Abstract
Conventional HIV gene therapy approaches are based on engineering HIV target cells that are non-permissive to viral replication. However, expansion of gene-modified HIV target cells has been limited in patients. Alternative genetic strategies focus on generating gene-modified producer cells that secrete antiviral proteins (AVPs). The secreted AVPs interfere with HIV entry, and, therefore, they extend the protection against infection to unmodified HIV target cells. Since any cell type can potentially secrete AVPs, hematopoietic and non-hematopoietic cell lineages can function as producer cells. Secretion of AVPs from non-hematopoietic cells opens the possibility of using a genetic approach for HIV prevention. Another strategy aims at modifying cytotoxic T cells to selectively target and eliminate infected cells. This review provides an overview of the different genetic approaches for HIV treatment and prevention.
Keywords: HIV, genetic therapies, antiviral proteins
Main Text
The human immunodeficiency virus type-1 (HIV) is the causative agent of AIDS. Worldwide, over 36.7 million individuals are living with HIV and over 2 million new infections occur annually.1 Drug-based antiretroviral therapy (ART) and pre-exposure prophylaxis (PrEP) help to manage and prevent HIV infection, respectively. Limitations of ART and PrEP include the need for lifelong adherence, the emergence of drug-resistant virus strains, drug-associated short- and long-term toxicities, the direct costs of the drugs, and the indirect costs such as regular doctor visits. Despite major advances in biomedical research, a cure or reliable vaccine remains elusive. The viral reservoir, a pool of long-lived infected CD4+ T cells and macrophages established during the acute phase of infection, represents a major obstacle to a cure. The reservoir is much larger than initially anticipated, and it is unclear whether complete elimination of the infected cells is possible. The development of an HIV vaccine is hindered by the genetic diversity of HIV and the ability of the virus to evade immune responses. While naturally occurring antibodies with broad and potent HIV neutralization activity have been identified, they only appear in a minority of patients, take years to develop, and contain a high degree of somatic mutations. Therefore, it remains questionable whether such antibodies can be elicited by conventional vaccination strategies.
Conventional gene therapy strategies focus on generating an immune system that is resistant to HIV in order to suppress viral replication in the absence of ART. The case of the Berlin patient is generally seen as a proof of principle that replacing a patient’s immune system with genetically resistant cells can result in a functional cure. Timothy Ray Brown was infected with HIV and diagnosed with acute myeloid leukemia. For the treatment of his leukemia, he received five rounds of chemotherapy, two rounds of total-body radiation, two rounds of immunosuppressive therapy, and two allogeneic bone marrow transplants from a donor with a naturally occurring mutation in the CCR5 gene (CCR5Δ32).2 After the treatment, his entire immune system was replaced with CCR5-negative donor cells (100% chimerism), and he was cured of HIV and leukemia.3 However, due to the risks associated with the treatment and the difficulty in finding matched donors with the CCR5Δ32 mutation, this procedure is not amenable for the treatment of a larger population.
Using genetic approaches to secrete antiviral proteins (AVPs) that interfere with HIV entry represents an alternative strategy to control HIV replication. Proof of principle that the administration of recombinant AVPs can suppress viral replication has been provided in a clinical trial and in a pre-clinical macaque model. In the clinical trial, twice daily infusions of soluble CD4 (sCD4) resulted in sustained suppression of viremia.4 In the pre-clinical model, infected animals were infused with a combination of two antibodies. Upon a single administration, viremia was suppressed for 3–5 weeks in chronically infected animals, and subsequent administrations prevented virus rebound.5 Since almost any cell type can be modified to secrete AVPs, hematopoietic and non-hematopoietic cells can serve as producer cells for the secreted AVPs. Strategies using gene-modified T cells or hematopoietic stem and/or progenitor cells (HSPCs) require ex vivo gene modification, and they should mainly be used for therapeutic purposes. Liver and muscle are highly vascularized and can be directly modified in vivo. Since in vivo gene modification is non-invasive and less complex than ex vivo gene therapy, liver- or muscle-directed genetic modification could be used for therapy and prevention.
Another approach to control HIV replication focuses on engineering CD8+ T cells that can recognize and kill infected cells. While initial clinical trials were disappointing, the recent successes of modifying CD8+ T cells to kill cancer cells have rekindled the interest in using retargeted CD8+ T cells to eliminate HIV-positive cells. This review provides an overview of the different genetic approaches.
Conventional HIV Gene Therapy Approaches
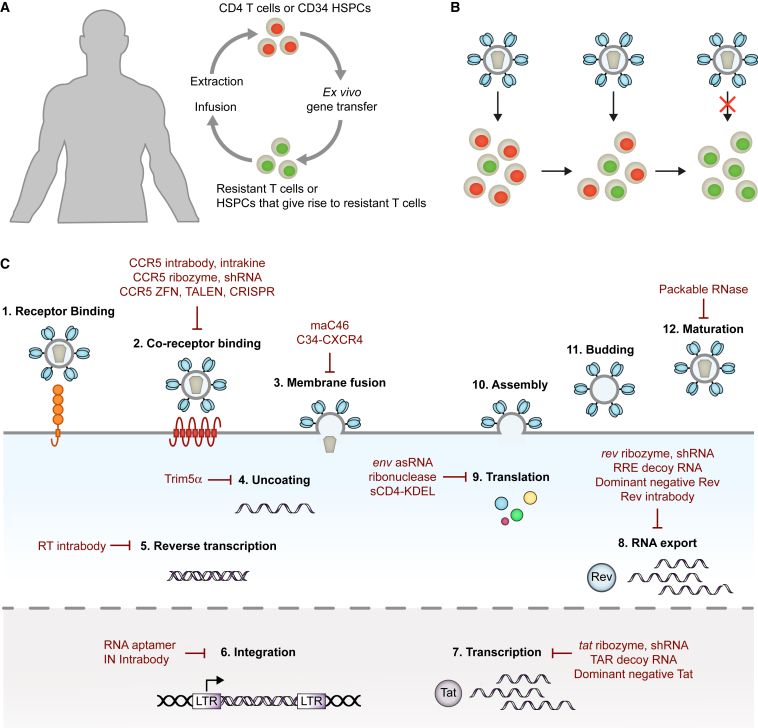
Conventional HIV gene therapy approaches focus on rendering HIV target cells non-permissive to viral replication. To this end, CD4+ T cells or CD34+ HSPCs are extracted from a patient, genetically modified ex vivo to express one or multiple antiviral genes, and infused into the same patient (Figure 1A).
Figure 1.
Conventional HIV Gene Therapy
(A) Ex vivo gene delivery. Autologous CD4+ T cells or CD34+ HSPCs are genetically modified ex vivo using a suitable vector. The gene-modified cells are infused back into the patient. (B) Positive selection of gene-modified HIV target cells. HIV replicates in susceptible HIV target cells (red). Gene-modified cells (green) are resistant to infection and accumulate to therapeutically relevant levels. (C) The HIV replication cycle and examples of gene therapeutics. RT, HIV reverse transcriptase; IN, HIV integrase.
HSPCs are usually not infected by HIV, but they give rise to lymphoid progenitors that migrate from the bone marrow to the thymus, where T cell differentiation and thymic education occur. The de novo development of T cells predominantly takes place before adolescence. In adults, the size of the thymus is decreased and the contribution of HSPCs to T cell homeostasis declines. Instead, T cell numbers are largely maintained through the division of T cells outside of the central lymphoid organs, such as CD4+ stem memory T cells (TSCMs). However, thymic output increases again in the first year after an HSPC transplant, resulting in the production of T cells with a new T cell receptor (TCR) repertoire. Therefore, gene-modified HSPCs and CD4+ T cells have the potential to give rise to new gene-modified HIV target cells.
Following infusion, mixed populations of gene-modified and unmodified cells coexist in the patient. Ideally, the gene-modified HIV target cells would have a survival advantage over unmodified cells and replace the unmodified HIV target cell population over time, resulting in an immune system that is resistant to HIV (Figure 1B).
Examples of HIV Gene Therapeutics
The antiviral gene products tested to date can generally be classified into RNA-based and protein-based therapeutics. They interfere with various stages of the HIV replication cycle by targeting viral factors or by targeting cellular factors that are essential for viral replication but dispensable for the host (Figure 1C). The steps of HIV entry are receptor binding, co-receptor binding, and membrane fusion. CD4 serves as the receptor, while CXCR4 or CCR5 usually function as a co-receptor. Receptor binding and co-receptor binding are mediated by the HIV envelope (Env) protein gp120, which is difficult to target because of its high variability and the inaccessibility of conserved sites within gp120. Targeting the receptor, CD4, also proves difficult due to the central role CD4 plays in the immune system. Similarly, CXCR4 is essential during embryonic development and plays an important role in the tissue recruitment of immune cells in adults.6 However, individuals born with a naturally occurring mutation in the CCR5 gene (CCR5Δ32) are apparently healthy.7 Consequently, multiple approaches have been developed to reduce CCR5 on the surface of HIV target cells. Cytokines and antibody derivatives with endoplasmic reticulum retention signal (intrakines and intrabodies, respectively) were designed to retain CCR5 inside gene-modified cells.8, 9, 10, 11 Ribozymes and small hairpin RNAs (shRNAs) have been utilized to target CCR5 mRNA,12, 13, 14, 15 while genome-editing enzymes, such as zinc-finger nucleases (ZFNs),16, 17 transcription activator-like effector nucleases (TALENs),18, 19 and CRISPR-associated protein-9 nuclease (Cas9) have been employed to introduce mutations into the CCR5 gene.20, 21, 22
Following engagement of CD4 and the co-receptor, HIV Env gp41 facilitates the fusion of the viral and cellular membranes. In the absence of target cell binding, gp41 is not accessible because it is shielded by gp120.23, 24 It contains highly conserved sequences, such as the heptad repeats.25, 26 The membrane-associated fusion inhibitor C46 (maC46) comprises a transmembrane domain and a short peptide derived from the heptad repeat 2 of gp41. Expression of the gene encoding maC46 results in high concentrations of maC46 on the surface of gene-modified cells, where it binds to the heptad repeat 1 of gp41, thereby locking gp41 in a fusion inactive state.27 In a similar approach, the fusion inhibitor C34 was fused to CXCR4 (C34-CXCR4).28 Although C34-CXCR4 confers protection to gene-modified HIV target cells, it is not clear to what extent expression of CXCR4 affects the natural function of these cells.
Once entry takes place, the viral capsid proteins are lost in the cytoplasm of host cells, a process that is defined as uncoating. The alpha isoform of tripartite motif-containing protein 5 (TRIM5α) is a natural retrovirus restriction factor, and it inhibits uncoating of retroviruses by interacting with the capsid proteins.29 While human TRIM5α is ineffective against HIV, rhesus macaque TRIM5α efficiently protects cells from HIV infection.30, 31 Expression of a chimeric human and rhesus macaque TRIM5α has been shown to protect HIV target cells from infection.32, 33
Following uncoating and reverse transcription, the HIV genome integrates into transcriptionally active sites of host chromosomes. Recently, an RNA aptamer has been identified that targets the HIV integrase.34 Aptamers are short DNA or RNA oligonucleotides with a stable three-dimensional conformation that enables them to bind to molecular targets with high specificity and affinity. In an elegant approach, the integrase-targeting RNA aptamer, incorporated as the terminal loop of an shRNA, was expressed by gene-modified HIV target cells, and it inhibited HIV replication in cell cultures.34
Other approaches have focused on inhibiting HIV gene expression in infected cells. The two HIV regulatory proteins, transactivator of transcription (Tat) and regulator of expression of virion proteins (Rev), have been targeted because they are expressed early during replication and are essential for viral gene expression. Tat interacts with the transactivation response (TAR) element present at the 5-prime end of nascent HIV transcripts and greatly enhances transcription of the integrated HIV genome,35, 36 while Rev binds to the Rev response element (RRE) of unspliced and singly spliced HIV mRNAs and promotes their export from the nucleus to the cytoplasm.37 Ribozymes and shRNAs have been designed to target a site in the overlapping open reading frames of tat and rev RNA.38, 39 TAR and RRE decoy RNAs mimic their natural counterpart and act by occupying the RNA-binding sites of Tat and Rev.39, 40 Additionally, trans-dominant negative Rev mutant proteins, e.g., RevM10, have been used to disrupt Rev function by binding and inactivating the wild-type protein.41, 42, 43 In another approach, long antisense RNAs were developed to bind to the HIV mRNA encoding the viral Env glycoproteins.44 The enzyme double-stranded RNA (dsRNA)-dependent adenosine deaminase recognizes the dsRNA complexes and converts adenosines to inosines, which are recognized as guanines and thus cause mutations.45 Infection of cells expressing the env antisense RNAs (VRX496) results in the accumulation of mutations in HIV Env proteins and the production of severely attenuated progeny viruses.44 MazF is an endoribonuclease derived from Escherichia coli that cleaves adenine-cytosine-adenine (ACA) in single-stranded RNA. While MazF is not specific for HIV, HIV transcripts are rich in ACA sequences, and MazF expression has been shown to reduce HIV infection in target cells.46
In a different approach, a recombinase has been developed that recognizes a conserved region in the long terminal repeats (LTRs) located on either side of the provirus DNA. The recombinase excises the provirus from the gene-modified HIV-infected cells, thereby curing them from infection.47 Similarly, the CRISPR/Cas9 system has been used to excise HIV provirus DNA from infected cells.48 However, HIV was shown to rapidly develop resistance to excision by Cas9.49, 50 The high mutation rate of HIV is indeed a major challenge for HIV gene therapies, as the antiviral gene products must confer lifelong protection to HIV target cells. While CCR5 is a cellular target with a low chance to mutate, HIV can switch co-receptor usage from CCR5 to CXCR4,51 and half of all circulating HIV isolates are capable of utilizing CXCR4 as a co-receptor. One possibility to prevent escape from inhibition is to combine different antiviral genes, similar to combination ART. For example, a vector encoding the tat/rev shRNA, a TAR decoy RNA, and a CCR5 ribozyme (rHIV7-shI-TAR-CCR5RZ) has been developed to inhibit HIV entry and gene expression,39 while a vector encoding maC46 and a CCR5 shRNA (Cal-1) has been designed to inhibit two different steps of entry.52
Summary of Clinical Trial Results
Based on promising results obtained in vitro, several antiviral genes advanced to testing in clinical trials (Table 1). All clinical trials completed to date have shown that the approaches are safe and that long-term engraftment of gene-modified cells can be achieved in patients. In some trials, modest clinical benefits were documented. For example, a trend toward a lower viral load was observed in patients receiving infusions of T cells modified to express the antisense RNA VRX496.53 While antisense-mediated genetic pressure on HIV was demonstrated, no enrichment of gene-modified cells occurred.53 Other trials have reported a modest survival advantage for gene-modified cells during ongoing viral replication. When autologous T cells modified with a control vector or a vector encoding RevM10 were infused into the same patient, RevM10-expressing cells had a survival advantage over cells modified with the control vector. Similarly, following infusion of autologous T cells modified with the CCR5 ZFN, preferential survival of gene-modified cells in comparison to unmodified cells was demonstrated during a scheduled ART treatment interruption. In trials based on the infusion of HSPCs modified with RevM10 or rHIV7-shI-TAR-CCR5RZ, increases in the level of gene-modified cells were observed during phases of viremia. However, the effects were transient and no clear accumulation of gene-modified cells to therapeutically relevant levels has been observed in any trial to date. There are multiple possible transgene-specific and general explanations for the lack of expansion.
