Abstract
We had experienced 117 Japanese Fabry patients (72 males and 45 females) from 1977 to 2006, and then we generated an improved Fabry analysis system in 2007 and have found 196 ones (95 males and 101 females) since then. In this study, we summarized the data of the patients and tried to elucidate the molecular and biochemical characteristics of Japanese Fabry patients. Gene analysis revealed various GLA mutations, including missense mutations (56.5%, 48 types); nonsense mutations (15.9%, 13 types); deletions (12.6%, 13 types); splicing defects (10.1%, 6 types); insertions (1.0%, 2 types), and insertions/deletions (0.5%, 1 type), in the patients that were tested. Amino acid substitutions resulting from the missense mutations found in the classic form patients tended to be localized in the core of the GLA protein, and those in the later-onset ones in the peripheral region. The most commonly identified pathogenic mutations are c.888G > A (p.M296I), c.936 + 919G > A, c.679C > T (p.R227X), c.335G > A (p.R112H), c.334C > T (p.R112C), and c.902G > A (p.R301Q). Among them, c.888G > A (p.M296I) is unique to Japanese Fabry patients. On the other hand, c.936 + 919G > A is a variant that has been frequently detected in Taiwan Chinese Fabry patients, and c.335G > A (p.R112H) in various countries. These are found in later-onset patients, and c.679C > T (p.R227X) and c.334C > T (p.R112C) classic ones. c.902G > A (p.R301Q) is found in both classic and later-onset form patients. A possible functional polymorphism, c.196G > C (p.E66Q), was identified in 0.4% of the subjects who underwent high-risk screening. The biochemical findings including leukocyte α-galactosidase A activity, plasma globotriaosylsphingosine level and urinary globotriaosylceramide in the individual phenotypic groups well reflected the phenotypic differences in this disease. The results will be useful for understanding the basis of Fabry disease in Japan.
Keywords: Fabry disease, α-Galactosidase A, Globotriaosylceramide, Globotriaosylsphingosine, Gene mutation
Highlights
-
•
The characteristics of Japanese Fabry patients were elucidated.
-
•
p.M296I unique to Japanese Fabry patients was commonly identified.
-
•
The biochemical findings well reflected the phenotypic differences.
1. Introduction
Fabry disease (OMIM 301500) is an X-linked genetic disorder with reduced or deficient activity of α-galactosidase A (GLA, EC 3. 2. 1. 22) resulting in accumulation of glycolipids, predominantly globotriaosylceramide (Gb3) and globotriaosylsphingosine (Lyso-Gb3), in organs and body fluids. This disease shows a wide clinical spectrum, and the clinical manifestations and findings are influenced by the GLA mutation and gender of a patient [1,2]. Males with the “classic form” Fabry disease exhibit pain in the peripheral extremities, angiokeratomas, hypohidrosis, corneal opacities, and bowel disturbance during childhood or adolescence, and develop renal, cardiac and cerebrovascular involvement, which influence their prognosis, in adulthood. “Later-onset form” of Fabry males are characterized by relatively milder clinical manifestations limited to the heart and/or kidneys in adulthood without any symptoms in childhood. “Fabry females” exhibit quite heterogeneous disease expression from asymptomatic to severe, depending on random X-chromosomal inactivation [[1], [2], [3], [4]].
Through medical and technological development, enzyme replacement therapy (ERT) and chaperone therapy are now available [[5], [6], [7]], and neonatal and high-risk screening is performed for early diagnosis and early treatment of Fabry patients [[8], [9], [10], [11], [12], [13]]. Accordingly, an effort to elucidate the characteristics of Fabry patients in various countries and areas has been made [[11], [12], [13], [14], [15], [16]]. For example, in Japan, many case reports and results of small cohort studies have been published [[17], [18], [19], [20]]. However, there has been few comprehensive studies of large numbers of Japanese Fabry patients. In this study, we tried to summarize the molecular and biochemical data for a lot of Japanese Fabry patients diagnosed in our laboratory, and to elucidate the characteristics of Japanese Fabry patients in order to understand the basis of this disease in Japan.
2. Materials and methods
2.1. Study design
In this study, we examined the phenotype, genotype, and biochemical characteristics of Japanese Fabry patients who had been diagnosed in our laboratory, and summarized the results.
We generated an improved Fabry analysis system including assaying of GLA activity in serum and leukocytes, GLA gene analysis, and detection of urinary Gb3 for diagnosis of Fabry disease in the present institute in 2007, and thereby found Japanese Fabry patients, answering requests from clinicians in various fields. Measurement of the plasma lyso-Gb3 concentration has been included in the Fabry analysis system since 2009. So, up to the present, we have found 196 Fabry patients (95 males and 101 females) in the “diagnosis of individual cases” for subjects strongly suspected of having Fabry disease, “high-risk screening” for males presenting any symptoms among the following, acroparesthesia, hypohidrosis, angiokeratomas, renal disorder, heart involvement and cerebrovascular disease, and “family diagnosis”. Then, we summarized the clinical, molecular and biochemical data, and compared them with the data for 117 Fabry patients (72 males and 45 females) that had been diagnosed by us in other hospitals before we moved to the present institute (1977–2006). Unfortunately, sufficient detailed information of the patients diagnosed before 2007 was not available because their records had been already discarded according to the hospitals' policy after a certain period of storage. However, the basic information on the genotype and phenotype of the patients was available.
This study was approved by the ethical committee of Meiji Pharmaceutical University, and was performed according to the ethical guidelines of the 1975 Declaration of Helsinki. Informed consent for this study was obtained from all the participants.
2.2. GLA gene analysis
Genomic DNA was extracted from blood samples of the Fabry patients with a Wizard genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's instructions. Seven exons and intron/exon boundaries of the GLA gene, and the specific intronic region containing c.936 + 919 were amplified by polymerase chain reaction (PCR) using appropriate primers described previously [21], and another pair, g.9331-F (5′-GCAGACCAAACGCCACATA-3′) and g.9331-R (5′-TGCGAGAGATACAGTCAAAGTC-3′). The PCR protocol consisted of 2 min at 94 °C, then 35 cycles of 15 s at 94 °C, 30 s at 55 °C, and 40 s at 68 °C, and finally 1 min at 68 °C, with KOD-plus DNA polymerase (Toyobo, Osaka, Japan). The amplified PCR fragments were directly sequenced.
