Abstract
Mutant KRAS drives glycolytic flux in lung cancer, potentially impacting aberrant protein glycosylation. Recent evidence suggests aberrant KRAS drives flux of glucose into the hexosamine biosynthetic pathway (HBP). HBP is required for various glycosylation processes, such as protein N- or O-glycosylation and glycolipid synthesis. However, its function during tumorigenesis is poorly understood. One contributor and proposed target of KRAS-driven cancers is a developmentally conserved epithelial plasticity program called epithelial-mesenchymal transition (EMT). Here we showed in novel autochthonous mouse models that EMT accelerated KrasG12D lung tumorigenesis by upregulating expression of key enzymes of the HBP pathway. We demonstrated that HBP was required for suppressing KrasG12D-induced senescence, and targeting HBP significantly delayed KrasG12D lung tumorigenesis. To explore the mechanism, we investigated protein glycosylation downstream of HBP and found elevated levels of O-linked β-N-acetylglucosamine (O-GlcNAcylation) posttranslational modification on intracellular proteins. O-GlcNAcylation suppressed KrasG12D oncogene-induced senescence (OIS) and accelerated lung tumorigenesis. Conversely, loss of O-GlcNAcylation delayed lung tumorigenesis. O-GlcNAcylation of proteins SNAI1 and c-MYC correlated with the EMT-HBP axis and accelerated lung tumorigenesis. Our results demonstrated that O-GlcNAcylation was sufficient and required to accelerate KrasG12D lung tumorigenesis in vivo, which was reinforced by epithelial plasticity programs.
Keywords: Oncology
Keywords: Cancer, Cellular senescence, Glycobiology
Introduction
In the presence of mutant oncogenes, such as KRASG12D, normal primary cells initiate intrinsic tumor suppressor fail-safe mechanisms, such as oncogene-induced senescence (OIS) or oncogene-induced apoptosis (OIA), in response to oncogenic cellular stress (1, 2). To progress toward full-blown malignancy, these premalignant cells must initiate programs to suppress OIS-OIA. We and others have previously shown that TWIST1, a major regulator of embryogenesis and epithelial-mesenchymal transition (EMT) can suppress OIS-OIA (3–9). However, the more general role of the EMT in suppressing mutant Kras-induced OIS has not been fully explored in vivo and the underlying mechanisms for this process are not well understood.
The EMT is a highly conserved embryonic development program that is hijacked by cancers to regulate cellular plasticity. Epithelial plasticity programs like EMT can be activated by many oncogenes, including mutant KRAS and a variety of contextual signals that cancer cells may experience in diverse tissue sites throughout the body (10–13). Recent studies have confirmed molecular and cellular markers of EMT in diverse established human solid tumors. Epithelial plasticity programs have important roles in therapeutic resistance, tumor recurrence, and metastatic progression (11, 14, 15), and have been associated with poor clinical outcomes in multiple tumor types (16).
The hexosamine biosynthetic pathway (HBP) is a sugar metabolism pathway incorporating metabolites from major metabolic processes in the cell, such as glutamine, acetyl coenzyme A, glucose, and uridine (17) to form the amino-sugar molecule end product (UDP-GlcNAc). UDP-GlcNAc provides a substrate for the posttranslational modifications (PTMs) of both intracellular plasma membrane and secretory proteins. UDP-GlcNAc can be conjugated to target proteins in several ways, resulting in N-linked and O-linked glycosylated products with some of the most complex occurring in the Golgi apparatus and endoplasmic reticulum (18–21). One O-linked modification is O-GlcNAcylation or the addition of UDP-GlcNAc on serine and threonine residues of cytosolic and nuclear proteins by the enzyme O-GlcNAc transferase (OGT). HBP glycosylation function as a major metabolic integration point influencing a range of processes involved in the cell cycle, metabolism, and stress (22, 23). Recent evidence suggests that mutant KRAS drives increased glucose flux into the HBP pathway (24); however, the function of the HBP during tumorigenesis is poorly understood.
In the current study, using in vitro and in vivo models, we demonstrated that the EMT induced the expression of HBP genes, which was required for suppressing KrasG12D-induced senescence, and that targeting the HBP significantly delayed KrasG12D-induced lung tumorigenesis as well as established Kras mutant lung tumors. We investigated protein glycosylation downstream of the HBP and found elevated levels of O-linked β-N-acetylglucosamine (O-GlcNAcylation) posttranslational modifications on intracellular proteins. O-GlcNAcylation promoted suppression of KrasG12D-induced senescence (OIS) and accelerated KrasG12D-induced lung tumorigenesis. O-GlcNAcylation of the proteins Snai1 and c-Myc correlated with the EMT-HBP axis and accelerated KrasG12D-induced lung tumorigenesis. Conversely, loss of O-GlcNAcylation delayed KrasG12D-induced lung tumorigenesis. Our results demonstrate, for what we believe is the first time, that O-GlcNAcylation was sufficient and required to accelerate KrasG12D-induced lung tumorigenesis, which was reinforced by epithelial plasticity programs such as EMT.
