Abstract
Enterovirus A71 (EV-A71) receptors that have been identified to date cannot fully explain the pathogenesis of EV-A71, which is an important global cause of hand, foot, and mouth disease and life-threatening encephalitis. We identified an IFN-γ–inducible EV-A71 cellular entry factor, human tryptophanyl-tRNA synthetase (hWARS), using genome-wide RNAi library screening. The importance of hWARS in mediating virus entry and infectivity was confirmed by virus attachment, in vitro pulldown, antibody/antigen blocking, and CRISPR/Cas9-mediated deletion. Hyperexpression and plasma membrane translocation of hWARS were observed in IFN-γ–treated semipermissive (human neuronal NT2) and cDNA-transfected nonpermissive (mouse fibroblast L929) cells, resulting in their sensitization to EV-A71 infection. Our hWARS-transduced mouse infection model showed pathological changes similar to those seen in patients with severe EV-A71 infection. Expression of hWARS is also required for productive infection by other human enteroviruses, including the clinically important coxsackievirus A16 (CV-A16) and EV-D68. This is the first report to our knowledge on the discovery of an entry factor, hWARS, that can be induced by IFN-γ for EV-A71 infection. Given that we detected high levels of IFN-γ in patients with severe EV-A71 infection, our findings extend the knowledge of the pathogenicity of EV-A71 in relation to entry factor expression upon IFN-γ stimulation and the therapeutic options for treating severe EV-A71–associated complications.
Keywords: Infectious disease, Microbiology
Keywords: Molecular pathology
Introduction
Enterovirus A71 (EV-A71) is a positive-sense, single-stranded RNA, nonenveloped virus that was first isolated in California, United States, in 1969 (1). It belongs to the human enterovirus A (EV-A) subspecies of the Enterovirus genus in the Picornaviridae family. EV-A71 is a major causative agent of hand, foot, and mouth disease (HFMD), which can be complicated by severe neurological diseases including aseptic meningitis, acute flaccid paralysis, and fatal neurogenic pulmonary edema (2). Severe EV-A71 outbreaks have been reported periodically throughout the world, particularly in the Asia-Pacific region. In addition to EV-A71, there are many other serotypes of enteroviruses that can cause a myriad of diseases ranging from self-limiting febrile exanthematous illness to fatal visceral disease (2). Most EV-A serotypes such as A6, A8, A10, and A16 are known to cause HFMD, herpangina, aseptic meningitis, and acute flaccid paralysis. Enterovirus B serotypes such as echoviruses 6, 11, 25, and 30 are especially known to cause infantile liver failure, myocarditis, pericarditis, pneumonia, encephalitis, and sudden infant death. Another serotype, enterovirus D68 (EV-D68), has caused recent epidemics of severe respiratory illness and fatal acute flaccid myelitis (3) in different regions of the United States and has also been associated with mortality and public health concerns (4, 5).
Currently identified EV-A71 receptors cannot fully explain the pathogenesis of EV-A71. Although a broad tissue tropism can be observed in EV-A71 infection in vitro, its replication capacity differs widely in various cell types. One of the determining factors is the expression of cognate entry factors on the host cell surface. Most EV-A71 studies to date have focused on 2 well-characterized receptors: human scavenger receptor class B member 2 (hSCARB2) and human P-selectin glycoprotein ligand 1 (hPSGL1) (6, 7). For hSCARB2, a previous study indicated that only a subset of serotype A enteroviruses that are closely related to EV-A71 are dependent on SCARB2 for infection (8). Receptor usage of hPSGL1 for infection is even more restrictive than that of hSCARB2, in which only a subset of EV-A71 strains can utilize hPSGL1 for cell entry, and its expression of hPSGL1 is primarily on hematopoietic cells, implying that it is less likely to be a critical receptor for EV-A71 (9). To date, knowledge of the expression patterns of both hSCARB2 and hPSGL1 cannot fully recapitulate the pathogenesis of EV-A71, including tissue tropism for viral replication and clinical manifestations (9). Other entry factors may exist and play a critical role in EV-A71 pathogenesis. In this study, we identified an IFN-γ–inducible cellular entry factor, human tryptophanyl-tRNA synthetase (hWARS), for EV-A71 using shRNA lentiviral library screening for human transcripts. We examined the functional role of hWARS in EV-A71 infection by experiments using in vitro virus attachment, pulldown, and antibody/antigen blocking, confirmed its function by CRISPR/Cas9 and an in vivo mouse model, and compared our findings with previously identified receptors including hSCARB2 and hPSGL1 (6, 7). We also studied the functional role of hWARS in other serotypes of human enteroviruses. The inducibility of hWARS in the disease progression of EV-A71 and potential treatment options for EV-A71 infections are also discussed.
Results
Identification of hWARS as an important host factor for productive EV-A71 infection.
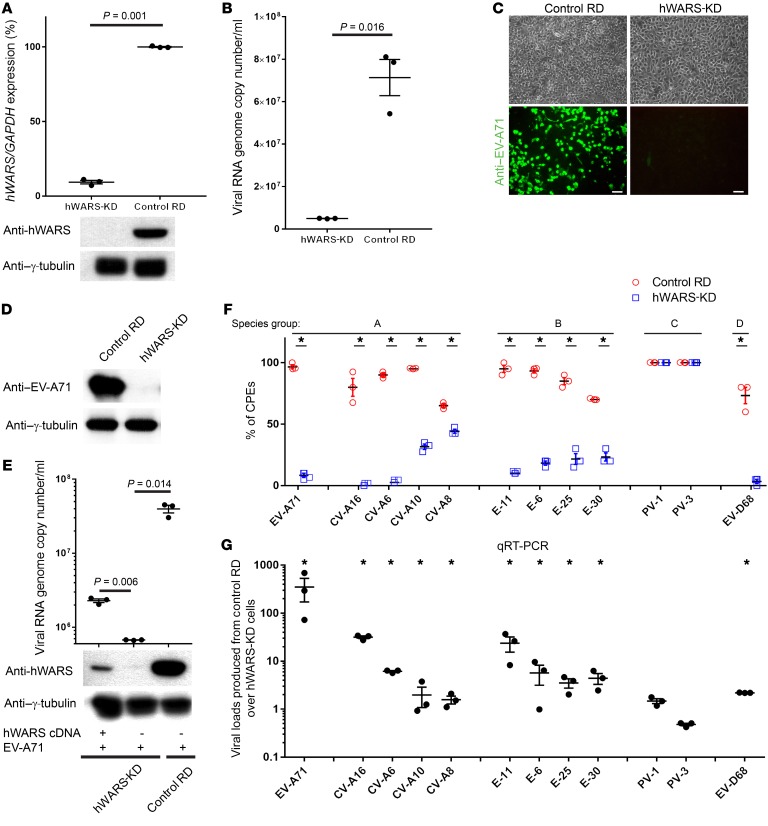
A lentiviral shRNA library targeting 54,509 human transcripts was transduced into 3 × 108 rhabdomyosarcoma (RD) cells, which are highly susceptible to EV-A71 infection. Cells carrying individual discrete shRNAs were challenged by a high titer of EV-A71. We anticipated that knockdown (KD) of a cellular gene that is critical for EV-A71 replication would halt the viral replication and hence protect the cells from the EV-A71–induced cytopathic effects (CPEs). Total RNAs from the pool of EV-A71–resistant cells were isolated, and the shRNAs were identified using an Affymetrix microarray, as described in our previous study (10). The data set containing the shRNA screening results has been deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE80407). We identified 118 candidate genes, the KD of which protected the cell clones from EV-A71–induced CPEs. The annotation and Ingenuity Pathway analyses of these 118 candidate genes are shown in Supplemental Table 1 and Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI99411DS1 Among these candidates, we discovered that silencing a human gene named hWARS could effectively protect cells against EV-A71–induced CPEs (Supplemental Figure 2, A and B). To establish the importance of hWARS in EV-A71 replication, we generated an independent RD cell clone that stably expressed hWARS shRNA. Successful KD of hWARS was confirmed by quantitative real-time PCR (qRT-PCR) and Western blot analyses (Figure 1A). Despite a significant reduction in hWARS expression, we observed no obvious deleterious effect on cell growth in these cells (Supplemental Figure 2, A and B). Under microscopic examination, hWARS-KD cells showed no CPEs compared with control RD cells after the challenge by high-titer EV-A71 (Supplemental Figure 2A). We further confirmed these results by modified MTT assays that measured the cellular metabolic activities. Compared with the control RD cells without EV-A71 infection, the modified MTT readouts of EV-A71–infected control RD cells showed a marked reduction throughout the 48 hours of EV-A71 infection period, suggesting that the KD of hWARS protected RD cells from EV-A71–induced CPEs (Supplemental Figure 2, A and B). Upon EV-A71 infection, we detected a significant reduction in viral RNA genome copy numbers in hWARS-KD cells compared with numbers in control RD cells (Figure 1B). Furthermore, both immunostaining and Western blot analyses revealed minimal expression of EV-A71 protein in EV-A71–inoculated hWARS-KD cells (Figure 1, C and D). Replenishment of hWARS by transfecting a hWARS hyperexpression plasmid into the hWARS-KD cells partially restored the susceptibility of the clones to EV-A71 (Figure 1E). Thus, our loss-of-function and gain-of-function experiments consistently supported the notion that hWARS is important for productive EV-A71 replication.
Figure 1. Generation of hWARS-KD cells and requirement of hWARS for EV-A71 infection.