2.3. Localization of the amino acid substitutions caused by Fabry missense mutations in the GLA structure
The amino acid substitutions caused by missense mutations found in the classic and later-onset Fabry patients were localized in the crystal structure of human GLA (PDB: 1R45) [22].
2.4. Measurement of GLA activity in serum and leukocytes
GLA activity in serum was fluorometrically measured for the first examination for high-risk screening [20]. Briefly, 20 μl of serum was mixed with 40 μl of a substrate solution comprising 5 nmol/L 4-methylumbelliferyl α-D-galactopyranoside (Calbiochem, La Jolla, CA) as a substrate and 117 nmol/L N-acetyl-D-galactosamine (Sigma-Aldrich, St. Louis, MO) as an inhibitor of α-galactosidase B (α-N-acetylgalactosaminidase) in 0.1 mol/L citrate-phosphate buffer, pH 4.6, in a 96-well plate. The mixture was incubated at 37 °C for 4 h, and then the reaction was stopped by adding 200 μL of 0.2 mol/L glycine buffer, pH 10.7. Then the released 4-methylumbelliferone (MU) was measured using a Wallac 1420 ARVO MX multi-label counter (PerkinElmer Inc., Waltham, MA) at excitation and emission wavelengths of 355 nm and 460 nm, respectively.
For measurement of GLA activity in leukocytes, 10 μl of a leukocyte homogenate (about 10 μg protein) was mixed with 40 μl of the substrate solution in a 1.5 mL micro-tube, followed by incubation at 37 °C for 30 min [20,23]. Then, the reaction was stopped by adding 950 μL of the stopping solution, and the released MU was measured using a spectrofluorometer (F2700; Hitachi Ltd., Tokyo, Japan) at excitation and emission wavelengths of 365 nm and 450 nm, respectively. Protein determination was performed with a Micro BCA protein assay kit (PIERCE, Rockford, IL), using bovine serum albumin as the standard.
2.5. Measurement of plasma Lyso-Gb3
Lyso-Gb3 in plasma was measured by means of liquid chromatography-tandem mass spectrometry (LC-MS/MS) according to the method described previously [24]. Various concentrations of Lyso-Gb3, ranging from 0.08 to 250 nmol/L, and stable-isotope labeled Lyso-Gb3 [25], which has one 13C and three deuteriums, were used as the standard and internal standard, respectively. For the LC, a LC system (Shimadzu, Kyoto, Japan) and an InertSustain C18 column (30 mm × 3.0 mm I.D., 5 μm: GL Science, Tokyo, Japan) were used. For detection of Lyso-Gb3, a LCMS-8040 triple quadrupole mass spectrometer (Shimadzu, Kyoto, Japan) equipped with an electrospray ionization interface was used in the positive-ion mode. The Multiple Reaction Monitoring (MRM) conditions were optimized with an automatic MRM optimization function. In the MRM mode, the following transitions were monitored: m/z 786.8 → 282.3 for Lyso-Gb3 and m/z 790.8 → 286.2 for the internal standard.
2.6. Detection of Gb3 in urine
Gb3 in urine was detected by thin-layer chromatography (TLC) followed by Gb3-staining with an anti-Gb3 antibody, according to the method described previously [26]. Briefly, 30 mL of urine was centrifuged, and the urinary sediment was obtained. Then, the glycolipid fraction including Gb3 of the urinary sediment was extracted with CHCl3/MeOH/water (2:1:0.1). The glycolipid fraction was evaporated to dryness and then diluted in CHCl3/MeOH/water (60:30:4.5), followed by TLC. For TLC, HPTLC silica gel 60 (Merck, Darmstadt, Germany) was used, and the separated Gb3 was immunologically stained with a hybridoma supernatant containing an anti-Gb3 mouse monoclonal antibody [27]. Purified Gb3 (Larodan AB, Solna, Sweden) was used as a standard.
2.7. Statistical analysis
Data for the various phenotypic groups are basically expressed as means ± standard deviation (SD) [n: number of trials]. The differences among the targeted groups were assessed by means of Welch's t-test, it being taken that there was a significant difference if p < .05. To examine the distribution of the cases in each group, dot plot analysis was performed.
3. Results
3.1. Phenotype of the Japanese Fabry patients
The phenotype of the Fabry patients diagnosed in our laboratory was examined. From 1977 to 2006, 117 patients (72 males and 45 females) with Fabry disease were diagnosed by means of measurement of leukocyte GLA activity, urinary Gb3 detection, and GLA gene analysis (1990~). When necessary, pathological examination was performed. Unfortunately, clinical information on 15 of the 72 male patients could not be obtained from the clinicians who requested the diagnosis. The phenotypes of the Fabry patients were determined according to their manifestations and family history as follows: Classic (C)/Later-onset (L)/Unknown: A group of Fabry males for which detailed clinical information are not available (U)/Female (F), 43/14/15/45. Since 2007, 196 Fabry patients (95 males and 101 females) have been found with the established analysis system, and their phenotypes were determined in the same way (C/L/U/F, 48/35/12/101). The ratios (percentages) of the patients developing heart disease, renal disorder, and cerebrovascular involvement as to the total were 52%, 51%, and 11%, respectively, in the male patients, and those being 32%, 31%, and 6%, respectively, in the female ones. When these two groups were compared, the number of patients diagnosed in a year and the ratios of the later-onset Fabry males and Fabry females as to the total Fabry patients apparently increased after 2007.