Results
Snai1 and Twist1 accelerated KrasG12D-induced lung tumorigenesis.
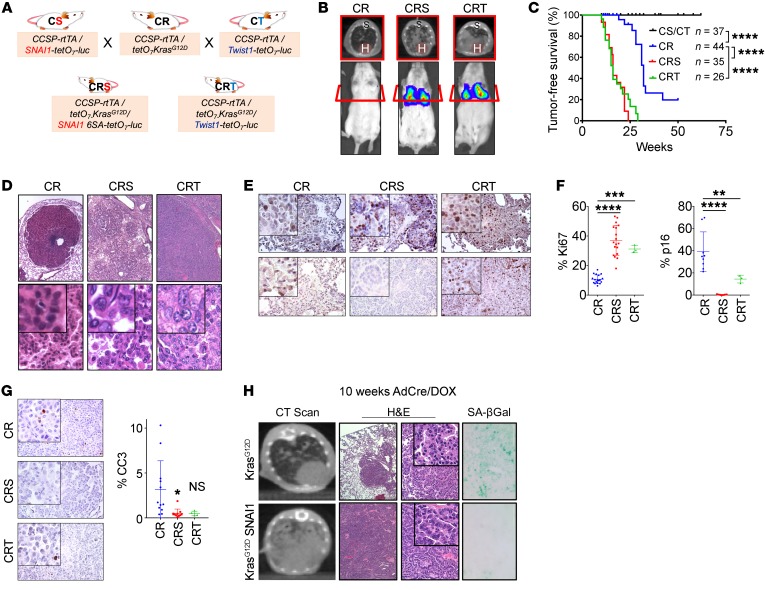
Two well-established EMT transcription factors (TFs) are SNAI1 and TWIST1, both of which are critical in development, aberrantly expressed in numerous cancers, and correlate with poor patient outcomes (25, 26). We generated novel lung-specific inducible mouse models coexpressing mutant KrasG12D and Snai1 (CRS) or Twist1 (CRT) (3) and compared them to mice expressing KrasG12D alone (CR) (Figure 1A and Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI94844DS1). Consistent with our previous work on TWIST1 (3), cone-beam computed tomography (CBCT) scans (Figure 1B) revealed that EMT TF-expressing CRS and CRT mice developed tumors twice as quickly (median 16 weeks) as CR mice overexpressing KrasG12D alone (median 32 weeks) and have significantly more tumors (Figure 1C and Supplemental Figure 1, C and D). Furthermore, while CR mice tend to develop adenomas, the EMT TF-expressing CRS and CRT mice developed adenocarcinomas (Figure 1D), which were dependent on the coexpression of KrasG12D and an EMT TF for tumor maintenance (Supplemental Figure 1E). The EMT TF overexpressing KrasG12D lung tumors also showed high proliferation, activated MAPK signaling, modestly decreased apoptosis, and low senescence relative to KrasG12D-alone tumors (Figure 1, E–H and Supplemental Figure 1, F and G, and ref. 3). To demonstrate EMT TF suppression of OIS, we generated Snai1- and Twist1-expressing (3) mouse embryonic fibroblast (MEF) cell lines and found EMT TF expression suppressed mutant Ras-induced OIS as shown by activated proliferation and reduced senescence-associated β-galactosidase (SA-βGal) staining (Supplemental Figure 1, H–J and ref. 3). Overall, EMT TFs suppressed OIS, accelerated lung tumorigenesis, and promoted lung tumor progression toward aggressive adenocarcinoma in KrasG12D-driven lung cells.
Figure 1. EMT transcription factors accelerated KrasG12D-induced lung tumorigenesis.
