Abstract
Atypical antipsychotics are highly effective antischizophrenic medications but their clinical utility is limited by adverse metabolic sequelae. We investigated whether upregulation of macrophage migration inhibitory factor (MIF) underlies the insulin resistance that develops during treatment with the most commonly prescribed atypical antipsychotic, olanzapine. Olanzapine monotherapy increased BMI and circulating insulin, triglyceride, and MIF concentrations in drug-naive schizophrenic patients with normal MIF expression, but not in genotypic low MIF expressers. Olanzapine administration to mice increased their food intake and hypothalamic MIF expression, which led to activation of the appetite-related AMP-activated protein kinase and Agouti-related protein pathway. Olanzapine also upregulated MIF expression in adipose tissue, which reduced lipolysis and increased lipogenic pathways. Increased plasma lipid concentrations were associated with abnormal fat deposition in liver and skeletal muscle, which are important determinants of insulin resistance. Global MIF-gene deletion protected mice from olanzapine-induced insulin resistance, as did intracerebroventricular injection of neutralizing anti–MIF antibody, supporting the role of increased hypothalamic MIF expression in metabolic dysfunction. These findings uphold the potential pharmacogenomic value of MIF genotype determination and suggest that MIF may be a tractable target for reducing the metabolic side effects of atypical antipsychotic therapy.
Keywords: Metabolism
Keywords: Schizophrenia
Introduction
Atypical antipsychotic medications have replaced conventional antipsychotics because of their superior therapeutic action and reduced risk of extrapyramidal sequelae such as tardive dyskinesia (1). Unfortunately, the clinical utility of these drugs is limited by the development of impaired glucose tolerance, insulin resistance, and obesity in some individuals (2). Olanzapine is a widely prescribed atypical antipsychotic that is especially efficacious for long-term treatment, although weight gain occurs in many treated patients and is a significant detriment to patient compliance (3).
The acute administration of olanzapine to healthy volunteers induces hypertriglyceridemia and insulin resistance, but little is known about the molecular mechanisms underlying these actions and whether central or peripheral regulatory pathways play a role (4). In rodents, a 30-day infusion of olanzapine induces obesity, whole-body insulin resistance, a reduction in whole-body energy expenditure, and increased lipid storage in liver (5). Long-term olanzapine administration also increases meal size and whole-body fat accumulation in rats (6, 7).
Macrophage migration inhibitory factor (MIF) is an immunoregulatory mediator that is widely expressed in tissues and contributes to the dysmetabolic effects of severe infection, including insulin resistance and reduced glucose uptake in adipose tissue (8). MIF promotes monocyte retention and activation within white adipose tissue (9), and increased MIF expression has been reported in patients with obesity or type 2 diabetes (10–12). In contrast, in cardiac muscle, MIF activates AMP-activated protein kinase (AMPK) to promote glucose utilization and protect against ischemic injury (13). MIF is encoded in a functionally polymorphic genetic locus, and MIF-dependent responses vary in individuals based on their allelic composition (14).
We performed a prospective longitudinal study in drug-naive and first-episode schizophrenic patients to examine whether MIF expression and MIF genotype influence the development of metabolic dysfunction in those receiving olanzapine monotherapy. We studied olanzapine administration in a genetically engineered mouse model to further elucidate these findings, and delineated auto- and paracrine actions for MIF that contribute to olanzapine-induced metabolic dysfunction.
Results
Olanzapine-induced insulin resistance and metabolic dysfunction is associated with increased plasma MIF concentration.
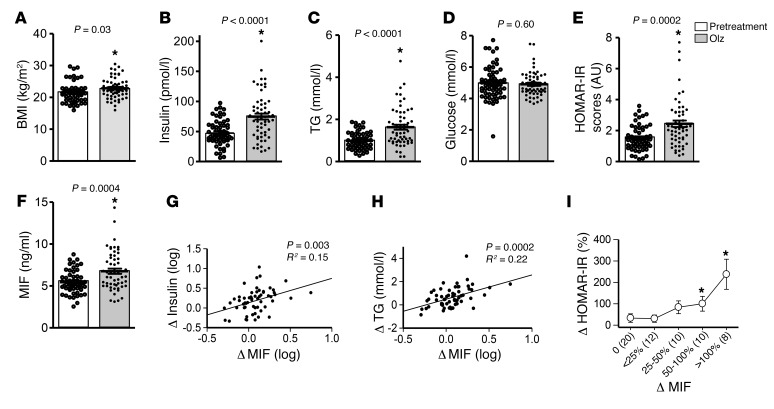
We characterized the metabolic response in 60 drug-naive and first-episode schizophrenic patients treated with olanzapine monotherapy for 2 months (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI93090DS1). As expected, olanzapine alleviated schizophrenia symptoms, as assessed by the Positive and Negative Syndrome Scale (PANSS) (Supplemental Figure 1). Increased BMI and both hyperinsulinemia and hypertriglyceridemia were evident after 8 weeks of treatment in these subjects (Figure 1, A–C), although fasting plasma glucose levels were unchanged (Figure 1D). The homeostasis model assessment insulin resistance (HOMA-IR) score, which is an index of metabolic dysfunction (10), also increased from a baseline level of 1.5 to 2.5 (Figure 1E). Notably, plasma MIF concentrations were increased following olanzapine treatment in these patients (Figure 1F) and these measurements correlated with the increment in hyperinsulinemia (Figure 1G), hypertriglyceridemia (Figure 1H), and insulin resistance scores (Figure 1I).
Figure 1. MIF is associated with the development of metabolic dysfunction in first-episode schizophrenia patients following olanzapine treatment.
Sixty first-episode patients with schizophrenia were studied for (A) BMI, (B) insulin, (C) triglyceride (TG), (D) glucose, and (F) plasma MIF before and after olanzapine monotherapy. HOMA-IR scores (E) were determined on the basis of plasma insulin and glucose values. The changes in MIF correlated with alterations in insulin (G), TG (H), and HOMA-IR scores (I) following olanzapine. Student’s t test (A–F), Pearson correlation (G, H), and 1-way ANOVA plus Tukey’s test (I) were used for data analysis. All t tests were 2-tailed. Mean ± SD.*P < 0.05 versus pretreatment.
Olanzapine increases MIF expression in a –794 CATT5–8 length-dependent manner.
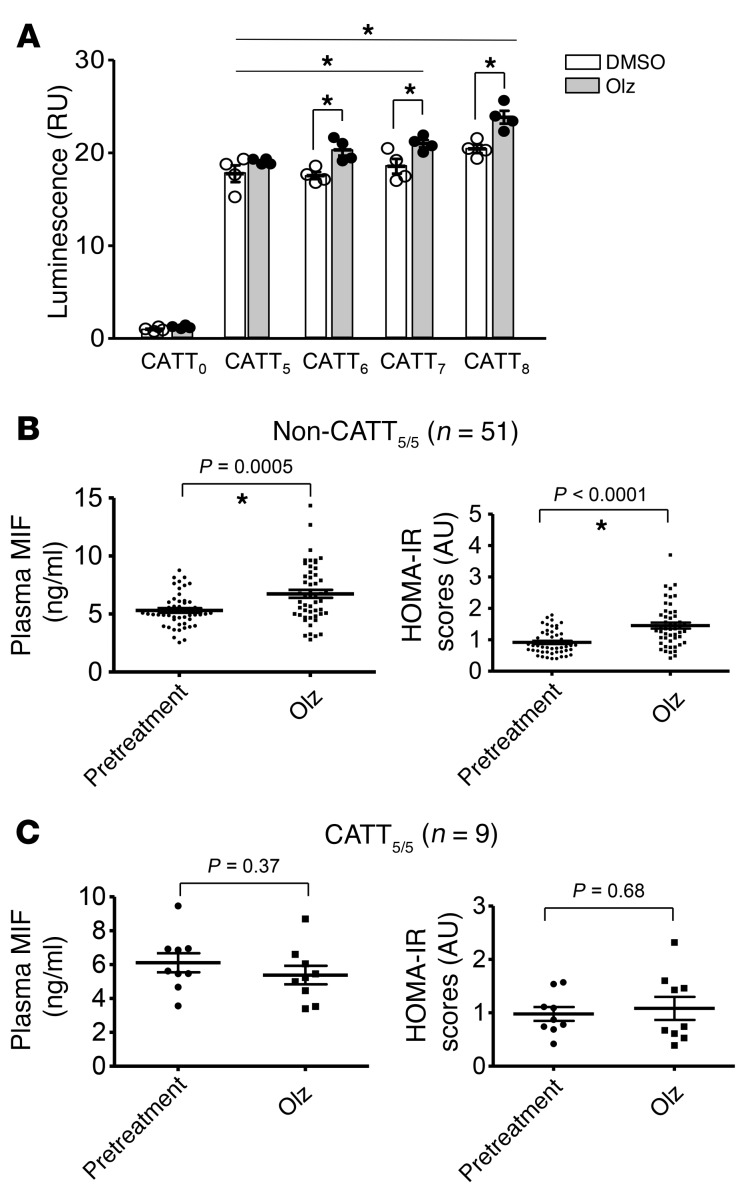
The human MIF gene (MIF) is characterized by a 4-nucleotide microsatellite repeat within the gene promoter (–794 CATT5–8, rs5844572) (14). Variant MIF alleles occur commonly in the population (minor allele frequency >5%) and microsatellite copy number is associated with increased mRNA expression such that the CATT5 repeat is a low-expression allele and the CATT6–8 repeats are higher expression alleles. We examined the impact of olanzapine on human MIF promoter activity by transfecting MIF promoter-luciferase fusion plasmids engineered to contain increasing lengths of the CATT microsatellite (CATT5–8) into HeLa cells, and measuring transcriptional activity. As shown in Figure 2A, olanzapine stimulated transcription in a CATT-length–dependent manner, with increased activity observed relative to vehicle for promoter constructs with more than 5 CATT repeats. These data suggest that olanzapine upregulates cellular MIF gene expression by influencing transcription at the –794 CATT5–8 polymorphic site.
