kg/m2, n (%) | 0.7 | ||
| ≤24.9 | 20 (58.8) | 1207 (56.4) | |
| 25–29.9 | 9 (26.5) | 506 (23.6) | |
| ≥30 | 5 (14.7) | 427 (20.0) | |
| Gestational age weeks, mean (SD) | 37.6 (2.5) | 39.1 (2.3) | <0.001 |
| Birth weight g, mean (SD) | 3204 (676) | 3393 (596) | 0.02 |
| Low birth weight, n (%) | 10 (17.9) | 243 (6.2) | 0.002 |
| SGA, n (%) | 10 (18.2) | 431 (11.6) | 0.1 |
BMI, body mass index; SGA, small for gestational age.
Data missing for NY state male births on maternal smoking: 8, alcohol: 10, race/ethnicity: 7, education: 38, BMI: 1776, gestational age: 182, SGA: 184. Among the missing SGA, two infants born <22 weeks gestation were not coded since the Kramer et al. reference cut-off points start at 22 weeks gestation.
Data missing for PUV patients on maternal education: 1, BMI: 22, gestational age: 1.
The microarray analysis of all 56 PUV samples resulted in a total of 4,879 autosomal PennCNV calls, 19 chromosome × PennCNV calls, and 2,045 autosomal cnvPartition calls. After applying the inclusion criteria reported in the methods, we identified 47 candidate CNVs (43 autosomal and 4 × chromosome CNVs) of interest in 32 different cases (19 cases with 1 CNV, 11 cases each with 2 CNVs, and two cases each with 3 CNVs) generated from the PennCNV analysis. All 43 autosomal CNVs were identified by both the cnvPartition and PennCNV algorithms (all cnvPartition calls had >83% overlap with PennCNV calls). Table II shows the 17 CNVs that met our in clusion criteria and were selected for follow-up as they were more biologically relevant based on the genes affected and/or were large in size. All 17 CNV calls were validated using qPCR. Supplemental Table SII shows 30 additional CNVs (26 autosomal CNVs and 4 × chromosome CNVs) that met our inclusion criteria, including not having significant overlap with catalogued CNVs, but were not selected for validation by qPCR.
TABLE II.
Seventeen Rare CNVs Identified and Validated in 15 Patients With Isolated Posterior Urethral Valves
| Locus | Genomic coordinates | Size (bps) | Type | Study subject ID | DECIPHER patients with overlapping CNVs & kidney & urinary tract anomalies | Gene(s)/transcript(s) |
|---|---|---|---|---|---|---|
| 1p35.1 | 33,543,972‥33,625,417 | 81,446 | Het Del | 5 | ADC; TRIM62 | |
| 1p34.1 | 46,557,405‥46,649,046 | 91,642 | Het Del | 7 | PIK3R3; RP11-322N21.2; RP4-533D7.5; TSPAN1 | |
| 1p31.1 | 82,947,889‥83,089,454 | 141,566 | Dupl | 12 | 266358: Hydronephrosis | Intergenic |
| 1q21.3 | 152,651,029‥153,284,782 | 633,754 | Dupl | 10 | C1orf68; KPRP; LCE1A; LCE1B; LCE1C; LCE1D; LCE1E; LCE1F; LCE2A; LCE2B; LCE4A; LCE6A; IVL; RP1-13P20.6; SMCP; SNORA31; SPRR1B; SPRR2A; SPRR2B; SPRR2D; SPRR2E; SPRR2F; LELP1; PRR9; RP1-140J1.1; LOR; PGLYRP3 | |
| 2p14–15 | 63,936,936‥64,110,691 | 173,756 | Dupl | 11 | ACA59; UGP2 | |
| 3p25.3 | 10,253,823‥10,332,468 | 78,646 | Dupl | 4 | 1876: Horseshoe kidney | GHRL; IRAK2; TATDN2 |
| 3q23 | 139,162,298‥140,231,134 | 1,068,837 | Dupl | 8 | 250665: Renal hypoplasia | 7SK; AC048346.1; CLSTN2; NMNAT3; RBP1; RBP2; RP11-13L2.2; RP11-166D18.1; RP11-31966.1; RP11-31966.3; RP11-442N1.1; RP11-442N1.2; RP11-68L1.1; RP11-68L1.2 |
| 3q28 | 191,907,021‥191,947,455 | 40,435 | Dupl/Tripl* | 9 | 256994: Hydronephrosis | FGF12 |
| 4q35.2 | 187,444,387‥187,593,959 | 149,573 | Dupl | 6 | 2384: Hypospadias, Micropenis 267301: Hypospadias | FAT1; MTNR1A |
| 6p22.3 | 16,446,643‥16,853,607 | 406,965 | Dupl | 3 | AL137003.1; ATXN1; RP1-151F17.1 | |
| 6q24.1 | 140,776,223‥142,206,247 | 1,430,025 | Dupl | 13 | 288262: Hydronephrosis | 7SK; AL356137.1; RP11-551A11.1; RP11-63E9.1; RP3-46062.2 |
| 12p13.2 | 10,874,088‥11,029,515 | 155,428 | Dupl | 14 | 266262: Hydronephrosis | CSDA; PRR4; RP13-81N3.2; TAS2R10; TAS2R7; TAS2R8; TAS2R9 |
| 16p13.12 | 13,938,976‥14,372,873 | 433,898 | Dupl | 2 | 264947: Hypospadias | CTA-276F8.2; CTD-2135D7.2; CTD-2135D7.3; CTD-2135D7.4; CTD-2135D7.5; ERCC4; MKL2; YRNA |