(A and B) Breeding schematic and bioluminescence imaging and CT scans of CR, CRS, and CRT mice. H, heart; S, spine. (C) Kaplan-Meier curve for tumor-free survival. The number of mice in each group is indicated. ****P < 0.0001, log-rank test. (D) H&E staining of representative mouse lung tumors in B at approximately 40 weeks. Original magnifications: top ×40, bottom ×200, insets ×400. (E–G) IHC of Ki67, p16, and cleaved caspase 3 (CC3) in CR, CRS, and CRT tumors with quantification. Positive staining was quantified in 5 different fields in each section. CR: n = 4, CRS: n = 5, CRT: n = 3. Original magnifications ×100, insets ×200. (H) CT scan, H&E, and SA-βGal staining of representative mouse lung tumor tissues from KrasG12D (LSL) or KrasG12D/SNAI1 (CS-LSL) mice. Original magnifications: second column ×4, third column ×100, insets ×200, last column ×200. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by 1-way ANOVA and Dunnett’s multiple comparisons test in F and G.
Snai1- and Twist1-induced expression of rate limiting enzymes of the HBP pathway.
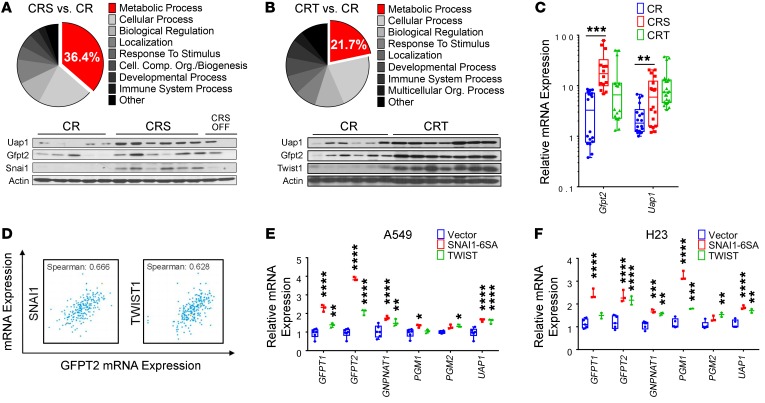
To identify mechanisms of EMT TF-mediated suppression of OIS, we used mRNA microarray analysis to identify differentially expressed genes between CR versus CRS and CR versus CRT lung tumors (Supplemental Table 1 and Supplemental Table 2). Gene Set Enrichment Analysis (GSEA) revealed a number of gene sets related to the EMT, senescence, and mutant KRAS-driven lung cancer (Supplemental Figure 1G and Supplemental Table 3). Interestingly, using gene ontology pathway analysis, we identified that one-third and one-fifth of the differentially expressed genes were involved in “metabolic biological processes,” in CR versus CRS and CR versus CRT models, respectively (Figure 2, A and B). Seahorse metabolic analyses confirmed that EMT TF-expressing non–small cell lung cancer (NSCLC) cells, A549 and PC9, exhibited elevated glucose utilization (Supplemental Figure 2, A and B). Interestingly, we found that our mouse lung tumors overexpressing Snai1 or Twist1 correlated with increased expression of critical genes in the HBP, a versatile energy-sensing pathway downstream of glycolysis. Specifically, EMT TFs correlated with elevated expression of the enzyme catalyzing the rate-limiting step (GFPT2) and final enzymatic step (UAP1) of the HBP as confirmed by qPCR and Western blot in both EMT TF mouse models (Figure 2, C–E) and in patient lung adenocarcinoma data from cBIO and The Cancer Genome Atlas (TCGA) (Figure 2F and Supplemental Figure 2C). In addition, both the transcriptomic profile of mesenchymal stem cells (Supplemental Figure 2D) and a cancer “mesenchymal metabolic signature” (MMS) (27) showed upregulation of GFPT2 and UAP1. GFPT2 was the top metabolism gene significantly overexpressed in tumorigenic KRASG12V-driven human lung epithelial cells (28). To verify this EMT-HBP axis, we overexpressed SNAI1 and TWIST1 in human mutant KRAS-driven NSCLC cell lines and found that HBP genes were similarly overexpressed (Figure 2, G and H). Additionally, knockdown models of SNAI1 in NSCLC cells and CRS lung tumor moribund mice without doxycycline (DOX) showed direct correlation of EMT with HBP genes (Supplemental Figure 2, E and F). Together, these data suggested a novel EMT-HBP axis in NSCLC cells in vitro, in patient lung cancer samples, and in our in vivo EMT TF-accelerated KrasG12D lung tumorigenesis mouse models.
Figure 2. EMT-induced expression of rate-limiting enzymes of the HBP.