Figure 2. MIF promoter activity is regulated in a –794 CATT length-dependent manner and is associated with olanzapine treatment–related alterations in plasma MIF levels and insulin resistance.
(A) HeLa cells cotransfected with MIF promoter-luciferase reporter plasmids bearing 0, 5, 6, 7, or 8 CATT repeats were treated with olanzapine (Olz, 5 mM) or vehicle (DMSO) for 18 hours and MIF promoter activity was determined by luciferase activity relative to a cotransfected pRL-β actin vector. In the schizophrenic patients (n = 60), plasma MIF levels and HOMA-IR scores were measured before and after 2 months of olanzapine treatment. DNA was extracted from whole-blood samples and the MIF gene was sequenced for determination of –794 CATT5–8 polymorphism (rs5844572). Mean ± SD; *P < 0.05. (B) High-expression –794 non-CATT5/5 (non-CATT5/5) genotypes (n = 51) are associated with posttreatment increases in plasma MIF levels and HOMA-IR scores. (C) The low-expression –794 CATT5/5 genotype (CATT5/5) (n = 9) is associated with protection from metabolic dysfunction. One-way ANOVA plus Tukey’s test or 2-tailed Student’s t test was performed for the data analysis. Mean ± SD; *P < 0.05 versus pretreatment.
Olanzapine’s metabolic side effects, but not therapeutic efficacy, correlate with high-expression MIF alleles.
Previous studies have linked high-expression MIF alleles with increased MIF plasma levels and clinical severity of various inflammatory disorders (14). Thus, patients were classified into low- or high-expresser MIF genotypes based on the presence of a –794 CATT5/5 genotype (CATT5/5 = lowest MIF expresser) or a non-794 CATT5/5 genotype (non-CATT5/5 = higher MIF expresser) (13, 14). The high expresser MIF genotype group (non-CATT5/5) exhibited increased plasma MIF, HOMA-IR scores, and BMI as well as increased plasma levels of insulin and triglyceride, and decreased HDL, when compared with the pretreatment group (Figure 2B and Supplemental Table 2). By contrast, in the CATT5/5 patients, olanzapine treatment did not alter plasma MIF levels, HOMA-IR scores, BMI, or other measured metabolic indicators (i.e., insulin, glucose, triglycerides, HDL, and total cholesterol) (Figure 2C and Supplemental Table 3). Importantly, MIF genotype did not influence psychiatric treatment response as assessed by improvement in PANSS measurements (Supplemental Figure 2). These data indicate that MIF genetic variants influence the development of olanzapine-induced metabolic side effects but not its therapeutic action.
MIF modulates obesity and food intake behavior by AMPK in the hypothalamus following olanzapine treatment.
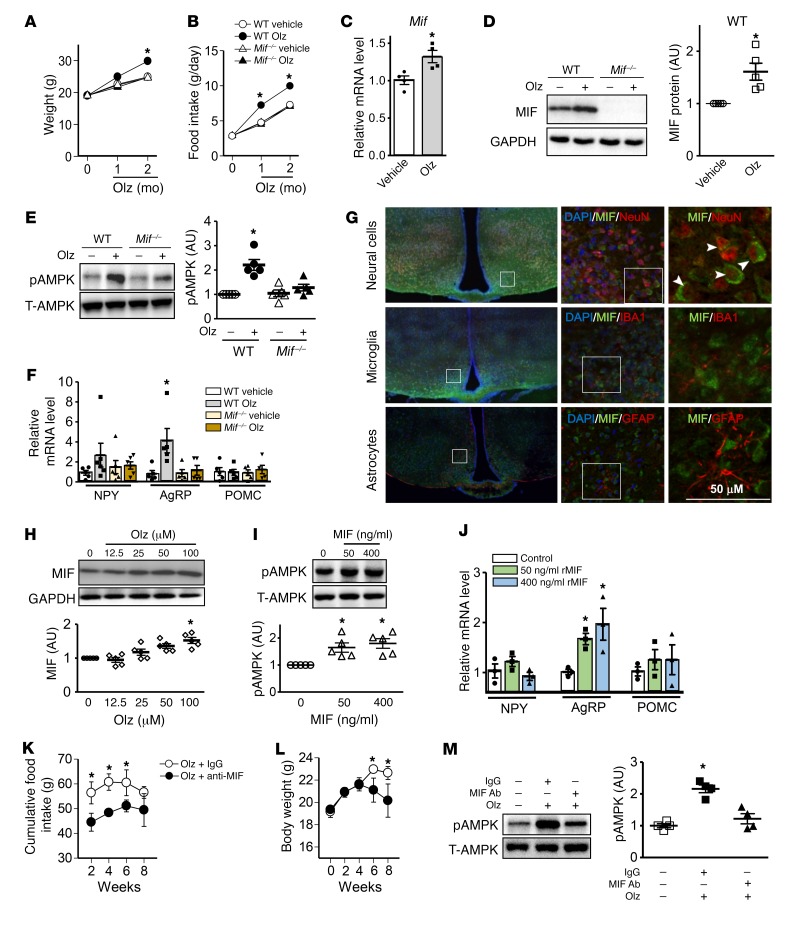
Olanzapine-induced adiposity and body weight gain is probably associated with hyperphagia and/or a reduction in basal energy expenditure (15–19). In order to delineate the mechanism(s) underlying MIF’s role in the hyperphagic action of olanzapine, we treated WT or global MIF-gene deficient (Mif–/–) C57BL/6 mice with olanzapine following a standard protocol (20, 21). We performed 2 studies with Mif–/– mice: the first employed WT nonlittermate controls (Figure 3) and the second employed WT littermate controls (Supplemental Figure 10). The results, which were similar in both studies, showed an increase in body weight in WT mice after 2 months of olanzapine treatment and a corresponding early increment in food intake at 1 and 2 months of treatment (Figure 3, A and B; Supplemental Figure 10, A and B). In contrast, Mif–/– mice were fully resistant to olanzapine-induced hyperphagia and weight gain. We hypothesized that olanzapine might influence MIF expression in the hypothalamus, which has a central role in appetite control. We found that both MIF mRNA and protein expression levels were increased in the hypothalamus of treated mice (Figure 3, C and D). MIF is known to increase AMP-kinase (AMPK) activation in heart muscle (13), and increased activation of AMPK in the hypothalamus increases appetite (22). Olanzapine treatment significantly increased AMPK phosphorylation at its activating Thr172 site in hypothalamic tissue from WT mice, but this effect was partially attenuated in Mif–/– mice (Figure 3E; Supplemental Figure 10C). Several neuropeptides act within the hypothalamus to influence food intake. During fasting, the expression of neuropeptide Y (NPY) and Agouti-related protein (AgRP) increases, and the expression of proopiomelanocortin (POMC) decreases (23). Following 2 months of olanzapine treatment, the hypothalamic expression of AgRP but not NPY or POMC was upregulated by olanzapine treatment in WT mice, but not in Mif–/– animals (Figure 3F; Supplemental Figure 10D).
Figure 3. MIF modulates food intake and obesity by activating AMPK in the hypothalamus following olanzapine treatment.