| 17q23.1–23.2 | 58,113,570‥60,325,665 | 2,212,096 | Het Del | 1 | 276114: Hypospadias 287822: Hypospadias | AC002994.1; AC008158.1; AC018628.1; AC079005.1; APPBP2; BCAS3; BRIP1; C17orf64; C17orf82; CA4;CTD-2319I12.1; HEATR6; INTS2; MED13; NACA2; PPM1D; RP11-15E18.1; SCARNA20; TBX2; TBX4; U6; USP32; Y_RNA; snoU13 |
| 18q21.31 | 56,018,908‥56,110,489 | 91,582 | Dupl | 8 | NEDD4L | |
| 20q13.2–13.31 | 52,395,038‥56,373,484 | 3,978,447 | Het Del | 15 | 251098: Hydronephrosis 278520: Male hypogonadism | AC005220.3; AC006076.1; AL117380.1; AL117380.2; AL121914.1; AL133232.1; AURKA; BCAS1; BMP7; C20orf43; CASS4; CBLN4; CSTF1; CTCFL; CYP24A1; DOK5; FAM209A; FAM209B; FAM210B; GCNT7; MC3R; MTRNR2L3; PCK1; PFDN4; PMEPA1; RAE1; RBM38; RN5S487; RP11-380D15.3; RP11-560A15.3; RP11-560A15.4; RP4-718J7.4; RP4-800J21.3; RP4-813D12.2; RP4-813D12.3; RP5-1010E17.1; RP5-1010E17.2; RP5-843L14.1; RP5-897D18.1; SPO11; TFAP2C; U3; U4atac; U6; YRNA; ZBP1; snoU13 |
| 21q22.11 | 31,523,734‥31,545,654 | 21,921 | Het Del | 14 | 286121: Hypospadias, Micropenis 288278: Micropenis | CLDN17 |
Het Del, heterozygous deletion; Dupl, duplication; Tripl, Triplication.
Bolded genes are highlighted in the figures and/or discussion section.
Coordinates (hg19) predicted using PennCNV.
CNV predicted as duplication by both algorithms was determined to be copy-number 4–5 (most likely a triplication) via qPCR.
The 17 CNVs that were validated (Table II) ranged in size from 21 Kb to 3.9 Mb and were found in 15 different cases. These included: 1 intergenic duplication, 1 duplication intersecting at least one GENCODE transcript, five heterozygous deletions intersecting genes, and 10 duplications intersecting genes; one CNV predicted to be a duplication by both algorithms was determined to be copy-number 4–5 via qPCR (most likely a triplication and henceforth described as a duplication/triplication). In screening 181 controls via qPCR for the rare variants listed in Table II, we found no controls with the same copy-number as that of our cases across all probes spanning each region. However, one control subject carried a duplication at both probe locations in the 21q22.11 region, whereas our case carried a deletion. In silico screening against 1928 controls, none of the controls harbored an autosomal CNV at the same locus with the same copy-number as our cases; however, there was major overlap (>50% of the region is overlapped by a CNV in a control sample) with one heterozygous deletion in the chromosome 2 region that was duplicated in patient ID 11. For the × chromosome calls (Supplemental Table SII), there were two controls that overlapped the CNV at 92 Mb (one deletion and one duplication; the case had a deletion), one control that overlapped the CNV at 112 Mb (a deletion; the case had a duplication), five controls that overlapped the CNV at the 149 Mb (two deletions and three duplications; the case had a duplication), and zero controls that significantly overlapped the duplication at 153 Mb. Therefore, CNVs at each of these four chromosome × loci were very rare, with a minor allele frequency less than of 0.002.
The rare CNVs found in individual cases included: a 3.9 Mb deletion at 20q13.2–13.31 that comprises the coding region of bone morphogenic protein 7 (BMP7) (Fig. 1); a 40 Kb duplication/triplication at 3q28 that overlaps the fibroblast growth factor 12 (FGF12) gene (Fig. 2); a 91 Kb heterozygous deletion at 1p34.1 that includes phosphoinositide-3-kinase regulatory subunit 3 (PIK3R3) and tetraspanin-1 (TSPAN1); a 149 Kb duplication at 4q35.2 that includes FAT atypical cadherin 1 (FAT1) gene; and a large heterozygous deletion 2.2 Mb at 17q23.1–23.2 that includes T-box 2 (TBX2) and T-box 4 (TBX4) (Fig. 3). We also identified two large duplications 1 Mb at 3q23 and 1.4 Mb at 6q24.1, neither of which encompasses genes of known function in growth and development.