Table 1.
HIV Gene Therapy Clinical Trials
| Gene | Vector | Target Cell | Phase | Status | Reference or ClinicalTrials.gov Trial |
|---|---|---|---|---|---|
| RNA-Based Approaches | |||||
| U5 ribozyme and pol ribozyme | GV | CD4 | I | completed | 203 |
| tat and tat/rev ribozyme | GV | CD34 | I | completed | 38 |
| tat/vpr ribozyme (Rz2/Oz1) | GV | CD4 | I | completed | 204, 205 |
| GV | CD34 | I | completed | 206 | |
| GV | CD34 | II | completed | 75 | |
| RRE decoy RNA | GV | CD34 | I | completed | 40 |
|
env antisense RNA (VRX-496) |
LV | CD4 | I | completed | 44 |
| LV |
CD4 |
II |
active |
53 |
|
| Protein-Based Approaches | |||||
| Dominant-negative Rev (RevM10) | GP | CD4 | I and II | completed | 41 |
| GV | CD4 | I and II | completed | 42 | |
| GV | CD34 | I | completed | 43 | |
| Dominant-negative Rev (TdRev) | GV | CD34 | I | completed | 207 |
| Membrane-anchored fusion inhibitor (maC46) | GV | CD4 | I | completed | 56 |
| CXCR4-anchored fusion inhibitor (C34-CXCR4) | LV | CD4 | I | recruiting | NCT03020524 |
| CCR5 CRISPR/Cas9 | NA | CD34 | NA | recruiting | NCT03164135 |
| CCR5 zinc-finger nuclease (SB-728) | Ad | CD4 | I | completed | 63 |
| Ad | CD4 | I | completed | NCT01044654a | |
| Ad | CD4 | I and II | completed | NCT01252641b | |
| Ad | CD4 | I and II | completed | NCT01543152c | |
| mRNA | CD4 | I | active | NCT02388594 | |
| mRNA | CD4 | I and II | active | NCT02225665 | |
| mRNA | CD34 | I | recruiting | NCT02500849 | |
| MazF endoribonuclease | LV | CD4 | I | active | NCT01787994 |
| Combination Approaches | |||||
| TAR antisense RNA, dominant-negative Rev | GV | CD4 | I | completed | 208 |
| CCR5 shRNA, TRIM5α, TAR decoy RNA | LV | CD34 | I and II | recruiting | NCT02797470 |
| tat/rev shRNA, TAR decoy RNA, CCR5 ribozyme | LV | CD34 | I | completed | 39 |
| (rHIV7-shI-TAR-CCR5RZ) | LV | CD34 | I | active | NCT01961063 |
| LV | CD34 | I | recruiting | NCT02337985 | |
| CCR5 shRNA, maC46 (Cal-1) | LV | CD4 CD34 |
I and II | completed | NCT01734850 |
| Cal-1 plus MGMT(P140K) | LV | CD34+ | I | recruiting | NCT02343666 |
GV, gammaretroviral vector; LV, lentiviral vector; Ad, adenoviral vector; GP, gold particles; CD4, CD4+ T cells; CD34, CD34+ HSPCs.
T cell dose escalation, patients with ART failure, and patients with heterozygote CCR5 delta-32 mutation.
Patients with ART failure.
Cyclosphosphamide dose escalation.
Transgene-Specific Issues
In specific cases, expression of the transgenes could have reduced the engraftment or repopulation potential of gene-modified cells, thereby giving a survival disadvantage over unmodified cells. Additionally, it is also possible that transgene expression was reduced or lost over time due to promoter-related issues. In other cases, the lack of expansion could have been related to the potency of the antiviral genes. A follow-up study has compared the antiviral effect of the tat/rev shRNA, VRC496, and maC46.54 In pure cultures of gene-modified HIV target cells, a strong protective effect was observed for all three antiviral genes. In contrast, upon infection of mixed cultures consisting of unmodified and genetically modified HIV target cells, only cells expressing maC46 had a strong survival advantage over unmodified cells. One possible explanation for the strong survival advantage conferred by maC46 could be that maC46 targets HIV before integration occurs. Once integrated, proviral genes will continuously be transcribed, which could facilitate escape from inhibition or result in reduced fitness of the infected cell. Mathematical modeling predicts that only antiviral genes that inhibit HIV replication before virus integration can provide a significant therapeutic benefit.55
However, there was no evidence of positive selection when autologous CD4+ T cells expressing maC46 were infused into HIV-positive individuals who had drug-resistant virus and incomplete suppression of viremia despite ART.56 Instead, the number of gene-modified cells rapidly declined within a week after infusion. Since maC46 is derived from viral proteins, immune responses could have contributed to the elimination of cells expressing maC46. Indeed, patients had pre-existing antibodies to maC46, but there was no clear indication that antibody-dependent cell-mediated cytotoxicity (ADCC) or other immune responses were responsible for the observed decline.56 Therefore, other factors could have been responsible for the result.
General Obstacles
Conventional HIV gene therapy approaches require HIV replication for the positive selection of gene-modified cells. The largest obstacle to this approach is probably the indirect cytopathic effect of HIV replication that leads to the destruction of uninfected cells. The immune system in untreated patients is in a hyperactive state. Systemic inflammation and chronic immune activation contribute to the dysregulation of T cell homeostasis,57 resulting in an unfavorable environment for engraftment and expansion. Even patients on ART who achieve suppression of viral replication have increased inflammation in comparison to uninfected individuals. Furthermore, infected cells secrete various HIV proteins that can induce apoptosis in uninfected bystander cells, e.g., the HIV accessory protein negative factor (Nef) fulfills multiple intracellular functions, but extracellular Nef was also detected in the plasma of half of HIV-positive individuals on ART with undetectable viral load58 and can trigger apoptosis of uninfected CD4+ T cells.59 Additionally, soluble HIV Env gp120 and antibodies targeting soluble gp120 are commonly found in the blood of HIV-positive individuals. Binding of soluble gp120 to CD4 and CCR5 or CXCR4 on uninfected cells can also induce apoptosis,60, 61 while anti-gp120 antibodies recognize soluble gp120 bound to surface CD4 and mediate the destruction of these cells by ADCC.62 Therefore, low levels of HIV replication could already impair the engraftment and expansion of gene-modified cells, even when the cells are highly resistant to HIV. Other general obstacles include but are not limited to slower replication of less aggressive HIV isolates that may not lead to the efficient selection of gene-modified cells, the replenishment of unmodified CD4+ T cells by unmodified HSPCs and/or CD4+ T cells, and long-lived HIV reservoirs such as CD4+ TSCMs and macrophages that harbor replication-competent HIV in patients.
The Level of Gene Modification
The level of gene modification that can be achieved in patients plays a central role for the success of gene therapy. Since conventional HIV gene therapy strategies rely on HIV replication to select gene-modified cells, the initial size of the gene-modified HIV target cell population has to be large enough to offset the negative effects of viral replication in the unmodified target cell population. The exact threshold has not been determined and may vary for different approaches, but pre-clinical and ongoing clinical studies have given valuable insights about the required level of gene modification.
For T cell-based therapies, the highest level of gene modification has been observed in the trial testing the CCR5 ZFN, in which autologous CCR5-modified CD4+ T cells were infused into patients on ART with undetectable viral load (below 20 HIV genome copies/mL).63 Aside from one patient who had a natural mutation in one CCR5 allele prior to gene modification, patient 203 had the highest level of CCR5-modified cells (close to 30% of the CD4+ T cell compartment) at the beginning of a scheduled treatment interruption period at 4 weeks post-infusion.63 Clonal analysis of CCR5-modified cells indicated that approximately 33% of cells underwent bi-allelic modification.63 Therefore, almost 10% of the CD4+ T cells of patient 203 were completely resistant at the beginning of the treatment interruption. During the treatment interruption, a transient survival advantage was evident for gene-modified cells, but no clinical benefits were observed, e.g., the viral load returned to historic set points and CD4+ T cell counts declined drastically.63 Similar results were observed in other patients with lower levels of gene modification.63
To increase engraftment of CCR5-modified T cells, pre-treatment with low doses of the chemotherapeutic agent cyclophosphamide is currently under investigation (ClinicalTrials.gov: NCT01543152). Low doses of cyclophosphamide have primarily been used to promote anti-tumor immunity by selectively depleting immunosuppressive regulatory T cells and stimulating the expansion of effector T cells,64, 65 but they were also shown to promote proliferation of adoptively transferred CD4+ T cells, which in turn enhanced tumor-specific CD8+ T cell responses.65 Preliminary results from the trial showed a cyclophosphamide dose-dependent increase in the number of engrafting CD4+ T cells.66 The effect was maximal at a dose of 1 g/m2 body surface area, resulting in an ∼2.5-fold increase in comparison to the previous trial without cyclophosphamide.63, 66 Although no accumulation of CCR5-modified cells was reported, 6 of 9 patients pre-conditioned with cyclophosphamide doses between 1 and 1.5 g/m2 demonstrated control of viremia (viral load below 10,000 RNA copies/mL) during an extended ART interruption for 14–26 months.66, 67 Interestingly, control of viremia was associated with increased HIV-specific CD8+ T cell responses and higher levels of CCR5-modified CD4+ TSCMs.67 These results spark many questions, e.g., was doubling the number of engrafting cells enough to reach clinically significant levels of gene-modified cells; did cyclophosphamide pre-treatment promote the engraftment of longer-lived T cell subsets; or was the combination of cyclophosphamide and infusion of unmodified T cells sufficient to induce HIV-specific immune responses, as has been seen in anti-tumor therapies? The final results for this trial as well as future trials in which unmodified T cells are infused after cyclophosphamide conditioning will offer further insights.
For all HSPC-based trials, the level of gene modification that was achieved in patients was extremely low (below 1%; Table 2). There are different possible explanations for this. Early trials were performed with vectors derived from gammaretroviral viruses, such as the murine leukemia virus (MLV). MLV vectors require cell division for integration, as they need the breakdown of the nuclear membrane, and therefore they can only integrate successfully into dividing cells.68 As HSPCs are more quiescent in culture, growth factor cocktails are necessary to induce cell division of HSPCs, which can lead to a reduced engraftment potential.69 For example, the protocol for the transduction of HSPCs with a retroviral vector encoding RevM10 required 5 days of ex vivo culture, with a standard growth factor cocktail consisting of thrombopoietin, stem cell factor, and Flt3 ligand (Table 2). Under these conditions, engraftment of HSPCs is drastically reduced in comparison to HSPCs that were just cultured for 1–2 days.70 Additionally, pre-conditioning regimens could have influenced the engraftment of HSPCs.
Table 2.
Completed HSPC-Based HIV Gene Therapy Trials
| Antiviral Gene (Vector) | ART | GF | E | E-GM (%) | C | I-GM (%) | Outcome |
|---|---|---|---|---|---|---|---|
| tat and tat/rev ribozyme (GV) | yes | NA | NA | NA | NMA | <1 | safe38 |
| MA | |||||||
| tat/vpr ribozyme (GV) | yes | no | 3 | 54 | NMA | <1 | safe, overall lower viral load and higher CD4+ T cells counts75 |
| RRE decoy RNA (GV) | yes | IL-3 | 3 | 17 | NMA | <1 | safe and survival advantage over cells modified with a control vector40 |
| SCF | |||||||
| RevM10 (GV) | yes | SCF | 5 | 24 | NMA | <1 | safe and survival advantage over cells modified with a control vector43 |
| TPO | |||||||
| Flt3L | |||||||
| TdRev (GV) | yes | IL-3 | 4 | 80 | RI | <1 | safe207 |
| GM-CSF | |||||||
| Flt3L | |||||||
| tat/rev shRNA, TAR decoy RNA, CCR5 ribozyme (LV) | yes | SCF | 2 | 18 | MA | <1 | safe and survival advantage over unmodified cells39 |
| TPO | |||||||
| Flt3L |
GV, gammaretroviral vector; LV, lentiviral vector; ART, antiretroviral therapy; GF, growth factors; E, ex vivo culture period in days; E-GM, ex vivo level of gene modification; C, conditioning regimen; MA, myeloablative; NMA, non-myeloablative; RI, reduced intensity; I-GM, in vivo level of gene modification.
In general, myeloablative (MA)-, reduced intensity (RI)-, or non-MA (NMA)-conditioning regimens can be used prior to HSPC transfer.71 MA conditioning completely depletes hematopoietic stem cells and their progeny. Accordingly, MA conditioning would substantially reduce the existing HIV reservoir72 and potentially promote the highest level of engraftment. However, MA conditioning is associated with life-threatening toxicities, and it is only an option in a small group of patients who require high-dose chemotherapy for the treatment of other life-threatening diseases, such as HIV-associated lymphoma.73 RI conditioning has a reduced toxicity while still promoting engraftment of HSPCs, and it could be an option for a larger group of patients.74 NMA conditioning is the least toxic, but it also mediates the lowest level of engraftment in the setting of autologous HSPC transplants.74 Since some trials have used NMA conditioning, e.g., a phase II trial based on the infusion of HSPCs modified to express a tat/vpr ribozyme,75 this could have contributed to the low level of gene modification. In comparison to gammaretroviral vectors, lentiviral vectors transduce HSPCs more efficiently because of their inherent ability to integrate into non-dividing cells. One trial has examined the engraftment of HSPCs modified with the lentiviral vector rHIV7-shI-TAR-CCR5RZ.39 Although MA conditioning and short ex vivo transduction protocols were used, very low levels of gene modification were observed. This can at least partially be explained by the fact that the study protocol required co-infusion of an HSPC batch that was only minimally manipulated, e.g., no ex vivo culture period.
A clinical trial for the treatment of a genetic disease reported gene modification levels of ∼15% after lentiviral vector-mediated gene modification of autologous HSPCs and MA conditioning.76 The question remains whether there are factors aside from the cyptopathic effects of HIV replication that may affect gene transfer in HIV-positive individuals. All completed HSPC-based trials to date have used gammaretroviral or lentiviral vectors, and patients were on ART prior to the extraction of HSPCs (Table 2). While viral load was not completely suppressed in the majority of patients, some ART regimens could have reduced the efficacy of gammaretroviral and lentiviral vector-mediated gene transfer. For example, a nucleoside reverse-transcriptase inhibitor (azidothymidine) efficiently inhibited gammaretroviral murine leukemia virus (MLV) infections.77 In another study, pigtailed macaques were infected with simian HIV (SHIV) and treated with ART consisting of an integrase inhibitor (raltegravir), a nucleoside reverse-transcriptase inhibitor (emtricitabine), and a nucleotide reverse-transcriptase inhibitor (tenofovir).78 Autologous macaque HSPCs contained detectable levels of all three drugs, even when ART was temporarily interrupted prior to HSPC extraction. Following transduction with lentiviral vectors, an accumulation of non-integrated vector genomes was detected, which resulted in a much lower long-term engraftment of gene-modified cells in comparison to gene-modified HSPCs from treatment-naive animals.78 Therefore, ART regimens may need to be adjusted well in advance of retrovirus-mediated gene transfer. The infusion of CD4+ T cells or HSPCs modified with the lentiviral vector Cal-1 into patients without ART was investigated in a phase I trial, which recently was completed (ClinicalTrials.gov: NCT01734850). In another trial, HSPCs electroporated with mRNAs encoding CCR5 ZFNs were infused into patients on ART with viremia suppressed below 20 RNA copies/mL (ClinicalTrials.gov: NCT02500849). The results from these trials will provide further information about the impact of ART on lentiviral vector-mediated gene transfer and/or to what extent ongoing viral replication interferes with the engraftment of cells.