WT and MIF knockout (Mif–/–) C57BL/6 mice were administered vehicle or 3 mg/kg olanzapine per day for 2 months. (A) Body weight and (B) food intake were monitored. Quantification of (C) MIF transcript levels by qPCR, (D) MIF protein, and (E) AMPK levels (pAMPK: Thr172 phosphorylated AMPK; T-AMPK: total AMPK) in homogenates of murine hypothalamic tissue after 2 months of treatment. (F) Transcript levels of NPY, AgRP, and POMC measured by qPCR in hypothalamic homogenates from mice treated with or without olanzapine for 2 months. Immunostaining was performed in the hypothalamus from WT mice. The staining for neural cells (NeuN, top panels), microglia (IBA1, middle panels), and astrocytes (GFAP, bottom panels) is red, while the staining for MIF is green (G). Arrow and arrowheads represent costaining of MIF and NeuN in neural cells. Isolated hypothalamic cells were treated with increasing concentrations of olanzapine for 24 hours, after which (H) MIF protein content was measured by Western blot. (I) Total and phospho-AMPK levels in isolated hypothalamic cells were evaluated following 24-hour MIF stimulation in vitro. (J) NPY, AgRP, and POMC gene expression measured in hypothalamic cells following 24 hours of MIF treatment. IgG or anti–MIF monoclonal antibody (2 μg/day) was administered i.c.v. by an osmotic pump to WT mice treated with olanzapine for 2 months. The cumulative food intake (K) and body weight gain (L) were subsequently evaluated. The hypothalamic tissues were collected for AMPK measurements (M). The pAMPK and T-AMPK Western blots are from parallel gels run contemporaneously on identical samples. For each animal group, n = 4–6. A, B, K, and L were analyzed by multivariate (2-way) ANOVA and the rest of the data were analyzed by 2-tailed Student’s t test or 1-way ANOVA. Mean ± SE in A, B, K, and L; mean ± SD in other panels; *P < 0.05 versus vehicle in C–F, versus other groups in A and B, versus control in H–J, versus IgG group in K–M. Olz: olanzapine.
We performed MIF costaining with specific markers of neural cells (NeuN), microglia cells (IBA1), and astrocytes (GFAP) in the WT hypothalamus to identify the location of MIF expression. We found that MIF colocalized primarily with neural cells rather than astrocytes or microglia cells (Figure 3G), suggesting that neural cells are likely the major source of MIF in hypothalamus. To further delineate the effects of olanzapine on MIF expression in the hypothalamus, hypothalamic neural cells were isolated and incubated with olanzapine for 24 hours (Supplemental Figure 3). We found that olanzapine upregulated MIF expression in a dose-dependent manner (Figure 3H) and that recombinant MIF treatment increased AMPK phosphorylation (Figure 3I) and AgRP expression in cultured hypothalamic neural cells in vitro (Figure 3J). Notably, the MIF receptor CD74 also was observed by immunostaining to be expressed in the hypothalamus (Supplemental Figure 4). These data indicate a role for olanzapine in augmenting MIF expression in the hypothalamus and suggest that centrally expressed MIF may have a paracrine/autocrine effect to stimulate downstream AMPK phosphorylation, leading to an increase in feeding behavior by upregulating AgRP expression.
To determine whether hypothalamic MIF mediates olanzapine-induced hyperphagia in vivo, we administered a neutralizing anti–MIF monoclonal antibody intracerebroventricularly (i.c.v.) to WT mice and then treated them with olanzapine for 2 months. Anti–MIF antibody treatment significantly inhibited olanzapine-induced food intake and subsequent body weight gain after 6–8 weeks (Figure 3, K and L) without influencing peripheral MIF levels (Supplemental Figure 5). Anti–MIF antibody also attenuated olanzapine-induced hypothalamic AMPK activation (Figure 3M). In contrast, a single dose of recombinant MIF (1.38 μg, i.c.v.) acutely stimulated hyperphagia (Supplemental Figure 6). These data support the conclusion that hypothalamic MIF modulates food intake behavior and body weight gain during olanzapine treatment by stimulating the AMPK pathway.
Olanzapine augments MIF release from adipocytes, which contributes to increased plasma MIF levels.
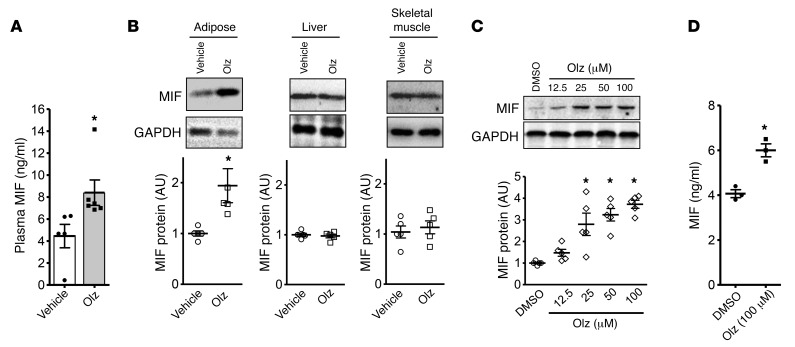
Consistent with our observations in humans with high MIF expression alleles, olanzapine treatment of WT mice increased plasma MIF levels (Figure 4A). MIF is expressed by the parenchymal cells of the liver, muscle, and adipose tissue (24). To better understand the potential tissue origins of the elevated plasma MIF levels observed during olanzapine therapy, we measured MIF protein expression in murine tissues after 2 months of treatment. MIF content was increased in adipose tissue, but not in liver or skeletal muscle (Figure 4B). Olanzapine also increased intracellular MIF content in cultured 3T3-L1 adipocytes (Figure 4C) as well as MIF release into conditioned medium (Figure 4D). Of note, olanzapine increased IL-1β and IL-6 gene expression and release in WT but not in Mif–/– mouse adipose tissue (Supplemental Figure 7), suggesting that olanzapine induction of a proinflammatory adipocyte phenotype is also MIF dependent. These latter data are consistent with observations that MIF mediates the inflammatory activation of adipose tissue and insulin resistance in the LDL-receptor knockout (Ldlr–/–) model of accelerated atherosclerosis (9).
Figure 4. Olanzapine increases plasma MIF levels and MIF expression in adipose tissue.
WT and MIF knockout (Mif–/–) C57BL/6 mice were administered vehicle or 3 mg/kg olanzapine per day for 2 months. Plasma MIF concentrations in WT mice with or without olanzapine treatment were measured by ELISA (A). Adipose, liver, and skeletal muscle tissues were subsequently isolated and (B) tissue MIF content was evaluated by Western blot. Olanzapine stimulation for 24 hours induced a dose-dependent increase in intracellular MIF protein levels in fully differentiated 3T3-L1 adipocytes (C) and induced MIF release into conditioned medium (D) (DMSO was employed as vehicle). Values are mean ± SD. A 2-tailed Student’s t test was used for data analysis in A, B, and D and 1-way ANOVA followed by Tukey’s test was used in C. Mean ± SD; *P < 0.05 versus vehicle or control in A, B, and C, and versus DMSO in D. For each animal group, n = 4–5 in A and B; n = 5 in C, and n = 3 in D.
Olanzapine attenuates lipolysis and increases lipogenesis in adipose tissue via MIF.
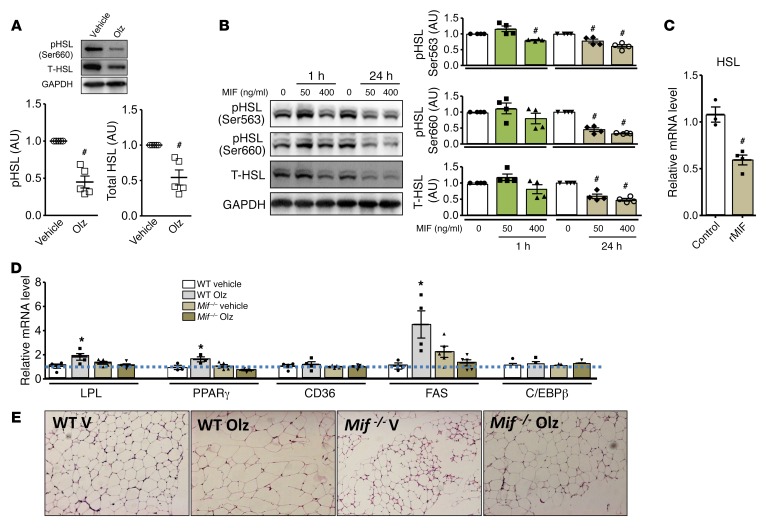
Chronic olanzapine treatment induces adipocyte hypertrophy (17). Hormone sensitive lipase (HSL) is the primary mechanism responsible for lipolysis in adipocytes and its activity is inhibited by insulin-mediated Akt phosphorylation (25). MIF reduces phosphorylation of Akt by inhibiting insulin-dependent phosphorylation of insulin receptor substrate 1 (IRS-1) and its association with the p85 regulatory subunit of phosphoinositide 3-kinase (8). We observed that the increase in MIF protein expression in adipose tissue after olanzapine treatment was also associated with a decrease in Akt phosphorylation (Supplemental Figure 8A), with diminished downstream phosphorylation of HSL (Figure 5A). We noted that total HSL protein expression was also reduced in adipose tissue from olanzapine-treated mice (Figure 5A). Additional in vitro experiments showed that the addition of recombinant MIF to cultured adipocytes reduced total and phosphorylated HSL protein content as well as HSL mRNA levels (Figure 5, B and C), indicating that MIF might inhibit lipolysis through a direct effect to inhibit both HSL expression and activation.
Figure 5. MIF downregulates lipolysis and upregulates adipogenesis following olanzapine treatment.