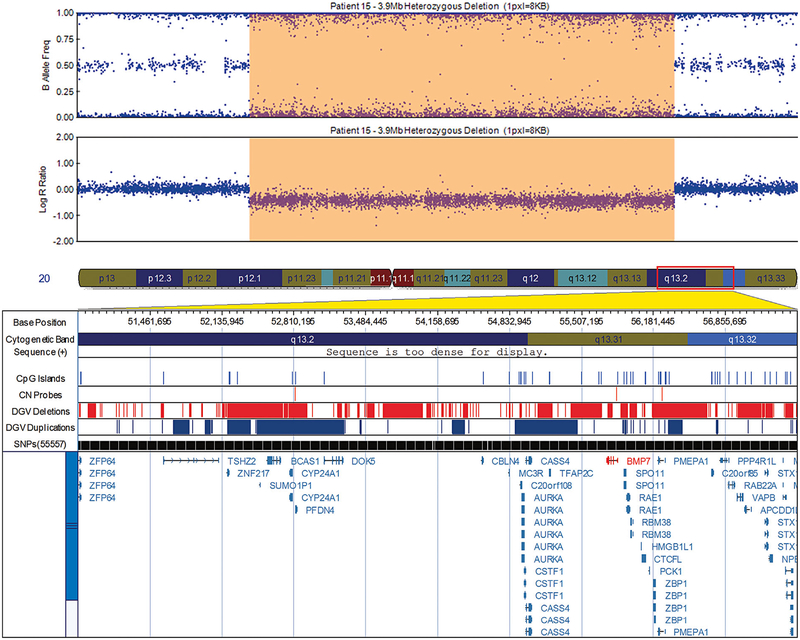
FIG. 1.
Visualization of the 3.9 Mb deletion in patient 15 at 20q13.2–13.31 encompassing bone morphogenic protein 7 (BMP7), exported from the Illumina Genome Viewer. The top panel depicts B-allele frequency (ratio of minor to major alleles) and the bottom panel depicts the logR ratio data (signal intensity). Tracks provided by Illumina show cytobands, CpG islands and the location of SNPs on the array. Custom tracks were created to display the location targeted by copy number assays used to validate CNVs (“CN probes”), and copy number losses (“DGV Deletions,” shown in red) and gains (“DGV Duplications,” shown in blue), both of which were downloaded from the DGV2 database (2014-10-16 version). A subset of genes/transcripts overlapping CNVs are listed below the panels. Genes mentioned in the tables/text are in red. CNV calls made using the pennCNV algorithm are highlighted (heterozygous deletions in orange and duplications are in blue). hg19 coordinates shown. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmga].
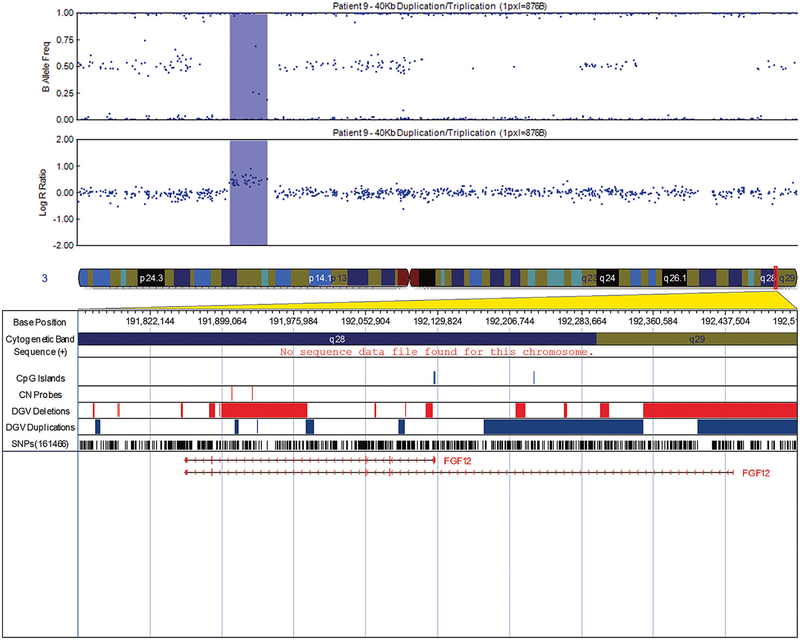
FIG. 2.
Visualization of the 40 Kb duplication/triplication in patient 9 at 3q28 that overlaps the fibroblast growth factor 12 (FGF12) gene, exported from the Illumina Genome Viewer. The top panel depicts B-allele frequency (ratio of minor to major alleles) and the bottom panel depicts the logR ratio data (signal intensity). Tracks provided by Illumina show cytobands, CpG islands and the location of SNPs on the array. Custom tracks were created to display the location targeted by copy number assays used to validate CNVs (“CN probes”), and copy number losses (“DGV Deletions,” shown in red) and gains (“DGV Duplications,” shown in blue), both of which were downloaded from the DGV2 database (2014-10-16 version). A subset of genes/transcripts overlapping CNVs are listed below the panels. Genes mentioned in the tables/text are in red. CNV calls made using the pennCNV algorithm are highlighted (heterozygous deletions in orange and duplications are in blue). hg19 coordinates shown. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmga].