3.2. Fabry patients diagnosed by “high-risk screening”, “diagnosis of individual cases”, and “family diagnosis”
“High-risk screening” was performed for 9106 males presenting any of the clinical manifestations indicating the possibility of Fabry disease, regardless of the severity of the disease, according to the method described previously [20]. As the first examination of the test, serum GLA activity was measured. The results of the preliminary study revealed that the serum GLA activity in apparently normal subjects was 4.5 ± 1.7 nmol/h/mL [n = 3963], and the cut-off point was determined to be 1.5 nmol/h/mL (mean – 1.75SD). Of all the patients who underwent the first examination of the” high-risk screening” test, 91 showed a positive reaction, and thus the second examination including measurement of leukocyte GLA activity, GLA gene analysis, plasma lyso-Gb3 measurement, and detection of urinary Gb3 (and pathological examination, when necessary) was performed for them. The results revealed that 38, 38, and 15 of them were Fabry patients, subjects having a possible functional polymorphism, c.196G > C (p.E66Q), and non-Fabry cases having sufficient leukocyte GLA activity, respectively. Fabry patients accounted for 0.4% of the males subjected to the “high-risk screening”, and subjects harboring c.196G > C (p.E66Q) were identified at the same incidence (0.4%). On the other hand, 148 subjects, who had been strongly suspected of having Fabry disease from their specific clinical manifestations and findings, received the “diagnosis of individual cases”, 83 (56%; 45 males and 38 females) being found to have Fabry disease. Following the “high-risk screening” and “diagnosis of individual cases”, “family diagnosis” was performed for 96 subjects, and 75 Fabry patients (78%; 12 males and 63 females) were found among them. The number of Fabry patients in each phenotypic group and the specialty of the clinicians who requested the diagnosis are shown in Table 1. The results revealed that cardiologists and nephrologists followed by pediatricians were involved in the diagnosis of Fabry patients, and many female patients were found on the “family diagnosis”.
Table 1.
Fabry patients found in our laboratory and the specialities of the clinicians who requested the diagnosis (2007~).
| Male |
Female |
Total | ||||
|---|---|---|---|---|---|---|
| Clinician fields | High-risk screening | Diagnosis of individual cases | Family diagnosis | Diagnosis of individual cases | Family diagnosis | |
| Cardiologists | 11 | 22 | 5 | 14 | 24 | 76 |
| Nephrologists | 21 | 9 | 4 | 11 | 25 | 70 |
| Pediatricians | 4 | 14 | 2 | 12 | 11 | 43 |
| Neurologists | 2 | 0 | 1 | 0 | 3 | 6 |
| Dermatologists | 0 | 0 | 0 | 1 | 0 | 1 |
| Total | 38 | 45 | 12 | 38 | 63 | 196 |
| 95 | 101 | |||||
3.3. GLA gene mutations
GLA gene analysis was performed for 207 patients from which informed consent was obtained since 1990. The information on individual cases is shown in our fabry-database (http://fabry-database.org/). Eighty-three pathological gene mutations including 18 that have never been registered in the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/) were identified in 200 cases: missense mutations, 117 cases (56.5%, 48 types); nonsense mutations, 33 cases (15.9%, 13 types); deletions, 26 cases (12.6%, 13 types); splicing defects, 21 cases (10.1%, 6 types); insertions, 2 cases (1.0%, 2 types), and insertions/deletions, 1 case (0.5%, 1 type) (Table 2). There are seven cases (five males and two females, 3.4%) for which no pathological gene mutations could be found in either the exonic regions or intron/exon boundaries of the GLA gene. All of the five male patients exhibited almost deficient GLA activity and typical pathological changes in the biopsied heart tissues, and the two female patients moderately decreased GLA activity and increased plasma Lyso-Gb3 concentration. In one of the two female patients, over-excretion of urinary Gb3 was found. Gross alterations, nonsense mutations, and splicing defects except for c.936 + 919 G > A [28] were identified in the classic Fabry patients. However, missense mutations comprising the majority of Fabry mutations were found in both the classic and later-onset Fabry patients.
Table 2.
GLA gene mutations identified in our laboratory (1990~).
| Missense mutations (48 types, 117 cases) |
| c.47 T > G (p.L16R), c.58G > C (p.A20P), c.104G > A (p.G35E), c.109G > A (p.A37T), c.110C > T (p.A37V), c.124A > G (p.M42 V), c.128G > T (p.G43 V), c.139 T > C (p.W47R), c.207C > A (p.F69 L), c.288G > A (p.M96I), c.334C > T (p.R112C), c.335G > A (p.R112H), c.337 T > A (p.F113I), c.338 T > C (p.F113S), c.425G > A (p.C142Y), c.437C > G (p.P146R), c.440G > A (p.G147E), c.443G > A (p.S148 N), c.496C > G (p.L166 V), c.548G > A (p.G183D), c.584G > T (p.G195 V), c.595G > A (p.V199 M), c.607G > A (p.E203K), c.623 T > G (p.M208R), c.628C > T (p.P210S), c.629C > T (p.P210L), c.644A > G (p.N215S), c.668G > T (p.C223F), c.688G > A (p.A230T), c.725 T > A (p.I242N), c.725 T > C (p.I242T), c.758 T > C (p.I253T), c.835C > G (p.Q279E), c.888G > A (p.M296I), c.899 T > C (p.L300P), c.902G > A (p.R301Q), c.950 T > C (p.I317T), c.960 T > G (p.N320 K), c.982G > A (p.G328R), c.1025G > A (p.R342Q), c.1061 T > A (p.I354K), c.1065C > A (p.N355 K), c.1078G > A (p.G360S), c.1124G > A (p.G375E), c.1133G > T (p.C378F), c.1208 T > C (p.L403S), c.1244 T > C (p.L415P), c.1250 T > C (p.L417P) |
| Nonsense mutations (13 types, 33 cases) |
| c.131G > A (p.W44X), c.242G > A (p.W81X), c.607G > T (p.E203X), c.612G > A (p.W204X), c.679C > T (p.R227X), c.707G > A (p.W236X), c.734G > A (p.W245X), c.772G > T (p.G258X), c.901C > T (p.R301X), c.1019G > A (p.W340X), c.1034C > G (p.S345X), c.1095 T > G (p.Y365X), c.1196G > A (p.W399X) |
| Deletions (13 types, 26 cases) |
| c.85delG, c.125_137del13, c.167_172del6, c.370delG, c.409delG, c.718_719delAA, c.733delT, c.1033-1034delTC, c.1072_1074delGAG, c.1177_1185del9, c.1235_1236delCT, c.1277_1278delAA, IVS2_IVS4 |
| Splicing defects (6 types, 21 cases) |
| c.548-1G > A, c.639 + 1G > A, c.936 + 919G > A, c.640-1G > T, c.802-1G > C, c.999 + 1G > T |
| Insertions (2 types, 2cases) |
| c.723_724insT, c.906_935dup30 |
| Insertions/Deletions (1 type, 1 case) |
| c.1067_1082del16ins9 |
The mutations in Gothic (18 types) are variants that have never been registered in the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/).