(A) mRNA and protein levels of the hWARS in hWARS-KD cells were measured by qRT-PCR and Western blot analyses. Control shRNA–transfected RD cells were included for comparison. Viral RNA genome copy numbers and EV-A71 protein levels of the EV-A71–infected hWARS-KD and control RD cells were determined by (B) qRT-PCR, (C) immunostaining (scale bars: 100 μm), and (D) Western blot analyses. (E) Transfection of hWARS cDNA (lane 1) or empty vector (lane 2) into hWARS RD cells followed by EV-A71 challenge. Viral RNA genome copy numbers and EV-A71 protein levels of the transfected cells were determined by qRT-PCR and Western blot analyses. EV-A71–infected control RD cells (lane 3) were included for comparison. (F) Differential CPEs of enterovirus serotypes in hWARS-KD and control RD cells. (G) Measurement of viral loads produced from supernatants of control RD cells over that of hWARS-KD cells. Data represent the mean ± SEM. All means were derived from 3 independent experiments. *P < 0.05, by 2-tailed Student’s t test (A, B, F, and G). The significance in E (determined by 2-tailed Student’s t test) for multiple group comparisons has been adjusted to 0.025 by Bonferroni’s correction. Endogenous γ-tubulin was detected as a loading control for all Western blot analyses. Anti–EV-A71 and anti-hWARS antibodies were used to detect EV-A71 and hWARS proteins. Data in A, C, D, and E are representative of 3 independent experiments.
Besides EV-A71, we tested the requirement of hWARS in RD cells for viral replication by other common human enterovirus isolates of different serotypes, including group A, B, C, and D human enteroviruses. Intriguingly, we found that KD of hWARS expression altered the virus-induced CPEs and viral loads in cells infected with many of the examined serotypes of enterovirus species (Figure 1F). Except for serotypes poliovirus 1 (PV-1) and PV-3, belonging to enterovirus C, all other tested serotypes belonging to enteroviruses A, B, and D, including EV-A71, coxsackievirus A16 (CV-A16), CV-A6, echovirus 11 (E-11), E-6, E-25, E-30, and EV-D68, showed a significant decrease in CPEs in hWARS-KD RD cells compared with control RD cells (Figure 1F). We also found that hWARS-KD RD cells produced fewer viruses in many of the tested serotypes of enterovirus species (Figure 1G). Overall, our results suggest that hWARS may play an important role in viral infection by a broad spectrum of serotypes of enterovirus species.
hWARS acts in the early stage of EV-A71 infection.
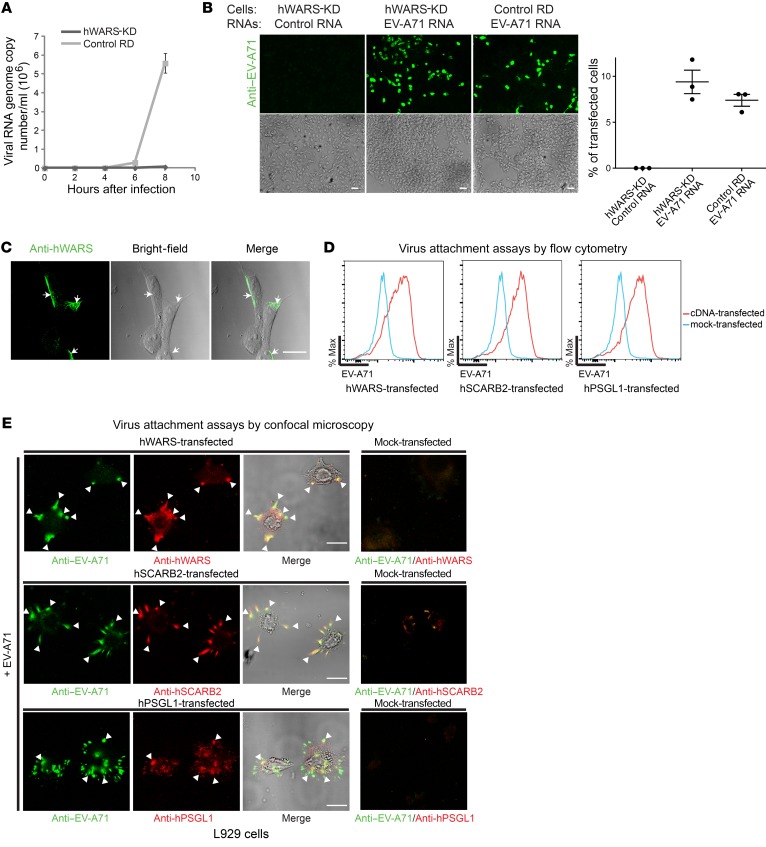
Although hWARS was first recognized as an aminoacyl-tRNA synthetase catalyzing the attachment of tryptophan onto cognate tRNA, its additional roles in angiogenesis, cytoskeletal reorganization, and shear stress–responsive gene expression have also been reported (11–16). Because EV-A71 has a positive-stranded RNA genome that can serve as a template for the synthesis of both viral proteins and RNAs, we anticipated that both processes would be affected if hWARS plays a role in EV-A71 replication. Indeed, we found that both viral RNAs and proteins were significantly reduced in hWARS-KD cells (Figure 1, B, G, and C, D, respectively). Consistent with a previous study on the replication kinetics of EV-A71 (17), our results showed an increase in viral RNA production as early as 6 hours after infection in RD cells. However, this was completely abrogated in hWARS-KD cells (Figure 2A). To identify the point of interaction between EV-A71 and hWARS, we transfected purified EV-A71 RNAs into hWARS-KD RD cells and control RD cells. We detected similar levels of cytoplasmic expression of viral proteins in both cells, indicating that the downstream machinery of EV-A71 replication remained intact (Figure 2B and Supplemental Figure 3A).
Figure 2. hWARS affects early EV-A71 replication.
(A) Amount of EV-A71 RNA in hWARS-KD and control RD cells during early infection. (B) Immunostained EV-A71 protein in hWARS-KD cells transfected with control RNA or purified EV-A71 RNA and control RD cells transfected with EV-A71 RNA. Histogram shows the estimated percentages of transfected cells. (C) Confocal microscopy showing endogenous hWARS expression. RD cells were stained with mouse monoclonal anti-hWARS antibodies (green). Also shown are the bright-field image and the overlapping image of hWARS protein. Arrows indicate positively stained hWARS. (D) Flow cytometric assays of EV-A71 virus attachment to hWARS, hSCARB2, and hPSGL1. High-titer EV-A71 was incubated with nonpermissive L929 cells overexpressing hWARS, hSCARB2, and hPSGL1, separately, at 4oC for 2 hours, and then washed to remove unbound viruses before fixation for flow cytometric analyses. EV-A71–inoculated mock-transfected L929 cells were included as a negative control. Max, maximum. (E) Assays of EV-A71 virus attachment to hWARS, hSCARB2, and hPSGL1 by confocal microscopy. Immunostaining for EV-A71 (green), hWARS (red), hSCARB2 (red), and hPSGL1 (red) was performed. Arrowheads indicate the colocalization (yellow) of EV-A71 and surface proteins. Immunostaining for mock-transfected L929 cells was also included as a negative control. Data in A, B, and D represent the mean ± SEM of 3 independent experiments. Images shown in B, D, and E are representatives of 3 independent experiments. Scale bars: 80 μm.
If hWARS is important for EV-A71 entry, it should be possible to detect the surface expression of hWARS. To examine endogenous hWARS expression on an intact plasma membrane by confocal microscopy, we stained RD cells with anti-hWARS antibodies under nonpermeabilized conditions and observed a sharp and polarized spindle-like pattern of hWARS concentrated at the cell periphery (Figure 2C). The membrane association of hWARS was further demonstrated in the orthogonal view of the anti-hWARS–stained RD cells treated with 0.1% Triton X-100 (Supplemental Video 1). Although many cellular factors have been shown to modulate the efficiency of EV-A71 entry (18, 19), EV-A71 pathogenesis is generally attributed to the tissue expression patterns of 2 known cellular receptors: hSCARB2 and hPSGL1 (6, 7). Consistent with previous findings on the capacity of EV-A71 to bind to cellular receptors (20), our virus attachment assays by flow cytometry in nonpermeabilized transfected L929 cells also showed an increased capacity of EV-A71 to bind to the surface of hSCARB2- and hPSGL1-transfected L929 cells when compared with mock-transfected L929 cells (Figure 2D). Notably, L929 cells transfected with hWARS cDNA also showed a similarly strong EV-A71–binding capacity (Figure 2D). Confocal microscopic analyses further revealed the surface colocalization of EV-A71 on hWARS, hSCARB2, and hPSGL1 in transfected mouse L929 (Figure 2E) and human RD cells (Supplemental Figure 3B). These results suggest that hWARS is expressed on the cell surface to mediate direct interaction with EV-A71 for cell entry.
Direct interaction between hWARS and EV-A71 is required for host cell entry.
Next, we compared EV-A71 infectivity in L929 cells expressing hWARS, hSCARB2, and hPSGL1, respectively. We detected similar levels of viral protein in cell lysates harvested from EV-A71–inoculated L929 cells expressing either hWARS or hSCARB2, suggesting that both surface proteins could effectively sensitize nonpermissive cells to EV-A71 infection (Figure 3A). Despite the high binding capacity of hPSGL1 and EV-A71 observed in our virus attachment assays (Figure 2, D and E), we detected minimal to no EV-A71 viral proteins in cell lysates harvested from hPSGL1-expressing L929 cells (Figure 3A). Direct interaction between hWARS and EV-A71 was confirmed by immunoprecipitation of hWARS followed by pulldown of EV-A71. Endogenous hWARS from RD cells and overexpressed hWARS from hWARS-transfected L929 cells were first immunoprecipitated using resin-coupled anti-hWARS–specific antibodies. Pulldown assays were then performed by inoculating the immunocomplex with EV-A71. Our results showed that both endogenous hWARS (lane 2) and overexpressed hWARS (lane 4) were sufficient to pull down EV-A71 (Figure 3B). Next, we tested three EV-A71 clinical isolates for their ability to bind to resin beads coupled with immune complexes containing the hWARS protein and anti-hWARS antibodies. Consistently, the results showed that all of the tested clinical isolates were highly enriched in an immune complex containing both hWARS protein and anti-hWARS antibodies, but they did not bind to anti-hWARS antibodies alone (Figure 3C). The functional importance of hWARS for EV-A71 replication was further demonstrated by antibody blocking of surface hWARS. Various amounts of anti-hWARS antibodies were administered to RD cells prior to EV-A71 infection. We observed dose-dependent reductions of the viral loads and viral proteins in cells treated with increasing concentrations of anti-hWARS antibodies, suggesting that the anti-hWARS antibodies could hinder EV-A71 infection (Figure 3D; see Supplemental Figure 4A for treatment of the control antibodies). Reciprocally, we saturated the surface antigens of EV-A71 by preincubating the viruses with soluble recombinant hWARS proteins prior to infection. The results were in line with the findings of previous antibody-blocking experiments, in that reduced infectivity was also observed when the viruses were pretreated with increasing amounts of soluble recombinant hWARS proteins (Figure 3E; see Supplemental Figure 4B for treatment of the soluble control protein).