Consistent with clinical trials for non-infectious diseases, 20% gene modification has been achieved in uninfected macaques after MA conditioning and infusion with autologous HSPCs modified with the clinical grade lentiviral vector Cal-1 encoding maC46 and a CCR5 shRNA.79 Following infection with SHIV, an accumulation of gene-modified cells, controlled viremia (viral load under 10,000 RNA copies/mL), and increased CD4+ T cell counts in the range of healthy subjects were observed.80 The study suggests that a level of gene modification of ∼20% is sufficient to achieve a therapeutic benefit in HSPC-based approaches. However, due to the associated risks, MA conditioning is not an option for the majority of patients. The question remains whether this level of gene modification can be achieved in patients undergoing RI or even NMA conditioning, where the infused cells are competing with existing cells.81 The impact of RI-conditioning regimens is currently under investigation in multiple trials (Table 3). For example, a clinical trial investigating the effect of RI conditioning on the engraftment of HSPCs modified with the lentiviral vector rHIV7-shI-TAR-CCR5RZ is currently recruiting. RI conditioning is also being tested for HSPCs modified with the CCR5 ZFN or HSPCs modified with Cal-1.
Table 3.
Ongoing Clinical HSPC-Based HIV Gene Therapy Trials
| Antiviral Gene | ART prior to HSPCT | VL prior to HSPCT (gc/mL) | Gene Transfer | Conditioning |
|---|---|---|---|---|
| CCR5 CRISPR/Cas9 | yes | <40 | RNP nucleofection | NA |
| CCR5 ZFN | yes | <20 | mRNA electroporation | busulfan |
| maC46 | no | 5,000–100,000 | lentiviral vector | busulfan |
| CCR5 shRNA | ||||
| maC46 | yes | <50 | lentiviral vector | BCNU, O6-benzylguanine |
| CCR5 shRNA | ||||
| MGMT(P140K) | ||||
| tat/rev shRNA | yes | NA | lentiviral vector | BCNU, cytarabine, etoposide, melphalan |
| TRIM5α | ||||
| TAR decoy RNA | ||||
| tat/rev shRNA | yes | NA | lentiviral vector | R-EPOCH (rituximab, etoposide, prednisone, vincristine Sulfate, cyclophosphamide, doxorubicin hydrochloride) |
| TAR decoy RNA | ||||
| CCR5 ribozyme |
ART, antiretroviral therapy; HSPCT, HSPC transplant; RNP, ribonucleoprotein; NA, not available; gc, HIV RNA genome copies.
In Vivo Selection of Gene-Modified Cells
To further boost the level of gene modification, vectors expressing drug resistance genes are currently under investigation. Rendering gene-modified cells resistant to chemotherapeutic agents enables the in vivo selection of gene-modified cells. For example, the P140K mutant of O6-alkylguanine-DNA-methyltransferase, MGMT(P140K), was used to confer resistance to the chemotherapeutic agent bis-chloroethylnitrosourea (BCNU), which is toxic to hematopoietic stem cells.82, 83, 84 MGMT(P140K) selection of gene-modified HSPCs was safe and feasible in clinical trials in glioblastoma patients.85, 86 Proof of principle has also been demonstrated in a pre-clinical macaque model of HIV infection. After transplantation of autologous gene-modified HSPCs expressing maC46 and MGMT(P140K), the level of gene modification was 4%–5%.84 Several rounds of BCNU treatment resulted in 20% stable gene marking in one animal and 60% in another animal.79 The macaques were subsequently infected with SHIV. During the acute phase of infection, comparable levels of virus replication were observed in macaques infused with control cells or with gene-modified cells expressing maC46. During this phase, a clear survival advantage of gene-modified CD4+ T cells expressing maC46 was evident, resulting in an accumulation of gene-modified cells to a level of 90% of total CD4+ T cells. After the acute phase, the viral load dropped in all animals, but significantly lower levels were observed in macaques infused with gene-modified cells expressing maC46 up to 196 days post-infection. Interestingly, the drop in viral load was accompanied by an increase of unmodified CD4+ T cells in macaques treated with HSPCs expressing maC46.
MGMT(P140K) has also been integrated into other vectors. For example, a lentiviral vector encoding multiple anti-HIV RNAs and MGMT(P140K) was used to transduce human HSPCs.87 Following injection into immunodeficient mice, three rounds of BCNU treatment resulted in overall reduced engraftment of human cells, but the percentage of gene-modified cells in the bone marrow and spleen were enriched by 15-fold, while the frequency of gene-modified CD4+ T cells was increased by 3-fold in comparison to unselected controls.87 Therefore, in vivo selection may represent a solution to increase gene modification in patients without the need for MA conditioning. For example, RI conditioning with BCNU and temozolomide was sufficient to select autologous HSPCs expressing MGMT(P140K) in a dog model.88 The ability to select gene-modified cells in patients has a vast potential to increase the efficacy of gene therapy approaches. One trial is currently recruiting patients with HIV-associated lymphoma to test HSPCs modified with the Cal-1 vector that also encodes MGMT(P140K). However, since chemotherapy can have severe side effects, the safety and feasibility of this method will have to be carefully determined for ART-suppressed HIV-positive individuals who do not require chemotherapy otherwise.89
Genetic Strategies Using Secreted Anti-HIV Proteins
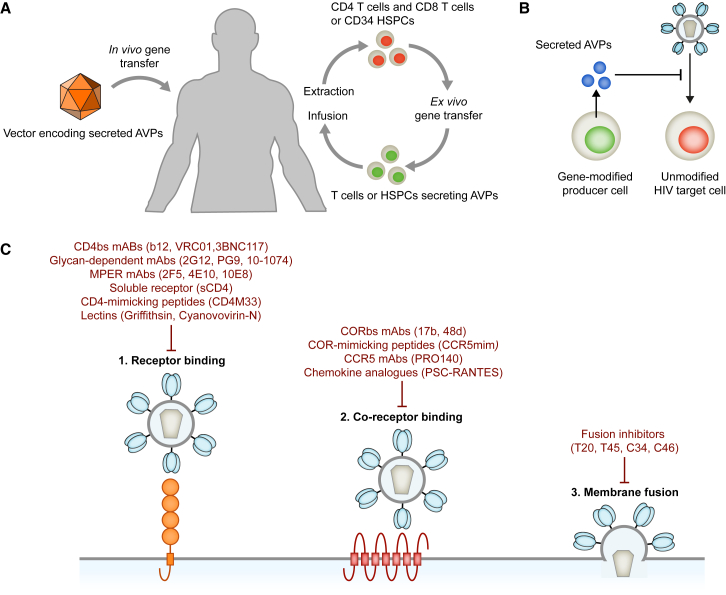
Alternative genetic strategies focus on generating producer cells that secrete AVPs. Intuitively, cells of the immune system are optimally suited for the production of secreted AVPs. Autologous CD4+ T cells, CD8+ T cells, or HSPCs can be modified ex vivo to secrete AVPs. The gene-modified hematopoietic cells are present at the sites of viral replication, e.g., lymphatic tissues, which can result in high local concentrations of the secreted AVPs. Especially gene-modified HSPCs can generate an armada of AVP-secreting cells, as the majority of myeloid and lymphoid cell lineages can serve as producers for the secreted AVPs. Since nearly any cell type can serve as a producer cell, it is also possible to modify non-hematopoietic cells to secrete AVPs. The liver and muscle are highly vascularized organs that can be genetically modified in vivo by injecting the appropriate vectors directly into the target organ (Figure 2A). AVPs secreted from gene-modified liver or muscle cells can enter the bloodstream and reach sites of viral replication via circulation. Ideally, AVPs secreted from gene-modified HIV target and/or non-target cells would accumulate to therapeutically relevant concentrations and lead to a systemic inhibition of viral replication (Figure 2B).
Figure 2.
Genetic Strategies Based on Secreted AVPs
(A) Genes encoding secreted AVPs can be delivered by ex vivo gene therapy. Alternatively, suitable vectors can be injected directly into the target tissue, such as muscle. (B) Gene-modified producer cells secrete the AVPs into their surrounding. The secreted AVPs inhibit HIV entry by binding to HIV Env present on virus particles and infected cells or by binding to cellular receptors, such as CCR5. (C) HIV entry and examples of protein-based entry inhibitors.
Secreted AVPs have to inhibit HIV by binding to extracellular targets. A multitude of protein-based entry inhibitors that interfere with one or multiple steps of the HIV entry process by binding to HIV Env on virus particles or proteins on the surface of target cells has been identified (Figure 2C). For example, receptor binding has been inhibited by soluble CD4 (sCD4)4, 90 as well as monoclonal antibodies (mAbs) targeting the CD4-binding site (CD4bs)91, 92 or glycan-dependent epitopes across HIV Env.93 Chemokine analogs,94 co-receptor-mimicking peptides,95 and mAbs targeting CCR596, 97 or the co-receptor-binding site98, 99 have been utilized to inhibit binding of gp120 to the co-receptor, while fusion inhibitors have been developed to prevent the function of gp41.100, 101 To overcome the limitations of single inhibitors, several inhibitors have also been combined, e.g., sCD4 was fused to a co-receptor-mimicking peptide or fusion inhibitors.102, 103, 104, 105 Aside from inhibiting HIV entry, several AVPs have the advantage that they can also reduce the cytopathic effects of HIV replication and mediate the elimination of infected cells. For example, antibodies and sCD4 can neutralize free gp120 in the serum of HIV-positive individuals or gp120 on the surface of infected cells. Additionally, antibodies bound to cell surface gp120 can mediate ADCC,106 while sCD4 binding to cell surface gp120 enables binding of co-receptor-binding site (CORbs) antibodies present in the serum of infected individuals, which can also mediate ADCC.107 The following two sections will provide examples of AVPs secreted from hematopoietic and non-hematopoietic cell lineages.
Secretion of AVPs from Hematopoietic Cell Lineages
Multiple studies have investigated secretion of AVPs from HIV target cells and HSPCs. This section provides examples of AVPs secreted from hematopoietic cell lineages. Fusion inhibitors are highly potent entry inhibitors that have been approved for clinical use.100 However, they need to be injected twice daily because of their relatively short half-life. To provide a continuous supply of fusion inhibitors, HIV target cells were engineered to secrete the fusion inhibitor C46.108 While gene-modified T lymphoid PM1 and Jurkat cells secreted significant quantities of C46, C46 secretion from gene-modified primary CD4+ T cells was below the limit of detection, and only partial inhibition of HIV replication was observed. In another in vitro study, secretion of a single-chain variable fragment (scFv) of PRO140 was investigated.109 The CCR5-targeting mAb PRO140 is currently being tested in phase III clinical trials (ClinicalTrials.gov: NCT02483078 and NCT02859961). Phase II trials have shown that once weekly injections of PRO140 mediate a significant reduction in viral load.96, 97 While the scFv PRO140 mediated protection of unmodified cells, the scFv was less potent than the parental mAb.109
In vivo studies have demonstrated more encouraging results. Two similar studies have investigated the secretion of the glycan-targeting mAb 2G12 or the CD4bs-targeting antibody b12, which were two of the most potent anti-HIV mAbs available at that time.110, 111 Immunodeficient mice were transplanted with gene-modified HSPCs expressing one of the mAbs. Following engraftment, blood samples contained low levels of either mAb (Table 4). At 1 week after infection, mice expressing 2G12 had significantly reduced viral loads in comparison to control mice; viral loads at later time points were not reported. Mice expressing b12 had higher levels of CD4+ T cells and a lower percentage of infected cells after infection, but viral load data were not reported.
Table 4.
Examples of Genetic Strategies Using Secreted AVPs
| Secreted AVP | Target | Vector | Promoter | Target Cells | Model System | sAVP Concentration | Outcome |
|---|---|---|---|---|---|---|---|
| Soluble receptor (sCD4) | gp120 | LV | EF1α | CD34 | hu-mice | 100 ng/mL | control of HIV replication after infection119 |
| CD4bs | |||||||
| mAb b12-IgA | gp120 | LV | MHa | CD34 | hu-mice | 10 ng/mL | partial protection against infection111 |
| CD4bs | |||||||
| mAb 2G12 | gp120 | LV | PGM | CD34 | hu-mice | 40 ng/mL | partial protection against infection110 |
| glycans | |||||||
| mAb b12 | gp120 | AAV2 | CMV (HC) | muscle | hu-mice | 8 μg/mL | NA123 |
| CD4bs | EF1α (LC) | ||||||
| mAb VRC07G54W | gp120 | AAV8 | CASIb | muscle | hu-mice | 100 μg/mL | protection against infection124 |
| CD4bs | |||||||
| mAb 10-1074 | gp120 | AAVc | thyroglobulin | muscle | hu-mice | 200 mg/mL | suppression of HIV replication after infection126 |
| glycans | |||||||
| Simian scFv-IgG1 (4L6 or 5L7) | gp120 | AAV1 | CMV | muscle | macaques | 1–270 μg/mL | immunogenic; protection against infection dependent on lower immune responses against the secreted AVP127 |
| NA | |||||||
| Simian mAb (4L6 and 5L7) | gp120 | AAV1 | CMV | muscle | macaques | 1–270 μg/mL | immunogenic; partial protection against infection128 |
| NA | |||||||
| Siminized mAB VRC07 | gp120 | AAV8 | β-actin | muscle | macaques | 66 μg/mL | immunogenic; protection against infection when immune suppressant were used129 |
| CD4bs | |||||||
| Rhesus sCD4-IgG1 | gp120 | AAV1 | CMV | muscle | macaques | 3–10 μg/mL | immunogenic; protection against infection dependent on lower immune responses against the secreted AVP127 |
| CD4bs | |||||||
| Rhesus eCD4-Ig | gp120 | AAV2 | CMV | muscle | macaques | 17–77 μg/mL | less immunogenic than mAbs; protection against infection130 |
| CD4bs CORbs |
hu-mice, humanized mice.
B cell-specific promoter containing Eμ enhancer and MAR upstream of human μ heavy-chain promoter.
A combination of the CMV enhancer, the chicken β-actin promoter, and the ubiquitin enhancer region flanked by a splice donor and splice acceptor.
Serotype not specified.
Since mAbs are highly specific for their target and HIV Env is highly variable, HIV can easily escape antibody binding.23, 112, 113, 114 In contrast, HIV cannot escape binding to sCD4 without losing its ability to bind to cellular CD4,115, 116 but initial clinical trials with sCD4 proved disappointing.117 Follow-up studies showed that the majority of clinical isolates require higher sCD4 concentrations for inhibition than initially anticipated.118 Subsequent clinical trials demonstrated that twice daily administration of sCD4 could completely neutralize cell-free HIV.4, 90 In proof-of-principle studies, we have modified human HSPCs with a lentiviral vector expressing sCD4.102 Although sCD4 has a much shorter half-life than antibodies, we detected 10-fold higher concentrations of sCD4 in the peripheral blood of immunodeficient mice engrafted with gene-modified HSPCs in comparison to the values reported for b12 (Table 4). This was probably due to the use of different promoters. Expression of sCD4 was under the control of the EF1α promoter, which was equally active in B cells and T cells, while b12 was expressed from a B cell-specific promoter. Directly after infection, viral load and percentages of CD4+ T cells were similar between the control and sCD4 groups.119 In control mice, viral load continued to increase and the percentage of CD4+ T cells decreased.119 In contrast, viral load continuously decreased and the percentage of CD4+ T cells remained stable in mice expressing sCD4, indicating that constitutive expression and secretion of an entry inhibitor can control HIV replication in vivo.119
After autologous T cell gene therapy, up to 30% of total CD4+ T cells can consist of gene-modified cells.63 The ratio of gene-modified producer cells to HIV target cells could be further increased if gene-modified CD8+ T cells were co-infused. Therefore, secreted AVPs would have to be identified that can suppress virus replication at this level of T cell gene marking. For HSPC gene therapy approaches, the required level of gene marking would likely be lower because all hematopoietic lineages would serve as producer cells, including B cells and monocytes, which would drastically increase the ratio of gene-modified producer cells to unmodified HIV target cells.