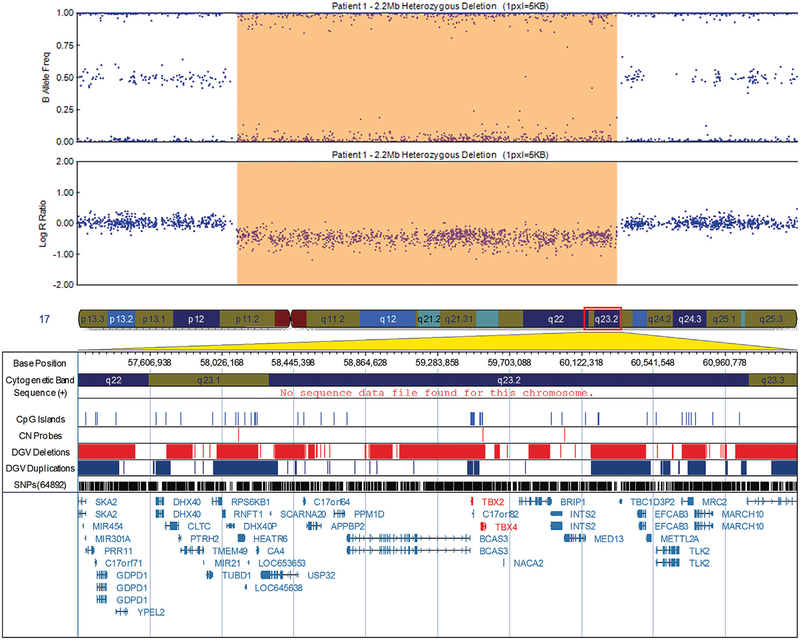
FIG. 3.
Visualization of the 2.2 Mb deletion in patient 1 at 17q23.1–23.2 that includes T-box 2 (TBX2) and TBX4, exported from the Illumina Genome Viewer. The top panel depicts B-allele frequency (ratio of minor to major alleles) and the bottom panel depicts the logR ratio data (signal intensity). Tracks provided by Illumina show cytobands, CpG islands and the location of SNPs on the array. Custom tracks were created to display the location targeted by copy number assays used to validate CNVs (“CN probes”), and copy number losses (“DGV Deletions,” shown in red) and gains (“DGV Duplications,” shown in blue), both of which were downloaded from the DGV2 database (2014-10-16 version). A subset of genes/transcripts overlapping CNVs are listed below the panels. Genes mentioned in the tables/text are in red. CNV calls made using the pennCNV algorithm are highlighted (heterozygous deletions in orange and duplications are in blue). hg19 coordinates shown. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmga].
Of the 30 CNVs reported in Supplemental Table SII not selected for validation, 12 duplications and three heterozygous deletions intersected genes, four heterozygous deletions and two duplications intersected at least one GENCODE transcript, and six heterozygous deletions and three duplications were intergenic.
DISCUSSION
The exact mechanism which causes PUV is unknown. Some have suggested that PUV represents an anomalous insertion of the mesonephric duct into the urogenital sinus and others have suggested that PUV represents remnants of the cloacal membrane [Krishnan et al., 2006]. More recent studies support the concept of a persistent urogenital membrane as the etiology of PUV and call for well-designed studies that aid in the understanding of the embryological origin of PUV [Krishnan et al., 2006]. Our study has identified genes in CNVs that possibly contribute to development of PUV. In this first genome-wide population based study of CNVs in isolated PUV, we identified several novel candidate gene regions that have not been previously reported including a 3.9 Mb deletion that includes BMP7, a 2.2 Mb deletion that involves TBX2 and TBX4, a deletion that involves PIK3R3 and TSPAN1, a duplication/triplication that involves FGF12, and a duplication encompassing FAT1.
Among the regions we identified as potentially causal is a large heterozygous deletion of 3.9 Mb on chromosome 20 harboring many genes, several of which are overlapped by CNVs carried by subjects with hydronephrosis, a common condition linked to PUV, in the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER). Among the deleted genes in this region is BMP7 which belongs to a family of secreted signaling molecules that are expressed early in embryogenesis and have a potential role in early development [Solloway and Robertson, 1999]. In late embryogenesis of the mouse urogenital system, BMP7 has been shown to be expressed in the urethra and its loss resulted in arrest in cloacal septation and severe anomalies in morphogenesis of the genital urethra and mesenchyme [Wu et al., 2009]. Several of the other genes in this region play a role in multiple types of cancer [Xu et al., 2003; Cox et al., 2006; Hobaus et al., 2013]. We are not, however, aware of any reports linking patients with PUV to increased cancer incidence in later life.
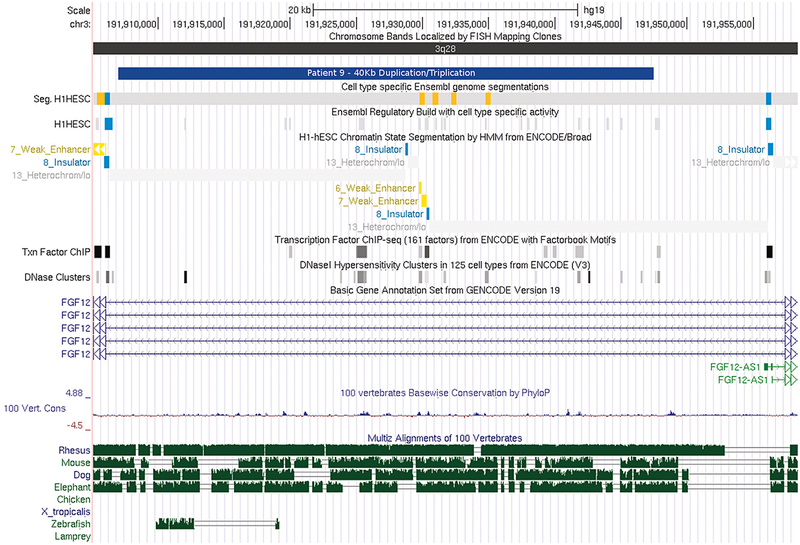
Among the other identified CNVs is a duplication/triplication in FGF12, a member of the fibroblast growth factor (FGF) family. The duplication/triplication is intronic, but does overlap transcription factors and a regulatory region (UCSC Browser; Fig. 4). To the best of our knowledge, there are no data examining the clinical significance of this duplication/triplication. FGF family members play many important roles in development. These include pre-gastrulation embryonic growth, patterning the central nervous system, and formation, growth, and shaping of several organs and tissues [Goldfarb, 1996]. FGF12 has been shown to be expressed in male reproductive organs including the ductus deferens, a derivative of the mesonephric duct [Mouse Genome Database]. This overlaps with one of the theories on PUV etiology namely, the abnormal insertion of the mesonephric duct into the urogenital sinus. The 40 Kb duplication/triplication in FGF12 also overlaps with a deletion in a subject with hydro-nephrosis in the DECIPHER database.