WT mice were treated with olanzapine (Olz, 3 mg/kg) or vehicle control for 2 months, and adipose tissue was isolated for the measurement of hormone-sensitive lipase (HSL) activation (A) by Western blotting. MIF (50 ng/ml and 400 ng/ml for 1 to 24 hours of treatment) was added to cultured 3T3-L1 adipocytes and (B) phospho- and total-HSL evaluated by Western blot, and (C) HSL mRNA by qPCR (400 ng/ml MIF). The GAPDH Western blot shown in B is from a parallel gel run contemporaneously on identical samples. (D) Adipose transcript levels of lipoprotein lipase (LPL), PPARγ, CD36, and fatty acid synthase (FAS) were quantified by qPCR in WT or Mif–/– mice treated for 2 months with olanzapine or vehicle. (E) H&E staining of representative adipose tissue sections (n = 5 examined per experimental group) was performed to evaluate adipocyte hypertrophy in WT and Mif–/– mice following 2 months of treatment with olanzapine (Olz) or vehicle (V). Original magnification, ×40. A 2-tailed Student’s t test or 1-way ANOVA plus Tukey’s test was used for statistical analysis. Mean ± SD; *P < 0.05 shows an increase versus vehicle or control; #P < 0.05 shows a reduction versus vehicle or control; n = 4–5 for each animal group.
We next investigated whether the induction of MIF expression by olanzapine might also increase lipogenesis in adipose tissue. Examination of transcript levels for genes that regulate lipogenesis revealed that olanzapine increased mRNA encoding lipoprotein lipase (LPL), peroxisome proliferator-activated receptor-γ (PPARγ), and fatty acid synthase (FAS) in adipose tissue from WT but not Mif–/– mice (Figure 5D). The expression levels of CD36, which functions in fatty acid uptake, was unchanged (Figure 5D). These data indicate that olanzapine upregulates lipogenesis by promoting fatty acid synthesis and storage in a MIF-dependent manner. Histologic examination of adipose tissue supported this conclusion by revealing hypertrophied adipocytes in olanzapine-treated WT but not Mif–/– mice (Figure 5E).
Olanzapine increases peripheral lipid accumulation and insulin resistance by a Mif-dependent pathway.
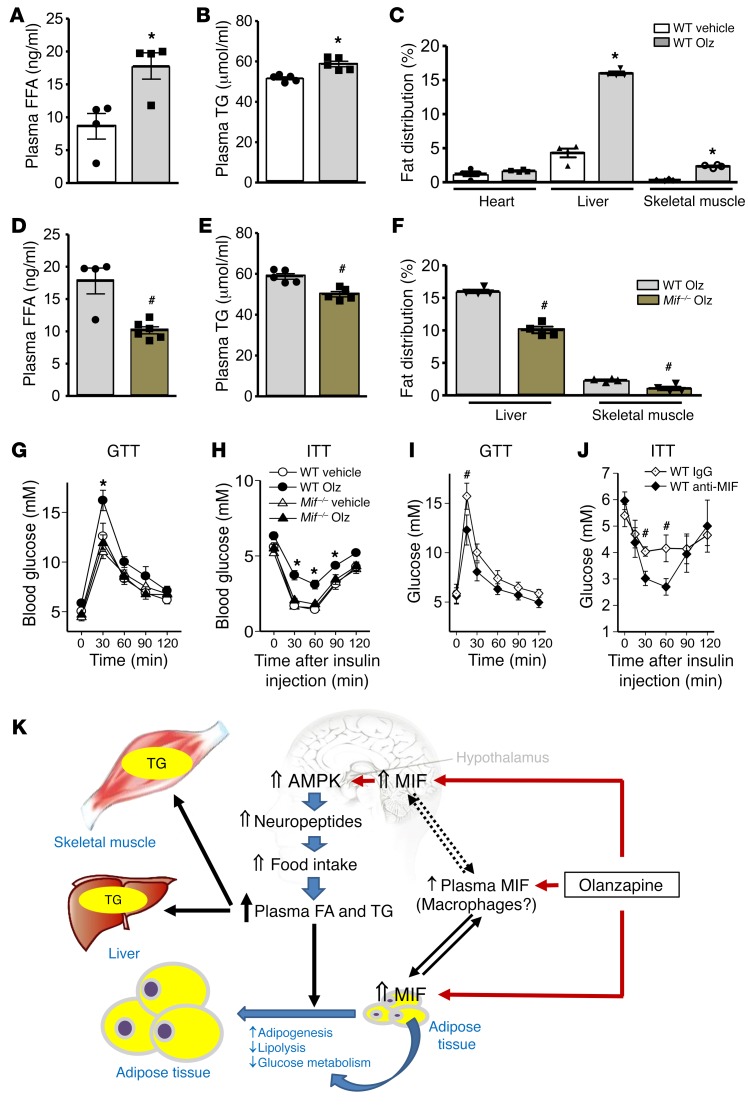
Consistent with our observations in human subjects with high MIF expression alleles, olanzapine treatment of WT mice increased food intake and plasma lipid levels (Figure 3B and Figure 6, A and B). To assess whether the elevation in plasma lipids was associated with an increase in fat content within nonlipid storage organs such as the heart, skeletal muscle, and liver, we determined fat distribution by quantitative MRI imaging. As shown in Figure 6C and Supplemental Figure 10E, olanzapine treatment led to an increase in lipid accumulation in liver and skeletal muscle but not in heart. These sequelae of olanzapine treatment were significantly attenuated in Mif–/– mice (Figure 6, D and E), suggesting that olanzapine-induced hyperlipidemia and peripheral lipid accumulation are MIF dependent.
Figure 6. MIF regulates abnormal lipid storage in liver and skeletal muscle, which may contribute to the occurrence of whole-body insulin resistance.
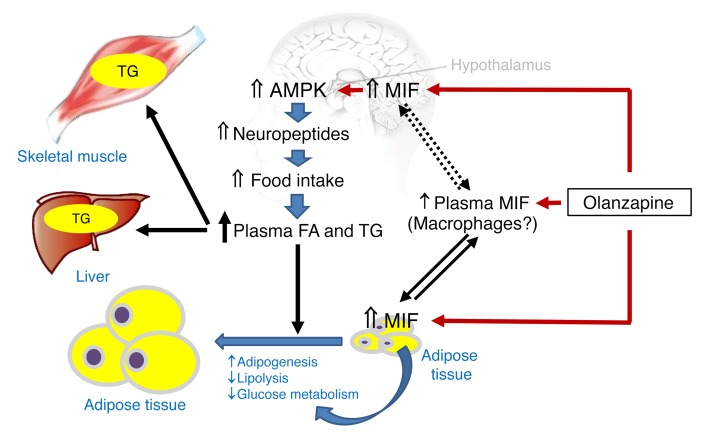
(A) Plasma free fatty acid (FFA) and (B) triglyceride (TG) levels were measured in blood samples collected from vehicle- or olanzapine-treated WT mice. (C) Fat distribution in heart, liver, and skeletal muscle was quantified in mice by MRI scanning using the T2*-IDEAL water-fat decomposition method (1). Plasma FFA (D) and TG (E) levels were quantified in olanzapine-treated WT and Mif–/– mice. Panel F shows the comparison of fat distribution in liver and skeletal muscle between WT and Mif–/– mice following 2 months of olanzapine treatment. In a separate experiment, intraperitoneal glucose tolerance (G) and insulin tolerance (H) tests (GTT, ITT) were performed following 2 months of olanzapine treatment in WT or Mif–/– mice. Glucose and insulin tolerance tests (I, J) in mice that received IgG or anti–MIF monoclonal antibody (2 μg/day) i.c.v. by an osmotic pump, accompanied with olanzapine for 2 months as in Figure 3, I and J. Mean ± SE shown in G–I; mean ± SD in the other panels. *P < 0.05 versus vehicle in A–C; #P < 0.05 versus WT Olz in D–F; *P < 0.05 versus other groups in G and H; #P < 0.05 versus IgG group in I and J. Data in A–F were analyzed by Student’s t test and G–J were analyzed by multivariate ANOVA. For each animal group, n = 4–5. Olz: olanzapine; Anti-MIF: anti–MIF antibody. (K) Schematic diagram for a proposed mechanism of olanzapine-induced metabolic dysfunction. Olanzapine induces MIF expression in the hypothalamus, which upregulates AMPK phosphorylation and food intake. In peripheral tissues, olanzapine stimulates MIF expression and release from adipose tissue, which may contribute to circulating levels of MIF. In adipose tissue, MIF also mediates glucose metabolism, adipogenesis, and adipolysis, leading to adiposity. Hyperlipidemia contributes to increased TG storage in liver and skeletal muscle, leading to insulin resistance in peripheral tissues.
After 2 months of olanzapine treatment, WT mice showed evidence of significant insulin resistance as assessed during intraperitoneal glucose and insulin tolerance tests (Figure 6, G and H; Supplemental Figure 10F and G). In contrast, Mif–/– mice were protected from these effects, suggesting that MIF is an obligatory modulator of olanzapine-induced insulin resistance. Preserved insulin sensitivity in Mif–/– mice following olanzapine was associated with less lipid accumulation in liver and skeletal muscle than in WT mice (Figure 6F; Supplemental Figure 10E). Fat deposition in liver or skeletal muscle is known to blunt insulin signaling in these tissues, and is a critical determinant of whole-body insulin resistance (26). In order to determine whether MIF had direct effects on insulin signaling in these tissues, we performed additional in vitro experiments with liver or skeletal muscle cells following 24-hour treatment with recombinant MIF, but did not find evidence for a direct effect of MIF on insulin-stimulated Akt phosphorylation (Supplemental Figure 9).