The most commonly identified six mutations, c.888G > A (p.M296I), c.936 + 919G > A, c.679C > T (p.R227X), c.335G > A (p.R112H), c.334C > T (p.R112C), and c.902G > A (p.R301Q), and brief information on them are presented in Table 3. The c.888G > A (p.M296I), c.936 + 919G > A, and c.335 G > A (p.R112H) mutations are thought to be essentially associated with the later-onset form of Fabry disease, and c.679C > T (p.R227X) and c.334C > T (p.R112C) with the classic ones. The c.902G > A (p.R301Q) mutation was found in both a classic Fabry patient and two later-onset ones.
Table 3.
Most common GLA gene mutations and the phenotypes of Fabry patients harboring them found in our laboratory (1990~).
| Phenotype |
||||||
|---|---|---|---|---|---|---|
| No. of cases | No. of families | Male |
Female | |||
| Classic | Later-onset | Unknown | ||||
| c.888G > A (p.M296I) | 12 | 7 | 0 | 6 | 2 | 4 |
| c.936 + 919G > A | 9 | 6 | 0 | 4 | 2 | 3 |
| c.679C > T (p.R227X) | 9 | 3 | 2 | 0 | 0 | 7 |
| c.335G > A (p.R112H) | 8 | 6 | 0 | 5 | 0 | 3 |
| c.334C > T (p.R112C) | 8 | 7 | 5 | 0 | 0 | 3 |
| c.902G > A (p.R301Q) | 8 | 3 | 1 | 2 | 0 | 5 |
3.4. Distribution of the amino acid substitutions caused by missense mutations in the GLA structure
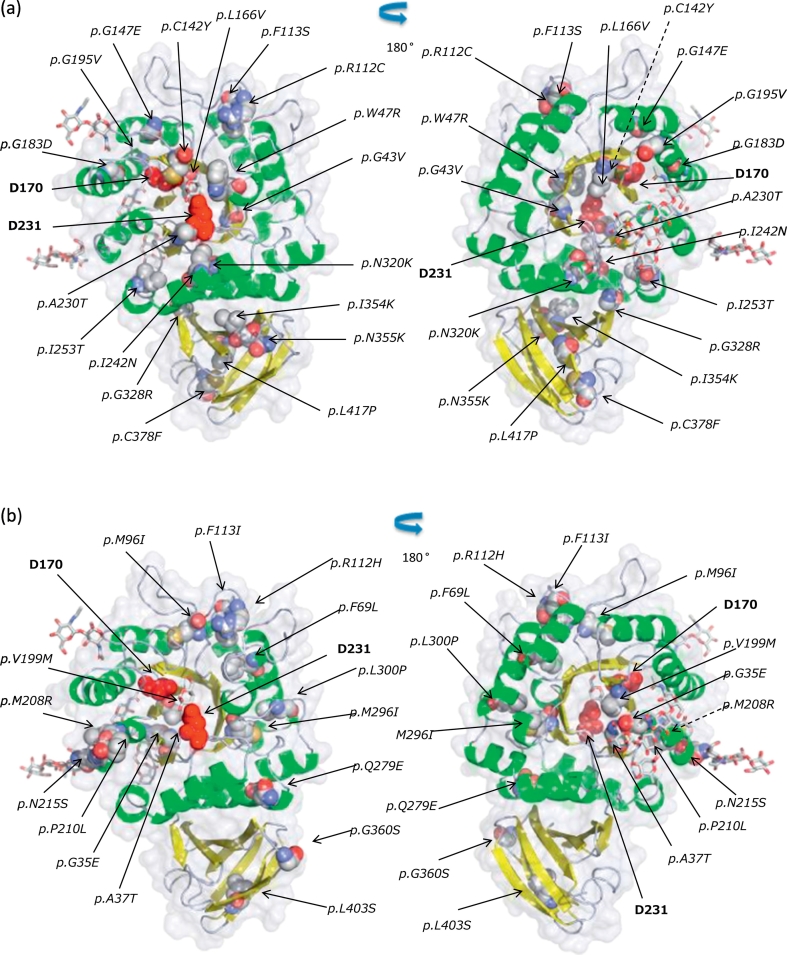
According to the crystallographic structure of human GLA, the enzyme subunit is composed of two domains: an N-terminal (β/α)8-barrel domain and a C-terminal antiparallel β-sheet one [22]. We determined the location of the amino acid substitutions caused by missense mutations identified in the classic (Fig. 1a) and later-onset (Fig. 1b) Fabry patients, except for those found in both the classic and later-onset Fabry males and those in Fabry females only. Although the amino acid substitutions found in the classic Fabry patients were broadly distributed over the GLA structure, many of them were localized in the core of the GLA protein. On the other hand, most of those found in the later-onset ones were localized in the peripheral region.
Fig. 1.