(A and B) Enrichment for genes of metabolic process using gene ontology on differentially expressed genes between EMT TF and non–EMT TF–driven mouse tumors. (C–E) qPCR (C) and Western blot (D and E) for the HBP genes Gftp2 and Uap1 in lung tumors (CR: n = 6, CRS: n = 6, CRT: n = 7 mice per genotype). 18s rRNA was used as internal control for all qPCR assays, and CRS and CRT values were normalized to CR. (F) Correlation of EMT TFs SNAI1/TWIST1 and HBP gene GFPT2 in lung adenocarcinoma patient samples from cBIO database. (G and H) Expression of HBP genes in NSCLC cell lines stably infected with SNAI1 or TWIST1. Quantitative PCR was run in triplicate from 2 independent experiments. Unless stated, error bars indicate mean ± SD. P values were derived from 2-way ANOVA and Sidak’s multiple comparisons test in C, G, and H. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The HBP is required for EMT-mediated suppression of OIS.
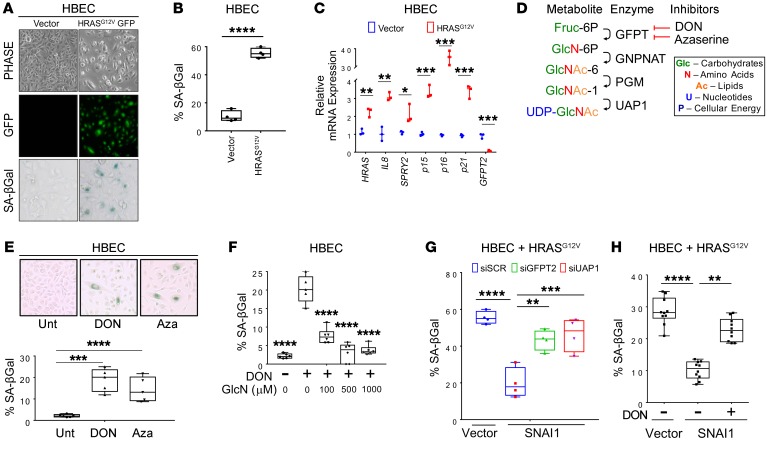
To determine the requirement of the HBP for senescence, we made use of the non–cancer human bronchial epithelial cell line (HBEC3-KT) (29) that underwent OIS upon HRASG12V transduction (Figure 3, A–C). Genetic and pharmacological inhibition of the HBP (Figure 3D) in HBEC3-KTs induced markers of senescence and decreased cell counts (Figure 3E and Supplemental Figure 3, A–E). As the glutamine antagonist 6-diazo-5-oxo-L-norleucine (DON) is not a specific inhibitor of GFPT2, we also demonstrated that senescence was specific to the HBP with a metabolite rescue experiment: addition of downstream metabolites of the HBP rescued DON-induced senescence (Figure 3F and Supplemental Figure 3, F and G), suggesting the HBP is a critical pathway controlling cellular senescence. We also found that SNAI1 could suppress HRASG12V-mediated OIS in HBEC3-KT cells (Figure 3, G and H), in alignment with previous work with Twist1-expressing MEFs (3). Genetic (Figure 3G and Supplemental Figure 3H) and pharmacological (Figure 3H and Supplemental Figure 3I) inhibition of the HBP abrogated the ability of EMT TF expression to suppress HRASG12V-mediated OIS in our SNAI1-overexpressing HBEC-3KT model.
Figure 3. The HBP is required for EMT-mediated suppression of oncogene-induced senescence (OIS).
(A–C) HBECs infected with HRASG12D were stained by counting SA-βGal–positive cells (A), and quantified (B) using qPCR of senescence marker gene expression (C). Original magnification in A is ×200. Quantitative PCR was run in triplicate from 2 independent experiments for C. (D) Illustration of the metabolites, enzymatic steps, and the inhibitors of the HBP pathway. (E) SA-βGal staining in HBECs upon 48-hour pharmacological inhibition of the HBP with 50 μM 6-diazo-5-oxo-L-nor-leucine (DON) and Azaserine (Aza). Original magnification ×200. (F) SA-βGal quantification of HBECs rescued with glucosamine (GlcN). (G and H) SNAI1-mediated OIS suppression was assessed with genetic (G) or pharmacological (H) inhibition of the HBP. More than 500 cells were counted for SA-βGal quantification in each experiment. Data are representative of 3 independent experiments. Unless stated, error bars indicate mean ± SD. P values were derived from 1-way ANOVA and Dunnett’s multiple comparisons test in E–H. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine if the EMT TF expression directly impacts the HBP and OIS in vivo, we crossed our inducible Snai1-expressing mouse line (CS) with the conditionally activated Lox-Stop-Lox-KrasG12D (LSL) (30) to generate a novel triple transgenic CS-LSL mouse line (Supplemental Figure 4A). In support of our findings in the CRS mouse model (Figure 1), CS-LSL mice generated more rapidly growing, less senescent, and more aggressive tumors compared with LSL mice (Figure 1H). With this model, we were able to inactivate expression of Snai1 alone in the lungs while maintaining expression of KrasG12D, and found that inactivation of Snai1 promoted tumor stasis and elevated levels of p53 (Supplemental Figure 4, B–E). Furthermore, without Snai1, mRNA expression of the HBP genes diminished (Supplemental Figure 4D). Thus, EMT-mediated suppression of mutant RAS-induced OIS required the HBP in vitro, and Snai1 directly modulated the HBP in vivo.