Since olanzapine did not appear to increase MIF expression in liver and skeletal muscle (Figure 4B), and MIF did not directly induce insulin resistance in liver or muscle cells (Supplemental Figure 9), we further tested whether olanzapine-induced MIF expression in the hypothalamus might be the primary mechanism responsible for insulin resistance during treatment with this drug. We found that i.c.v. administration of a neutralizing anti–MIF antibody not only decreased olanzapine-induced weight gain and hyperphagia (Figure 3, K and L) but also reduced insulin resistance (Figure 6, I and J). Thus, the overall evidence indicates that MIF stimulation of food intake during olanzapine therapy contributes to weight gain, increased circulating triglycerides, and abnormal lipid accumulation in liver and skeletal muscle, leading to the development of insulin resistance (Figure 6K).
Discussion
Atypical antipsychotics are often prescribed for many years to patients with schizophrenia. With long-term use, the associated metabolic side effects of these agents significantly increase the risk of heart disease (2, 27). The present study identifies a genetic susceptibility to olanzapine-induced metabolic dysfunction that is based on high genotypic MIF expression. Our findings support the pharmacogenomic value of MIF genotype determination and also suggest that increased MIF expression is an important pathogenic mechanism that might be a tractable target for reducing the metabolic side effects of atypical antipsychotic therapy.
We studied 60 first-episode and treatment-naive schizophrenia patients after a 2-month course of olanzapine therapy. Our results show that the susceptibility to the development of increased BMI, hyperinsulinemia, and hypertriglyceridemia was related to the patient’s plasma MIF levels and in turn with MIF genotype. Metabolic side effects did not develop in subjects with the lowest MIF expresser CATT5/5 genotype, suggesting that MIF genotype determination may be useful in the risk stratification of patients for the adverse metabolic effects of olanzapine. Since MIF expression may vary across a spectrum of possible CATT copy number genotypes (e.g., 5/6, 5/7, 5/8, 6/6, 6/7, 6/8, 7/7, 7/8), a further study in a larger population would be necessary to better define risk among combinations of these allelic variants. MIF genotype did not influence olanzapine treatment efficacy during the present 2-month study; however, longer term investigations would be required to rule out this possibility. It might be noted that olanzapine-induced metabolic effects occur independent of the presence of mental illness (28), and prior studies have shown that healthy volunteers receiving acute administration of olanzapine also develop glucose intolerance and hypertriglyceridemia (4).
The elevated metabolic parameters (i.e., body mass, hyperinsulinemia, hypertriglyceridemia) observed in the patient cohort were recapitulated in WT mice administered olanzapine. Our experimental studies provide compelling evidence that MIF indeed plays an important pathogenic role in the metabolic side effects associated with olanzapine treatment. MIF-deficient mice are metabolically comparable to WT mice at baseline, with similar body mass and tissue fat distribution (8, 29). They demonstrated little evidence of olanzapine-induced metabolic dysfunction during therapy, with lower circulating plasma triglycerides and free fatty acids, less lipid accumulation in liver and skeletal muscle, and less insulin resistance compared with WT mice during olanzapine treatment.
Olanzapine induces greater weight gain than most other second-generation antipsychotics, which appears to be due to its stimulating action on appetite and food intake (3). We demonstrated increased food intake in olanzapine-treated WT mice, similar to prior findings in olanzapine-treated rats (15). Furthermore, we found that MIF levels were increased in the hypothalamus as a consequence of treatment and that genetic deletion of MIF abolished this hyperphagic effect of olanzapine. Importantly, neutralizing hypothalamic MIF by local administration of anti–MIF antibody inhibited olanzapine-induced hyperphagia and insulin resistance. Thus, these data support the hypothesis that olanzapine-induced hyperphagia, weight gain, and insulin-resistance are mediated at least in part by increased hypothalamic MIF expression and action.
The present study also helps to elucidate the mechanism for how olanzapine-induced hypothalamic MIF expression stimulates an increase in appetite. Olanzapine treatment increased the phosphorylation of hypothalamic AMPK, which has a known effect to stimulate feeding by augmenting expression of the NPY and AgRP neuropeptides (22). Hypothalamic AMPK activation has been implicated in olanzapine-induced hyperphagia (30, 31), but the mechanism(s) linking olanzapine and AMPK were previously not known. Our results indicate that increased MIF expression plays an important role in the activation of AMPK, based on attenuation of AMPK phosphorylation by the i.c.v. administration of a neutralizing MIF antibody during olanzapine treatment. We previously demonstrated that MIF activates AMPK in heart muscle, where it has an important role in the response to ischemia (13). However, the current data indicate that an olanzapine-MIF-AMPK-AgRP axis has an important role in the metabolic dysfunction associated with therapy.
Elevated circulating MIF levels are associated with obesity and type 2 diabetes in various populations (32, 33), and prior studies support a strong positive correlation between circulating MIF levels and BMI (11). MIF was first discovered in immune cells as a cytokine released from preformed storage pools in response to stimulation (34). MIF also is widely expressed in metabolically active parenchymal tissues including adipose tissue, the heart, and the liver (35). Adipose tissue has been implicated as the major source of the high circulating plasma MIF levels observed in subjects with a high BMI (36, 37). We found that olanzapine increased adipose MIF expression in vivo and induced MIF release from cultured adipocytes, consistent with the possibility that the increase in plasma MIF observed in human subjects administered olanzapine originated at least in part from adipose tissue (35).
Our results also demonstrate a novel role for MIF in mediating fat storage in adipose tissue. Systemic fatty acid metabolism reflects the balance of lipogenesis and lipolysis. Free fatty acids are generated during lipolysis by HSL, which is regulated by protein kinase A–dependent serine phosphorylation (25). Protein kinase A in turn is responsive to intracellular cAMP levels that are modulated by Akt signaling (38). Our results in adipose tissue indicate that increased MIF expression inhibits Akt signaling and leads to a reduction in HSL phosphorylation. HSL protein content also was reduced, which may further decrease lipolysis. A decrease in the expression of HSL has been reported to occur during olanzapine treatment (7). Our data suggest a distinct MIF-dependent action to downregulate HSL protein expression and phosphorylation following olanzapine treatment.
Lipogenesis relies on several enzymes and transcription factors, including lipoprotein lipase, fatty acid synthase, and PPARγ. Lipoprotein lipase catalyzes fatty acid release from circulating lipoproteins and facilitates lipid uptake into adipocytes. The transcription factor PPARγ is highly expressed in adipose tissue and its activation leads to an increase in triglyceride formation. Olanzapine increases fatty acid uptake into adipose tissue in rodents (17) and we observed that it upregulates lipoprotein lipase and PPARγ gene expression in WT but not in MIF-deficient mice. In contrast, the expression of the major fatty acid transporter CD36 was not affected, suggesting that the olanzapine-mediated increase in lipid uptake occurs independent of this transporter. The expression of fatty acid synthase, which catalyzes de novo synthesis of long-chain fatty acids, also was upregulated by olanzapine in a MIF-dependent manner. Collectively, the observation that Mif–/– mice are resistant to olanzapine-induced increases in lipoprotein lipase, PPARγ, and fatty acid synthase expression positions MIF as a central regulator of these effects. These results are in line with the finding that Mif–/– mice maintain normal body weight and adipocyte size during olanzapine treatment.
Our results also indicate that olanzapine-induced upregulation of inflammatory factors, including IL-6, IL-1β, and TNFα, in adipose tissue is also MIF dependent. Insulin resistance exhibits features of a chronic inflammatory state (39) and is accompanied by expression of inflammatory cytokines such as IL-6, IL-1β, and TNFα (10). MIF is an upstream regulator of the innate response, and genetic MIF deficiency is associated with a reduction in the inducible expression of these cytokines (40). Human subjects who are low MIF expressers have reduced responses to microbial stimuli and show protection from the inflammatory complications of infections or autoimmunity (14, 41, 42). Clinical studies support a relationship between plasma MIF levels and impaired glucose tolerance and type 2 diabetes, and these associations were statistically more significant than with C-reactive protein or IL-6 (10). Thus, the precise role of MIF-induced adipocyte inflammation in olanzapine-induced insulin resistance is of interest and requires further evaluation.