Structure of human GLA and positions of the amino acid substitutions resulting from the missense mutations identified in the classic (a) and later-onset (b) Japanese Fabry patients. The backbone is displayed as a ribbon model, and the ligand and sugars as a stick model. The amino acids involved in the substitutions and the catalytic residues (D170 and D231) are indicated as a CPK model and red spheres, respectively. Front view (left) and back view (right). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. GLA activity in leukocytes
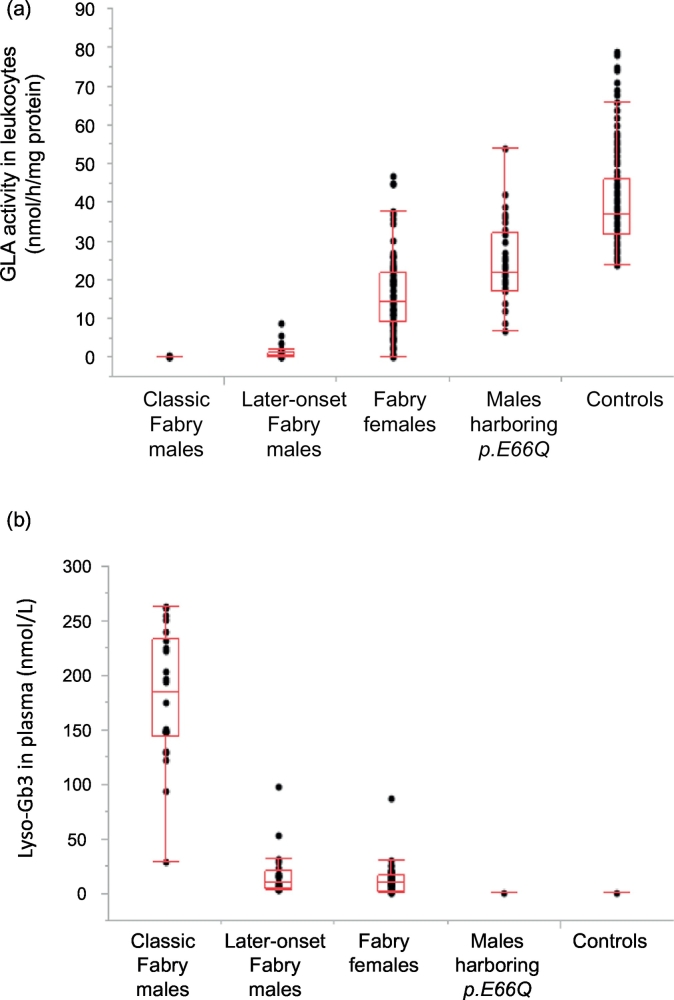
The leukocyte GLA activities in the different phenotypic groups are shown in Fig. 2a. The average leukocyte GLA activities in the classic Fabry males, later-onset Fabry males, Fabry females, males harboring c.196G > C (p.E66Q), and control subjects were 0.13 ± 0.14 [n = 37], 1.4 ± 2.2 [n = 29], 17 ± 10 [n = 90], 24 ± 10 [n = 38], and 41 ± 12 nmol/h/mg protein [n = 136], respectively. Both the classic and later-onset Fabry males exhibited apparently lower GLA activity than the control group (p < .01, for the classic group vs. the control one, and the later-onset group vs. the control one), although slight residual activity was found in the later-onset Fabry males. Fabry females showed a wide range of GLA activity, the average value being about half of the control mean. Approximately 1/3 of them could not be differentiated from the controls. The subjects with c.196G > C (p.E66Q) exhibited about 59% (24–65%) of the control mean.
Fig. 2.
Leukocyte GLA activity (a) and plasma Lyso-Gb3 concentration (b) in the classic Fabry males, later-onset Fabry males, Fabry females, males harboring c.196G > C (p.E66Q), and controls. A box plot shows the distribution of cases in each group.
3.6. Lyso-Gb3 in plasma
The plasma Lyso-Gb3 concentrations of Fabry patients have been measured since 2009, and the results are summarized in Fig. 2b. The average plasma Lyso-Gb3 levels in the classic Fabry males, later-onset Fabry males, Fabry females, males harboring c.196G > C (p.E66Q), and control subjects were 177 ± 61 [n = 24], 16 ± 20 [n = 25], 12 ± 13 [n = 48], 0.38 ± 0.13 [n = 17], and 0.37 ± 0.11 [n = 34] nmol/L, respectively. The average plasma Lyso-Gb3 concentration in the classic Fabry males was significantly higher than that in the control subjects (p < .01), and those in the later-onset Fabry males and Fabry females were lower than that in the classic Fabry males (p < .01, for the later-onset group vs. the classic one, and the female one vs. the classic one), but apparently higher than that in the controls (p < .01, for the later-onset group vs. the control one, and the female one vs. the control one). Among the Fabry patients, subjects harboring c.888G > A (p.M296I) or c.335G > A (p.R112H) exhibited relatively low plasma Lyso-Gb3 concentrations (c.888G > A (p.M296I), hemizygotes: 5.3 ± 2.3 [n = 3] (2.9–7.4) nmol/L, heterozygotes: 1.3 ± 0.5 [n = 3] (0.87–1.8) nmol/L; and c.335G > A (p.R112H), hemizygotes: 5.4 ± 2.6 [n = 5] (3.2–9.9) nmol/L, heterozygotes: 0.96 ± 0.83 [n = 3] (0.42–1.9) nmol/L) compared with the others, as previously reported [23,25]. Especially in the heterozygotes, the plasma Lyso-Gb3 level was near the control value, although it was a little bit higher than that in the controls. On the other hand, there was no statistical difference in the plasma Lyso-Gb3 concentration between the males with c.196G > C (p.E66Q) and the controls (p > .05).
3.7. Gb3 in urine
Detection of urinary Gb3 by means of TLC followed by immunostaining has been performed using available samples from 30 classic Fabry males, 30 later-onset Fabry males, and 80 Fabry females since 2007. All of the classic Fabry males were Gb3+ (positive reaction showing over-excretion of Gb3 in urine). The numbers of later-onset Fabry males who were Gb3+, ± (pseudopositive reaction), and – (negative reaction) were 22, 4, and 4, respectively. In the Fabry females examined, the numbers of subjects who were Gb3+, ±, and – were 47, 13, and 20, respectively. All of the 47 control subjects examined were Gb3–.
4. Discussion
The number of Fabry patients found in a year apparently increased from 4.0 (1977–2006) to 16.3 (2007~). Widening of clinicians' knowledge on Fabry disease and advances of diagnostic technology after the introduction of ERT have been helpful for finding such patients. As to Fabry females, many of them have been found on “family diagnosis”, suggesting the importance of genetic counselling. Furthermore, when we consider that many Fabry patients exhibited cardiac and/or renal symptoms and have been found due to requests for diagnosis from cardiologists and nephrologists, an effort not to miss such patients and diagnose them in the early stage of the disease is required.