Secretion of AVPs from Non-hematopoietic Cell Lineages
In vivo gene therapy of non-hematopoietic cells is less invasive and complex than ex vivo gene therapy of hematopoietic cells. Therefore, secretion of AVPs from non-hematopoietic cell lineages could theoretically be used for HIV treatment and prevention. Adeno-associated virus (AAV)-derived vectors can be used to directly modify cells in vivo. Several studies have used AAV vectors to transfer genes encoding secreted AVPs to muscle cells. Although AAV vectors are generally non-integrating, the vector genomes persist for long times because skeletal muscle cells do not undergo mitosis.120 Instead, damaged cells are repaired or replaced by satellite cells.120 The average lifespan of skeletal muscle cells in a human adult is 15.1 years.121 AAV vectors encoding mAb b12 were injected into the muscle of immunodeficient mice.122 Following vector administration, the maximal concentration of antibody that bound to HIV gp120 was 8 μg/mL; in vivo inhibition of HIV replication was not examined.
In a similar study, significantly higher levels of the mAb b12 (over 100 μg/mL) were observed after AAV vector-mediated gene transfer into the muscle tissue of mice.123 The same study also showed that the newer and more potent CD4bs mAb VRC01 accumulated to similar levels in mice. Mice capable of secreting b12 or VRC01 were engrafted with human CD4+ T cells, and they were protected from intravenous HIV challenge. In a follow-up study, AAV vector-mediated expression of the mAb VRC07G54W protected humanized mice from multiple mucosal HIV challenges, while incomplete protection was observed with b12 or VRC01.124 The mAb VRC07G54W was chosen because VRC07 is a more potent clonal variant of VRC01, and introducing a G54W mutation into the heavy-chain complementarity-determining region 2 of VRC07 further improved neutralization breadth and potency.125
Even higher mAb levels were observed in another study examining the ability of the glycan-targeting mAb 10-1074 to suppress HIV replication after infection was established.126 While mAb 10-1074 has a lower neutralization breadth than newer CD4bs-targing antibodies, such as VRC01, lower concentrations of mAb 10-1074 are necessary for efficient inhibition.93 Humanized mice were infected with HIV and treated with ART consisting of one integrase inhibitor and two nucleotide reverse-transcriptase inhibitors.126 However, ART seemed to interfere with AAV transduction as ART-treated mice had significantly lower antibody expression after AAV injection, which was attributed to the presence of nucleotide analogs in ART-treated animals. Therefore, treatment was switched to infusions of mAb 10-1074 followed by injection of the AAV vector encoding the mAb 10-1074. After cessation of mAb 10-1074 infusions, vector-mediated expression resulted in antibody levels of 200 mg/mL. The widely different levels of mAbs observed in the different studies were probably due to the use of different promoters (Table 4). Importantly, expression of mAb 10-1074 maintained suppression of viral load in six of seven mice.
Multiple studies also examined secretion of AVPs in macaques. Three AAV vectors were used for intramuscular injection in nine macaques (three animals for each vector).127 Two vectors encoded scFvs (4L6 or 5L7) derived from anti-simian immunodeficiency virus (SIV) antibodies fused to the crystallizable fragment (Fc) region of IgG1 (termed immunoadhesins). The third vector encoded rhesus sCD4-immunoglobulin G (IgG)1. Serum concentrations of the transgenes varied from 3 to 190 μg/mL. After injection with SIVmac316, four of six macaques receiving AAV vectors encoding immunoadhesins and two of three macaques receiving vectors encoding sCD4-IgG1 were completely protected from infection. Infection in the three remaining animals correlated with detectable immune responses to the transgenes. Hoping that full-length rhesus-derived antibodies would not elicit an immune response, AAV vectors were designed that encoded 4L6 and 5L7 mAbs that only contained rhesus-derived sequences. However, immune responses to the antibodies were still observed in nine of twelve macaques that were intramuscularly injected with the vectors.128 Similarly, immunosuppression was required to sustain expression of a simianized form of mAb VRC07 after intramuscular injection of AAV vectors.129
Recently, AAV vector-mediated delivery of sCD4-IgG1 fused to a co-receptor-mimicking peptide (eCD4-Ig) was tested in macaques.130 sCD4 has the unique ability to induce conformational changes in HIV Env, resulting in the exposure of the normally shielded co-receptor-binding site. The fusion protein eCD4-Ig was highly active against isolates that were relatively resistant to multiple CD4bs-targeting mAbs, but it requires co-expression of a tyrosine protein sulfotransferase to promote sulfation of the co-receptor-mimicking peptide.130 Concentrations of rhesus eCD4-Ig reached 17 to 77 μg/mL in blood samples of macaques, which were protected from multiple SHIV-AD8-EO challenges. Although eCD4-Ig also caused immune responses, the fusion protein was less immunogenic in macaques in comparison to four anti-HIV mAbs. Although no significant immune responses were reported in clinical trials based on the infusion of the mAbs VRC01, 3BNC117, and 10-1074, the macaque studies illustrate that immune responses to secreted AVPs could represent a significant obstacle to AAV-mediated gene transfer in humans. Two clinical trials are currently testing delivery of the glycan-targeting mAb PG9 (ClinicalTrials.gov: NCT01937455) or VRC07 (ClinicalTrials.gov: NCT03374202) via AAV vector gene therapy, and they will provide further insights about immune responses to the secreted AVPs.
In a different approach, AAV vectors encoding a fusion protein consisting of the scFv of human mAb b12 and the Fc region of IgG1 were used to transduce human vaginal epithelial cells (VECs).131 Gene-modified VECs secreted ∼12 μg/mL b12 immunoadhesin. In experiments utilizing VECs and transwell plates, secreted b12 immunoadhesin inhibited HIVBaL transfer. Local expression of secreted AVPs may also enhance the efficacy over secreted AVPs expressed at distant sites, such as muscle tissue, but further studies would be necessary to determine if there is a benefit in local expression of secreted AVPs for HIV prophylaxis.
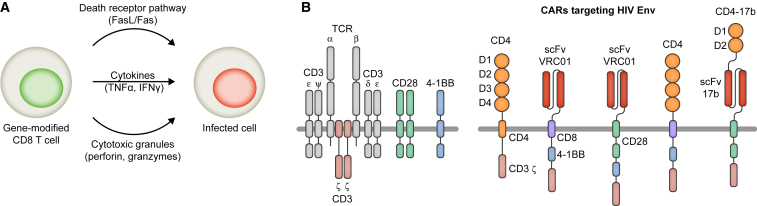
Gene Therapy Using Engineered CD8+ T Cells
Reprogramming CD8+ T cells to recognize and kill HIV-infected cells represents another genetic approach to treat HIV-positive individuals (Figure 3A). To that end, CD8+ T cells or HSPCs are modified ex vivo to express HIV-specific TCRs or chimeric antigen receptors (CARs). For example, a naturally occurring HIV Gag-specific TCR (A2-SL9) is associated with lower viremia in HIV-positive individuals.132, 133 Gene-modified CD8+ T cells expressing A2-SL9 reduced HIV infection in a mouse model of HIV infection. Phage display was used to isolate an A2-SL9 variant with enhanced affinity, and a clinical trial was initiated to test its safety and efficacy (ClinicalTrials.gov: NCT00991224).134 However, the trial was cancelled because off-target effects resulting in severe adverse effects were observed in other trials testing affinity-enhanced TCRs.135
Figure 3.
Gene Therapy Using Engineered CD8+ T Cells
(A) CD8+ T cells are modified to express HIV-specific TCRs or CARs. Upon recognition of an infected cell, gene-modified CD8+ T cells mediate the destruction of the infected cell. (B) Examples of HIV-specific CARs. The structure of the TCR/CD3 complex with co-stimulatory receptors CD28 and 4-1BB is shown in the left panel. The structure of HIV-specific CARs is depicted in the right panel.
In another approach, CD4 was used to engineer an HIV-specific CAR. The 4 extracellular domains and the transmembrane domain of CD4 were covalently linked to the zeta subunit of the CD3 TCR, which is the cytoplasmic domain involved in signal transduction and T cell activation (Figure 3B).136 Expression of the CD4 CAR enabled CD8+ T cells to selectively kill infected cells and suppress viral replication in vitro.137 The CD4 CAR was subsequently tested in phase I and II clinical trials.138, 139, 140 While no differences in viral load were detected, decreases in infected peripheral blood mononucleocytes as well as HIV DNA isolated from rectal mucosa were observed. Interestingly, a long-term follow-up study showed that CAR T cells were detected in 98% of samples tested for at least 11 years after infusion.141 Despite the persistence of the gene-modified cells, no clinical benefit was evident, indicating that a significant challenge for HIV CAR-T cell therapy is the requirement for persistent activity.142 Factors that can influence the expansion, persistence, and activity of adoptively transferred T cells include but are not limited to the ex vivo culture conditions of the T cell product, the patient pre-conditioning regimens, the development of T cell exhaustion, immune responses, and the design of CARs.143
To create more potent HIV-specific CARs, a series of novel mAbs with broad and potent neutralization activity was converted to scFvs and linked to different transmembrane domains as well as cytoplasmic co-stimulatory domains and the CD3 zeta subunit (Figure 3B).144, 145 The novel CARs expressed on CD8+ T cells mediated specific proliferation and killing in response to HIV-infected cells, and one of them is entering clinical trials (ClinicalTrials.gov: NCT03240328); but, as mentioned earlier, HIV can easily escape mAb-mediated inhibition.
Since HIV cannot escape binding to CD4 without a significant loss of fitness, multiple approaches have been tested to improve CD4 CARs. In one study, optimization of the vector backbone, promoter, transmembrane domain, and signaling domains resulted in significantly more potent CD4 CARs in comparison to the original construct and CARs based on anti-HIV mAbs (Figure 3B).146 However, since the extracellular domains of CD4 CARs are identical to CD4, expression of CD4 CARs renders CD8+ T cells susceptible to HIV infection.146 Additionally, bispecific CARs consisting of the first two extracellular domains of CD4 linked to an scFv of the CORbs-targeting mAb 17b were designed to improve potency and block infection of gene-modified CD8+ T cells (Figure 3B).147 In a combination approach, CD4 CARs were co-expressed with antiviral genes that protect cells from infection. For example, trends toward a lower viral load and increased expansion of gene-modified cells were observed in macaques that were infused with autologous HSPCs modified to express maC46 and CD4 CARs in comparison to control animals that were infused with HSPCs expressing maC46 and CD4 CARs with a deleted signaling domain.148 However, it is of note that multiple fatal cases were observed after the infusion of gene-modified CAR-T cells.149, 150 The severe adverse events, such as neurologic toxicity and cytokine release syndrome, were not related to the vector that was used to transfer the gene encoding the CAR, but rather to the effects of expressing CARs on T cells.
Gene Transfer Vectors and the Safety of Genetic Strategies
Early clinical trials used vectors derived from gammaretroviruses. MLV vectors were used to genetically modify T cells and HSPCs. However, MLV vectors preferentially integrate near the transcriptional start sites of proto-oncogenes, and their promoter and/or enhancer regions in the viral LTRs can modulate proto-oncogene expression, resulting in monoclonal expansion of genetically modified cells.151, 152, 153 Consequently, severe adverse effects, such as leukemia or myelodysplasia, were observed in several patients treated with autologous HSPCs that were modified with MLV vectors.154, 155, 156 The safety risk seems to be specific to the use of HSPCs, as no adverse effects have been reported in clinical trials based on the MLV-mediated expression of therapeutic genes in autologous T cells.157 MLV-based vectors were also used in multiple clinical trials testing HIV gene therapy approaches based on the modification of HSPCs (Table 1). While no severe adverse effects were detected in these trials, MLV vectors are not used for the modification of HSPCs anymore because of their genotoxicity.
Self-inactivating (SIN) MLV vectors have deletions in their LTRs that inactivate the viral promoter and/or enhancer after integration. In these vectors, transgene expression is under the control of an internal promoter. SIN MLV vectors have an improved safety profile, but their clinical use was initially limited because they could not be produced at high titers.158 The low titer was largely due to transcriptional interference of the MLV promoter and/or enhancer in the 5′ LTR with the internal promoter, which reduced the production of full-length vector RNA during vector particle production in the packaging cells.159 This problem was overcome by substituting the MLV promoter region in the 5′ LTR with another promoter to drive expression of the full-length vector RNA.159, 160 Their safety and effectiveness are currently being investigated in clinical trials for the treatment of X-linked severe combined immunodeficiency (SCID).161 However, direct integration into regulatory elements of proto-oncogenes still poses a safety risk that is not addressed by the SIN design.
In contrast to MLV vectors, lentiviral vectors do not preferentially integrate near transcription start sites. Instead, the vector genome integrates randomly into transcriptionally active regions of chromosomes.162 Furthermore, the SIN design could be applied to lentiviral vectors without causing a significant loss of titer.163, 164 SIN lentiviral vectors have been tested extensively in clinical trials. The data accumulated to date suggest that lentiviral vectors have an exceptional safety profile. No vector-associated severe adverse events were reported in any trial to date. In one patient, monoclonal expansion of a gene-modified cell was observed, but the expansion was transient and was not associated with any malignancy.165 In all other trials, polyclonal development of gene-modified HSPCs and their progeny was observed. Based on the safety and effectiveness of lentiviral vector-mediated expression of therapeutic genes, a gene therapy using autologous HSPCs has recently been approved for the treatment of adenosine deaminase (ADA)-SCID in Europe.166 Furthermore, lentiviral vector-mediated expression of anti-CD19 CARs in T cells has recently been approved for the treatment of B cell malignancies. To date, one trial using a combination lentiviral vector expressing three anti-HIV genes has been completed (Table 2), and four trials based on the modification of HSPCs with lentiviral vectors are planned or ongoing (Table 3).
The sleeping beauty transposon system represents a non-viral method of integrating antiviral genes into the chromosomes of human cells. The antiviral gene flanked by terminal inverted repeats (TIRs) is introduced into cells as part of so-called DNA minicircle vectors. Co-transfer of mRNAs encoding the transposase enzyme results in the excision of the antiviral gene from the minicircles and random integration into the host chromosome. An advantage of the transposon system is the reduced production cost in comparison to viral delivery methods. In a proof-of-principle study, the sleeping beauty transposon system was used to deliver genes encoding small interfering RNAs (siRNAs) targeting CXCR4 or CCR5 into HIV-sensitive cell lines.167 However, further improvements may be necessary to increase the transduction efficacy of repopulating HSPCs.168 Ongoing clinical trials using the sleeping beauty transposon system are testing the sleeping beauty transposon system for CAR-T cell engineering to treat patients with B cell malignancies.