High-fat diet feeding studies have shown that MIF expression increases in adipocytes and attenuates insulin signaling and glucose uptake by interfering with the phosphorylation of IRS-1 (8, 43). Our signal transduction studies in WT and Mif–/– adipocytes suggest that olanzapine modulates insulin-stimulated Akt activation and reduces glucose uptake and subsequent metabolism in a MIF-dependent fashion in adipose tissue (Supplemental Figure 8, B–D). In contrast, MIF does not attenuate insulin-stimulated Akt phosphorylation in liver and skeletal muscle cells, suggesting that circulating MIF may not have a direct effect on insulin signaling in liver and skeletal muscle. However, MIF may have an indirect effect to cause insulin resistance in peripheral tissues. Olanzapine treatment in mice increased plasma FA and TG concentrations, and the increased PPARγ expression observed in adipose tissue may promote expansion of fat mass that is a compensatory mechanism to exclude fat from liver and muscle and modulate insulin resistance. Nevertheless, this upregulation of PPARγ is not sufficiently compensatory to hyperlipidemia. Persistent elevation in lipid levels therefore leads to TG deposition in liver and skeletal muscle, observed by MRI imaging, which likely contributed to the whole-body insulin resistance observed in this and prior studies (17). MIF gene–deficient mice did not develop increased fat stores in the liver and muscle and were protected from the insulin resistance that was observed in the olanzapine-treated WT mice.
In conclusion, this study identifies a genetic susceptibility to olanzapine-induced metabolic dysfunction that is based on high genotypic MIF expression. In addition, increased MIF expression is a critical pathogenic mechanism in the metabolic derangements observed after olanzapine administration in patients with schizophrenia. Olanzapine increases MIF expression in the murine hypothalamus, leading to a hyperphagic response via activation of AMPK and AgRP expression. The increased lipid uptake exceeded the storage capacity of adipose tissue, leading to abnormal fat deposition in liver and skeletal muscle and the development of insulin resistance (Figure 6K).
These results also have clinical implications for schizophrenic patients treated with olanzapine. Determination of an individual’s MIF genotype may be useful to assess risk for adverse metabolic sequelae associated with olanzapine treatment. Pharmacologic MIF inhibition, which is presently under clinical evaluation for inflammatory diseases, could minimize metabolic side effects that detract from an otherwise highly effective schizophrenia therapy.
Methods
Human studies.
Inpatients were recruited at the Shanghai Mental Health Center, Shanghai Jiao Tong University School of Medicine, from October 2010 to October 2015. All subjects received the Mini-International Neuropsychiatric Interview (MINI), and demographic data including age, sexual phenotype, and BMI were collected (Supplemental Table 1). The symptoms of schizophrenic patients were evaluated with PANSS by 2 experienced psychiatrists (interrater reliability, Kappa = 0.86). First-episode and drug-naive Chinese Han population patients with ages ranging from 18 to 50 years old who met the DSM-IV criteria for schizophrenia and had a total PANSS score of at least 60 were recruited for this study. Patients who smoked, drank alcohol, were substance dependent, had severe medical illness (e.g., cancer, diabetes) or other CNS conditions, or who were pregnant were excluded from participation, leaving a total of 60 patients with a mean age of 28.0 ± 6.6 years (30 males and 30 females) for study. Qualifying patients were subjected to olanzapine monotherapy for 60 days, starting with an initial dose of 5 mg daily and gradually increasing to 15–20 mg daily. Benzhexol hydrochloride tablets were permitted if patients experienced extrapyramidal symptoms. BMI and PANSS scores were assessed in patients at 2 time points: at the first diagnosis of schizophrenia and following 60 days of olanzapine therapy.
Analysis of MIF and metabolic indicators.
Peripheral venous blood was collected from patients in the fasting state between 7:00 am to 9:00 am at the time of diagnosis, then again at 1 and 2 months after initiation of olanzapine therapy. Plasma glucose, insulin, and lipid levels were determined in the Clinical Laboratory at Shanghai Mental Health Center, which was certified by the China National Accreditation Service for Conformity Assessment (CNAS). Plasma MIF level was determined with an ELISA method according to the protocol from R&D Systems. DNA was extracted from whole-blood samples and the MIF gene was sequenced for determination of –794 CATT5–8 polymorphism (rs5844572) (44). Data were obtained and analyzed in a blinded fashion.
MIF promoter assay.
Human HeLa cells were transfected with 500 ng pGL-3 CATT plasmids (CATT0, CATT5, CATT6, CATT7, CATT8) together with 10 ng pRL-β actin plasmids in 24-well plates (5 × 104 cells/well) using Lipofectamine 3000 Transfection reagent (Invitrogen) (43). Cells were incubated for 6 hours and then treated with 5 μM olanzapine or DMSO for 18 hours. After cellular lysis, MIF promoter activity was determined by Dual-Luciferase Reporter Assay (Promega) and signals normalized in relation to the Renilla luciferase activity.
Data analysis and statistical testing of human studies.
Statistical analysis was performed with the Statistical Package for Social Sciences (SPSS, version 17.0). A 1-sample Kolmogorov-Smirnov test was used to examine normal data distribution. Demographic data were analyzed using χ2, t test, and 1-way ANOVA as indicated in the text. If the data were not normally distributed, they were transformed into normal distribution by using natural logarithms or analyzed by nonparametrical statistical analysis (Whitney-Mann U test). Paired t test was performed to analyze plasma MIF in pre- and posttreatment groups. All significance levels were 2-tailed, and a P value of less than 0.05 was considered statistically significant.
Experimental animals.
Female C57BL/6 mice (8 weeks of age) were used in the current study, congruous with previous studies that explored olanzapine-induced metabolic dysfunction (45). Mif–/– mice were created and maintained on a pure C57BL/6 (Charles River) background, as previously described (21), and together with WT controls (Charles River) were maintained at the Shanghai Key Laboratory of Psychotic Disorders at Shanghai Mental Health Center. Mice were housed in individually ventilated cages with an artificial 12-hour light/dark cycle at room temperature (20–25°C) and fed with a medium fat diet (metabolizable energy, 17.5 kJ/g), which resembles a typical human diet consisting of 14% protein, 31% lipid, and 54% carbohydrate (46). Mice were treated with 3 mg/kg olanzapine (MilliporeSigma) or vehicle (DMSO) for 2, 4, and 8 weeks as previously described (20, 47).
MIF antibody i.c.v. infusion by mini-pump.
A cannula was placed sterilely in the third ventricle (1.8 mm caudal to bregma, 5.0 mm ventral to the sagittal sinus) as previously described (48) to administer neutralizing antibodies for the purpose of studying the effect of MIF in the hypothalamic feeding pathway. Anti–MIF antibody (2 μg/day, clone NIHIII.D9ll) or isotypic IgG1 control was injected via a mini–osmotic pump implanted in a subcutaneous pocket (Alzet model 2002).
Metabolic studies.
Mice were fasted for 6 hours and fasting blood glucose levels were determined from tail venous blood; 1 mg/g body glucose or 0.5 U/kg insulin then was injected i.p. and blood glucose values were obtained at 30, 60, 90, and 120 minutes. Fat composition in liver, heart, and skeletal muscle (gastrocnemius) was determined by MRI scanning and the T2*-IDEAL water-fat decomposition method (49). Glucose uptake into adipocytes was measured as previously described (8). Briefly, differentiated 3T3-L1 cells were incubated with 0.5 μCi/ml of 2-deoxy-[3H] glucose for 10 minutes and lysed with 50 mM NaOH for the measurement of radioactivity. Following olanzapine treatment for 2 months, visceral adipose tissue was isolated from WT and Mif–/– mice and H&E staining was performed.
Antibodies and MIF reagents.
Antibodies against phospho-AMPK (Thr172, catalog 2535), phospho-Akt (Ser473, catalog 9271), phospho-HSL (Ser660 and Ser563, catalog 4126 and 4139), AMPK (catalog 2535), Akt (catalog 9272), HSL (catalog 4107), and JNK (catalog 9252) were from Cell Signaling Technology, and the anti–phospho-JNK antibody (V7931) was obtained from Promega. A previously described anti–MIF monoclonal antibody (NIHIII.D.9) was employed to measure MIF protein expression (50). Recombinant mouse MIF was purified from a high-yield E. coli expression system by fast protein liquid chromatography (FPLC) followed by C8 chromatography to remove endotoxin (51). Mouse MIF concentrations were measured by a 1-step sandwich ELISA as previously described (8).
Immunofluorescence.
Immunofluorescence staining was performed on 8-week-old WT C57BL/6 mouse brain sections (30 μm), which were blocked with 3% donkey serum in 0.3% Triton X-100 for 1 hour at room temperature and incubated for 24 hours at 4°C with primary antibodies (MIF antibody 1:100; NeuN antibody 1:500, Millipore; IBA1 antibody 1:500, Millipore; GFAP antibody 1:1,000, Millipore). Following the incubation with primary antibodies, the sections were washed 3 times for 30 minutes with PBS and then incubated for 1 hour at room temperature with FITC- or/and CY3-conjugated secondary antibody. Finally, the images were obtained with an Olympus confocal microscope.
Cell culture studies.
Hypothalamic cells were isolated as previously described (52). Briefly, hypothalamic tissue was dissected from 14-day-old mouse embryos (C57BL/6 strain), washed in PBS, cut into small fragments, dispersed into single cells, and cultured in Ham F10 medium plus 15% heat-inactivated horse serum, 2.5% fetal calf serum, 1.5% of 200 mM l-glutamine, penicillin (50 IU/ml), and streptomycin (50 μg/ml). 3T3-L1 adipocytes were cultured and differentiated as previously described (53). Before all experiments, cells were briefly serum-starved in DMEM 0.2% bovine serum albumin (BSA) for 2 hours.