FIG. 4.
Visualization of the transcription factors and regulatory regions overlapped by the 40 Kb duplication/triplication in the intron of fibroblast growth factor 12 (FGF12) gene, exported from the UCSC genome browser. hg19 coordinates shown. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmga].
The 91 Kb deletion at 1p34.1 spans exonic regions of both PIK3R3 and TSPAN1 genes. The mouse expresses PIK3R3 in the urinary system at embryonic day 14.5 [Mouse Genome Database] which is equivalent to Carnegie stage 20 or week 8 following ovulation in humans, the time during which the urogenital system begins to develop. Additionally, both PIK3R3 and TSPAN1 have a medium to high protein localization score in the urinary bladder of adult humans [Uhlen et al., 2015]. PIK3R3 belongs to a family of lipid lipases responsible for coordinating a diverse range of cell functions including proliferation, cell survival, and cell migration [Genecards]. Proteins encoded by the TSPAN1 gene are involved in signal transduction events regulating cell development, activation, growth, and motility [Genecards]. These genes have not been investigated previously for involvement in genitourinary system development or malformations.
The 149 Kb duplication on chromosome 4q35.2 involves several FAT1 exons. FAT1 is highly expressed in a number of fetal epithelia, including the lower urinary tract. It facilitates cell to cell adhesion and is likely essential in developmental processes and cell communication [GUDMAP].
Other CNVs identified in a separate patient included a large 2.2 Mb deletion on chromosome 17q23.1–23.2 that involves TBX2 and TBX4, a phylogenetically conserved family of genes involved in the regulation of developmental processes [Showell et al., 2004]. Both TBX2 and TBX4 are expressed in the male urethra [Mouse Genome Database]. To our knowledge, this is the first study to report urinary system involvement in the phenotypical spectrum of chr17q23.1–q23.2 deletion syndrome [Ballif et al., 2010].
Both of the above regions on chromosomes 4q and 17q as well as regions on chromosomes 16q13.12 and 21q22.11 have been associated with hypospadias. Although reports of hypospadias occurring in conjunction with PUV are rare [Bhagat et al., 2008; Carvell and Mulik, 2013; Ranawaka and Dickson, 2013], several gene variants, including BMP7, that contribute to early urethral development have been shown to be associated with hypospadias risk in humans [Carmichael et al., 2013].
Few studies have investigated associations between gene variants and PUV. One recent study demonstrated an association between PUV and two renin-angiotensin system gene polymorphisms including angiotensin converting enzyme insertion/deletion (ACE I/D) and angiotensin type 2 receptor (AT2R A1332G) [Laksmi et al., 2010]. Specifically, a significantly higher frequency of the AT2R GG genotype was found among PUV patients as compared to control subjects. Both the ACE DD and the AT2R GG genotypes were also associated with more severe renal disease in PUV patients [Laksmi et al., 2010]. ACE DD genotype has been implicated in affecting the activity of the Renin-Angiotensin System while the AT2R polymorphism has been associated with abnormal splicing resulting in shorter length mRNA and defective protein synthesis [Laksmi et al., 2010]. While we did not investigate gene polymorphisms in the etiology of our PUV cases, we can effectively rule out large (>20 Kb) CNVs including ACE as a common cause of PUV.
We identified only one previous study by Caruana et al. that has examined CNVs in PUV in a group of patients [Caruana et al., 2015]. In a cohort of 178 patients with congenital anomalies of the kidney and urinary tract, 29 were reported to have PUV with or without other associated anomalies. Of the 18 subjects in whom CNVs with a threshold ≥200 Kb were identified, those diagnosed with PUV were the most common (n = 7; six had isolated PUV and one had PUV, hypothyroidism, and a left hydrocele) [Caruana et al., 2015]. None of our CNVs overlapped those reported in the Caruana et al. study. Possible explanations include different demographics and the inclusion of renal anomalies in the Caruana study which was an exclusionary criterion for our study. We also conducted a higher resolution than Caruana et al. considering CNVs ≥20 Kb irrespective of whether they were intergenic regions allowing us to identify additional, potentially important CNVs.