Figure 3. hWARS interacts directly with EV-A71.
(A) cDNA-transfected L929 cells were inoculated with high-titer EV-A71 (TCID50/ml = 190,000). Forty-eight hours after inoculation, the cell lysates were harvested for Western blot analyses using specific antibodies. Mock-transfected L929 and EV-A71–infected RD cells were included as positive and negative controls, respectively. (B) Immunoprecipitation (IP) of hWARS from RD or hWARS-transfected L929 cells followed by pulldown of EV-A71. Endogenous hWARS from RD cells (lane 2) and overexpressed hWARS from hWARS-transfected L929 cells (lane 4) were first separately immunoprecipitated by anti-hWARS antibodies. The complexes were then inoculated with EV-A71 overnight at 4oC. After washing away unbound viruses, the complexes were dissociated by anti–EV-A71 antibodies for Western blot (WB) analyses. Asterisks indicate nonspecific bands. (C) Pulldown of EV-A71 clinical isolates by recombinant hWARS protein. Three different clinical EV-A71 isolates were inoculated with recombinant hWARS protein immunocomplexes and detected using Western blot analyses as described in B. (D) Anti-hWARS antibody blockage of EV-A71 infection. Surface hWARS on RD cells was blocked with anti-hWARS antibodies for 1 hour before EV-A71 infection. Virus production in conditioned supernatants and virus protein expression in infected cell lysates are shown. (E) Saturation of EV-A71 virions by recombinant hWARS protein (ranging from 18.8 to 75.2 nM). EV-A71 was preincubated with recombinant hWARS protein before challenging the RD cells. Virus production and viral protein expression were measured as described in D. (F) Western blot analyses of EV-A71, hWARS, and hSCARB2 proteins in RD CRISPR/Cas9 (CRISPR) cell clones. Endogenous γ-tubulin was used as a loading control. The r value represents the Pearson’s correlation coefficient. Data in A–G are representative of 3 independent experiments, and data in D and E represent the mean ± SEM.
We further investigated the requirement of hWARS for effective EV-A71 infection by inactivating the expression of hWARS and hSCARB2 in RD cells using the CRISPR/Cas9 system (Figure 3F). However, we were unable to examine EV-A71 infectivity in hWARS CRISPR/Cas9–edited (CRISPR) RD cells, because the hWARS CRISPR RD cells exhibited slow-growing and lethal phenotypes (Supplemental Figure 5A). In contrast, no obvious deleterious growth effect was observed in hSCARB2 CRISPR cells. Upon EV-A71 infection, we detected lower levels of EV-A71 protein in hSCARB2 CRISPR RD cells compared with levels in the control RD cells (Figure 3F). Notably, variable levels of EV-A71 protein were observed in hSCARB2 CRISPR clones (clone 2–5) with undetectable hSCARB2 expression (Figure 3F), which prompted us to assess whether additional factor(s) contribute to the cells’ susceptibility to EV-A71. We performed Western blot analyses to determine whether hWARS was uniformly expressed in these hSCARB2 CRISPR clones. Interestingly, we found that hWARS expression levels were positively correlated with those of the EV-A71 protein detected in these hSCARB2 CRISPR clones (Figure 3F), suggesting that the expression of hWARS played an important role in the cells’ susceptibility to EV-A71 in the absence of hSCARB2. Among the different hSCARB2 CRISPR RD clones, clone 1, which expressed the highest levels of both hSCARB2 and hWARS, showed the greatest susceptibility to EV-A71 (Figure 3F). Apparently, EV-A71 may require hWARS expression for effective cell entry.
IFN-γ induces the expression and membrane translocation of hWARS, which subsequently sensitizes cells to infection.
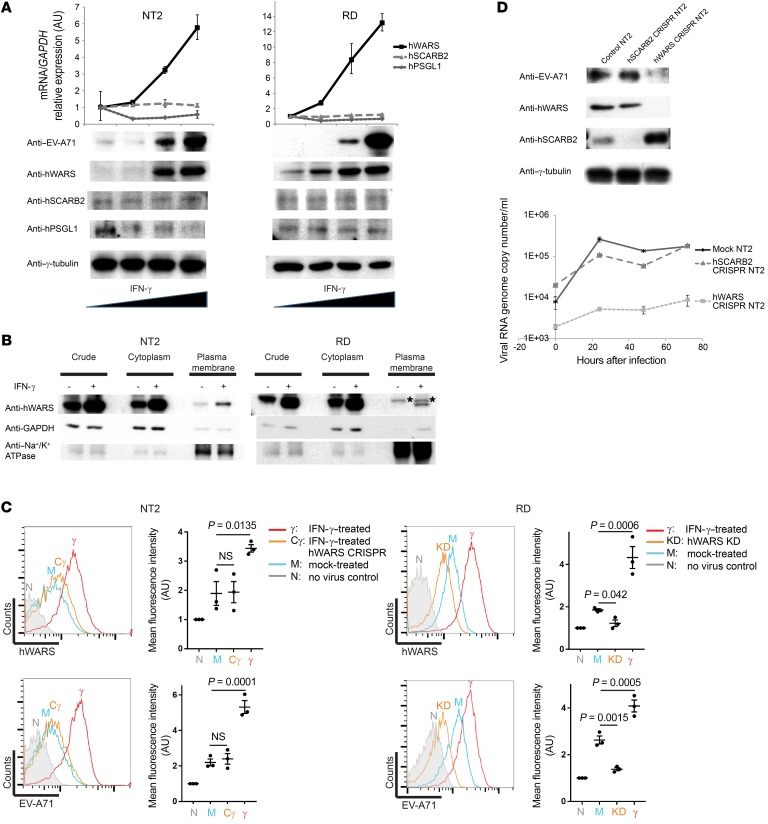
hWARS was previously shown to be upregulated by IFN-γ in several epithelial and fibroblast cell lines (21). We hypothesized that IFN-γ production elicited by innate immunity during primary infection could induce the expression of hWARS in certain cell types. To test this, we selected a human neuron-committed teratocarcinoma cell line (NT2), primary human neuron stem cells (HNSCs), and primary human bronchial epithelial cells (HBEpCs), because neuronal and pulmonary cells represent the most physiologically relevant cell models for studying EV-A71, which is known to be associated with severe neurological diseases and pulmonary edema. NT2 is known to be poorly susceptible to EV-A71 infection under unstimulated conditions. We stimulated NT2 cells, as well as RD cells with different doses of IFN-γ. Under low-titer infection conditions (50% tissue culture infectious dose [TCID50] per milliliter = 1), a small percentage of NT2 and RD cells were infected by EV-A71, as shown by the weak detection of viral protein using Western blot analysis (Figure 4A, lanes 1 and 5). When cells were cultured in the presence of IFN-γ, mimicking cytokine production as part of the host response to EV-A71 infection, we detected enhanced protein and mRNA expression of hWARS in an IFN-γ–dependent manner (Figure 4A; see Supplemental Figure 6, A and B for the expression of hWARS in response to treatment of proinflammatory cytokines in HNSCs and HBEpCs, respectively). More interestingly, we also observed plasma membrane translocation of hWARS upon IFN-γ stimulation (Figure 4B). The surface expression of hWARS in mock- and IFN-γ–treated NT2 and RD cells was further examined using flow cytometric analyses under nonpermeable conditions. We detected an increase in the surface expression of hWARS in both IFN-γ–treated cell types compared with their mock-treated counterparts (Figure 4C; top). Remarkably, the induced surface expression of hWARS correlated with an increase in the capacity of EV-A71 to bind to the IFN-γ–treated cells (Figure 4C; bottom).
Figure 4. Enhanced EV-A71 infection by IFN-γ–induced hWARS expression.
(A) Effect of IFN-γ on EV-A71 infection. NT2 and RD cells treated with different doses of IFN-γ (0, 10, 50, and 100 U/ml) were infected with low-titer EV-A71 (TCID50/ml = 1). Forty-eight hours after infection, mRNAs and proteins of the treated cells were harvested for qRT-PCR and Western blot analyses, respectively. Endogenous GAPDH and γ-tubulin were used as loading controls. (B) Induced expression and plasma membrane translocation of hWARS after treatment with IFN-γ. NT2 and RD cells were treated with 100 U/ml IFN-γ (+) or mock treated (–) for 72 hours, followed by subcellular fractionation. hWARS protein expression in the cytoplasm and plasma membrane fractions was detected using anti-hWARS antibodies. Total crude extracts were included. Cytosolic GAPDH and the plasma membrane protein sodium potassium ATPase (Na+/K+ ATPase) were detected to confirm the cytoplasmic and plasma membrane fractions, respectively. The asterisks indicate nonspecific bands. (C) Flow cytometric analyses of the surface expression of hWARS and EV-A71 attachment to NT2 and RD cells. (D) Western blot analyses of EV-A71, hWARS, and hSCARB2 proteins in the NT2 CRISPR cell clones. Endogenous γ-tubulin was used as a loading control. A time-course infection experiment monitoring the viral RNA genome within the NT2 CRISPR cell clones after the EV-A71 challenge was conducted using qRT-PCR. Data in A and D represent the mean ± SEM of 3 independent experiments. Results shown in A–D are representative of 3 independent experiments. Statistical significance was determined by 1-way ANOVA with Bonferroni’s post hoc test.