Inhibition of HBP in mutant KRAS lung cancer cells resulted in senescence.
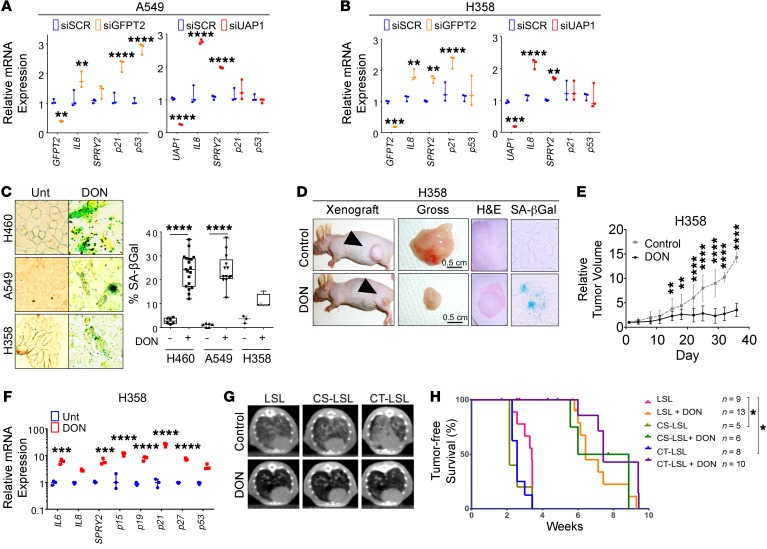
Genetic knockdown of GFPT2 and UAP1 in NSCLC cells revealed increased markers of senescence and reduced clonogenic potential (Figure 4, A and B and Supplemental Figure 5A, and Supplemental Figure 6, A and B). Conversely, GFPT2 overexpression increased clonogenic potential (Supplemental Figure 6C). Pharmacological inhibition of the HBP with low concentrations of DON (<10 μM) in NSCLC cell lines showed decreased cell viability, decreased clonogenicity, increased senescence morphology, and elevated molecular markers of senescence and apoptosis, whereas normal HBEC3-KT cells were seemingly unaffected (Figure 4C and Supplemental Figure 5, B–F). KRAS mutant H358 (Figure 4, D–F and Supplemental Figure 5, G–I) and A549 (Supplemental Figure 5, J–M) flank tumor moribund athymic nude mice were treated with DON (20 mg/kg/week) and treatment resulted in smaller tumors, reduced tumor growth, reduced gross tumor weight, and increased markers of senescence over 5 weeks without significantly impacting body weight (Supplemental Figure 5I). DON treatment also inhibited autochthonous lung tumorigenesis in the CS-LSL and CT-LSL mouse tumors without impacting body weight (Figure 4, G and H and Supplemental Figure 5, N and O). Therefore, targeting the HBP either genetically or pharmacologically in lung cancer inhibited early lung tumorigenesis and had therapeutic effects on established tumors that resulted in senescence and/or cell death.
Figure 4. Targeting the HBP in mutant KRAS lung cancer cells results in senescence.
(A and B) qPCR for senescence marker genes in NSCLC cell lines following 48 hours of GFPT2 and UAP1 siRNA knockdown. qPCR was run in triplicate from 3 independent experiments. (C) SA-βGal staining of NSCLC cell lines treated with ≤10 μM DON for 48 hours. More than 500 cells were counted for SA-βGal quantification in each experiment. Data are representative of 3 independent experiments. Original magnification ×400. (D) Tumor appearance and SA-βGal staining. (E) Quantified tumor volume (control: n = 10 tumors; DON: n = 14 tumors; original magnifications: H&E ×40, SA-Gal ×400). (F) qPCR for senescence marker gene expression of H358 NSCLC cell line xenograft in athymic nude mice treated with 20 mg/kg/week DON versus vehicle for 5 weeks. (G and H) Representative CT scans 6 weeks post AdCre (G) and tumor-free survival Kaplan-Meier curves of LSL, CS-LSL, and CT-LSL mice. Mice were treated with DON 20 mg/kg/3 days. The number of mice in each group is indicated (H). Unless stated, error bars indicate mean ± SD. P values were derived from 2-way ANOVA and Sidak’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
O-GlcNAcylation is required to suppress senescence.