Site-specific integration is currently under extensive investigation to reduce the risk of insertional mutagenesis.169, 170, 171 For example, the CRISPR/Cas9 system was combined with AAV vectors to achieve site-specific integration of the donor DNA into the beta-globin (HBB) and CCR5 gene.171 Ribonucleoprotein complexes (RNPs) containing single-guide RNAs (sgRNAs) and Cas9 were electroporated into HSPCs directly, followed by the application of AAV vectors containing donor DNA with regions homologous to the respective gene.171 While site-specific integration was observed, the repopulation potential of HSPCs with integrated genes was very low, indicating that further optimization of the protocol is necessary.171 Furthermore, off-target editing effects are a general concern when genome-editing enzymes are used. For instance, substantial off-target activity was observed in a different study when unoptimized sgRNAs were used to target the HBB and CCR5 genes.172 Interestingly, anti-CRISPR/Cas9 proteins isolated from bacteriophages have been shown to reduce off-target effects without significantly diminishing on-target activity.173 Inclusion of inducible suicide genes in the expression cassette represents an option to remove gene-modified hematopoietic cells in case of severe adverse effects caused by integrated vector DNA.174, 175
AAV vectors are generally non-integrating, although integration has been observed in some instances.176 In one study, the development of hepatic tumors was observed following AAV vector-mediated gene therapy in mice, and integrated AAV vector sequences were found in tumors.177 A major concern for using AAV vectors is immune responses against the viral capsid proteins. Multiple AAV serotypes have been discovered. Serotypes 1–9 are among the most commonly used ones. AAV serotypes 1, 4, and 7–9 were found in nonhuman primates, while AAV serotypes 2, 3, 5, and 6 were isolated from humans.178, 179 In a clinical trial for the treatment of hemophilia B via liver-directed gene therapy, six patients were infused through the hepatic artery with AAV2 vectors that mediated liver-specific expression of human Factor IX (FIX).180 However, FIX expression was transient and the decline was accompanied by elevated liver transaminases, suggesting that gene-modified liver cells were destroyed. Follow-up experiments indicated that cellular immune responses against AAV2 capsid proteins caused the disappearance of gene-modified hepatocytes and elevated transaminase levels. Immune responses against capsid proteins also limited the effectiveness of a similar liver-directed gene therapy using AAV8 vectors for the expression of FIX, but a transient application of immunosuppressive drugs halted the destruction of gene-modified cells.181 A gene therapy based on the intramuscular injection of an AAV1 vector (Alipogene Tiparvovec) has recently been approved for the treatment of lipoprotein lipase deficiency in Europe.182, 183 In a clinical study, the immune responses after Alipogene Tiparvovec administration were investigated.184 Five patients were maintained on immune suppressants from shortly before Alipogene Tiparvovec administration until 12 weeks after administration. Humoral and cellular immune responses against AAV1 capsid proteins were detected at various time points during and after immunosuppression. However, no major negative impact on clinical efficacy or safety was observed.184
In the above-mentioned clinical studies, no immune responses to the transgene products were observed. Furthermore, the administration of recombinant protein-based HIV entry inhibitors or antibodies was well tolerated in clinical trials, and no immune responses that adversely affected the treatment were detected.4, 90, 92, 93, 97, 117, 185, 186, 187, 188, 189 However, immune responses to anti-HIV mAbs were often detected in pre-clinical macaque models, and they reduced the effectiveness of the approach as mentioned in the section above. Clinical trials in humans will shed more light on immune responses to AAV vector transgene products.
Transgenes expressed by hematopoietic cells also have the potential to elicit immune responses. While infusion of autologous gene-modified T cells expressing maC46 did not elicit a de novo immune response in a clinical trial,56 there are reports of humoral and cellular immune responses to transgenes expressed by autologous gene-modified hematopoietic cells when they were infused without MA conditioning.190, 191, 192 No immune responses to transgenes are usually observed when gene-modified hematopoietic cells are infused after MA conditioning, suggesting that a state of tolerance is induced.79, 193, 194, 195, 196, 197, 198
The Cost of Genetic Strategies
According to an analysis from 2015, the estimated lifetime cost of drugs used for ART is ∼$400,000.199 Combining the cost of ART with additional direct and indirect costs, such as doctors’ visits and the impact on labor productivity, the cost per infected individual was estimated to be ∼$1 million.199 PrEP was estimated to cost ∼$450,000 per prevented infection, and it was found to be cost saving in comparison to ART.
A T cell-based therapy for the treatment of pediatric and young adult patients with acute lymphoblastic leukemia is priced at $475,000.200 The price for an approved HSPC-based gene therapy for the treatment of ADA-SCID is ∼$660,000,201 while the cost of an AAV-based approach approved for the therapy of lipoprotein lipase deficiency is up to ∼$1.2 million.183 The high cost of the AAV therapy is largely due to the need for ∼20 intramuscular injections of AAV administered in one procedure to reach therapeutic serum concentrations of lipoprotein lipase (∼60 ng/mL).183, 202 It is clear that the price of genetic approaches needs to be reduced in order to become an option for a larger population. However, a genetic therapy would only be needed once in an optimal scenario. While the initial cost of such a therapy is high, it has the potential to be cost saving in comparison to lifelong ART.
Conclusions
Pre-clinical studies have demonstrated that multiple genetic approaches can be used to control viremia in the absence of ART. To translate these results into the clinic, it will be important for all approaches to develop protocols that achieve consistently high levels of gene modification in patients. However, a combination of the different approaches, e.g., rendering HIV target cells resistant and enabling them to secrete AVPs, might be necessary in order to achieve full suppression of viremia.
Utilizing a genetic strategy based on secreted AVPs for HIV prophylaxis is an intriguing concept. Pre-clinical studies have shown the feasibility of this approach. However, whether AVPs can accumulate in high enough concentrations in humans remains to be answered. Local expression of secreted AVPs may be a possibility to increase AVP concentrations at the site of action. Additionally, the optimization of AAV vectors is required to increase the efficacy and significantly lower the cost of strategies utilizing AAV vectors. Due to the recent successes of gene therapy, it is foreseeable that technical and financial barriers will be further reduced, and genetic approaches for HIV therapy and prophylaxis could become an option for a larger population.
Acknowledgments
This research is funded by a grant from Canadian Foundation for AIDS Research (grant number 503531).
References
- 1.UNAIDS. (2016). Global AIDS update. Joint United Nations Programme on HIV/AIDS. http://www.unaids.org/sites/default/files/media_asset/global-AIDS-update-2016_en.pdf. [PubMed]
- 2.Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K., Schneider T., Hofmann J., Kücherer C., Blau O. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 3.Allers K., Hütter G., Hofmann J., Loddenkemper C., Rieger K., Thiel E., Schneider T. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 4.Schacker T., Collier A.C., Coombs R., Unadkat J.D., Fox I., Alam J., Wang J.P., Eggert E., Corey L. Phase I study of high-dose, intravenous rsCD4 in subjects with advanced HIV-1 infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995;9:145–152. [PubMed] [Google Scholar]
- 5.Shingai M., Nishimura Y., Klein F., Mouquet H., Donau O.K., Plishka R., Buckler-White A., Seaman M., Piatak M., Jr., Lifson J.D. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee S., Behnam Azad B., Nimmagadda S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 2014;124:31–82. doi: 10.1016/B978-0-12-411638-2.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samson M., Libert F., Doranz B.J., Rucker J., Liesnard C., Farber C.M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 8.Yang A.G., Bai X., Huang X.F., Yao C., Chen S. Phenotypic knockout of HIV type 1 chemokine coreceptor CCR-5 by intrakines as potential therapeutic approach for HIV-1 infection. Proc. Natl. Acad. Sci. USA. 1997;94:11567–11572. doi: 10.1073/pnas.94.21.11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang A.G., Zhang X., Torti F., Chen S.Y. Anti-HIV type 1 activity of wild-type and functional defective RANTES intrakine in primary human lymphocytes. Hum. Gene Ther. 1998;9:2005–2018. doi: 10.1089/hum.1998.9.14-2005. [DOI] [PubMed] [Google Scholar]
- 10.Schroers R., Davis C.M., Wagner H.J., Chen S.Y. Lentiviral transduction of human T-lymphocytes with a RANTES intrakine inhibits human immunodeficiency virus type 1 infection. Gene Ther. 2002;9:889–897. doi: 10.1038/sj.gt.3301711. [DOI] [PubMed] [Google Scholar]
- 11.Swan C.H., Bühler B., Steinberger P., Tschan M.P., Barbas C.F., 3rd, Torbett B.E. T-cell protection and enrichment through lentiviral CCR5 intrabody gene delivery. Gene Ther. 2006;13:1480–1492. doi: 10.1038/sj.gt.3302801. [DOI] [PubMed] [Google Scholar]
- 12.Nazari R., Ma X.Z., Joshi S. Inhibition of human immunodeficiency virus-1 entry using vectors expressing a multimeric hammerhead ribozyme targeting the CCR5 mRNA. J. Gen. Virol. 2008;89:2252–2261. doi: 10.1099/vir.0.2008/001222-0. [DOI] [PubMed] [Google Scholar]
- 13.Bai J., Gorantla S., Banda N., Cagnon L., Rossi J., Akkina R. Characterization of anti-CCR5 ribozyme-transduced CD34+ hematopoietic progenitor cells in vitro and in a SCID-hu mouse model in vivo. Mol. Ther. 2000;1:244–254. doi: 10.1006/mthe.2000.0038. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu S., Kamata M., Kittipongdaja P., Chen K.N., Kim S., Pang S., Boyer J., Qin F.X., An D.S., Chen I.S. Characterization of a potent non-cytotoxic shRNA directed to the HIV-1 co-receptor CCR5. Genet. Vaccines Ther. 2009;7:8. doi: 10.1186/1479-0556-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson J.S., Walker J., Nolta J.A., Bauer G. Specific transduction of HIV-susceptible cells for CCR5 knockdown and resistance to HIV infection: a novel method for targeted gene therapy and intracellular immunization. J. Acquir. Immune Defic. Syndr. 2009;52:152–161. doi: 10.1097/QAI.0b013e3181b010a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt N., Wang J., Kim K., Friedman G., Wang X., Taupin V., Crooks G.M., Kohn D.B., Gregory P.D., Holmes M.C., Cannon P.M. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez E.E., Wang J., Miller J.C., Jouvenot Y., Kim K.A., Liu O., Wang N., Lee G., Bartsevich V.V., Lee Y.L. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mock U., Machowicz R., Hauber I., Horn S., Abramowski P., Berdien B., Hauber J., Fehse B. mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res. 2015;43:5560–5571. doi: 10.1093/nar/gkv469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamin R., Berges B.K., Solis-Leal A., Igbinedion O., Strong C.L., Schiller M.R. TALEN gene editing takes aim on HIV. Hum. Genet. 2016;135:1059–1070. doi: 10.1007/s00439-016-1678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 21.Kang H., Minder P., Park M.A., Mesquitta W.T., Torbett B.E., Slukvin I.I. CCR5 Disruption in Induced Pluripotent Stem Cells Using CRISPR/Cas9 Provides Selective Resistance of Immune Cells to CCR5-tropic HIV-1 Virus. Mol. Ther. Nucleic Acids. 2015;4:e268. doi: 10.1038/mtna.2015.42. [DOI] [PubMed] [Google Scholar]
- 22.Xu L., Yang H., Gao Y., Chen Z., Xie L., Liu Y., Liu Y., Wang X., Li H., Lai W. CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Mol. Ther. 2017;25:1782–1789. doi: 10.1016/j.ymthe.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 24.Stewart-Jones G.B., Soto C., Lemmin T., Chuang G.Y., Druz A., Kong R., Thomas P.V., Wagh K., Zhou T., Behrens A.J. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 26.Holguín A., De Arellano E.R., Soriano V. Amino acid conservation in the gp41 transmembrane protein and natural polymorphisms associated with enfuvirtide resistance across HIV-1 variants. AIDS Res. Hum. Retroviruses. 2007;23:1067–1074. doi: 10.1089/aid.2006.0256. [DOI] [PubMed] [Google Scholar]
- 27.Melikyan G.B., Egelhofer M., von Laer D. Membrane-anchored inhibitory peptides capture human immunodeficiency virus type 1 gp41 conformations that engage the target membrane prior to fusion. J. Virol. 2006;80:3249–3258. doi: 10.1128/JVI.80.7.3249-3258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leslie G.J., Wang J., Richardson M.W., Haggarty B.S., Hua K.L., Duong J., Secreto A.J., Jordon A.P., Romano J., Kumar K.E. Potent and Broad Inhibition of HIV-1 by a Peptide from the gp41 Heptad Repeat-2 Domain Conjugated to the CXCR4 Amino Terminus. PLoS Pathog. 2016;12:e1005983. doi: 10.1371/journal.ppat.1005983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grütter M.G., Luban J. TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr. Opin. Virol. 2012;2:142–150. doi: 10.1016/j.coviro.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayah D.M., Sokolskaja E., Berthoux L., Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 31.Stremlau M., Owens C.M., Perron M.J., Kiessling M., Autissier P., Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 32.Anderson J., Akkina R. Human immunodeficiency virus type 1 restriction by human-rhesus chimeric tripartite motif 5alpha (TRIM 5alpha) in CD34(+) cell-derived macrophages in vitro and in T cells in vivo in severe combined immunodeficient (SCID-hu) mice transplanted with human fetal tissue. Hum. Gene Ther. 2008;19:217–228. doi: 10.1089/hum.2007.108. [DOI] [PubMed] [Google Scholar]