Other identified studies included a case report of PUV with chronic renal disease in a child with mild intellectual disability carrying a homozygous 16p13.11 duplication [Houcinat et al., 2015], which we did not replicate in this study. As several of the CNV regions that we identified have been previously associated with renal anomalies, despite the different embryonic origins for the kidney and the urethra, we also examined studies with reported CNVs in anomalies of the kidney and urinary tract. In one study of pediatric patients with chronic kidney disease due to congenital and non-congenital abnormalities of the kidney and urinary tract, none of the reported CNVs overlapped ours [Verbitsky et al., 2015]. In another study of individuals with renal hypodysplasia, a region of one of our duplicated CNVs (chromosome 16p11, study subject 24) that was not selected for validation, overlapped a duplicated region of the study’s reported CNVs [Sanna-Cherchi et al., 2012]. There is a potential that the CNV overlap between studies might differ given the breakpoint estimate inaccuracies.
Our study had some limitations common to CNV investigations. There is some inherent degree of uncertainty when examining catalogued CNVs in databases due to technical and platform limitations, stringency of the parameters used to impute CNVs, and whether the imputed CNVs were validated [Duclos et al., 2011]. Additionally, while hospitals and physicians are required to report birth defect cases, there is the possibility of under-ascertainment of PUV cases and PUV-associated birth defects in the CMR. Our birth prevalence estimate of 1 in 9,500 male live-births however, is similar to a recent study of 1 in 11,000 live-births [Lloyd et al., 2013]. Several strengths should also be noted. Our study is one of the largest to examine genetics of isolated PUV in a population-based setting. While we restricted our case definition to isolated PUV in an attempt to identify CNVs common to a more homogeneous group of cases, none of the CNVs analyzed occurred in more than one case. Given the population-based nature of our study, we were also able to examine the demographics of this group of infants and we noted lower gestational age and birth weight in comparison with a random sample of NYS births, a finding that is common among infants with genetic syndromes [Boghossian et al., 2012]. Our study is the second to provide data demonstrating that CNVs can reliably be detected from DNA extracted from DBS without whole-genome amplification. We recently reported using DBS to detect 20 rare CNVs, including a deletion of BMP2, in the etiology of classic heterotaxy [Rigler et al., 2015]. At this point, diagnosis using microarrays is unlikely to be implemented clinically given the genetic heterogeneity in the cases and the uncertainty associated with interpretation.
Although we deliberately excluded PUV cases with associated defects, we surprisingly detected several CNV regions that have been previously associated with renal anomalies. This finding suggests that there might be a common genetic pathway shared between PUV and renal anomalies. Our finding of previously unreported novel CNVs in PUV suggests that genetic factors may play a larger role, in as many as 57% (32/56) of cases, than previously understood. Follow-up studies might include sequencing genes in the regions we identified to determine whether point mutations in candidate genes also play a role in PUV etiology.
Supplementary Material
ACKNOWLEDGMENTS
We thank Natalie Weir at the Minnesota Core Laboratories and the staff at the Biomedical Genomics Center Facility at the University of Minnesota for microarray genotyping; April J. Atkins, Emily C. McGrath, and Adam C. Gearhart at the Wadsworth Center, New York State Department of Health for laboratory and technical assistance; Sandra D. Richardson at the Congenital Malformations Registry, New York State Department of Health for data management; and Dr. Karl G. Hill with the Social Development Research Group (5R01DA024411) at the University of Washington for generously sharing population B allele frequency and GC content files for PennCNV software. We also thank Dr. Hill with the Social Development Research Group and Drs. Ken C. Winters, George M. Realmuto, and Gerald J. August with the study on Youth Drug Abuse, ADHD and Related Disorders (5R01DA012995) for providing population allele frequencies for the CNVs nominated in this study. This study makes use of data generated by the DECIPHER Consortium. A full list of centers who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the DECIPHER project was provided by the Wellcome Trust. Those who carried out the original analysis and collection of the data bear no responsibility for the further analysis or interpretation of it. Some data used for comparison in this manuscript were obtained from the ISCA Consortium database (www.iscaconsortium.org), which generates this information using NCBI’s database of genomic structural variation (dbVar, www.ncbi.nlm.nih.gov/dbvar/), study nstd37. Samples and associated phenotype data were provided by ISCA Consortium member laboratories.
Grant sponsor: Intramural Research Program of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development; Grant numbers: HHSN275201100001I, HHSN27500005; Grant sponsor: NICHD; Grant number: N01-DK-73431; Grant sponsor: National Human Genome Research Institute.
Footnotes
Conflict of interest: None.