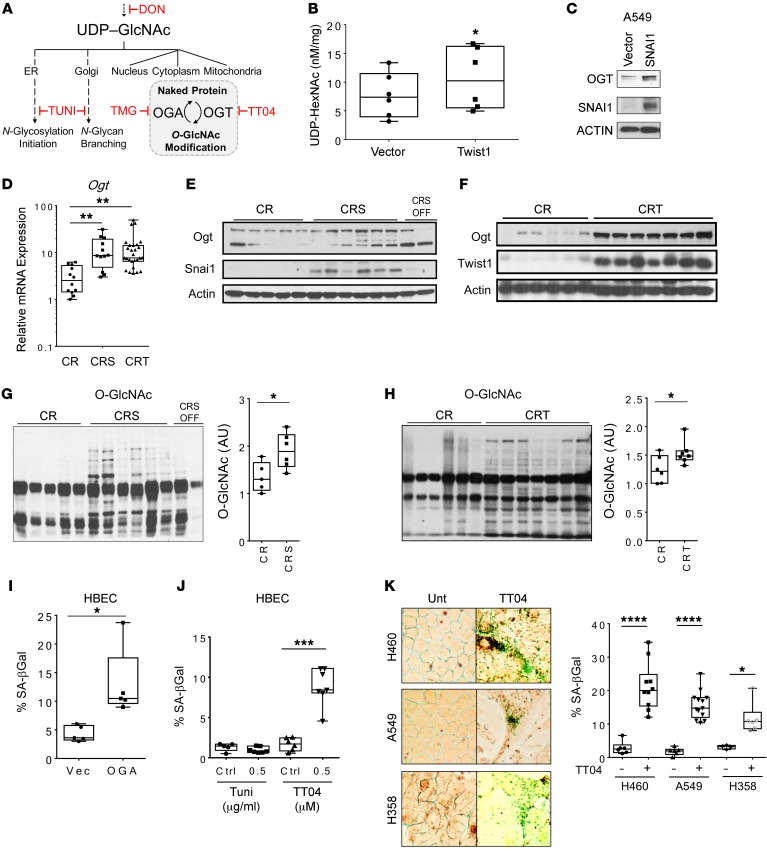
The HBP produces the nucleotide sugar UDP-GlcNAc, which can be utilized in multiple pathways, including both N-linked and O-linked protein glycosylations (Figure 5A), which regulate protein stability and activity (31–33). We performed reversed-phase HPLC to measure UDP-GlcNAc levels in isogenic human NSCLC cell lines (A549, H358, and H460) with and without Twist1 overexpression. Twist1 overexpression in aggregate showed increased levels of UDP-HexNAc (UDP-GlcNAc and downstream metabolite UDP-GalNAc), suggesting increased HBP flux (Figure 5B). O-GlcNAcylation is a single sugar modification found on proteins in the nucleus, cytoplasm, and mitochondria (34), and has previously been shown to stabilize SNAI1 in the nucleus (35). The O-GlcNAc modification (34, 36) is added and removed by just 2 enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), respectively (Figure 5A). In our models overexpressing EMT TFs, we found that the OGT levels were increased in both human NSCLC cells (Figure 5C) and mouse tumor tissues (Figure 5, D–F). Correspondingly, the total O-GlcNAcylation level was also directly correlated with EMT TF expression in mouse lung tumor tissues, NSCLC cells, and MEFs (Figure 5, G and H, Supplemental Figure 7, and Supplemental Figure 8, A–D).
Figure 5. O-GlcNAcylation is required to suppress senescence.