- 33.Anderson J.S., Javien J., Nolta J.A., Bauer G. Preintegration HIV-1 inhibition by a combination lentiviral vector containing a chimeric TRIM5 alpha protein, a CCR5 shRNA, and a TAR decoy. Mol. Ther. 2009;17:2103–2114. doi: 10.1038/mt.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang K.M., Castanotto D., Li H., Scherer L., Rossi J.J. Incorporation of aptamers in the terminal loop of shRNAs yields an effective and novel combinatorial targeting strategy. Nucleic Acids Res. 2018;46:e6. doi: 10.1093/nar/gkx980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kao S.Y., Calman A.F., Luciw P.A., Peterlin B.M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 36.Feng S., Holland E.C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 37.Felber B.K., Hadzopoulou-Cladaras M., Cladaras C., Copeland T., Pavlakis G.N. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michienzi A., Castanotto D., Lee N., Li S., Zaia J.A., Rossi J.J. RNA-mediated inhibition of HIV in a gene therapy setting. Ann. N Y Acad. Sci. 2003;1002:63–71. doi: 10.1196/annals.1281.008. [DOI] [PubMed] [Google Scholar]
- 39.DiGiusto D.L., Krishnan A., Li L., Li H., Li S., Rao A., Mi S., Yam P., Stinson S., Kalos M. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohn D.B., Bauer G., Rice C.R., Rothschild J.C., Carbonaro D.A., Valdez P., Hao Ql., Zhou C., Bahner I., Kearns K. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood. 1999;94:368–371. [PubMed] [Google Scholar]
- 41.Woffendin C., Ranga U., Yang Z., Xu L., Nabel G.J. Expression of a protective gene-prolongs survival of T cells in human immunodeficiency virus-infected patients. Proc. Natl. Acad. Sci. USA. 1996;93:2889–2894. doi: 10.1073/pnas.93.7.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranga U., Woffendin C., Verma S., Xu L., June C.H., Bishop D.K., Nabel G.J. Enhanced T cell engraftment after retroviral delivery of an antiviral gene in HIV-infected individuals. Proc. Natl. Acad. Sci. USA. 1998;95:1201–1206. doi: 10.1073/pnas.95.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Podsakoff G.M., Engel B.C., Carbonaro D.A., Choi C., Smogorzewska E.M., Bauer G., Selander D., Csik S., Wilson K., Betts M.R. Selective survival of peripheral blood lymphocytes in children with HIV-1 following delivery of an anti-HIV gene to bone marrow CD34(+) cells. Mol. Ther. 2005;12:77–86. doi: 10.1016/j.ymthe.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 44.Levine B.L., Humeau L.M., Boyer J., MacGregor R.R., Rebello T., Lu X., Binder G.K., Slepushkin V., Lemiale F., Mascola J.R. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. USA. 2006;103:17372–17377. doi: 10.1073/pnas.0608138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.George C.X., Gan Z., Liu Y., Samuel C.E. Adenosine deaminases acting on RNA, RNA editing, and interferon action. J. Interferon Cytokine Res. 2011;31:99–117. doi: 10.1089/jir.2010.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamoto M., Chono H., Kawano Y., Saito N., Tsuda H., Inoue K., Kato I., Mineno J., Baba M. Sustained inhibition of HIV-1 replication by conditional expression of the E. coli-derived endoribonuclease MazF in CD4+ T cells. Hum. Gene Ther. Methods. 2013;24:94–103. doi: 10.1089/hgtb.2012.131. [DOI] [PubMed] [Google Scholar]
- 47.Karpinski J., Hauber I., Chemnitz J., Schäfer C., Paszkowski-Rogacz M., Chakraborty D., Beschorner N., Hofmann-Sieber H., Lange U.C., Grundhoff A. Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity. Nat. Biotechnol. 2016;34:401–409. doi: 10.1038/nbt.3467. [DOI] [PubMed] [Google Scholar]
- 48.Ebina H., Misawa N., Kanemura Y., Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z., Pan Q., Gendron P., Zhu W., Guo F., Cen S., Wainberg M.A., Liang C. CRISPR/Cas9-Derived Mutations Both Inhibit HIV-1 Replication and Accelerate Viral Escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 50.Wang G., Zhao N., Berkhout B., Das A.T. CRISPR-Cas9 Can Inhibit HIV-1 Replication but NHEJ Repair Facilitates Virus Escape. Mol. Ther. 2016;24:522–526. doi: 10.1038/mt.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffmann C. The epidemiology of HIV coreceptor tropism. Eur. J. Med. Res. 2007;12:385–390. [PubMed] [Google Scholar]
- 52.Wolstein O., Boyd M., Millington M., Impey H., Boyer J., Howe A., Delebecque F., Cornetta K., Rothe M., Baum C. Preclinical safety and efficacy of an anti-HIV-1 lentiviral vector containing a short hairpin RNA to CCR5 and the C46 fusion inhibitor. Mol. Ther. Methods Clin. Dev. 2014;1:11. doi: 10.1038/mtm.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tebas P., Stein D., Binder-Scholl G., Mukherjee R., Brady T., Rebello T., Humeau L., Kalos M., Papasavvas E., Montaner L.J. Antiviral effects of autologous CD4 T cells genetically modified with a conditionally replicating lentiviral vector expressing long antisense to HIV. Blood. 2013;121:1524–1533. doi: 10.1182/blood-2012-07-447250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimpel J., Braun S.E., Qiu G., Wong F.E., Conolle M., Schmitz J.E., Brendel C., Humeau L.M., Dropulic B., Rossi J.J. Survival of the fittest: positive selection of CD4+ T cells expressing a membrane-bound fusion inhibitor following HIV-1 infection. PLoS ONE. 2010;5:e12357. doi: 10.1371/journal.pone.0012357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Laer D., Hasselmann S., Hasselmann K. Impact of gene-modified T cells on HIV infection dynamics. J. Theor. Biol. 2006;238:60–77. doi: 10.1016/j.jtbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 56.van Lunzen J., Glaunsinger T., Stahmer I., von Baehr V., Baum C., Schilz A., Kuehlcke K., Naundorf S., Martinius H., Hermann F. Transfer of autologous gene-modified T cells in HIV-infected patients with advanced immunodeficiency and drug-resistant virus. Mol. Ther. 2007;15:1024–1033. doi: 10.1038/mt.sj.6300124. [DOI] [PubMed] [Google Scholar]
- 57.Février M., Dorgham K., Rebollo A. CD4+ T cell depletion in human immunodeficiency virus (HIV) infection: role of apoptosis. Viruses. 2011;3:586–612. doi: 10.3390/v3050586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferdin J., Goričar K., Dolžan V., Plemenitaš A., Martin J.N., Peterlin B.M., Deeks S.G., Lenassi M. Viral protein Nef is detected in plasma of half of HIV-infected adults with undetectable plasma HIV RNA. PLoS ONE. 2018;13:e0191613. doi: 10.1371/journal.pone.0191613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lenassi M., Cagney G., Liao M., Vaupotic T., Bartholomeeusen K., Cheng Y., Krogan N.J., Plemenitas A., Peterlin B.M. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic. 2010;11:110–122. doi: 10.1111/j.1600-0854.2009.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahr B., Robert-Hebmann V., Devaux C., Biard-Piechaczyk M. Apoptosis of uninfected cells induced by HIV envelope glycoproteins. Retrovirology. 2004;1:12. doi: 10.1186/1742-4690-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perfettini J.L., Castedo M., Roumier T., Andreau K., Nardacci R., Piacentini M., Kroemer G. Mechanisms of apoptosis induction by the HIV-1 envelope. Cell Death Differ. 2005;12(Suppl 1):916–923. doi: 10.1038/sj.cdd.4401584. [DOI] [PubMed] [Google Scholar]
- 62.Veillette M., Coutu M., Richard J., Batraville L.A., Dagher O., Bernard N., Tremblay C., Kaufmann D.E., Roger M., Finzi A. The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-infected individuals. J. Virol. 2015;89:545–551. doi: 10.1128/JVI.02868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madondo M.T., Quinn M., Plebanski M. Low dose cyclophosphamide: Mechanisms of T cell modulation. Cancer Treat. Rev. 2016;42:3–9. doi: 10.1016/j.ctrv.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 65.Scurr M., Pembroke T., Bloom A., Roberts D., Thomson A., Smart K., Bridgeman H., Adams R., Brewster A., Jones R. Low-Dose Cyclophosphamide Induces Antitumor T-Cell Responses, which Associate with Survival in Metastatic Colorectal Cancer. Clin. Cancer Res. 2017;23:6771–6780. doi: 10.1158/1078-0432.CCR-17-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu R., DeJesus E., Hawkins T., Mitsuyasu R.T., Wang S., Lee G., Tang W., Ando D. Cytoxan enhancement of SB-728-T engraftment: A strategy to improve anti-HIV Response. Top Antivir Med. 2015;23:179. [Google Scholar]
- 67.Ando D., Blick G., Lalezari J., Hsu R., DeJesus E., Hawkins T., Parks D., Wang S., Tang W., Mitsuyasu R. A dose escalation study of cyclophophamide (CTX) to enhance SB-728-T engraftment. Mol Ther. 2015;23:S11. [Google Scholar]
- 68.Maetzig T., Galla M., Baum C., Schambach A. Gammaretroviral vectors: biology, technology and application. Viruses. 2011;3:677–713. doi: 10.3390/v3060677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aggarwal R., Lu J., Pompili V.J., Das H. Hematopoietic stem cells: transcriptional regulation, ex vivo expansion and clinical application. Curr. Mol. Med. 2012;12:34–49. doi: 10.2174/156652412798376125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiekmeijer A.S., Pike-Overzet K., Brugman M.H., Salvatori D.C., Egeler R.M., Bredius R.G., Fibbe W.E., Staal F.J. Sustained Engraftment of Cryopreserved Human Bone Marrow CD34(+) Cells in Young Adult NSG Mice. Biores. Open Access. 2014;3:110–116. doi: 10.1089/biores.2014.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bacigalupo A., Ballen K., Rizzo D., Giralt S., Lazarus H., Ho V., Apperley J., Slavin S., Pasquini M., Sandmaier B.M. Defining the intensity of conditioning regimens: working definitions. Biol. Blood Marrow Transplant. 2009;15:1628–1633. doi: 10.1016/j.bbmt.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mavigner M., Watkins B., Lawson B., Lee S.T., Chahroudi A., Kean L., Silvestri G. Persistence of virus reservoirs in ART-treated SHIV-infected rhesus macaques after autologous hematopoietic stem cell transplant. PLoS Pathog. 2014;10:e1004406. doi: 10.1371/journal.ppat.1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diaconescu R., Flowers C.R., Storer B., Sorror M.L., Maris M.B., Maloney D.G., Sandmaier B.M., Storb R. Morbidity and mortality with nonmyeloablative compared with myeloablative conditioning before hematopoietic cell transplantation from HLA-matched related donors. Blood. 2004;104:1550–1558. doi: 10.1182/blood-2004-03-0804. [DOI] [PubMed] [Google Scholar]
- 74.Atilla E., Ataca Atilla P., Demirer T. A Review of Myeloablative vs Reduced Intensity/Non-Myeloablative Regimens in Allogeneic Hematopoietic Stem Cell Transplantations. Balkan Med. J. 2017;34:1–9. doi: 10.4274/balkanmedj.2017.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mitsuyasu R.T., Merigan T.C., Carr A., Zack J.A., Winters M.A., Workman C., Bloch M., Lalezari J., Becker S., Thornton L. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat. Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Bougnères P., Schmidt M., Kalle C.V., Fischer A., Cavazzana-Calvo M., Aubourg P. Lentiviral hematopoietic cell gene therapy for X-linked adrenoleukodystrophy. Methods Enzymol. 2012;507:187–198. doi: 10.1016/B978-0-12-386509-0.00010-7. [DOI] [PubMed] [Google Scholar]
- 77.Koh Y., Matreyek K.A., Engelman A. Differential sensitivities of retroviruses to integrase strand transfer inhibitors. J. Virol. 2011;85:3677–3682. doi: 10.1128/JVI.02541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Younan P.M., Peterson C.W., Polacino P., Kowalski J.P., Obenza W., Miller H.W., Milless B.P., Gafken P., DeRosa S.C., Hu S.L., Kiem H.P. Lentivirus-mediated Gene Transfer in Hematopoietic Stem Cells Is Impaired in SHIV-infected, ART-treated Nonhuman Primates. Mol. Ther. 2015;23:943–951. doi: 10.1038/mt.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Younan P.M., Polacino P., Kowalski J.P., Peterson C.W., Maurice N.J., Williams N.P., Ho O., Trobridge G.D., Von Laer D., Prlic M. Positive selection of mC46-expressing CD4+ T cells and maintenance of virus specific immunity in a primate AIDS model. Blood. 2013;122:179–187. doi: 10.1182/blood-2013-01-482224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peterson C.W., Haworth K.G., Burke B.P., Polacino P., Norman K.K., Adair J.E., Hu S.L., Bartlett J.S., Symonds G.P., Kiem H.P. Multilineage polyclonal engraftment of Cal-1 gene-modified cells and in vivo selection after SHIV infection in a nonhuman primate model of AIDS. Mol. Ther. Methods Clin. Dev. 2016;3:16007. doi: 10.1038/mtm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Uchida N., Weitzel R.P., Evans M.E., Green R., Bonifacino A.C., Krouse A.E., Metzger M.E., Hsieh M.M., Donahue R.E., Tisdale J.F. Evaluation of engraftment and immunological tolerance after reduced intensity conditioning in a rhesus hematopoietic stem cell gene therapy model. Gene Ther. 2014;21:148–157. doi: 10.1038/gt.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lohrmann H.P., Lepp K.A., Schreml W. 5-Fluorouracil, 1,3-bis(2-chloroethyl)-1-nitrosourea, and 1-(2-chloroethyl)-3-(4-methylcyclohexyl)-1-nitrosourea: effect on the human granulopoietic system. J. Natl. Cancer Inst. 1982;68:541–547. [PubMed] [Google Scholar]
- 83.Zielske S.P., Reese J.S., Lingas K.T., Donze J.R., Gerson S.L. In vivo selection of MGMT(P140K) lentivirus-transduced human NOD/SCID repopulating cells without pretransplant irradiation conditioning. J. Clin. Invest. 2003;112:1561–1570. doi: 10.1172/JCI17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beard B.C., Trobridge G.D., Ironside C., McCune J.S., Adair J.E., Kiem H.P. Efficient and stable MGMT-mediated selection of long-term repopulating stem cells in nonhuman primates. J. Clin. Invest. 2010;120:2345–2354. doi: 10.1172/JCI40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adair J.E., Beard B.C., Trobridge G.D., Neff T., Rockhill J.K., Silbergeld D.L., Mrugala M.M., Kiem H.P. Extended survival of glioblastoma patients after chemoprotective HSC gene therapy. Sci. Transl. Med. 2012;4:133ra57. doi: 10.1126/scitranslmed.3003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adair J.E., Johnston S.K., Mrugala M.M., Beard B.C., Guyman L.A., Baldock A.L., Bridge C.A., Hawkins-Daarud A., Gori J.L., Born D.E. Gene therapy enhances chemotherapy tolerance and efficacy in glioblastoma patients. J. Clin. Invest. 2014;124:4082–4092. doi: 10.1172/JCI76739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chung J., Scherer L.J., Gu A., Gardner A.M., Torres-Coronado M., Epps E.W., Digiusto D.L., Rossi J.J. Optimized lentiviral vectors for HIV gene therapy: multiplexed expression of small RNAs and inclusion of MGMT(P140K) drug resistance gene. Mol. Ther. 2014;22:952–963. doi: 10.1038/mt.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gori J.L., Beard B.C., Ironside C., Karponi G., Kiem H.P. In vivo selection of autologous MGMT gene-modified cells following reduced-intensity conditioning with BCNU and temozolomide in the dog model. Cancer Gene Ther. 2012;19:523–529. doi: 10.1038/cgt.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reithmeier T., Graf E., Piroth T., Trippel M., Pinsker M.O., Nikkhah G. BCNU for recurrent glioblastoma multiforme: efficacy, toxicity and prognostic factors. BMC Cancer. 2010;10:30. doi: 10.1186/1471-2407-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schacker T., Coombs R.W., Collier A.C., Zeh J.E., Fox I., Alam J., Nelson K., Eggert E., Corey L. The effects of high-dose recombinant soluble CD4 on human immunodeficiency virus type 1 viremia. J. Infect. Dis. 1994;169:37–40. doi: 10.1093/infdis/169.1.37. [DOI] [PubMed] [Google Scholar]