Up to now, >900 GLA mutations have been reported in the world (HGMD, http://www.hgmd.cf.ac.uk/). In this study, we performed GLA gene analysis for 207 Fabry patients and identified 83 mutations in 200 (96.6%) of them. We did not find any pathological mutations in another seven cases (3.4%) on the ordinary gene analysis. Pathological changes of the GLA gene may exist somewhere in the region that the present gene analysis did not cover in these cases, i.e., the regulation area of the GLA gene. Of the GLA mutations identified in Japanese Fabry patients, the most frequently identified variants were missense ones; followed by nonsense ones, deletions, splicing defects, insertions, and insertions/deletions. It is difficult to clearly determine the relationship between the genotype and phenotype [29,30]. However, regarding the Japanese Fabry patients examined in this study, nonsense mutations, gene rearrangements, and most splicing defects except for c.936 + 919G > A were found in the classic form patients. On the other hand, missense mutations were identified in both the classic and later-onset patients.
To determine the locations of amino acid substitutions caused by missense mutations in the GLA structure, we performed structural analysis. The amino acid residues of which substitutions associated with the later-onset Fabry disease tended to be located near or on the molecular surface, and those with the classic form near or in the core of the GLA molecule. The substitution of buried amino acids is thought to cause serious conformational changes and to severely affect the stability of the enzyme molecule and/or the catalytic site.
The six commonly found mutations in Japanese Fabry patients account for 26% of all subjects who underwent the gene analysis.
c.888G > A (p.M296I) is a mutation unique to Japanese (fabry-database, http://fabry-database.org/) [31]. M296 is located in the core region of the GLA molecule, and it is predicted that the amino acid substitution causes a very small structural change in a restricted region from the core to the surface, and that the active site is not affected [23]. Male patients harboring this mutation have residual GLA activity. The plasma Lyso-Gb3 level in the patients is relatively lower than those in the classic and other later-onset Fabry males, but higher than that in the healthy subjects [23,25]. The patients with this mutation exhibit later-onset and mild clinical manifestations [31].
The c.936 + 919 G > A mutation is an intronic variant causing aberrant alternative splicing. Newborn screening in Taiwan has revealed a surprisingly high frequency of Fabry patients with this mutation [[11], [12], [13]]. In Japan, this mutation has been found in Fabry patients who live in the south-eastern area of the country. The patients with this mutation have residual GLA activity and exhibit a moderately increased plasma Lyso-Gb3 concentration. They presented the later-onset form of Fabry disease, and this mutation is predicted to have spread widely in the eastern Asia.
c.679C > T (p.R227X) is a nonsense mutation, leading to premature termination of protein translation of GLA. In the Fabry males with this mutation, GLA activity was almost completely deficient, and the plasma Lyso-Gb3 concentration was significantly elevated. This mutation has been frequently identified in various countries of the world (fabry-database, http://fabry-database.org/).
The c.334C > T (p.R112C) and c.335G > A (p.R112H) mutations are missense ones resulting in different amino acid substitutions at the same residue on GLA [32]. Both have been identified in various countries (http://fabry-database.org/). R112 is located on the loop comprising the barrel domain of the GLA structure, being near the surface of the molecule, and both the amino acid substitutions do not affect the active site [23]. This structural change is thought to affect the stability of the GLA protein. The GLA activity in the patients with c.334C > T (p.R112C) was almost completely deficient and the plasma Lyso-Gb3 level was very high. The males harboring this mutation exhibited the classic Fabry disease. On the other hand, the males with c.335G > A (p.R112H) exhibited residual GLA activity, and their plasma Lyso-Gb3 concentrations were almost the same as in the males with c.888G > A (p.M296I) [23,25].
The males harboring c.902G > A (p.R301Q) exhibited clinical symptoms of both the classic and the later-onset Fabry disease (http://fabry-database.org/). Their plasma lyso-Gb3 levels were considerably increased.
c.196G > C (p.E66Q) has been frequently identified in Koreans (approximately 1% of the Korean population) [33]. This variant was found in 0.4% of the Japanese subjects that underwent the “high-risk screening” test in this study. Their GLA activity level was 59% of the control mean, and there was no statistical difference in the plasma Lyso-Gb3 concentration between the cases with c.196G > C (p.E66Q) and the healthy subjects, strongly suggesting that this is a functional polymorphism. The evidence that no pathological findings specific to Fabry disease have been observed in biopsied tissues supports it [34].
The biochemical data including leukocyte GLA activity, plasma Lyso-Gb3 concentration, and urinary Gb3 excretion in the individual phenotypic groups well reflected the phenotypic differences in this disease.
In conclusion, we elucidated the molecular and biochemical characteristics of Japanese Fabry patients. The results will be useful for understanding the basis of Fabry disease in Japan.
Acknowledgments
Acknowledgements
We thank the many doctors involved in this study for providing of the patients' information and Ms. M. Tanaka for typing the manuscript. This work was partly supported by JPS KAKENHI Grant Number JP18K15071.
Financial disclosures
Hitoshi Sakuraba has received grants and personal fees from Dainippon Sumitomo Pharma Co., Ltd. and Sanofi Japan Co., outside of the submitted work.
Tadayasu Togawa has received grants from Sanofi Japan Co., outside of the submitted work.
Takahiro Tsukimura received JPS KAKENHI Grant Number JP18K15071.
Toshie Tanaka, Tomoko Otsuka, Atsuko Sato, Tomoko Shiga, Seiji Saito, and Kazuki Ohno have nothing to disclose.
Conflict of interest
Although Kazuki Ohno is an employee of Catalyst Inc., he has no competing interest.
We declare that none of the authors have any competing interests.
Human and animal rights
This study was approved by the ethical committee of Meiji Pharmaceutical University, and was performed according to the ethical guidelines of the 1975 Declaration of Helsinki. Informed consent for this study was obtained from all the participants.
Author contributions
Conceived and designed the study: Hitoshi Sakuraba, Tadayasu Togawa, Takahiro Tsukimura. Performed the experiments: Takahiro Tsukimura, Toshie Tanaka, Tomoko Ohtsuka, Atsuko Sato, Tomoko Shiga, Seiji Saito, Kazuki Ohno. Wrote the paper: Hitoshi Sakuraba.