(A) Schematic of UDP-GlcNAc incorporation into N-linked and O-linked glycosylation with inhibitors (red). (B) Nucleotide sugar analysis with reversed-phase HPLC in several lung cancer cell lines overexpressing TWIST1 led to increased levels of O-HexNAc. Aggregate UDP-HexNAc level in A549, H460 and H358 cells with or without TWIST1 overexpression. Experiment was repeated 3 times. P = 0.0313, 2-sided Welch t test. (C) OGT protein level in A549 cells with overexpression of SNAI1. (D) Ogt mRNA level and (E and F) OGT protein levels by Western blot in lung tumor tissues from CR, CRS, and CRT mice (CR: n = 5, CRS: n = 6, CRT: n = 7 mice, same samples and actin/Snai1/Twist1 blot membrane as in Figure 2, D and E). (G and H) Global O-GlcNAcylation of proteins by Western blot in lung tumor tissues from CR, CRS, and CRT mice (CR: n = 5, CRS: n = 6, CRT: n = 7 mice). (I) SA-βGal staining following 48-hour attenuation of O-GlcNAcylation via overexpression of OGA. (J) SA-βGal staining following pharmacological inhibition of N-glycosylation (TUNI) compared with OGT inhibition (TT04). (K) SA-βGal staining of TT04-treated NSCLC cell lines. More than 500 cells were counted for SA-βGal quantification in each experiment. Data are representative of 3 independent experiments. Original magnification ×400. Error bars indicate mean ± SD. Unless stated, P values were derived from an unpaired, 2-tailed Student’s t test in B, G, H, and I; 1-way ANOVA and Dunnett’s multiple comparisons test were performed in D and J; 2-way ANOVA and Sidak’s multiple comparisons test were performed in K. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We performed a variety of genetic and pharmacologic methods to manipulate O-GlcNAcylation in normal and cancer cells to assess effects on senescent phenotypes. Genetic ablation of Ogt reduced cell viability upon conditional Ogt knockout in MEFs over 5 days (Supplemental Figure 9A). Reduction of O-GlcNAcylation in HBEC3-KT cells via genetic overexpression of OGA (Figure 5I) and pharmacological inhibition with OGT inhibitor TT04 (Figure 5J) promoted senescence, whereas inhibition of N-glycosylation with tunicamycin (TUNI) had no effect on senescence (Figure 5J, Supplemental Figure 10). Requirement of O-GlcNAcylation was assessed by crossing the LSL mouse line with a Cre-conditional Ogtfl/fl mouse line (37). Ex vivo knockout of Ogt in isolated type II pneumocytes from LSL-Ogtfl/Y mice resulted in more senescent cells, which could be partially rescued by the OGA inhibitor thiamet-G (TMG) (Supplemental Figure 9B). Genetic knockdown of OGT in A549 cells reduced NSCLC clonogenicity (Supplemental Figure 9C) while reducing O-GlcNAcylation in A549 cells in vitro with overexpression of OGA also led to cellular senescence (Supplemental Figure 9D). Additionally, treatment with the OGT inhibitor TT04 increased SA-βGal–positive staining and promoted senescent cellular morphology in other NSCLC cells (Figure 5K). Importantly, TMG treatment of LSL mice resulted in elevated protein O-GlcNAcylation and decreased expression of several genes associated with senescence in vivo (Supplemental Figure 9E). These data indicate that EMT TFs and the sugar PTM O-GlcNAcylation promoted each other in a feed-forward loop to regulate senescence in vitro and in vivo.
O-GlcNAcylation is sufficient and required for acceleration of KrasG12D-induced lung tumorigenesis.
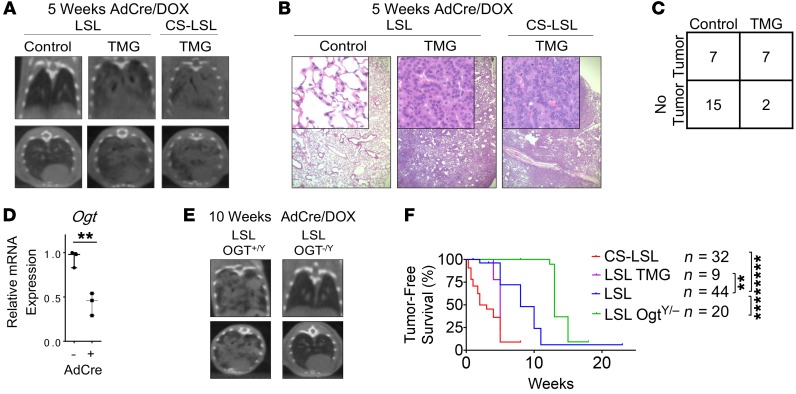
We next determined the importance of O-GlcNAcylation for KrasG12D-induced lung tumorigenesis by using established and novel autochthonous mouse models. O-GlcNAcylation was sufficient to significantly accelerate KrasG12D-induced lung tumorigenesis as shown by pharmacological elevation of O-GlcNAcylation with TMG treatment in the LSL model (median 5 weeks; Figure 6, A–C). We then generated LSL-Ogtfl/fl mice that simultaneously express KrasG12D with concomitant knockout Ogt specifically in the lung epithelial cells upon intranasal administration of Adeno-CMV-Cre (AdCre) virus (Figure 6, D and E). Genetic knockout of Ogt in vivo reduced O-GlcNAc levels, but did not induce obvious cell death in mouse lung tissues (Supplemental Figure 11 and Supplemental Figure 12). However, male LSL-Ogtfl/Y mice demonstrated that O-GlcNAcylation was required for KrasG12D-induced lung tumorigenesis as demonstrated by a significant delay in lung tumor development (median 15 weeks; Figure 6F). Overall, data from these autochthonous mouse models revealed O-GlcNAcylation was necessary and sufficient for KrasG12D-induced lung tumorigenesis.