Next, we studied the effect of hWARS on EV-A71 infection in NT2 cells. The expression of hWARS in NT2 cells was first targeted using the CRISPR/Cas9 system followed by EV-A71 infection under IFN-γ–stimulated infection conditions. In contrast to CRISPR RD cells, we found that reduced expression of hWARS in NT2 cells did not result in obvious growth defects (Supplemental Figure 5B). Interestingly, we observed a marked decrease in EV-A71 protein levels in hWARS CRISPR NT2 cells when compared with levels in the control NT2 cells (Figure 4D). As a control, hSCARB2 CRISPR NT2 cells were also generated, and their EV-A71 infectivity was examined in parallel. Consistent with the CRISPR RD cell data (Figure 3F), suppression of the hSCARB2 in NT2 cells did not result in a detectable change in EV-A71 infectivity when compared with control NT2 cells (Figure 4D). The high responsiveness of hWARS expression in neuron-committed NT2 cells upon IFN-γ treatment and the requirement of hWARS for effective EV-A71 infection could indicate the existence of previously neglected cell types that could support enterovirus replication under certain conditions and may relate to EV-A71 pathogenesis.
hWARS sensitizes mouse cells to EV-A71 in vivo.
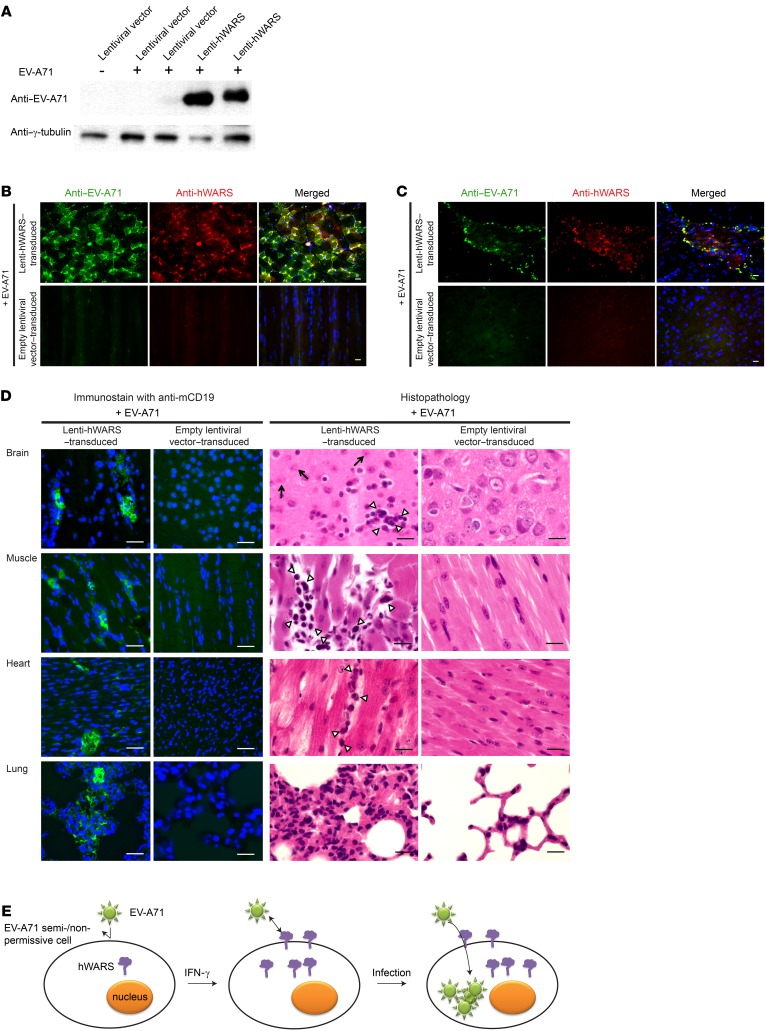
To confirm the functional importance of hWARS for EV-A71 infection in vivo, we developed a mouse model that overexpressed hWARS using a lentiviral vector (lenti-) expression system. We confirmed successful gene delivery by detection of hWARS expression in lenti-hWARS–transduced L929 cells (Supplemental Figure 7). Next, we transduced neonatal BALB/c mice with 106 copies of lenti-hWARS or an empty lentiviral vector via simultaneous intraperitoneal, intracerebral, and subcutaneous injections. hWARS was allowed to be overexpressed for 5 days before the mice were challenged with EV-A71. As early as 5 days after EV-A71 inoculation, we detected the presence of EV-A71 proteins and RNAs in the skeletal muscles and brains of mice transduced with lenti-hWARS (Figure 5, A–C). In contrast, viral proteins were almost undetectable in the control mice transduced with the empty lentiviral vector followed by EV-A71 inoculation. Pathologically, we observed interstitial infiltration of inflammatory cells into various organs including the brain, muscle, heart, and lungs (Figure 5D; left). We also observed the degeneration of neurons in brain tissue, which is consistent with the ataxia and paralysis phenotype of the EV-A71–inoculated lenti-hWARS–transduced mice (Figure 5D; right). These findings recapitulate the neurological symptoms associated with EV-A71 infection in humans (Supplemental Figure 8 and Supplemental Video 2). We observed no abnormality in EV-A71–inoculated lentiviral vector–transduced mice (Supplemental Video 3 and Figure 5D; right, mock-infected brain tissue).
Figure 5. EV-A71 infection in mouse cells overexpressing hWARS.
(A) Western blot analyses of EV-A71 protein in the skeletal muscles of EV-A71–challenged neonatal BALB/c mice pretransduced with empty lentiviral vectors (lanes 2 and 3) or a lentiviral vector expressing hWARS (lanes 4 and 5). Two representative samples were included in each category. A noninfected empty lentiviral vector (lane 1) was included as a control. Representative images of muscle (B) and brain (C) tissues from hWARS-transduced mice. Tissues from EV-A71–infected mice were immunostained with anti–EV-A71 (green) and anti-hWARS antibodies (red), respectively. The tissues from empty lentiviral vector–transduced mice inoculated with EV-A71 served as negative controls. DAPI-stained nuclei (blue) are also shown in the merged images. (D) Histopathology of the EV-A71–inoculated hWARS-transduced mice on day 5 after infection. Interstitial infiltrations of lymphocytes in various organs were detected using anti-CD19 antibodies (green). The nuclei were stained with DAPI (blue). H&E staining showed the inflammatory infiltrates of mononuclear cells (arrowheads) and degenerated neuronal cells (arrows). (E) Schematic representation of an IFN-γ–inducible cell entry model for EV-A71 infection. EV-A71 semi- or nonpermissive cells show low or no surface expression of hWARS in a quiescent state (left). The proinflammatory response leading to the production of IFN-γ can induce the expression and membrane translocation of hWARS. This in turn sensitizes poorly susceptible cells to EV-A71 infection by facilitating hWARS–EV-A71 interaction (middle). As a result, EV-A71 can be internalized into the cells, and infection occurs. The images shown in B–D are representative of 3 independent experiments. Scale bars: 30 μm.
Although mice are known to be semi-/nonpermissive to EV-A71 infection, qRT-PCR measurement of the mouse homolog of WARS (mWARS) revealed that its expression pattern was highly correlated with the tissue tropism and pathogenesis of EV-A71, in which relatively high levels of mWARS expression were observed in the intestine, lungs, and liver (Supplemental Figure 9A). Notably, under a quiescent state, high expression levels of mWARS in the brain were detected, which was also correlated with neurological complications of EV-A71 infection (Supplemental Figure 9A). In line with the results obtained in human cell line models, the present results also show that the expression of mWARS can be induced by IFN-γ in murine L929 cells (Supplemental Figure 9B).
Discussion
We have identified a new entry factor, hWARS, for EV-A71 infection, which is an important causative agent of HFMD and can result in severe neurologic complications leading to paralysis or death. EV-A71 is able to enter host cells using different cellular receptors, resulting in broad tissue tropism. Previous EV-A71 studies have focused on 2 receptors: hPSGL1 and hSCARB2. hPSGL1, a leukocyte-specific membrane protein, is involved in leukocyte interactions with vascular endothelium (22). The absolute requirement of hPSGL1 for EV-A71 cell entry is not known, because it is primarily expressed in hematopoietic cells, and it did not support all of the EV-A71 strains tested for infection (7). hSCARB2, in contrast, has been proposed as the major receptor for systemic EV-A71 infection, given its relatively robust expression in the lysosomes and endosomes of many cell types (6, 23), and served as a receptor for all of the tested EV-A71 strains (8). Yamayoshi et al. previously reported that other enteroviruses, including CV-A7, CV-A14, and CV-A16, which are highly related to EV-A71, can use hSCARB2 for infections (8). In this study, we compared the functional role of hWARS with that of hSCARB2 and hPSGL1. First, we generated hWARS-KD and hWARS CRISPR cell lines and examined the requirement of hWARS for EV-A71 infection (Figure 1, A–G and Figure 4F). Upon EV-A71 inoculation, we observed a significant reduction in virus replication and CPEs in hWARS-KD cells compared with the corresponding parental cells (Figure 1, F and G). Generally, a higher amount of virus production results in greater CPEs. However, factors including the virus strain and the infected cell type could contribute to the variability of CPEs. Previous studies have suggested that different enterovirus-specific proteins play regulatory roles in virus-induced CPEs (24). Host-specific factors that regulate apoptotic and cytokine/chemokine production pathways may also modulate CPEs and virus production independently (25, 26). We measured both the virus-induced CPEs (Figure 1F) and the relative amount of virus production (Figure 1G) in control RD and hWARS-KD RD cells. Our results showed that cells protected from virus-induced CPEs in the hWARS-KD cells produced fewer viruses in hWARS-KD cells than did their control counterparts. The results are consistent with the positive correlation between the level of virus production and the degree of the CPE on infected cells. We were consistently unable to detect viral protein in either hWARS-KD or hWARS CRISPR cells after EV-A71 challenge (Figure 1, C and D, and Figure 4D). Recently, CRISPR/Cas9 technology has proven useful in identifying new host cell entry factors or receptors for virus entry, such as the anthrax toxin receptor 1 for the Seneca Valley virus (27). Therefore, CRISPR/Cas9 was also used in this study to confirm the role of hWARS for EV-A71 cell entry. Overall, our loss-of-function experiments using shRNA and CRISPR/Cas9 consistently showed that hWARS is important for EV-A71 infection.