- 91.Ledgerwood J.E., Coates E.E., Yamshchikov G., Saunders J.G., Holman L., Enama M.E., DeZure A., Lynch R.M., Gordon I., Plummer S., VRC 602 Study Team Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin. Exp. Immunol. 2015;182:289–301. doi: 10.1111/cei.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Caskey M., Klein F., Lorenzi J.C., Seaman M.S., West A.P., Jr., Buckley N., Kremer G., Nogueira L., Braunschweig M., Scheid J.F. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caskey M., Schoofs T., Gruell H., Settler A., Karagounis T., Kreider E.F., Murrell B., Pfeifer N., Nogueira L., Oliveira T.Y. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 2017;23:185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hartley O., Gaertner H., Wilken J., Thompson D., Fish R., Ramos A., Pastore C., Dufour B., Cerini F., Melotti A. Medicinal chemistry applied to a synthetic protein: development of highly potent HIV entry inhibitors. Proc. Natl. Acad. Sci. USA. 2004;101:16460–16465. doi: 10.1073/pnas.0404802101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dorfman T., Moore M.J., Guth A.C., Choe H., Farzan M. A tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J. Biol. Chem. 2006;281:28529–28535. doi: 10.1074/jbc.M602732200. [DOI] [PubMed] [Google Scholar]
- 96.Jacobson J.M., Lalezari J.P., Thompson M.A., Fichtenbaum C.J., Saag M.S., Zingman B.S., D’Ambrosio P., Stambler N., Rotshteyn Y., Marozsan A.J. Phase 2a study of the CCR5 monoclonal antibody PRO 140 administered intravenously to HIV-infected adults. Antimicrob. Agents Chemother. 2010;54:4137–4142. doi: 10.1128/AAC.00086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jacobson J.M., Thompson M.A., Lalezari J.P., Saag M.S., Zingman B.S., D’Ambrosio P., Stambler N., Rotshteyn Y., Marozsan A.J., Maddon P.J. Anti-HIV-1 activity of weekly or biweekly treatment with subcutaneous PRO 140, a CCR5 monoclonal antibody. J. Infect. Dis. 2010;201:1481–1487. doi: 10.1086/652190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thali M., Moore J.P., Furman C., Charles M., Ho D.D., Robinson J., Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sullivan N., Sun Y., Sattentau Q., Thali M., Wu D., Denisova G., Gershoni J., Robinson J., Moore J., Sodroski J. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wild C.T., Shugars D.C., Greenwell T.K., McDanal C.B., Matthews T.J. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eron J.J., Gulick R.M., Bartlett J.A., Merigan T., Arduino R., Kilby J.M., Yangco B., Diers A., Drobnes C., DeMasi R. Short-term safety and antiretroviral activity of T-1249, a second-generation fusion inhibitor of HIV. J. Infect. Dis. 2004;189:1075–1083. doi: 10.1086/381707. [DOI] [PubMed] [Google Scholar]
- 102.Fetzer I., Gardner M.R., Davis-Gardner M.E., Prasad N.R., Alfant B., Weber J.A., Farzan M. eCD4-Ig Variants That More Potently Neutralize HIV-1. J. Virol. 2018;92 doi: 10.1128/JVI.02011-17. e02011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lu L., Pan C., Li Y., Lu H., He W., Jiang S. A bivalent recombinant protein inactivates HIV-1 by targeting the gp41 prehairpin fusion intermediate induced by CD4 D1D2 domains. Retrovirology. 2012;9:104. doi: 10.1186/1742-4690-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Falkenhagen A., Ameli M., Asad S., Read S.E., Joshi S. A novel gene therapy strategy using secreted multifunctional anti-HIV proteins to confer protection to gene-modified and unmodified target cells. Gene Ther. 2014;21:175–187. doi: 10.1038/gt.2013.70. [DOI] [PubMed] [Google Scholar]
- 105.Falkenhagen A., Joshi S. Further Characterization of the Bifunctional HIV Entry Inhibitor sCD4-FIT45. Mol. Ther. Nucleic Acids. 2017;7:387–395. doi: 10.1016/j.omtn.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bruel T., Guivel-Benhassine F., Amraoui S., Malbec M., Richard L., Bourdic K., Donahue D.A., Lorin V., Casartelli N., Noël N. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat. Commun. 2016;7:10844. doi: 10.1038/ncomms10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ferrari G., Pollara J., Kozink D., Harms T., Drinker M., Freel S., Moody M.A., Alam S.M., Tomaras G.D., Ochsenbauer C. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 2011;85:7029–7036. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Egerer L., Volk A., Kahle J., Kimpel J., Brauer F., Hermann F.G., von Laer D. Secreted antiviral entry inhibitory (SAVE) peptides for gene therapy of HIV infection. Mol. Ther. 2011;19:1236–1244. doi: 10.1038/mt.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Falkenhagen A., Ameli M., Asad S., Read S.E., Joshi S. Gene Therapy Using a Secreted Single Chain Variable Fragment Targeting CCR5 to Inhibit HIV Infection. J. Antivir. Antiretrovir. 2013;5:085–091. [Google Scholar]
- 110.Joseph A., Zheng J.H., Chen K., Dutta M., Chen C., Stiegler G., Kunert R., Follenzi A., Goldstein H. Inhibition of in vivo HIV infection in humanized mice by gene therapy of human hematopoietic stem cells with a lentiviral vector encoding a broadly neutralizing anti-HIV antibody. J. Virol. 2010;84:6645–6653. doi: 10.1128/JVI.02339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hur E.M., Patel S.N., Shimizu S., Rao D.S., Gnanapragasam P.N., An D.S., Yang L., Baltimore D. Inhibitory effect of HIV-specific neutralizing IgA on mucosal transmission of HIV in humanized mice. Blood. 2012;120:4571–4582. doi: 10.1182/blood-2012-04-422303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moore P.L., Sheward D., Nonyane M., Ranchobe N., Hermanus T., Gray E.S., Abdool Karim S.S., Williamson C., Morris L. Multiple pathways of escape from HIV broadly cross-neutralizing V2-dependent antibodies. J. Virol. 2013;87:4882–4894. doi: 10.1128/JVI.03424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lynch R.M., Wong P., Tran L., O’Dell S., Nason M.C., Li Y., Wu X., Mascola J.R. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J. Virol. 2015;89:4201–4213. doi: 10.1128/JVI.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dingens A.S., Haddox H.K., Overbaugh J., Bloom J.D. Comprehensive Mapping of HIV-1 Escape from a Broadly Neutralizing Antibody. Cell Host Microbe. 2017;21:777–787.e4, e774. doi: 10.1016/j.chom.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brighty D.W., Rosenberg M., Chen I.S., Ivey-Hoyle M. Envelope proteins from clinical isolates of human immunodeficiency virus type 1 that are refractory to neutralization by soluble CD4 possess high affinity for the CD4 receptor. Proc. Natl. Acad. Sci. USA. 1991;88:7802–7805. doi: 10.1073/pnas.88.17.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Orloff S.L., Kennedy M.S., Belperron A.A., Maddon P.J., McDougal J.S. Two mechanisms of soluble CD4 (sCD4)-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) infectivity and their relation to primary HIV-1 isolates with reduced sensitivity to sCD4. J. Virol. 1993;67:1461–1471. doi: 10.1128/jvi.67.3.1461-1471.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schooley R.T., Merigan T.C., Gaut P., Hirsch M.S., Holodniy M., Flynn T., Liu S., Byington R.E., Henochowicz S., Gubish E. Recombinant soluble CD4 therapy in patients with the acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. A phase I-II escalating dosage trial. Ann. Intern. Med. 1990;112:247–253. doi: 10.7326/0003-4819-112-4-247. [DOI] [PubMed] [Google Scholar]
- 118.Daar E.S., Ho D.D. Relative resistance of primary HIV-1 isolates to neutralization by soluble CD4. Am. J. Med. 1991;90(4A):22S–26S. doi: 10.1016/0002-9343(91)90407-o. [DOI] [PubMed] [Google Scholar]
- 119.Falkenhagen A., Singh J., Asad S., Leontyev D., Read S., Zúñiga-Pflücker J.C., Joshi S. Control of HIV Infection In Vivo Using Gene Therapy with a Secreted Entry Inhibitor. Mol. Ther. Nucleic Acids. 2017;9:132–144. doi: 10.1016/j.omtn.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Collins C.A., Partridge T.A. Self-renewal of the adult skeletal muscle satellite cell. Cell Cycle. 2005;4:1338–1341. doi: 10.4161/cc.4.10.2114. [DOI] [PubMed] [Google Scholar]
- 121.Spalding K.L., Bhardwaj R.D., Buchholz B.A., Druid H., Frisén J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 122.Lewis A.D., Chen R., Montefiori D.C., Johnson P.R., Clark K.R. Generation of neutralizing activity against human immunodeficiency virus type 1 in serum by antibody gene transfer. J. Virol. 2002;76:8769–8775. doi: 10.1128/JVI.76.17.8769-8775.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Balazs A.B., Chen J., Hong C.M., Rao D.S., Yang L., Baltimore D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2011;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Balazs A.B., Ouyang Y., Hong C.M., Chen J., Nguyen S.M., Rao D.S., An D.S., Baltimore D. Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med. 2014;20:296–300. doi: 10.1038/nm.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Diskin R., Scheid J.F., Marcovecchio P.M., West A.P., Jr., Klein F., Gao H., Gnanapragasam P.N., Abadir A., Seaman M.S., Nussenzweig M.C., Bjorkman P.J. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Horwitz J.A., Halper-Stromberg A., Mouquet H., Gitlin A.D., Tretiakova A., Eisenreich T.R., Malbec M., Gravemann S., Billerbeck E., Dorner M. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc. Natl. Acad. Sci. USA. 2013;110:16538–16543. doi: 10.1073/pnas.1315295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Johnson P.R., Schnepp B.C., Zhang J., Connell M.J., Greene S.M., Yuste E., Desrosiers R.C., Clark K.R. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 2009;15:901–906. doi: 10.1038/nm.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fuchs S.P., Martinez-Navio J.M., Piatak M., Jr., Lifson J.D., Gao G., Desrosiers R.C. AAV-Delivered Antibody Mediates Significant Protective Effects against SIVmac239 Challenge in the Absence of Neutralizing Activity. PLoS Pathog. 2015;11:e1005090. doi: 10.1371/journal.ppat.1005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saunders K.O., Wang L., Joyce M.G., Yang Z.Y., Balazs A.B., Cheng C., Ko S.Y., Kong W.P., Rudicell R.S., Georgiev I.S. Broadly Neutralizing Human Immunodeficiency Virus Type 1 Antibody Gene Transfer Protects Nonhuman Primates from Mucosal Simian-Human Immunodeficiency Virus Infection. J. Virol. 2015;89:8334–8345. doi: 10.1128/JVI.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gardner M.R., Kattenhorn L.M., Kondur H.R., von Schaewen M., Dorfman T., Chiang J.J., Haworth K.G., Decker J.M., Alpert M.D., Bailey C.C. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature. 2015;519:87–91. doi: 10.1038/nature14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Abdel-Motal U.M., Sarkis P.T., Han T., Pudney J., Anderson D.J., Zhu Q., Marasco W.A. Anti-gp120 minibody gene transfer to female genital epithelial cells protects against HIV-1 virus challenge in vitro. PLoS ONE. 2011;6:e26473. doi: 10.1371/journal.pone.0026473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brander C., Hartman K.E., Trocha A.K., Jones N.G., Johnson R.P., Korber B., Wentworth P., Buchbinder S.P., Wolinsky S., Walker B.D., Kalams S.A. Lack of strong immune selection pressure by the immunodominant, HLA-A*0201-restricted cytotoxic T lymphocyte response in chronic human immunodeficiency virus-1 infection. J. Clin. Invest. 1998;101:2559–2566. doi: 10.1172/JCI2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kan-Mitchell J., Bisikirska B., Wong-Staal F., Schaubert K.L., Bajcz M., Bereta M. The HIV-1 HLA-A2-SLYNTVATL is a help-independent CTL epitope. J. Immunol. 2004;172:5249–5261. doi: 10.4049/jimmunol.172.9.5249. [DOI] [PubMed] [Google Scholar]
- 134.Varela-Rohena A., Molloy P.E., Dunn S.M., Li Y., Suhoski M.M., Carroll R.G., Milicic A., Mahon T., Sutton D.H., Laugel B. Control of HIV-1 immune escape by CD8 T cells expressing enhanced T-cell receptor. Nat. Med. 2008;14:1390–1395. doi: 10.1038/nm.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Leibman R.S., Riley J.L. Engineering T Cells to Functionally Cure HIV-1 Infection. Mol. Ther. 2015;23:1149–1159. doi: 10.1038/mt.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Roberts M.R., Qin L., Zhang D., Smith D.H., Tran A.C., Dull T.J., Groopman J.E., Capon D.J., Byrn R.A., Finer M.H. Targeting of human immunodeficiency virus-infected cells by CD8+ T lymphocytes armed with universal T-cell receptors. Blood. 1994;84:2878–2889. [PubMed] [Google Scholar]
- 137.Yang O.O., Tran A.C., Kalams S.A., Johnson R.P., Roberts M.R., Walker B.D. Lysis of HIV-1-infected cells and inhibition of viral replication by universal receptor T cells. Proc. Natl. Acad. Sci. USA. 1997;94:11478–11483. doi: 10.1073/pnas.94.21.11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mitsuyasu R.T., Anton P.A., Deeks S.G., Scadden D.T., Connick E., Downs M.T., Bakker A., Roberts M.R., June C.H., Jalali S. Prolonged survival and tissue trafficking following adoptive transfer of CD4zeta gene-modified autologous CD4(+) and CD8(+) T cells in human immunodeficiency virus-infected subjects. Blood. 2000;96:785–793. [PubMed] [Google Scholar]
- 139.Walker R.E., Bechtel C.M., Natarajan V., Baseler M., Hege K.M., Metcalf J.A., Stevens R., Hazen A., Blaese R.M., Chen C.C. Long-term in vivo survival of receptor-modified syngeneic T cells in patients with human immunodeficiency virus infection. Blood. 2000;96:467–474. [PubMed] [Google Scholar]
- 140.Deeks S.G., Wagner B., Anton P.A., Mitsuyasu R.T., Scadden D.T., Huang C., Macken C., Richman D.D., Christopherson C., June C.H. A phase II randomized study of HIV-specific T-cell gene therapy in subjects with undetectable plasma viremia on combination antiretroviral therapy. Mol. Ther. 2002;5:788–797. doi: 10.1006/mthe.2002.0611. [DOI] [PubMed] [Google Scholar]