Expression analyses.
Transcript levels for the genes for NPY, AgRP, POMC, HSL, and MIF were measured by quantitative real-time reverse transcription PCR (qPCR) (22). Phosphorylation and total levels of AMPK, JNK, Akt, and HSL and total MIF content were evaluated by Western blot as previously described (54).
Statistical analysis of animal studies.
One-way ANOVA with Tukey’s post hoc tests were used to determine differences between group mean values. The level of statistical significance was set at P < 0.05.
Study approval.
The human study was approved by the IRB of Shanghai Mental Health Center and all human experiments were conducted in accordance with the Declaration of Helsinki. All study-related procedures were carried out with written informed consent. All mouse studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011), as well as the China National Standards for Laboratory Animal Quality and the Chinese Guidelines for Care and Use of Laboratory Animals.
Author contributions
DC, YP, and CZ conducted human data analysis and animal experiments. ZL, YS, and MX collected blood samples from human patients and performed MIF measurement. YQ, MW, JX, and WD participated in the animal experiments. GEK, KNS, LL, KJ, YX, LY, and RB contributed intellectually to data analysis and manuscript editing. MP prepared animal models and the MIF ELISAs. JL performed MIF promoter assay. MH performed the i.c.v. injection of recombinant MIF in rats. LY and RB provided overall scientific support for the research project. DQ designed and managed the research.
Supplementary Material
Acknowledgments
Our gratitude is expressed to the patients and their families for participation in this clinical study. This study was supported by grants to DC from the Ministry of Science and Technology Precision Medicine Project (2017YFC0909200), the National Science Foundation of China (NSFC, 81671336, 81271481, 81171266 and 81500976), and the National Science Foundation of Shanghai (NSFS, 06ZR14147, 10PJ1408800 and 11140900400); to YP from the Shanghai Sailing Program (14YF1413500); to RB (R01 AI042310 and AR049610) and LY (HL128069) from NIH; and to DQ from the Research and Development Corporation (RDC) Newfoundland and Labrador (RDC 5404.1980.102), the National Sciences and Engineering Research Council of Canada (NSERC, RGPIN-2017-04542), and the Canadian Institutes of Health Research (CIHR Project Grant PJT-156116).
Version 1. 10/08/2018
Electronic publication
Version 2. 11/01/2018
Print issue publication
Funding Statement
NSFC (National Science Foundation of China, 81671336<FF0C>81271481, 81171266 and 81500976), NSFS (National Science Foundation of Shanghai, 06ZR14147, 10PJ1408800 and 11140900400) and Ministry of Science and Technology Precision Medicine Project (2017YFC0909200) to Dr. Cui.
RO1 AI042310 and AR049610 to Dr. Bucala and HL128069 to Dr. Young
Shanghai Sailing Program (14YF1413500)
Research and Development Corporation (RDC) Newfoundland and Labrador (RDC 5404.1980.102), National Sciences and Engineering Research Council of Canada (NSERC: RGPIN-2017-04542) and Canadian Institutes of Health Research (CIHR Project Grant: PJT-156116) for Dr. Qi.
Footnotes
Conflict of interest: Yale University holds rights to patents that describe the potential utility of MIF inhibition and MIF genotype determination. RB has applied for a patent related to MIF modulators (9540322). RB and LL have applied for a patent related to the method of inhibiting binding or activity of MIF by administering a MIF antagonist (9221903). RB has applied for a patent related to MIF promoter polymorphism in inflammatory disease (9139877).
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(11):4997–5007.https://doi.org/10.1172/JCI93090.
Contributor Information
Donghong Cui, Email: manyucc@126.com.
Yanmin Peng, Email: ameepeng@gmail.com.
Chengfang Zhang, Email: callup1987@126.com.
Zezhi Li, Email: lizezhi1981@aliyun.com.
Yousong Su, Email: wintersus@126.com.
Yadan Qi, Email: yadanqi2000@yahoo.ca.
Mengjuan Xing, Email: xingmj0209@sina.com.
Jia Li, Email: Leejiasjtu@163.com.
Jinjie Xu, Email: sjmedical_xjj@163.com.
Meiti Wang, Email: 865573592@qq.com.
Wenhua Ding, Email: dingwenhua1234@163.com.
Marta Piecychna, Email: Marta.piecychna@yale.edu.
Lin Leng, Email: lin.leng@yale.edu.
Michiru Hirasawa, Email: michiru@mun.ca.
Kaida Jiang, Email: jiangkaida@aliyun.com.
Lawrence Young, Email: lawrence.young@yale.edu.
Yifeng Xu, Email: hyyyyb@gmail.com.
Dake Qi, Email: dake.qi@mun.ca.
Richard Bucala, Email: richard.bucala@yale.edu.
References
- 1.Muench J, Hamer AM. Adverse effects of antipsychotic medications. Am Fam Physician. 2010;81(5):617–622. [PubMed] [Google Scholar]
- 2.Newcomer JW. Second-generation (atypical) antipsychotics and metabolic effects: a comprehensive literature review. CNS Drugs. 2005;19(Suppl 1):1–93. doi: 10.2165/00023210-200519001-00001. [DOI] [PubMed] [Google Scholar]
- 3.Domecq JP, et al. Clinical review: Drugs commonly associated with weight change: a systematic review and meta-analysis. J Clin Endocrinol Metab. 2015;100(2):363–370. doi: 10.1210/jc.2014-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albaugh VL, Singareddy R, Mauger D, Lynch CJ. A double blind, placebo-controlled, randomized crossover study of the acute metabolic effects of olanzapine in healthy volunteers. PLoS ONE. 2011;6(8):e22662. doi: 10.1371/journal.pone.0022662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chintoh AF, Mann SW, Lam TK, Giacca A, Remington G. Insulin resistance following continuous, chronic olanzapine treatment: an animal model. Schizophr Res. 2008;104(1–3):23–30. doi: 10.1016/j.schres.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Davoodi N, Kalinichev M, Korneev SA, Clifton PG. Hyperphagia and increased meal size are responsible for weight gain in rats treated sub-chronically with olanzapine. Psychopharmacology (Berl) 2009;203(4):693–702. doi: 10.1007/s00213-008-1415-1. [DOI] [PubMed] [Google Scholar]
- 7.Minet-Ringuet J, et al. Alterations of lipid metabolism and gene expression in rat adipocytes during chronic olanzapine treatment. Mol Psychiatry. 2007;12(6):562–571. doi: 10.1038/sj.mp.4001948. [DOI] [PubMed] [Google Scholar]
- 8.Atsumi T, et al. The proinflammatory cytokine macrophage migration inhibitory factor regulates glucose metabolism during systemic inflammation. J Immunol. 2007;179(8):5399–5406. doi: 10.4049/jimmunol.179.8.5399. [DOI] [PubMed] [Google Scholar]
- 9.Verschuren L, et al. MIF deficiency reduces chronic inflammation in white adipose tissue and impairs the development of insulin resistance, glucose intolerance, and associated atherosclerotic disease. Circ Res. 2009;105(1):99–107. doi: 10.1161/CIRCRESAHA.109.199166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herder C, et al. Association of systemic concentrations of macrophage migration inhibitory factor with impaired glucose tolerance and type 2 diabetes: results from the Cooperative Health Research in the Region of Augsburg, Survey 4 (KORA S4) Diabetes Care. 2006;29(2):368–371. doi: 10.2337/diacare.29.02.06.dc05-1474. [DOI] [PubMed] [Google Scholar]
- 11.Dandona P, et al. Increased plasma concentration of macrophage migration inhibitory factor (MIF) and MIF mRNA in mononuclear cells in the obese and the suppressive action of metformin. J Clin Endocrinol Metab. 2004;89(10):5043–5047. doi: 10.1210/jc.2004-0436. [DOI] [PubMed] [Google Scholar]
- 12.Koska J, et al. mRNA concentrations of MIF in subcutaneous abdominal adipose cells are associated with adipocyte size and insulin action. Int J Obes (Lond) 2009;33(8):842–850. doi: 10.1038/ijo.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller EJ, et al. Macrophage migration inhibitory factor stimulates AMP-activated protein kinase in the ischaemic heart. Nature. 2008;451(7178):578–582. doi: 10.1038/nature06504. [DOI] [PubMed] [Google Scholar]
- 14.Sreih A, et al. Dual effect of the macrophage migration inhibitory factor gene on the development and severity of human systemic lupus erythematosus. Arthritis Rheum. 2011;63(12):3942–3951. doi: 10.1002/art.30624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper GD, Pickavance LC, Wilding JP, Halford JC, Goudie AJ. A parametric analysis of olanzapine-induced weight gain in female rats. Psychopharmacology (Berl) 2005;181(1):80–89. doi: 10.1007/s00213-005-2224-4. [DOI] [PubMed] [Google Scholar]
- 16.Goudie AJ, Smith JA, Halford JC. Characterization of olanzapine-induced weight gain in rats. J Psychopharmacol (Oxford) 2002;16(4):291–296. doi: 10.1177/026988110201600402. [DOI] [PubMed] [Google Scholar]