The importance of hWARS during EV-A71 infection was further demonstrated by gain-of-function experiments. As with hSCARB2, overexpression of hWARS sensitized nonpermissive mouse L929 cells to EV-A71 infection (Figure 3A). To facilitate effective cell entry, viruses need to bind to the cell-surface proteins. Our virus attachment assays analyzed by flow cytometry (Figure 2D and Figure 4C) and confocal microscopy (Figure 2, C and E) showed that EV-A71 can attach to cells transfected with hWARS, hSCARB2, or hPSGL1. In vitro pulldown experiments further showed that the direct interactions between EV-A71 and hWARS occurred not just in one particular isolate, but also in other clinical isolates (Figure 3C). We further examined the importance of hWARS in EV-A71 infection by suppressing the expression of hSCARB2 in permissive RD cells. Interestingly, we found that these hSCARB2 CRISPR RD clones expressing various amounts of hWARS had variable susceptibilities to EV-A71 (Figure 3F). The amount of EV-A71 proteins in these clones was positively correlated with the expression level of hWARS, indicating the important role of hWARS in EV-A71 infection, even in the absence of hSCARB2 (Figure 3F). One possible explanation for the heterogeneity of the CRISPR RD cells could be their hyperdiploid karyotypes (>50 chromosomes) (28), which makes the targeting of gene expression by CRISPR/Cas9 challenging. Next, we studied the effect of hWARS on EV-A71 infection by generating hWARS CRISPR RD cells using the CRISPR/Cas9 system. Surprisingly, the RD cells did not survive after the expression of hWARS was suppressed by CRISPR/Cas9 (Supplemental Figure 5A). This deleterious growth effect, however, was not observed in either hWARS CRISPR NT2 cells or hWARS-KD RD cells (Supplemental Figure 5A and Supplemental Figure 2, A and B). We speculate that the residual expression of hWARS in the hWARS-KD RD cells (~10% that of the parental RD cells) is sufficient to support cell survival. Because RD cells (doubling time = 0.97 ± 0.08 days; ref. 29) replicate faster than do NT2 cells (doubling time = 2.29 ± 0.21 days; ref. 30), higher hWARS expression may be required for RD cells to maintain their basal metabolic activity. The differential requirement of hWARS in various cell types, such as muscle cells and neuronal cells, might prevent the identification of important entry factors similar to hWARS in other EV-A71 studies. Overall, our gain-of-function experiments in nonpermissive mouse L929 cells and lentiviral vector–transduced mice suggest that overexpression of hWARS alone is sufficient to sensitize cells to enterovirus infection (Figure 3A and Figure 5, A–C, respectively). Furthermore, our shRNA and CRISPR/Cas9 loss-of-function experiments showed that the enterovirus infectivities were significantly reduced in hWARS-KD/CRISPR cells (Figure 1, A–G, and Figure 4D). Confirmation of hSCARB2 expression in these cells by Western blot analyses (Figure 3F and Figure 4D) further suggested that hWARS may function independently of hSCARB2 in mediating enterovirus entry.
hWARS is an IFN-γ–inducible entry factor for EV-A71 and is able to sensitize poorly susceptible neuronal cells to EV-A71 infection. Previous studies showed that hWARS could be upregulated by IFN-γ in several epithelial and fibroblast cell lines (21). In this study, we demonstrated the expression of hWARS in response to IFN-γ in both neuronal (NT2) and muscle (RD) cells. Unlike RD cells, which are highly susceptible to EV-A71 infection, the inclusion of poorly susceptible NT2 cells allowed us to further examine whether the expression of hWARS can sensitize NT2 cells to EV-A71 infection. As shown in Figure 4, A and C, enhanced expression of hWARS was detected in both cells treated with increasing doses of IFN-γ, whereas the expression of hSCARB2 and hPSGL1 was not enhanced by the addition of IFN-γ (Figure 4A). A significant increase in mRNA and protein levels of EV-A71 was confirmed by qRT-PCR and Western blot analyses, respectively, supporting the idea that enhanced expression of hWARS induced by IFN-γ could sensitize poorly susceptible NT2 cells to EV-A71 infection (Figure 4A). Although it is known that proinflammatory cytokines induced by the early innate immune response often help to control virus replication and prime humoral and cell-based adaptive immune responses to combat viral infection, our study provides the first evidence to our knowledge that high levels of proinflammatory cytokines such as IFN-γ can also trigger the expression of host factors that in turn favor viral infection. Notably, a previous study showed that a subset of proinflammatory cytokines, including IFN-γ, was elevated in proportion to the severity of EV-A71 infection (31). These clinical observations corroborate our present findings, which showed that IFN-γ may alter the susceptibility to EV-A71 of certain cell types, such as neuronal cells, causing severe neurological infections. Furthermore, 2 recent studies showed that deregulation of hWARS expression was linked to neurodegenerative disorders, in which perturbation of hWARS activity by competitive inhibition of tryptamine, a decarboxylated tryptophan analog, induced neurodegeneration accompanied by subcellular redistribution (32). Because we and others have shown that hWARS expression and subcellular localization can be modulated in response to IFN-γ stimulation (Figure 4, A and B), it is possible that the alteration of hWARS expression could contribute to disease progression in EV-A71 pathogenesis.
The role of hWARS in virus entry was confirmed in our mouse infection model, with clinical progression compatible with EV-A71 infections in humans, including those associated with severe neurological disorders. It is known that current in vivo animal models do not fully recapitulate human diseases caused by EV-A71 (9). In this study, we attempted to overexpress hWARS using a lentiviral vector expression system followed by EV-A71 challenge in neonatal mice. Phenotypically, on day 5 after challenge, we observed that these challenged lenti-hWARS–transduced mice developed ruffling of the fur and bilateral rear limb paralysis with marked difficulty in walking, compatible with symptoms of neurological disorders (Supplemental Video 2). These symptoms were in line with the histopathological results, which indicated a degeneration of neuronal cells (arrows) in brain tissue (Figure 5D; right; EV-A71–infected brain tissue). The cause of damage in the CNS could be due to the virus-induced cytokine effect during hippocampal infection, as confirmed by the presence of EV-A71 proteins (Figure 5C). Indeed, similar pathological changes caused by coxsackievirus B3 hippocampal infection have been documented (33). Most interestingly, a recent report on the identification of the EV-D68 receptor suggested that hippocampal neurons are susceptible to EV-D68 (34). It should be noted that induced expression of hWARS could be detected on the plasma membrane of hippocampal neurons in a disease model (32). The upregulated plasma membrane expression of hWARS in disease and IFN-γ–stimulated models supports the notion that EV-A71 might target a specific type of neuron through hWARS-mediated cell entry. Though we cannot draw any correlation between tissue expression of specific receptors and viral tissue tropism in the genetically engineered mouse model used in this study, further examination of brain tissue from deceased patients with enterovirus encephalitis is warranted.
Like other studies using mouse models to study EV-A71 infection (9), we detected high levels of EV-A71 protein in the skeletal muscles of the hWARS-transduced mice (Figure 5, A and B). Indeed, a mechanism called “provocation poliomyelitis” has been proposed for enteroviral encephalitis (35). The mechanism suggests that skeletal muscle injury can induce retrograde axonal transport of the enterovirus and thereby facilitate viral invasion of the CNS and the progression of spinal cord damage. Another nonmutually exclusive mechanism for the enterovirus neuroinvasion is the “Trojan horse” hypothesis (36). An elegant study performed by Mena et al. in 1999 showed that CV-B3 was able to infect and replicate in B cells (36). They proposed that the B cell infection not only facilitated the dissemination of the virus, but also mediated immune escape by hiding within mobile host cells. Our immunostaining data showed strong B cell infiltration into various organs of our challenged lenti-hWARS mouse model (Figure 5D, left). Future studies of the effect of hWARS on B cell enteroviral infection could provide new insights into EV-A71 dissemination.
We have proposed an IFN-γ–inducible cell entry model that may partly explain the severe clinical outcomes due to EV-A71 and other severe enteroviral infections in humans (Figure 5E). A strong proinflammatory response in patients with severe EV-A71 infections leads to the production of IFN-γ, which induces hWARS expression in cells. This in turn sensitizes semi- or nonpermissive cells such as neuronal cells to EV-A71 infection. Our proposed model may also explain the anecdotal success of intravenous Ig in treating severe EV-A71 infection through downmodulation of the cytokine storm, including IFN-γ and thus hWARS (37). Alternatively, potential treatment options for severe EV-A71–associated neurological complications include blocking the accessibility of hWARS by specific antibodies or the use of recombinant proteins.
Methods
Cell lines.
The NT2 (American Type Culture Collection [ATCC]; CRL1973), RD (ATCC; CCL-136) cell lines and their derivative cell clones were maintained in DMEM (Life Technologies) supplemented with 10% (v/v) heat-inactivated FCS (HyClone), 100 units/ml penicillin, and 100 μg/ml streptomycin (Life Technologies, Thermo Fisher Scientific). Cells were selected and maintained in the same medium as that described above, with the addition of 1 μg/ml puromycin. L929 cells (ATCC; CCL-1) were maintained in RPMI-1640 medium (Life Technologies, Thermo Fisher Scientific) supplemented with FCS and antibiotics. HBEpCs (Cell Applications Inc.; 502-05a) were cultured in Bronchial Epithelial Cell Growth Medium (Cell Applications Inc.; 511-500). HNSCs (Cell Applications Inc.; HS820-20f) were grown in neurosphere form and maintained in HNSC Growth Medium (Cell Applications Inc.; 813-250). To differentiate the HNSCs into neurons, 50,000 HNSCs were seeded into a 24-well plate well precoated with Poly-D-Lysine with Laminin Coating Solution (Cell Applications Inc.; 127-25). The HNSCs were allowed to differentiate in HNSC Differentiation Medium (Cell Applications Inc.; 813D-250) for 3 weeks before cytokine treatments. All cell lines used were confirmed to be free of mycoplasma contamination using the Plasmo Test (InvivoGen). The identities of RD cells and their derivatives were confirmed by the service provider PANGENIA for human short tandem repeat (STR) profiling, followed by matching with the ATCC database.
Mice.