References
- 1.Germain D.P. Fabry disease. Orphanet J. Rare Dis. 2010;5:30. doi: 10.1186/1750-1172-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branton M.H., Schiffmann R., Sabnis S.G., Murray G.J., Quirk J.M., Altarescu G., Goldfarb L., Brady R.O., Balow J.E., Austin H.A., III, Kopp J.B. Natural history of Fabry renal disease: influence of alpha-galactosidase A activity and genetic mutations on clinical course. Medicine (Baltimore) 2002;81:122–138. doi: 10.1097/00005792-200203000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Nance C.S., Klein C.J., Banikazemi M., Dikman S.H., Phelps R.G., McArthur J.C., Rodriguez M., Desnick R.J. Later-onset Fabry disease: an adult variant presenting with the cramp-fasciculation syndrome. Arch. Neurol. 2006;63:453–457. doi: 10.1001/archneur.63.3.453. [DOI] [PubMed] [Google Scholar]
- 4.MacDermot K.D., Holmes A., Miners A.H. Anderson-Fabry disease: clinical manifestations and impact of disease in a cohort of 60 obligate carrier females. J. Med. Genet. 2001;38:769–775. doi: 10.1136/jmg.38.11.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germain D.P., Charrow J., Desnick R.J., Guffon N., Kempf J., Lachmann R.H., Lemay R., Linthorst G.E., Packman S., Scott C.R., Waldek S., Warnock D.G., Weinreb N.J., Wilcox W.R. Ten-year outcome of enzyme replacement therapy with agalsidase beta in patients with Fabry disease. J. Med. Genet. 2015;52:353–358. doi: 10.1136/jmedgenet-2014-102797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck M., Hughes D., Kampmann C., Larroque S., Mehta A., Pintos-Morell G., Ramaswami U., West M., Wijatyk A., Giugliani R. Fabry Outcome Survey Study Group, long-term effectiveness of agalsidase alfa enzyme replacement in Fabry disease: a Fabry Outcome Survey analysis. Mol. Genet. Metab. Rep. 2015;3:21–27. doi: 10.1016/j.ymgmr.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Germain D.P., Hughes D.A., Nicholls K., Bichet D.G., Giugliani R., Wilcox W.R., Feliciani C., Shankar S.P., Ezgu F., Amartino H., Bratkovic D., Feldt-Rasmussen U., Nedd K., Sharaf E., Din U., Lourenco C.M., Banikazemi M., Charrow J., Dasouki M., Finegold D., Giraldo P., Goker-Alpan O., Longo N., Scott C.R., Torra R., Tuffaha A., Jovanovic A., Waldek S., Packman S., Ludington E., Viereck C., Kirk J., Yu J., Benjamin E.R., Johnson F., Lockhart D.J., Skuban N., Castelli J., Barth J., Barlow C., Schiffmann R. Treatment of Fabry's disease with the pharmacologic chaperone migalastat. N. Engl. J. Med. 2016;375:545–555. doi: 10.1056/NEJMoa1510198. [DOI] [PubMed] [Google Scholar]
- 8.Sista R.S., Wang T., Wu N., Graham C., Eckhardt A., Winger T., Srinivasan V., Bali D., Millington D.S., Pamula V.K. Multiplex newborn screening for Pompe, Fabry, Hunter, Gaucher, and Hurler diseases using a digital microfluidic platform. Clin. Chim. Acta. 2013;424:12–18. doi: 10.1016/j.cca.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaggl M., Lajic N., Heinze G., Voigtländer T., Sunder-Plassmann R., Paschke E., Fauler G., Sunder-Plassmann G., Mundigler G. Screening for Fabry disease by urinary globotriaosylceramide isoforms measurement in patients with left ventricular hypertrophy. Int. J. Med. Sci. 2016;13:340–346. doi: 10.7150/ijms.14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiffmann R., Hughes D.A., Linthorst G.E., Ortiz A., Svarstad E., Warnock D.G., West M.L., Wanner C., Participants Conference. Screening, diagnosis, and management of patients with Fabry disease: conclusions from a "Kidney Disease: improving Global Outcomes" (KDIGO) Controversies Conference. Kidney Int. 2017;91:284–293. doi: 10.1016/j.kint.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Hwu W.L., Chien Y.H., Lee N.C., Chiang S.C., Dobrovolny R., Huang A.C., Yeh H.Y., Chao M.C., Lin S.J., Kitagawa T., Desnick R.J., Hsu L.W. Newborn screening for Fabry disease in Taiwan reveals a high incidence of the later-onset GLA mutation c.936+919G>a (IVS4+919G>a) Hum. Mutat. 2009;30:1397–1405. doi: 10.1002/humu.21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang W.H., Niu D.M., Lu C.Y., Lin S.Y., Liu T.C., Chang J.G. Modulation the alternative splicing of GLA (IVS4+919G>A) in Fabry disease. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin H.Y., Chong K.W., Hsu J.H., Yu H.C., Shih C.C., Huang C.H., Lin S.J., Chen C.H., Chiang C.C., Ho H.J., Lee P.C., Kao C.H., Cheng K.H., Hsueh C., Niu D.M. High incidence of the cardiac variant of Fabry disease revealed by newborn screening in the Taiwan Chinese population. Circ. Cardiovasc. Genet. 2009;2:450–456. doi: 10.1161/CIRCGENETICS.109.862920. [DOI] [PubMed] [Google Scholar]
- 14.Spada M., Pagliardini S., Yasuda M., Tukel T., Thiagarajan G., Sakuraba H., Ponzone A., Desnick R.J. High incidence of later-onset Fabry disease revealed by newborn screening. Am. J. Hum. Genet. 2006;79:31–40. doi: 10.1086/504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serebrinsky G., Calvo M., Fernandez S., Saito S., Ohno K., Wallace E., Warnock D., Sakuraba H., Politei J. Late onset variants in Fabry disease: results in high risk population screenings in Argentina. Mol. Genet. Metab. Rep. 2015;4:19–24. doi: 10.1016/j.ymgmr.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vieitez I., Souto-Rodriguez O., Fernandez-Mosquera L., San Millan B., Teijeira S., Fernandez-Martin J., Martinez-Sanchez F., Aldamiz-Echevarria L.J., Lopez-Rodriguez M., Navarro C., Ortolano S. Fabry disease in the Spanish population: observational study with detection of 77 patients. Orphanet J. Rare. Dis. 2018;13:52. doi: 10.1186/s13023-018-0792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue T., Hattori K., Ihara K., Ishii A., Nakamura K., Hirose S. Newborn screening for Fabry disease in Japan: prevalence and genotypes of Fabry disease in a pilot study. J. Hum. Genet. 2013;58:548–552. doi: 10.1038/jhg.2013.48. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi M., Ohashi T., Sakuma M., Ida H., Eto Y. Clinical manifestations and natural history of Japanese heterozygous females with Fabry disease. J. Inherit. Metab. Dis. 2008;3:483–487. doi: 10.1007/s10545-007-0740-6. [DOI] [PubMed] [Google Scholar]