WEBSITES FOR DATA PRESENTATION
Database of Genomic Variants (DGV)
http://dgv.tcag.ca/dgv/app/home
DECIPHER
https://decipher.sanger.ac.uk/
GUDMAP Genitourinary Database Molecular Anatomy Project
The Human Gene Compendium
UCSC Genome Browser Home
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
REFERENCES
- Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Rosenfeld JA, Traylor RN, Gastier-Foster J, Thrush DL, Astbury C, Bartholomew D, McBride KL, Pyatt RE, Shane K, Smith WE, Banks V, Gallentine WB, Brock P, Rudd MK, Adam MP, Keene JA, Phillips JA 3rd, Pfotenhauer JP, Gowans GC, Stankiewicz P, Bejjani BA, Shaffer LG. 2010. Identification of a recurrent microdeletion at 17q23.1q23.2 flanked by segmental duplications associated with heart defects and limb abnormalities. Am J Hum Genet 86:454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat SK, Gopalakrishnan G, Kekre NS, Kumar S. 2008. Anterior and posterior urethral valves with subcoronal hypospadias: A rare association. J Pediatr Surg 43:e23–e25. [DOI] [PubMed] [Google Scholar]
- Boghossian NS, Horbar JD, Carpenter JH, Murray JC, Bell EF. 2012. Major chromosomal anomalies among very low birth weight infants in the Vermont Oxford Network. J Pediatr 160:774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael SL, Ma C, Choudhry S, Lammer EJ, Witte JS, Shaw GM. 2013. Hypospadias and genes related to genital tubercle and early urethral development. J Urol 190:1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana G, Wong MN, Walker A, Heloury Y, Webb N, Johnstone L, James PA, Burgess T, Bertram JF. 2015. Copy-number variation associated with congenital anomalies of the kidney and urinary tract. Pediatr Nephrol 30:487–495. [DOI] [PubMed] [Google Scholar]
- Carvell J, Mulik R. 2013. A case of hypospadias, anterior, and posterior urethral valves. J Surg Case Rep 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DG, Hankinson SE, Hunter DJ. 2006. Polymorphisms of the AURKA (STK15/Aurora Kinase) gene and breast cancer risk (United States). Cancer Causes Control 17:81–83. [DOI] [PubMed] [Google Scholar]
- Duclos A, Charbonnier F, Chambon P, Latouche JB, Blavier A, Redon R, Frebourg T, Flaman JM. 2011. Pitfalls in the use of DGV for CNV interpretation. Am J Med Genet Part A 155A:2593–2596. [DOI] [PubMed] [Google Scholar]
- Genecards: Weizmann Institute of Science. 2013. http://www.genecards.org/ Accessed on 17 February 2015.
- Goldfarb M 1996. Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev 7:311–325. [DOI] [PubMed] [Google Scholar]
- GUDMAP Genitourinary Database Molecular Anatomy Project. http://www.gudmap.org/ Accessed on 13 December 2014. [DOI] [PubMed]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ. 2012. GENCODE: The reference human genome annotation for the ENCODE Project. Genome Res 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobaus J, Hummel DM, Thiem U, Fetahu IS, Aggarwal A, Mullauer L, Heller G, Egger G, Mesteri I, Baumgartner-Parzer S, Kallay E. 2013. Increased copy-number and not DNA hypomethylation causes over-expression of the candidate proto-oncogene CYP24A1 in colorectal cancer. Int J Cancer 133:1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houcinat N, Llanas B, Moutton S, Toutain J, Cailley D, Arveiler B, Combe C, Lacombe D, Rooryck C. 2015. Homozygous 16p13.11 duplication associated with mild intellectual disability and urinary tract malformations in two siblings born from consanguineous parents. Am J Med Genet Part A 167:2714–2719. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. 2004. Detection of large-scale variation in the human genome. Nat Genet 36:949–951. [DOI] [PubMed] [Google Scholar]
- Illumina. Infinium genotyping data analysis. http://www.illumina.com/documents/products/technotes/technote_infinium_genotyping_data_analysis.pdf Accessed 3 March 2014.
- Indudhara R, Joseph DB, Perez LM, Diethelm AG. 1998. Renal transplantation in children with posterior urethral valves revisited: A 10-year followup. J Urol 160:1201–1203. [PubMed] [Google Scholar]
- Kramer MS, Platt RW, Wen SW, Joseph KS, Allen A, Abrahamowicz M, Blondel B, Bréart G. 2001. A new and improved population-based Canadian reference for birth weight for gestational age. Pediatrics 108:E35. [DOI] [PubMed] [Google Scholar]
- Krishnan A, de SA, Konijeti R, Baskin LS. 2006. The anatomy and embryology of posterior urethral valves. J Urol 175:1214–1220. [DOI] [PubMed] [Google Scholar]
- Laksmi NK, Khullar M, Kaur B, Ahuja M, Mahajan JK, Mittal BR, Bhattacharya A, Medhi B. 2010. Association of angiotensin converting enzyme and angiotensin type 2 receptor gene polymorphisms with renal damage in posterior urethral valves. J Pediatr Urol 6:560–566. [DOI] [PubMed] [Google Scholar]
- Lloyd JC, Wiener JS, Gargollo PC, Inman BA, Ross SS, Routh JC. 2013. Contemporary epidemiological trends in complex congenital genitouri-nary anomalies. J Urol 190:1590–1595. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Database (MGD). The Jackson Laboratory, Bar Harbor, Maine. http://www.informatics.jax.org Accessed on 25 October 2014.