Five-day-old female BALB/c mice were obtained from the Laboratory Animal Unit of the University of Hong Kong. The animals were housed in specific pathogen–free facilities under a 12-hour light/12-hour dark cycle, together with lactating mothers. All animal-related experiments were performed according to standard procedures.
Production of the shRNA library and stable clones.
A feline immunodeficiency virus–based shRNA library was produced, as we described previously (10). Briefly, 2 μg shRNA library in lentiviral constructs (SBI) were cotransfected with 10 μg pPACK packaging plasmid mix into HEK293T cells (ATCC; CRL-1573) using Lipofectamine and Plus reagents (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s protocol. Twenty-four and seventy-two hours after transfection, the conditioned culture media were collected to harvest the packaged viruses. GFP-packaged viruses were produced in parallel. The MOI of the viruses was estimated on the basis of the percentage of GFP-transduced RD cells (Supplemental Figure 10). The shRNA library packaged viruses (MOI = 0.1) were used to transduce 3 × 108 RD cells. After 8 hours of exposure to virus, cells were washed twice with PBS and resuspended in RPMI-1640 Medium (Life Technologies, Thermo Fisher Scientific). One week after transduction, the cells were selected for 3 weeks in growth medium containing 1 μg/ml puromycin. For independent validation, shRNA-expressing vector constructs (Mission shRNA) were purchased from MilliporeSigma. These shRNA-packaged viruses were generated in HEK293T cells using ViraPower Lentiviral Expression Systems (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s protocol.
shRNA-based screening.
shRNA-transduced RD cell clones were challenged with EV-A71 (TCID50/ml = 190,000). Four weeks after infection, small RNAs were harvested using a mirVana Small RNA Isolation Kit (Ambion, Thermo Fisher Scientific). cDNA synthesis and labeling were performed as described in the manufacturer’s protocol. Briefly, 5 μg total RNA was reverse transcribed into cDNAs in the presence of 10 μM cDNA synthesis GNF primer (5′-ATTTATTGTATCTGTGGGAGCCTC-3′), 100 mM dithiothreitol, 10 mM each dNTP and 1× RT buffer, and 200 U SuperScript III Reverse Transcriptase (Invitrogen, Thermo Fisher Scientific). The reaction mixtures were incubated at 42°C for 1 hour. The reaction was stopped by heat inactivation at 72°C for 5 minutes. Half of the reaction mixture was then transferred to a tube containing 1× PCR reaction buffer, 20 mM dNTP, 20 μM forward GNF primer (5′-TGCATGTCGCTATGTGTTCTGGGA-3′) and 20 μM reverse GNF primer (5′-ACAAAGCACTGGAAGCTATCGAA-3′), and Advantage HF2 Taq Polymerase (Clontech). PCR amplification of the shRNA target region was performed under the following conditions: (step 1) 94°C for 4 minutes; (step 2) 94°C for 30 seconds, and then 68°C for 1 minute; (step 3) step 2 was repeated for 20 cycles; and (step 4) 68°C for 3 minutes. An aliquot was made by transferring 1 μl of the first-round PCR products into 4 new tubes, each containing 1× PCR reaction buffer, 20 mM dNTP, 20 μM each NRev GNF universal primer (5′-AAAGAATGCTTATGGACGCTAGAA-3′) and NFwd-Bio primer (Biotin-5′CTTCCTGTCAGA-3′), and Advantage HF2 Taq Polymerase (Clontech). PCR for biotin labeling was performed under the following conditions: (step 1) 94°C for 2 minutes, 50°C for 2 minutes, and then 68°C for 1 minute; (step 2) 94°C for 30 seconds and then 68°C for 30 seconds; (step 3) step 2 was repeated for 18 cycles; and (step 4) 68°C for 3 minutes. The PCR products were then gel purified using a QIAquick PCR Purification Kit (QIAGEN) as described in the manufacturer’s protocol. After purification, the PCR product was treated with Lambda Exonuclease (New England BioLabs) at 37°C for 2 hours to remove the nonbiotinylated strand. The digested products were purified as described above and then quantified. Ten micrograms of the purified products was hybridized on the GeneChip Human Genome U133+2 Array (Affymetrix) using Affymetrix hybridization buffer followed by staining with streptavidin phycoerythrin (Molecular Probes). The image of the processed chip was captured with a scanner controlled by Affymetrix GCOS software according to the standard Affymetrix protocol. The signals were summarized for replicates with 2 or more replicate values above 100. The fold changes were determined on the basis of the ratio of the signals between the mock and infected samples. Data were deposited en toto in the NCBI’s GEO database (GEO GSE80407). Pathway analysis was performed using the Ingenuity Pathway Analysis program (QIAGEN).
qRT-PCR analyses.
Total RNA was isolated using a mirVana miRNA Isolation Kit (Ambion, Thermo Fisher Scientific). The RNAs were first quantified with a NanoDrop Spectrophotometer (ND-1000; Thermo Fisher Scientific). Total RNA (1 μg) was reverse transcribed as described above. qRT-PCR was performed as described previously (38), using FS Universal SYBR Green Master (Rox) Reaction Mix (Roche Life Science) with the temperature-cycling condition of 15 seconds at 95°C followed by 1 minute at 60°C for 55 cycles in a 7900 Real-Time PCR System (Applied Biosystems). The cDNAs of EV-A71–infected samples were quantified using 300 nM each of forward and reverse specific primers (5′-CCCCTGAATGCGGCTAATCC-3′ and 5′-ACACGGACACCCAAAGTAGT-3′) (39, 40). Forward and reverse specific primers (5′-GCCCCTGAATGCGGCTAAT-3′ and 5′-ATTGTCACCATAAGCAGYCA-3′) and a probe (5′-FAM- CGGACACCCAAAGTAGTCGGTTCCG –lABkFQ-3′) were used for the detection of different enterovirus serotype samples. A plasmid with the target 5′ noncoding region (NCR) sequence of 129 bp was used to generate the standard curve. Expression of the housekeeping gene GAPDH was also measured as an internal control using the following primers: (forward) 5′-TCACCACCATGGAGAAGGC-3′ and (reverse) 5′-GCTAAGCAGTTGGTGGTGCA-3′. The expression levels of host gene candidates were quantified using sequence-specific fluorescent DNA probes (Thermo Fisher Scientific; WARS; Hs00188259_m1). After reverse transcription, the cDNAs were added to a reaction mixture containing TaqMan Universal PCR Master Mix (Applied Biosystems) and TaqMan Gene Expression Assay Mix (inventoried for the corresponding genes; Applied Biosystems). qRT-PCRs were performed under the same conditions as those described above.
CPE measurement.
The CPEs of unfixed, unstained infected cells were measured using an optical microscope, with the condenser down and the iris diaphragm partly closed as described in “Cytopathic Effects of Viruses Protocols,” by Suchman and Blair (41). Total detachment of the monolayer cell was considered to be 100%. To determine the percentage of CPEs, the RD cell line and its derivative shRNA cell clones were inoculated with enterovirus at a viral titer MOI of 1. The virus-induced CPEs were monitored daily.
Confocal and fluorescence microscopic analyses.
RD cells, their derivative KD cells, and L929 cells were fixed in 4% paraformaldehyde with or without 0.1 % Triton X-100. After 1 hour of blocking with 3% BSA at room temperature, the cells were stained with anti–EV-A71 antibody (monoclonal antibody recognizing the amino acid sequence of VP2; MilliporeSigma; MAB979), and/or anti-hWARS antibodies (monoclonal antibody recognizing amino acids 50–150 of hWARS; Abcam; ab58054), and/or anti-CD19 (BD Pharmingen; 550284) for 1 hour at room temperature. Unbound antibodies were washed away with 6× PBS. Positively stained cells were detected by secondary IgG (H+L) antibodies conjugated with either Alexa Fluor 488 or Alexa Fluor 594 (Life Technologies, Thermo Fisher Scientific; A11001 and A11012, respectively) for 30 minutes at room temperature. After washing 6 times with PBS, the stained cells were mounted onto glass slides with VECTASHIELD mounting medium with DAPI (Vector Laboratories) and examined with a Leica TCS-NT microscope (Leica Microsystems) or a LSM700 confocal microscope (Zeiss). For the 2-color immunofluorescence experiments, samples were immunostained with an antibody mix that contained 2 primary antibodies generated from different species under the same conditions as those described above. Backgrounds generated from the secondary antibodies were accessed by immunostaining samples with isotype IgG antibodies instead of the cognate primary antibodies. The specificities of the primary antibodies were confirmed by immunostaining of mouse L929 cells transfected with cDNA that expressed human proteins. Mock-transfected L929 cells were also immunostained for comparison.
For animal tissue stainings, tissue sections were first deparaffinized and rehydrated, followed by treatment with Antigen Unmasking Solution (Vector Laboratories; H-3300) to retrieve the antigens according to the manufacturer’s instructions. Detection of antigens was performed under the same conditions as those described above.
Flow cytometric analyses.
RD cells, their derivative KD cells, and L929 cells were trypsinized followed by fixation in 4% paraformaldehyde. After washing with PBS to remove the fixative, the cells were stained with anti–EV-A71 antibodies (MilliporeSigma; MAB979) and/or anti-hWARS (Abcam; ab58054), anti-hSCARB2 (Abcam; ab16522), or anti-hPSGL1 (Abcam; ab66882) antibodies for 1 hour at room temperature. Unbound antibodies were washed away once with PBS. Positively stained cells were detected by secondary IgG (H+L) antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 594 (Life Technologies, Thermo Fisher Scientific; A11001 and A11012, respectively) for 30 minutes at room temperature. Flow cytometric analyses were performed using a BD LSR Fortessa Analyzer, and data were analyzed using FlowJo vX software (Tree Star).
Virus infection, antibody and antigen blocking, and virus attachment assays.
Information on the viruses used in this study can be found in Supplemental Table 2. Unless otherwise specified, EV-A71 infection was performed by first inoculating 5 × 105 cells with EV-A71 at a titer of TCID50/ml = 190,000 for 1 hour. Unbound viruses were removed by washing with 3× PBS. After replenishment of the cell growth medium containing 1% FCS, virus replication and CPEs were monitored at various time points after the infected cells were reincubated in 5% CO2 at 37°C.