- 19.Kubo T., Ochi Y., Baba Y., Hirota T., Tanioka K., Yamasaki N., Yoshimitsu M., Higuchi K., Takenaka T., Nakajima K., Togawa T., Tsukimura T., Sano S., Tei C., Sakuraba H., Kitaoka H. Prevalence and clinical features of Fabry disease in Japanese male patients with diagnosis of hypertrophic cardiomyopathy. J. Cardiol. 2017;69:302–307. doi: 10.1016/j.jjcc.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Doi K., Noiri E., Ishizu T., Negishi K., Suzuki Y., Hamasaki Y., Honda K., Fujita T., Tsukimura T., Togawa T., Saito S., Sakuraba H. High-throughput screening identified disease-causing mutants and functional variants of α-galactosidase A gene in Japanese male hemodialysis patients. J. Hum. Genet. 2012;57:575–579. doi: 10.1038/jhg.2012.68. [DOI] [PubMed] [Google Scholar]
- 21.Takata T., Okumiya T., Hayashibe H., Shimmoto M., Kase R., Itoh K., Utsumi K., Kamei S., Sakuraba H. Screening and detection of gene mutations in Japanese patients with Fabry disease by non-radioactive single-stranded conformation polymorphism analysis. Brain Dev. 1997;19:111–116. doi: 10.1016/s0387-7604(96)00486-x. [DOI] [PubMed] [Google Scholar]
- 22.Garman S.C., Garboczi D.N. The molecular defect leading to Fabry disease: structure of human alpha-galactosidase. J. Mol. Biol. 2004;337:319–335. doi: 10.1016/j.jmb.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 23.Tsukimura T., Nakano S., Togawa T., Tanaka T., Saito S., Ohno K., Shibasaki F., Sakuraba H. Plasma mutant α-galactosidase a protein and globotriaosylsphingosine level in Fabry disease. Mol. Genet. Metab. Rep. 2014;1:288–298. doi: 10.1016/j.ymgmr.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakuraba H., Togawa T., Tsukimura T., Kato H. Plasma lyso-Gb3: a biomarker for monitoring fabry patients during enzyme replacement therapy. Clin. Exp. Nephrol. 2018;22:843–849. doi: 10.1007/s10157-017-1525-3. [DOI] [PubMed] [Google Scholar]
- 25.Sueoka H., Ichihara J., Tsukimura T., Togawa T., Sakuraba H. Nano-LC-MS/MS for quantification of Lyso-Gb3 and its analogues reveals a useful biomarker for Fabry disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0127048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawashima I., Takeuchi I., Ohsawa M., Kotani M., Tajima Y., Inomata T., Izumi T., Sakuraba H. Phospholipid storage in the myocardium of a unique Japanese case of idiopathic cardiomyopathy. Clin. Chim. Acta. 2006;372:154–157. doi: 10.1016/j.cca.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Kotani M., Kawashima I., Ozawa H., Ogura K., Ariga T., Tai T. Generation of one set of murine monoclonal antibodies specific for globo-series glycolipids: evidence for differential distribution of the glycolipids in rat small intestine. Arch. Biochem. Biophys. 1994;310:89–96. doi: 10.1006/abbi.1994.1144. [DOI] [PubMed] [Google Scholar]
- 28.Ishii S., Nakao S., Minamikawa-Tachino R., Desnick R.J., Fan J.Q. Alternative splicing in the alpha-galactosidase a gene: increased exon inclusion results in the Fabry cardiac phenotype. Am. J. Hum. Genet. 2002;70:994–1002. doi: 10.1086/339431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dipple K.M., McCabe E.R.B. Phenotypes of patients with “simple” Mendelian disorders are complex traits: Thresholds, modifiers, and systems dynamics. Am. J. Hum. Genet. 2000;66:1729–1735. doi: 10.1086/302938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scriver C.P., Water P.J. Monogenic trait are not simple: Lessons learned from phenylketonuria. Trends Genet. 1999;15:267–272. doi: 10.1016/s0168-9525(99)01761-8. [DOI] [PubMed] [Google Scholar]
- 31.Mitobe S., Togawa T., Tsukimura T., Kodama T., Tanaka T., Doi K., Noiri E., Akai Y., Saito Y., Yoshino M., Takenaka T., Saito S., Ohno K., Sakuraba H. Mutant α-galactosidase a with M296I does not cause elevation of the plasma globotriaosylsphingosine level. Mol. Genet. Metab. 2012;107:623–626. doi: 10.1016/j.ymgme.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Saito S., Ohno K., Sakuraba H. Comparative study of structural changes caused by different substitutions at the same residue on α-galactosidase a. PLoS One. 2013;8 doi: 10.1371/journal.pone.0084267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee B.H., Heo S.H., Kim G.H., Park J.Y., Kim W.S., Kang D.H., Choe K.H., Kim W.H., Yang S.H., Yoo H.W. Mutations of the GLA gene in Korean patients with Fabry disease and frequency of the E66Q allele as a functional variant in Korean newborns. J. Hum. Genet. 2010;55:512–517. doi: 10.1038/jhg.2010.58. [DOI] [PubMed] [Google Scholar]
- 34.Togawa T., Tsukimura T., Kodama T., Tanaka T., Kawashima I., Saito S., Ohno K., Fukushige T., Kanekura T., Satomura A., Kang D.H., Lee B.H., Yoo H.W., Doi K., Noiri E., Sakuraba H. Fabry disease: biochemical, pathological and structural studies of the α-galactosidase a with E66Q amino acid substitution. Mol. Genet. Metab. 2012;105:615–620. doi: 10.1016/j.ymgme.2012.01.010. [DOI] [PubMed] [Google Scholar]