- 141.Scholler J., Brady T.L., Binder-Scholl G., Hwang W.T., Plesa G., Hege K.M., Vogel A.N., Kalos M., Riley J.L., Deeks S.G. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci. Transl. Med. 2012;4:132ra53. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hale M., Mesojednik T., Romano Ibarra G.S., Sahni J., Bernard A., Sommer K., Scharenberg A.M., Rawlings D.J., Wagner T.A. Engineering HIV-Resistant, Anti-HIV Chimeric Antigen Receptor T Cells. Mol. Ther. 2017;25:570–579. doi: 10.1016/j.ymthe.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guedan S., Posey A.D., Jr., Shaw C., Wing A., Da T., Patel P.R., McGettigan S.E., Casado-Medrano V., Kawalekar O.U., Uribe-Herranz M. Enhancing CAR T cell persistence through ICOS and 4-1BB costimulation. JCI Insight. 2018;3:96976. doi: 10.1172/jci.insight.96976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ali A., Kitchen S.G., Chen I.S.Y., Ng H.L., Zack J.A., Yang O.O. HIV-1-Specific Chimeric Antigen Receptors Based on Broadly Neutralizing Antibodies. J. Virol. 2016;90:6999–7006. doi: 10.1128/JVI.00805-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu B., Zou F., Lu L., Chen C., He D., Zhang X., Tang X., Liu C., Li L., Zhang H. Chimeric Antigen Receptor T Cells Guided by the Single-Chain Fv of a Broadly Neutralizing Antibody Specifically and Effectively Eradicate Virus Reactivated from Latency in CD4+ T Lymphocytes Isolated from HIV-1-Infected Individuals Receiving Suppressive Combined Antiretroviral Therapy. J. Virol. 2016;90:9712–9724. doi: 10.1128/JVI.00852-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Leibman R.S., Richardson M.W., Ellebrecht C.T., Maldini C.R., Glover J.A., Secreto A.J., Kulikovskaya I., Lacey S.F., Akkina S.R., Yi Y. Supraphysiologic control over HIV-1 replication mediated by CD8 T cells expressing a re-engineered CD4-based chimeric antigen receptor. PLoS Pathog. 2017;13:e1006613. doi: 10.1371/journal.ppat.1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu L., Patel B., Ghanem M.H., Bundoc V., Zheng Z., Morgan R.A., Rosenberg S.A., Dey B., Berger E.A. Novel CD4-Based Bispecific Chimeric Antigen Receptor Designed for Enhanced Anti-HIV Potency and Absence of HIV Entry Receptor Activity. J. Virol. 2015;89:6685–6694. doi: 10.1128/JVI.00474-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhen A., Peterson C.W., Carrillo M.A., Reddy S.S., Youn C.S., Lam B.B., Chang N.Y., Martin H.A., Rick J.W., Kim J. Long-term persistence and function of hematopoietic stem cell-derived chimeric antigen receptor T cells in a nonhuman primate model of HIV/AIDS. PLoS Pathog. 2017;13:e1006753. doi: 10.1371/journal.ppat.1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bonifant C.L., Jackson H.J., Brentjens R.J., Curran K.J. Toxicity and management in CAR T-cell therapy. Mol. Ther. Oncolytics. 2016;3:16011. doi: 10.1038/mto.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hartmann J., Schüßler-Lenz M., Bondanza A., Buchholz C.J. Clinical development of CAR T cells-challenges and opportunities in translating innovative treatment concepts. EMBO Mol. Med. 2017;9:1183–1197. doi: 10.15252/emmm.201607485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 152.Cattoglio C., Facchini G., Sartori D., Antonelli A., Miccio A., Cassani B., Schmidt M., von Kalle C., Howe S., Thrasher A.J. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- 153.Wang G.P., Berry C.C., Malani N., Leboulch P., Fischer A., Hacein-Bey-Abina S., Cavazzana-Calvo M., Bushman F.D. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115:4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 155.Howe S.J., Mansour M.R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M.H., Pike-Overzet K., Chatters S.J., de Ridder D. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stein S., Ott M.G., Schultze-Strasser S., Jauch A., Burwinkel B., Kinner A., Schmidt M., Krämer A., Schwäble J., Glimm H. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 157.Collins M., Thrasher A. Gene therapy: progress and predictions. Proc. Biol. Sci. 2015;282:20143003. doi: 10.1098/rspb.2014.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Volkova N.A., Fomina E.G., Smolnikova V.V., Zinovieva N.A., Fomin I.K. The U3 region of Moloney murine leukemia virus contains position-independent cis-acting sequences involved in the nuclear export of full-length viral transcripts. J. Biol. Chem. 2014;289:20158–20169. doi: 10.1074/jbc.M113.545855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schambach A., Mueller D., Galla M., Verstegen M.M., Wagemaker G., Loew R., Baum C., Bohne J. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13:1524–1533. doi: 10.1038/sj.gt.3302807. [DOI] [PubMed] [Google Scholar]
- 160.Zychlinski D., Schambach A., Modlich U., Maetzig T., Meyer J., Grassman E., Mishra A., Baum C. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol. Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 161.Hacein-Bey-Abina S., Pai S.Y., Gaspar H.B., Armant M., Berry C.C., Blanche S., Bleesing J., Blondeau J., de Boer H., Buckland K.F. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2014;371:1407–1417. doi: 10.1056/NEJMoa1404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Craigie R., Bushman F.D. HIV DNA integration. Cold Spring Harb. Perspect. Med. 2012;2:a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Miyoshi H., Blömer U., Takahashi M., Gage F.H., Verma I.M. Development of a self-inactivating lentivirus vector. J. Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zufferey R., Dull T., Mandel R.J., Bukovsky A., Quiroz D., Naldini L., Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Aiuti A., Roncarolo M.G., Naldini L. Gene therapy for ADA-SCID, the first marketing approval of an ex vivo gene therapy in Europe: paving the road for the next generation of advanced therapy medicinal products. EMBO Mol. Med. 2017;9:737–740. doi: 10.15252/emmm.201707573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tamhane M., Akkina R. Stable gene transfer of CCR5 and CXCR4 siRNAs by sleeping beauty transposon system to confer HIV-1 resistance. AIDS Res. Ther. 2008;5:16. doi: 10.1186/1742-6405-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Holstein M., Mesa-Nuñez C., Miskey C., Almarza E., Poletti V., Schmeer M., Grueso E., Ordóñez Flores J.C., Kobelt D., Walther W. Efficient Non-viral Gene Delivery into Human Hematopoietic Stem Cells by Minicircle Sleeping Beauty Transposon Vectors. Mol. Ther. 2018;26:1137–1153. doi: 10.1016/j.ymthe.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.De Ravin S.S., Reik A., Liu P.Q., Li L., Wu X., Su L., Raley C., Theobald N., Choi U., Song A.H. Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 2016;34:424–429. doi: 10.1038/nbt.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cradick T.J., Fine E.J., Antico C.J., Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shin J., Jiang F., Liu J.J., Bray N.L., Rauch B.J., Baik S.H., Nogales E., Bondy-Denomy J., Corn J.E., Doudna J.A. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 2017;3:e1701620. doi: 10.1126/sciadv.1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhou X., Dotti G., Krance R.A., Martinez C.A., Naik S., Kamble R.T., Durett A.G., Dakhova O., Savoldo B., Di Stasi A. Inducible caspase-9 suicide gene controls adverse effects from alloreplete T cells after haploidentical stem cell transplantation. Blood. 2015;125:4103–4113. doi: 10.1182/blood-2015-02-628354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yagyu S., Hoyos V., Del Bufalo F., Brenner M.K. An Inducible Caspase-9 Suicide Gene to Improve the Safety of Therapy Using Human Induced Pluripotent Stem Cells. Mol. Ther. 2015;23:1475–1485. doi: 10.1038/mt.2015.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Deyle D.R., Russell D.W. Adeno-associated virus vector integration. Curr. Opin. Mol. Ther. 2009;11:442–447. [PMC free article] [PubMed] [Google Scholar]
- 177.Donsante A., Miller D.G., Li Y., Vogler C., Brunt E.M., Russell D.W., Sands M.S. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 178.Gao G.P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gao G., Vandenberghe L.H., Alvira M.R., Lu Y., Calcedo R., Zhou X., Wilson J.M. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 181.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ylä-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol. Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ylä-Herttuala S. Glybera’s second act: the curtain rises on the high cost of therapy. Mol. Ther. 2015;23:217–218. doi: 10.1038/mt.2014.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ferreira V., Petry H., Salmon F. Immune Responses to AAV-Vectors, the Glybera Example from Bench to Bedside. Front. Immunol. 2014;5:82. doi: 10.3389/fimmu.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hodges T.L., Kahn J.O., Kaplan L.D., Groopman J.E., Volberding P.A., Amman A.J., Arri C.J., Bouvier L.M., Mordenti J., Izu A.E. Phase 1 study of recombinant human CD4-immunoglobulin G therapy of patients with AIDS and AIDS-related complex. Antimicrob. Agents Chemother. 1991;35:2580–2586. doi: 10.1128/aac.35.12.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jacobson J.M., Lowy I., Fletcher C.V., O’Neill T.J., Tran D.N., Ketas T.J., Trkola A., Klotman M.E., Maddon P.J., Olson W.C., Israel R.J. Single-dose safety, pharmacology, and antiviral activity of the human immunodeficiency virus (HIV) type 1 entry inhibitor PRO 542 in HIV-infected adults. J. Infect. Dis. 2000;182:326–329. doi: 10.1086/315698. [DOI] [PubMed] [Google Scholar]
- 187.Walmsley S., Henry K., Katlama C., Nelson M., Castagna A., Reynes J., Clotet B., Hui J., Salgo M., DeMasi R., Delehanty J. Enfuvirtide (T-20) cross-reactive glycoprotein 41 antibody does not impair the efficacy or safety of enfuvirtide. J. Infect. Dis. 2003;188:1827–1833. doi: 10.1086/379810. [DOI] [PubMed] [Google Scholar]
- 188.Jacobson J.M., Israel R.J., Lowy I., Ostrow N.A., Vassilatos L.S., Barish M., Tran D.N., Sullivan B.M., Ketas T.J., O’Neill T.J. Treatment of advanced human immunodeficiency virus type 1 disease with the viral entry inhibitor PRO 542. Antimicrob. Agents Chemother. 2004;48:423–429. doi: 10.1128/AAC.48.2.423-429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lynch R.M., Boritz E., Coates E.E., DeZure A., Madden P., Costner P., Enama M.E., Plummer S., Holman L., Hendel C.S., VRC 601 Study Team Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 2015;7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Riddell S.R., Elliott M., Lewinsohn D.A., Gilbert M.J., Wilson L., Manley S.A., Lupton S.D., Overell R.W., Reynolds T.C., Corey L., Greenberg P.D. T-cell mediated rejection of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients. Nat. Med. 1996;2:216–223. doi: 10.1038/nm0296-216. [DOI] [PubMed] [Google Scholar]
- 191.Stripecke R., Carmen Villacres M., Skelton D., Satake N., Halene S., Kohn D. Immune response to green fluorescent protein: implications for gene therapy. Gene Ther. 1999;6:1305–1312. doi: 10.1038/sj.gt.3300951. [DOI] [PubMed] [Google Scholar]
- 192.Rosenzweig M., Connole M., Glickman R., Yue S.P., Noren B., DeMaria M., Johnson R.P. Induction of cytotoxic T lymphocyte and antibody responses to enhanced green fluorescent protein following transplantation of transduced CD34(+) hematopoietic cells. Blood. 2001;97:1951–1959. doi: 10.1182/blood.v97.7.1951. [DOI] [PubMed] [Google Scholar]
- 193.Horn P.A., Morris J.C., Bukovsky A.A., Andrews R.G., Naldini L., Kurre P., Kiem H.P. Lentivirus-mediated gene transfer into hematopoietic repopulating cells in baboons. Gene Ther. 2002;9:1464–1471. doi: 10.1038/sj.gt.3301820. [DOI] [PubMed] [Google Scholar]
- 194.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Veres G., Schmidt M., Kutschera I., Vidaud M., Abel U., Dal-Cortivo L., Caccavelli L. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 195.Gaspar H.B., Cooray S., Gilmour K.C., Parsley K.L., Adams S., Howe S.J., Al Ghonaium A., Bayford J., Brown L., Davies E.G. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 2011;3:97ra79. doi: 10.1126/scitranslmed.3002715. [DOI] [PubMed] [Google Scholar]
- 196.Gaspar H.B., Cooray S., Gilmour K.C., Parsley K.L., Zhang F., Adams S., Bjorkegren E., Bayford J., Brown L., Davies E.G. Hematopoietic stem cell gene therapy for adenosine deaminase-deficient severe combined immunodeficiency leads to long-term immunological recovery and metabolic correction. Sci. Transl. Med. 2011;3:97ra80. doi: 10.1126/scitranslmed.3002716. [DOI] [PubMed] [Google Scholar]
- 197.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 199.Ouellet E., Durand M., Guertin J.R., LeLorier J., Tremblay C.L. Cost effectiveness of ‘on demand’ HIV pre-exposure prophylaxis for non-injection drug-using men who have sex with men in Canada. Can. J. Infect. Dis. Med. Microbiol. 2015;26:23–29. doi: 10.1155/2015/964512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dolgin E. Bringing down the cost of cancer treatment. Nature. 2018;555:S26–S29. doi: 10.1038/d41586-018-02483-3. [DOI] [PubMed] [Google Scholar]
- 201.Touchot N., Flume M. Early Insights from Commercialization of Gene Therapies in Europe. Genes (Basel) 2017;8:E78. doi: 10.3390/genes8020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rip J., Nierman M.C., Wareham N.J., Luben R., Bingham S.A., Day N.E., van Miert J.N., Hutten B.A., Kastelein J.J., Kuivenhoven J.A. Serum lipoprotein lipase concentration and risk for future coronary artery disease: the EPIC-Norfolk prospective population study. Arterioscler. Thromb. Vasc. Biol. 2006;26:637–642. doi: 10.1161/01.ATV.0000201038.47949.56. [DOI] [PubMed] [Google Scholar]
- 203.Wong-Staal F., Poeschla E.M., Looney D.J. A controlled, Phase 1 clinical trial to evaluate the safety and effects in HIV-1 infected humans of autologous lymphocytes transduced with a ribozyme that cleaves HIV-1 RNA. Hum. Gene Ther. 1998;9:2407–2425. doi: 10.1089/hum.1998.9.16-2407. [DOI] [PubMed] [Google Scholar]
- 204.Macpherson J.L., Boyd M.P., Arndt A.J., Todd A.V., Fanning G.C., Ely J.A., Elliott F., Knop A., Raponi M., Murray J. Long-term survival and concomitant gene expression of ribozyme-transduced CD4+ T-lymphocytes in HIV-infected patients. J. Gene Med. 2005;7:552–564. doi: 10.1002/jgm.705. [DOI] [PubMed] [Google Scholar]
- 205.Cooper D., Penny R., Symonds G., Carr A., Gerlach W., Sun L.Q., Ely J. A marker study of therapeutically transduced CD4+ peripheral blood lymphocytes in HIV discordant identical twins. Hum. Gene Ther. 1999;10:1401–1421. doi: 10.1089/10430349950018067. [DOI] [PubMed] [Google Scholar]
- 206.Amado R.G., Mitsuyasu R.T., Rosenblatt J.D., Ngok F.K., Bakker A., Cole S., Chorn N., Lin L.S., Bristol G., Boyd M.P. Anti-human immunodeficiency virus hematopoietic progenitor cell-delivered ribozyme in a phase I study: myeloid and lymphoid reconstitution in human immunodeficiency virus type-1-infected patients. Hum. Gene Ther. 2004;15:251–262. doi: 10.1089/104303404322886101. [DOI] [PubMed] [Google Scholar]
- 207.Kang E.M., de Witte M., Malech H., Morgan R.A., Phang S., Carter C., Leitman S.F., Childs R., Barrett A.J., Little R., Tisdale J.F. Nonmyeloablative conditioning followed by transplantation of genetically modified HLA-matched peripheral blood progenitor cells for hematologic malignancies in patients with acquired immunodeficiency syndrome. Blood. 2002;99:698–701. doi: 10.1182/blood.v99.2.698. [DOI] [PubMed] [Google Scholar]
- 208.Morgan R.A., Walker R., Carter C.S., Natarajan V., Tavel J.A., Bechtel C., Herpin B., Muul L., Zheng Z., Jagannatha S. Preferential survival of CD4+ T lymphocytes engineered with anti-human immunodeficiency virus (HIV) genes in HIV-infected individuals. Hum. Gene Ther. 2005;16:1065–1074. doi: 10.1089/hum.2005.16.1065. [DOI] [PubMed] [Google Scholar]