The neutralization antibody assays were performed by first diluting the anti-hWARS (Abcam; ab31536) or control antibodies in 500 μl DMEM. The antibody mix (250 μl) was then added into 250 μl DMEM containing 5 × 104 RD cells in a 24-well plate 1 hour before the cells were challenged with EV-A71 at a titer of TCID50/ml = 19,000, as described above. The conditioned media and the cell lysates were harvested 1 day after infection for TCID50 and Western blot analyses.
The antigen-blocking assays were performed by premixing EV-A71 (TCID50/ml = 19,000) with soluble recombinant hWARS or control protein in 500 μl DMEM at 4oC overnight with shaking. The next day, infection experiments were performed by replacing the culture media of 5 × 104 RD cells in a 24-well plate with the mixtures for infection, as mentioned above. The conditioned media and the cell lysates were harvested 1 day after infection for TCID50 and Western blot analyses.
L929 cells transfected with hWARS, hSCARB2, hPSGL1, or an empty plasmid control were mixed with high-titer EV-A71 (TCID50/ml = 190,000), separately, at 4°C for 2 hours. Unbound viruses were removed by washing 3 times with ice-cold PBS. The cells were then fixed in 4% paraformaldehyde for 15 minutes. After 1 hour of blocking with 3% BSA at room temperature, the cells were costained with anti–EV-A71 (MAB979; MilliporeSigma) and anti-hWARS (Abcam; ab58054), anti-hSCARB2 (Abcam; ab16522), or anti-hPSGL1 (Abcam; ab66882) antibodies, as described in Confocal and fluorescence microscopic analyses in Methods.
Western blot analyses.
Cell lysates or tissue extracts were resolved by SDS-PAGE. Proteins were transferred onto a polyvinylidene difluoride membrane by electroblotting (Hoefer) at a constant current of 150 mA overnight. Detection of antigens was performed using anti–EV-A71 (MAB979; MilliporeSigma), anti-hWARS (Abcam; ab58054), anti-hSCARB2 (Abcam; ab16522), and anti-hPSGL1 (Abcam; ab66882) antibodies. For loading controls, the membranes were stripped with Restore Western blot stripping buffer (Pierce, Thermo Fisher Scientific; 21059) before reprobing with anti–γ-tubulin (MilliporeSigma; T6557), anti-Na+/K+-ATPase (Abcam; ab76020), and anti-GAPDH (Cell Signaling Technology; 2118).
In vitro pulldown assays.
Recombinant hWARS proteins were incubated with EV-A71. Both recombinant hWARS and EV-A71 were mixed in the presence of 0.1% Tween-20 at 4°C overnight with shaking. The next day, the anti-hWARS antibodies (Abcam; ab58054) and agarose-conjugated secondary IgG (H+L) antibodies (Abcam; ab97025) were added to the mixture and allowed to incubate for an additional 2.5 hours. After washing 6 times with buffer containing 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0, agarose-bound proteins were fractionated by SDS-PAGE and detected by Western blotting using anti-hWARS (Abcam; ab58054) and anti–EV-A71 (MilliporeSigma; MAB979) antibodies.
Subcellular fractionation.
Subcellular fractionation of RD and NT2 cells was performed using the Plasma Membrane Protein Extraction Kit (Abcam). Briefly, 108 cells were harvested by scraping. The cells were washed once with 3 ml PBS and then lysed in 1 ml homogenizing buffer mix on ice by passing through a syringe 10 times. The supernatants were collected after sedimentation at 700 g for 10 minutes at 4°C. The cytoplasmic fraction (supernatant) and the total cellular membrane (pellet) fractions were further separated by centrifugation at 10,000 g for 30 minutes at 4°C. Plasma membrane proteins were extracted by resuspending the total cellular membrane pellet in 200 μl of the upper-phase solution and 200 μl of the lower-phase solution. The complex was incubated on ice for 5 minutes before centrifugation at 1,000 g for 5 minutes at 4°C. The upper-phase solution was collected, and the steps were repeated by adding 100 μl of the lower-phase solution. The upper-phase solution was collected and combined with the previously harvested upper-phase solution. The resultant solution was then diluted in 5 volumes of water and kept on ice for 5 minutes. The plasma membrane proteins were collected by centrifugation of the diluted upper-phase solution at 10,000 g for 10 minutes at 4°C, and then 0.5 % Triton X-100 in PBS was added to dissolve the plasma membrane protein pellets for Western blot analyses.
Animal infection model.
The hWARS-expressing lentiviruses were generated using ViraPower Lentiviral Expression Systems (Invitrogen, Thermo Fisher Scientific), as described above. The virus titers were first successfully demonstrated in L929 cells (Supplemental Figure 7). Viruses (1 × 106) were concentrated by ultracentrifugation and then resuspended in 100 μl PBS for intranasal, intravenous, and intraperitoneal injections under anesthesia using ketamine (100 mg/kg) and xylazine (10 mg/kg), as described previously (42). Five days after transduction, the 10-day-old mice were again challenged with EV-A71 (TCID50/ml = 10,000) using the same routes of delivery. Three mice from each group were sacrificed to harvest their organs five days after virus inoculation.
Generation of CRISPR/Cas9 cell clones.
To clone the sgRNA guide sequence, the lentiCRISPR v2 plasmid (Addgene plasmid no. 52961; a gift of Feng Zhang, MIT, Cambridge, Massachusetts, USA; ref. 43) was cut and dephosphorylated with FastDigest BsmBI and FastAP (Fermentas) at 37°C for 2 hours. Oligonucleotides for the hWARS sgRNA (5′-CACCGCCAGCACCTACCAGTAATCA-3′ and 5′-AAACTGATTACTGGTAGGTGCTGGC-3′) and hSCARB2 (5′-CACCGTGTAGACCAGAGTATCGAGA-3′ and 5′-AAACTCTCGATACTCTGGTCTACAC-3′) (IDT) were phosphorylated separately using polynucleotide kinase (Fermentas) at 37°C for 30 minutes and then annealed by heating to 95°C for 5 minutes and cooling to 25°C at 1.5°C/minute. The annealed oligonucleotides were then ligated into a gel-purified lentiCRISPR v2 plasmid at 25°C for 5 minutes using T7 ligase (Enzymatics).
The hWARS-lentiCRISPR or hSCARB2-lentiCRISPR plasmids were cotransfected with the packaging plasmids pMD2.G and psPAX2 (Addgene; plasmids 12259 and 12260) into 293T cells for lentivirus production. Briefly, the transfection complex containing 10 μg hWARS-lentiCRISPR/hSCARB2-lentiCRISPR plasmid, 5 μg pMD2.G, 7.5 μg psPAX2, 100 μl Plus Reagent (Life Technologies, Thermo Fisher Scientific), and 50 μl Lipofectamine 2000 (Life Technologies) was allowed to inoculate with the 293T cells for 6 hours. At 60 hours after transfection, the lentiviruses were harvested from the conditioned media by centrifugation at 1,811 g at 4°C for 10 minutes and then passed through a 0.45-μm low-protein–binding membrane (MilliporeSigma). The purified lentiviruses were then transduced into RD or NT2 cells. On day 3 after transduction, 1 μg/ml and 0.5 μg/ml puromycin (MilliporeSigma) were added to the transduced RD and NT2 cells, respectively. The transduced cells were serially diluted into 96-well plates for clonal expansion. The inactivation of targeted gene expression in these clones was confirmed by Western blot analyses.
Study approval.
The mouse experiments were approved by the Committee on the Use of Live Animals in Teaching and Research, Hong Kong University (CULATR 3630–15) and the Department of Health of the Government of the Hong Kong Special Administrative Region under the Animals (Control of Experiments) Ordinance, Chapter 340 (15-235 in DH/HA&P/8/2/3 Pt.71).
Statistics.
Unless otherwise specified, all data were evaluated using a 2-tailed, unequal variance Student’s t test, and 1-way ANOVA with Bonferroni’s post hoc test. P values of less than 0.05 were considered significant. The statistical significance threshold was adjusted by Bonferroni’s correction for multiple comparisons.
Author contributions
MLY and KYY conceived, designed, and supervised the study. MLY and LJ performed most of the experiments with the help of CCYY, KHC, JPC, JLLT, CZ, AJZ, and WMW. SKPL, PCYW, and JYCL provided samples and analyzed the data. MLY, JLLT, SRS, and KYY wrote the manuscript. MLY, JLLT, SKPL, PCYW, JFWC, DYJ, JLLT, KHK, SRS, and KYY provided advice and contributed to data analysis and manuscript preparation. MLY, DYJ, SRS, and KYY secured funding and did trouble-shooting. All authors read and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank members of the Centre for Genomic Sciences at the University of Hong Kong for their technical support with high-throughput RNA sequencing. We thank Helen Zhi from the Biostatistics and Clinical Research Methodology Unit (Li Ka Shing Faculty of Medicine, University of Hong Kong) for providing advice on biostatistics. We are grateful to the staff of the Department of Microbiology at the University of Hong Kong for facilitation of the study. This work was supported in part by the Shaw Foundation; Prof. Richard Yu and Mrs. Carol Yu; the Providence Foundation Ltd. (in memory of the late Lui Hac Minh); the Respiratory Virus Research Foundation; the Hong Kong University Foundation; Cheer Master Investments Ltd; the Hong Kong Health and Medical Research Fund (12110822); the Health and Medical Research Fund Commissioned Research on Control of Infectious Diseases (Phase III) (HKM-15-M02, HKM-15-M04); the Food and Health Bureau; the Government of the Hong Kong Special Administrative Region; and the Collaborative Research Fund (C7011-15R) and the Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases (Ministry of Education of China). This work was also supported in part by the High Level Hospital — Summit Program in Guangdong at The University of Hong Kong–Shenzhen Hospital. The sponsors had no role in the design or conduct of the study, in the collection, analysis or interpretation of data, or in the preparation, review or approval of the manuscript.
Version 1. 08/28/2018
In-Press Preview
Version 2. 10/15/2018
Electronic publication
Version 3. 11/01/2018
Print issue publication
Footnotes
Conflict of interest: MLY, JFWC, and KYY have filed a patent application claiming the use of hWARS as a model for the treatment of enterovirus infection (PCT/US2017/051391).