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner MM, Landrum MJ, Aken B, Ayling S, Baertsch R, Fernandez-Banet J, Cherry JL, Curwen V, Dicuccio M, Kellis M, Lee J, Lin MF, Schuster M, Shkeda A, Amid C, Brown G, Dukhanina O, Frankish A, Hart J, Maidak BL, Mudge J, Murphy MR, Murphy T, Rajan J, Rajput B, Riddick LD, Snow C, Steward C, Webb D, Weber JA, Wilming L, Wu W, Birney E, Haussler D, Hubbard T, Ostell J, Durbin R, Lipman D. 2009. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res 19:1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranawaka R, Dickson AP. 2013. Multiple urethral anomalies: Anterior urethral diverticulum, posterior urethral valves, and distal hypospadias. J Pediatr Surg 48:e5–e8. [DOI] [PubMed] [Google Scholar]
- Rigler SL, Kay DM, Sicko RJ, Fan R, Liu A, Caggana M, Browne ML, Druschel CM, Romitti PA, Brody LC, Mills JL. 2015. Novel copy-number variants in a population-based investigation of classic heterotaxy. Genet Med 17:348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra-Matiz CA, Isabelle JT, Biski CK, Duva SJ, Sweeney ML, Parker AL, Young AJ, Diantonio LL, Krein LM, Nichols MJ, Caggana M. 2013. Cost-effective and scalable DNA extraction method from dried blood spots. Clin Chem 59:1045–1051. [DOI] [PubMed] [Google Scholar]
- Sanna-Cherchi S, Kiryluk K, Burgess KE, Bodria M, Sampson MG, Hadley D, Nees SN, Verbitsky M, Perry BJ, Sterken R, Lozanovski VJ, Materna-Kiryluk A, Barlassina C, Kini A, Corbani V, Carrea A, Somenzi D, Murtas C, Ristoska-Bojkovska N, Izzi C, Bianco B, Zaniew M, Flogelova H, Weng PL, Kacak N, Giberti S, Gigante M, Arapovic A, Drnasin K, Caridi G, Curioni S, Allegri F, Ammenti A, Ferretti S, Goj V, Bernardo L, Jobanputra V, Chung WK, Lifton RP, Sanders S, State M, Clark LN, Saraga M, Padmanabhan S, Dominiczak AF, Foroud T, Gesualdo L, Gucev Z, Allegri L, Latos-Bielenska A, Cusi D, Scolari F, Tasic V, Hakonarson H, Ghiggeri GM, Gharavi AG. 2012. Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreuder MF, van der Horst HJ, Bokenkamp A, Beckers GM, van Wijk JA. 2008. Posterior urethral valves in three siblings: A case report and review of the literature. Birth Defects Res A Clin Mol Teratol 82:232–235. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O’Hara R, Casalunovo T, Conlin LK, D’Arcy M, Frackelton EC, Geiger EA, Haldeman-Englert C, Imielinski M, Kim CE, Medne L, Annaiah K, Bradfield JP, Dabaghyan E, Eckert A, Onyiah CC, Ostapenko S, Otieno FG, Santa E, Shaner JL, Skraban R, Smith RM, Elia J, Goldmuntz E, Spinner NB, Zackai EH, Chiavacci RM, Grundmeier R, Rappaport EF, Grant SF, White PS, Hakonarson H. 2009. High-resolution mapping and analysis of copy number variations in the human genome: A data resource for clinical and research applications. Genome Res 19:1682–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showell C, Binder O, Conlon FL. 2004. T-box genes in early embryogenesis. Dev Dyn 229:201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solloway MJ, Robertson EJ. 1999. Early embryonic lethality in Bmp5;BMP7 double mutant mice suggests functional redundancy within the 60A subgroup. Development 126:1753–1768. [DOI] [PubMed] [Google Scholar]
- Thomalla JV, Mitchell ME, Garett RA. 1989. Posterior urethral valves in siblings. Urology 33:291–294. [DOI] [PubMed] [Google Scholar]
- Trembath DG, Rijhsinghani A. 2002. Possible maternal inheritance of a common obstructive urinary tract anomaly. Report of a case of a woman with multiple urinary tract infections and two sons with posterior urethral valves. J Reprod Med 47:962–964. [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. 2015. Proteomics. Tissue-based map of the human proteome. Science 347:1260419. [DOI] [PubMed] [Google Scholar]
- Verbitsky M, Sanna-Cherchi S, Fasel DA, Levy B, Kiryluk K, Wuttke M, Abraham AG, Kaskel F, Köttgen A, Warady BA, Furth SL, Wong CS, Gharavi AG. 2015. Genomic imbalances in pediatric patients with chronic kidney disease. J Clin Invest 125:2171–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. 2007. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 17:1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S, Mir S, Schlingmann KP, Nurnberg G, Becker C, Kara PE, Ozkayin N, Konrad M, Nürnberg P, Schaefer F. 2005. Gene locus ambiguity in posterior urethral valves/prune-belly syndrome. Pediatr Nephrol 20:1036–1042. [DOI] [PubMed] [Google Scholar]
- Wu X, Ferrara C, Shapiro E, Grishina I. 2009. BMP7 expression and null phenotype in the urogenital system suggest a role in re-organization of the urethral epithelium. Gene Expr Patterns 9:224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LL, Shi Y, Petrovics G, Sun C, Makarem M, Zhang W, Sesterhenn IA, McLeod DG, Sun L, Moul JW, Srivastava S. 2003. PMEPA1, an androgen-regulated NEDD4-binding protein, exhibits cell growth inhibitory function and decreased expression during prostate cancer progression. Cancer Res 63:4299–4304. [PubMed] [Google Scholar]
- Yohannes P, Hanna M. 2002. Current trends in the management of posterior urethral valves in the pediatric population. Urology 60:947–953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.