- 17.Albaugh VL, et al. Olanzapine promotes fat accumulation in male rats by decreasing physical activity, repartitioning energy and increasing adipose tissue lipogenesis while impairing lipolysis. Mol Psychiatry. 2011;16(5):569–581. doi: 10.1038/mp.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ader M, et al. Metabolic dysregulation with atypical antipsychotics occurs in the absence of underlying disease: a placebo-controlled study of olanzapine and risperidone in dogs. Diabetes. 2005;54(3):862–871. doi: 10.2337/diabetes.54.3.862. [DOI] [PubMed] [Google Scholar]
- 19.Gothelf D, et al. Weight gain associated with increased food intake and low habitual activity levels in male adolescent schizophrenic inpatients treated with olanzapine. Am J Psychiatry. 2002;159(6):1055–1057. doi: 10.1176/appi.ajp.159.6.1055. [DOI] [PubMed] [Google Scholar]
- 20.Coccurello R, et al. Olanzapine (LY170053, 2-methyl-4-(4-methyl-1-piperazinyl)-10H-thieno[2,3-b][1,5] benzodiazepine), but not the novel atypical antipsychotic ST2472 (9-piperazin-1-ylpyrrolo[2,1-b][1,3]benzothiazepine), chronic administration induces weight gain, hyperphagia, and metabolic dysregulation in mice. J Pharmacol Exp Ther. 2008;326(3):905–911. doi: 10.1124/jpet.108.137240. [DOI] [PubMed] [Google Scholar]
- 21.Fingerle-Rowson G, et al. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc Natl Acad Sci U S A. 2003;100(16):9354–9359. doi: 10.1073/pnas.1533295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson KA, et al. Hypothalamic CaMKK2 contributes to the regulation of energy balance. Cell Metab. 2008;7(5):377–388. doi: 10.1016/j.cmet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Bertile F, Oudart H, Criscuolo F, Maho YL, Raclot T. Hypothalamic gene expression in long-term fasted rats: relationship with body fat. Biochem Biophys Res Commun. 2003;303(4):1106–1113. doi: 10.1016/S0006-291X(03)00481-9. [DOI] [PubMed] [Google Scholar]
- 24.Fingerle-Rowson G, et al. Regulation of macrophage migration inhibitory factor expression by glucocorticoids in vivo. Am J Pathol. 2003;162(1):47–56. doi: 10.1016/S0002-9440(10)63797-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi SM, et al. Insulin regulates adipocyte lipolysis via an Akt-independent signaling pathway. Mol Cell Biol. 2010;30(21):5009–5020. doi: 10.1128/MCB.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shulman GI. Cellular mechanisms of insulin resistance. J Clin Invest. 2000;106(2):171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones ME, et al. Risk of mortality (including sudden cardiac death) and major cardiovascular events in users of olanzapine and other antipsychotics: a study with the general practice research database. Cardiovasc Psychiatry Neurol. 2013;2013:647476. doi: 10.1155/2013/647476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koro CE, et al. Assessment of independent effect of olanzapine and risperidone on risk of diabetes among patients with schizophrenia: population based nested case-control study. BMJ. 2002;325(7358):243. doi: 10.1136/bmj.325.7358.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serre-Beinier V, et al. Macrophage migration inhibitory factor deficiency leads to age-dependent impairment of glucose homeostasis in mice. J Endocrinol. 2010;206(3):297–306. doi: 10.1677/JOE-09-0342. [DOI] [PubMed] [Google Scholar]
- 30.He M, Zhang Q, Deng C, Wang H, Lian J, Huang XF. Hypothalamic histamine H1 receptor-AMPK signaling time-dependently mediates olanzapine-induced hyperphagia and weight gain in female rats. Psychoneuroendocrinology. 2014;42:153–164. doi: 10.1016/j.psyneuen.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Claret M, et al. AMPK is essential for energy homeostasis regulation and glucose sensing by POMC and AgRP neurons. J Clin Invest. 2007;117(8):2325–2336. doi: 10.1172/JCI31516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vozarova B, et al. Plasma concentrations of macrophage migration inhibitory factor are elevated in Pima Indians compared to Caucasians and are associated with insulin resistance. Diabetologia. 2002;45(12):1739–1741. doi: 10.1007/s00125-002-0896-4. [DOI] [PubMed] [Google Scholar]
- 33.Kim H, et al. Elevated levels of macrophage migration inhibitory factor in women with metabolic syndrome. Horm Metab Res. 2011;43(9):642–645. doi: 10.1055/s-0031-1283150. [DOI] [PubMed] [Google Scholar]
- 34.Calandra T, et al. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377(6544):68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 35.Morrison MC, Kleemann R. Role of macrophage migration inhibitory factor in obesity, insulin resistance, type 2 diabetes, and associated hepatic co-morbidities: a comprehensive review of human and rodent studies. Front Immunol. 2015;6:308. doi: 10.3389/fimmu.2015.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skurk T, Herder C, Kräft I, Müller-Scholze S, Hauner H, Kolb H. Production and release of macrophage migration inhibitory factor from human adipocytes. Endocrinology. 2005;146(3):1006–1011. doi: 10.1210/en.2004-0924. [DOI] [PubMed] [Google Scholar]
- 37.Church TS, et al. Obesity, macrophage migration inhibitory factor, and weight loss. Int J Obes (Lond) 2005;29(6):675–681. doi: 10.1038/sj.ijo.0802942. [DOI] [PubMed] [Google Scholar]
- 38.Duncan RE, Ahmadian M, Jaworski K, Sarkadi-Nagy E, Sul HS. Regulation of lipolysis in adipocytes. Annu Rev Nutr. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lafontan M. Adipose tissue and adipocyte dysregulation. Diabetes Metab. 2014;40(1):16–28. doi: 10.1016/j.diabet.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell RA, et al. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc Natl Acad Sci U S A. 2002;99(1):345–350. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDevitt MA, et al. A critical role for the host mediator macrophage migration inhibitory factor in the pathogenesis of malarial anemia. J Exp Med. 2006;203(5):1185–1196. doi: 10.1084/jem.20052398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das R, et al. Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2013;110(32):E2997–E3006. doi: 10.1073/pnas.1301128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finucane OM, et al. Macrophage migration inhibitory factor deficiency ameliorates high-fat diet induced insulin resistance in mice with reduced adipose inflammation and hepatic steatosis. PLoS ONE. 2014;9(11):e113369. doi: 10.1371/journal.pone.0113369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baugh JA, et al. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun. 2002;3(3):170–176. doi: 10.1038/sj.gene.6363867. [DOI] [PubMed] [Google Scholar]
- 45.Albaugh VL, et al. Hormonal and metabolic effects of olanzapine and clozapine related to body weight in rodents. Obesity (Silver Spring) 2006;14(1):36–51. doi: 10.1038/oby.2006.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minet-Ringuet J, Even PC, Lacroix M, Tomé D, de Beaurepaire R. A model for antipsychotic-induced obesity in the male rat. Psychopharmacology (Berl) 2006;187(4):447–454. doi: 10.1007/s00213-006-0433-0. [DOI] [PubMed] [Google Scholar]
- 47.Coccurello R, Moles A. A murine model of atypical antipsychotic-induced weight gain and metabolic dysregulation. Curr Protoc Neurosci. 2010;Chapter 9:Unit9.33. doi: 10.1002/0471142301.ns0933s52. [DOI] [PubMed] [Google Scholar]
- 48.Gil SY, et al. Clusterin and LRP2 are critical components of the hypothalamic feeding regulatory pathway. Nat Commun. 2013;4:1862. doi: 10.1038/ncomms2896. [DOI] [PubMed] [Google Scholar]
- 49.Yu H, et al. Multiecho reconstruction for simultaneous water-fat decomposition and T2* estimation. J Magn Reson Imaging. 2007;26(4):1153–1161. doi: 10.1002/jmri.21090. [DOI] [PubMed] [Google Scholar]
- 50.Donnelly SC, et al. Regulatory role for macrophage migration inhibitory factor in acute respiratory distress syndrome. Nat Med. 1997;3(3):320–323. doi: 10.1038/nm0397-320. [DOI] [PubMed] [Google Scholar]
- 51.Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF) Biochemistry. 1994;33(47):14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 52.Poon K, Barson JR, Fagan SE, Leibowitz SF. Developmental changes in embryonic hypothalamic neurons during prenatal fat exposure. Am J Physiol Endocrinol Metab. 2012;303(3):E432–E441. doi: 10.1152/ajpendo.00238.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zebisch K, Voigt V, Wabitsch M, Brandsch M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal Biochem. 2012;425(1):88–90. doi: 10.1016/j.ab.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Qi D, et al. Cardiac macrophage migration inhibitory factor inhibits JNK pathway activation and injury during ischemia/reperfusion. J Clin Invest. 2009;119(12):3807–3816. doi: 10.1172/JCI39738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.