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(11):5163–5177.https://doi.org/10.1172/JCI99411.
See the related Commentary at Not your usual tRNA synthetase: hWARS serves as an enterovirus entry factor.
Contributor Information
Man Lung Yeung, Email: pmlyeung@hku.hk.
Lilong Jia, Email: justin86@hku.hk.
Kwok-Hung Chan, Email: chan.kwok.hung@hkucc.hku.hk.
Jian-Piao Cai, Email: caijuice@163.com.
Chaoyu Zhang, Email: abbyzcy@hku.hk.
Anna J. Zhang, Email: zhangajx@hkucc.hku.hk.
Wan-Man Wong, Email: louise33@hku.hk.
Kin-Hang Kok, Email: khkok@hku.hk.
Dong-Yan Jin, Email: dyjin@hku.hk.
Shin-Ru Shih, Email: srshih@mail.cgu.edu.tw.
Kwok-Yung Yuen, Email: kyyuen@hkucc.hku.hk.
References
- 1.Yip CC, Lau SK, Woo PC, Yuen KY. Human enterovirus 71 epidemics: what’s next? Emerg Health Threats J. 2013;6:19780. doi: 10.3402/ehtj.v6i0.19780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong SS, Yip CC, Lau SK, Yuen KY. Human enterovirus 71 and hand, foot and mouth disease. Epidemiol Infect. 2010;138(8):1071–1089. doi: 10.1017/S0950268809991555. [DOI] [PubMed] [Google Scholar]
- 3.Lau SK, et al. Enterovirus D68 infections associated with severe respiratory illness in elderly patients and emergence of a novel clade in Hong Kong. Sci Rep. 2016;6:25147. doi: 10.1038/srep25147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imamura T, Oshitani H. Global reemergence of enterovirus D68 as an important pathogen for acute respiratory infections. Rev Med Virol. 2015;25(2):102–114. doi: 10.1002/rmv.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson R. Outbreaks of enterovirus D68 continue across the USA. Lancet Respir Med. 2014;2(10):791. doi: 10.1016/S2213-2600(14)70190-0. [DOI] [PubMed] [Google Scholar]
- 6.Yamayoshi S, et al. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat Med. 2009;15(7):798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura Y, Shimojima M, Tano Y, Miyamura T, Wakita T, Shimizu H. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat Med. 2009;15(7):794–797. doi: 10.1038/nm.1961. [DOI] [PubMed] [Google Scholar]
- 8.Yamayoshi S, et al. Human SCARB2-dependent infection by coxsackievirus A7, A14, and A16 and enterovirus 71. J Virol. 2012;86(10):5686–5696. doi: 10.1128/JVI.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang YF, Yu CK. Animal models of enterovirus 71 infection: applications and limitations. J Biomed Sci. 2014;21:31. doi: 10.1186/1423-0127-21-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284(29):19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolova L, Sudomoina MA, Grigorieva A, Zinovieva OL, Kisselev LL. Cloning and nucleotide sequence of the structural gene encoding for human tryptophanyl-tRNA synthetase. Gene. 1991;109(2):291–296. doi: 10.1016/0378-1119(91)90624-K. [DOI] [PubMed] [Google Scholar]
- 12.Tzima E, Reader JS, Irani-Tehrani M, Ewalt KL, Schwartz MA, Schimmel P. Biologically active fragment of a human tRNA synthetase inhibits fluid shear stress-activated responses of endothelial cells. Proc Natl Acad Sci U S A. 2003;100(25):14903–14907. doi: 10.1073/pnas.2436330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakasugi K, et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci U S A. 2002;99(1):173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kise Y, et al. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat Struct Mol Biol. 2004;11(2):149–156. doi: 10.1038/nsmb722. [DOI] [PubMed] [Google Scholar]
- 15.Yang XL, Schimmel P, Ewalt KL. Relationship of two human tRNA synthetases used in cell signaling. Trends Biochem Sci. 2004;29(5):250–256. doi: 10.1016/j.tibs.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Otani A, et al. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc Natl Acad Sci U S A. 2002;99(1):178–183. doi: 10.1073/pnas.012601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, He YQ, Yi LN, Zan H, Kung HF, He ML. Viral kinetics of enterovirus 71 in human abdomyosarcoma cells. World J Gastroenterol. 2011;17(36):4135–4142. doi: 10.3748/wjg.v17.i36.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamayoshi S, Fujii K, Koike S. Receptors for enterovirus 71. Emerg Microbes Infect. 2014;3(7):e53. doi: 10.1038/emi.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee PH, et al. Enterovirus 71 virion-associated galectin-1 facilitates viral replication and stability. PLoS One. 2015;10(2):e0116278. doi: 10.1371/journal.pone.0116278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamayoshi S, Ohka S, Fujii K, Koike S. Functional comparison of SCARB2 and PSGL1 as receptors for enterovirus 71. J Virol. 2013;87(6):3335–3347. doi: 10.1128/JVI.02070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleckner J, Martensen PM, Tolstrup AB, Kjeldgaard NO, Justesen J. Differential regulation of the human, interferon inducible tryptophanyl-tRNA synthetase by various cytokines in cell lines. Cytokine. 1995;7(1):70–77. doi: 10.1006/cyto.1995.1009. [DOI] [PubMed] [Google Scholar]
- 22.Huang HI, Weng KF, Shih SR. Viral and host factors that contribute to pathogenicity of enterovirus 71. Future Microbiol. 2012;7(4):467–479. doi: 10.2217/fmb.12.22. [DOI] [PubMed] [Google Scholar]
- 23.Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13(3):137–145. doi: 10.1016/S0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 24.Harris KG, Coyne CB. Death waits for no man — does it wait for a virus? How enteroviruses induce and control cell death. Cytokine Growth Factor Rev. 2014;25(5):587–596. doi: 10.1016/j.cytogfr.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romanova LI, et al. Variability in apoptotic response to poliovirus infection. Virology. 2005;331(2):292–306. doi: 10.1016/j.virol.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 26.Schulte BM, et al. Cytokine and chemokine production by human pancreatic islets upon enterovirus infection. Diabetes. 2012;61(8):2030–2036. doi: 10.2337/db11-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miles LA, Burga LN, Gardner EE, Bostina M, Poirier JT, Rudin CM. Anthrax toxin receptor 1 is the cellular receptor for Seneca Valley virus. J Clin Invest. 2017;127(8):2957–2967. doi: 10.1172/JCI93472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front Oncol. 2013;3:183. doi: 10.3389/fonc.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh WE, Jr , et al. Human chromosome 11 contains two different growth suppressor genes for embryonal rhabdomyosarcoma. Proc Natl Acad Sci U S A. 1992;89(5):1755–1759. doi: 10.1073/pnas.89.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houldsworth J, Reuter VE, Bosl GJ, Chaganti RS. ID gene expression varies with lineage during differentiation of pluripotential male germ cell tumor cell lines. Cell Tissue Res. 2001;303(3):371–379. doi: 10.1007/s004410000340. [DOI] [PubMed] [Google Scholar]
- 31.Lin TY, Hsia SH, Huang YC, Wu CT, Chang LY. Proinflammatory cytokine reactions in enterovirus 71 infections of the central nervous system. Clin Infect Dis. 2003;36(3):269–274. doi: 10.1086/345905. [DOI] [PubMed] [Google Scholar]
- 32.Paley EL. Tryptamine-induced tryptophanyl-tRNAtrp deficiency in neurodifferentiation and neurodegeneration interplay: progenitor activation with neurite growth terminated in Alzheimer’s disease neuronal vesicularization and fragmentation. J Alzheimers Dis. 2011;26(2):263–298. doi: 10.3233/JAD-2011-110176. [DOI] [PubMed] [Google Scholar]
- 33.Feuer R, Mena I, Pagarigan RR, Harkins S, Hassett DE, Whitton JL. Coxsackievirus B3 and the neonatal CNS: the roles of stem cells, developing neurons, and apoptosis in infection, viral dissemination, and disease. Am J Pathol. 2003;163(4):1379–1393. doi: 10.1016/S0002-9440(10)63496-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei W, et al. ICAM-5/Telencephalin Is a Functional Entry Receptor for Enterovirus D68. Cell Host Microbe. 2016;20(5):631–641. doi: 10.1016/j.chom.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Gromeier M, Wimmer E. Mechanism of injury-provoked poliomyelitis. J Virol. 1998;72(6):5056–5060. doi: 10.1128/jvi.72.6.5056-5060.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mena I, Perry CM, Harkins S, Rodriguez F, Gebhard J, Whitton JL. The role of B lymphocytes in coxsackievirus B3 infection. Am J Pathol. 1999;155(4):1205–1215. doi: 10.1016/S0002-9440(10)65223-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang SM, et al. Modulation of cytokine production by intravenous immunoglobulin in patients with enterovirus 71-associated brainstem encephalitis. J Clin Virol. 2006;37(1):47–52. doi: 10.1016/j.jcv.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Yeung ML, et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat Microbiol. 2016;1:16004. doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yip CC, et al. Emergence of enterovirus 71 “double-recombinant” strains belonging to a novel genotype D originating from southern China: first evidence for combination of intratypic and intertypic recombination events in EV71. Arch Virol. 2010;155(9):1413–1424. doi: 10.1007/s00705-010-0722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yip CC, Lau SK, Lo JY, Chan KH, Woo PC, Yuen KY. Genetic characterization of EV71 isolates from 2004 to 2010 reveals predominance and persistent circulation of the newly proposed genotype D and recent emergence of a distinct lineage of subgenotype C2 in Hong Kong. Virol J. 2013;10:222. doi: 10.1186/1743-422X-10-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suchman E, Blair C. Cytopathic effects of viruses protocols. Microbe Library. American Society for Microbiology. https://web.archive.org/web/20120602171845/http://microbelibrary.org/component/resource/laboratory-test/2875-cytopathic-effects-of-viruses-protocols Archived from the original June 2, 2012. Accessed November 20, 2014.
- 42.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4(7):1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.