Figure 6. O-GlcNAcylation is sufficient and required for KrasG12D-induced lung tumorigenesis.
(A–C) O-GlcNAcylation was elevated pharmacologically with 1.5 mg/mL TMG in LSL mouse drinking water, and tumor burden was assessed by CT scans (A), H&E (B), and contingency table (C). The number of mice in each group was indicated. Fisher’s exact test P = 0.0439, 2-tailed. Top and bottom panels in A are representative coronal and axial sections from CT scan of the same mouse. Original magnification ×40; insets ×200 (B). (D–F) Ogt mRNA expression (D) and tumor burden via CT scan (E) at 10 weeks after intranasal administration of AdCre in Ogt knockout mice (n = 3 without AdCre, n = 3 with AdCre). (F) Tumor-free survival Kaplan-Meier curves of LSL mice (median 8 weeks post-AdCre) compared with CS-LSL (median 2 weeks), elevated O-GlcNAcylation mice (LSL-TMG median 5 weeks), and Ogt knockout mice (LSL-Ogt–/Y median 15 weeks). The number of mice in each group is indicated. Error bars indicate mean ± SD. Unless stated, P values were derived from unpaired, 2-tailed Student’s t test in D, and log-rank test in F. **P < 0.01, ****P < 0.0001.
EMT elevates O-GlcNAcylation to suppress senescence and stabilizes oncoproteins like c-Myc.
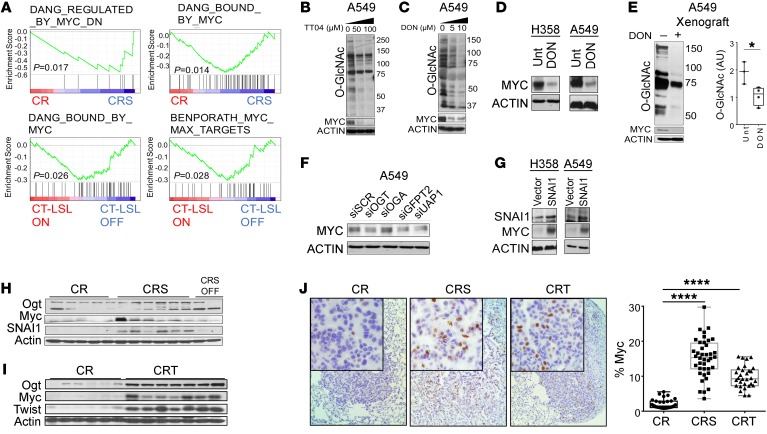
Having established that the EMT-HBP axis required O-GlcNAcylation to suppress senescence and accelerate KrasG12D-induced lung tumorigenesis, we then focused on O-GlcNAcylated candidates that have been shown to be critical for tumorigenesis. One promising target was the oncoprotein c-MYC, based on multiple studies that KrasG12D and MYC oncogenes can have collaborative effects during tumorigenesis in vivo (38–40) and c-MYC itself is stabilized by O-GlcNAcylation (41). We found Myc gene sets were overrepresented by GSEA in our EMT KrasG12D-driven lung cancer mouse models (Figure 7A). We then found that pharmacological inhibition of O-GlcNAcylation with TT04 or DON (Figure 7, B–E) or genetic knockdown (Figure 7F) resulted in a reduction of c-MYC in NSCLC cells in vitro and in vivo. Similarly, overexpression of EMT TFs in human and mouse NSCLC cells resulted in increased c-MYC levels in vitro (Figure 7G) and in vivo (Figure 7, H–J, and Supplemental Figure 13, A and B). Taken together, our data suggested that the EMT-HBP axis may suppress OIS through stabilizing oncogenes such as c-Myc, which have been shown previously to suppress mutant Kras-induced OIS.
Figure 7. EMT elevates O-GlcNAcylation stabilizing the oncoprotein c-Myc in KRAS mutant lung cancer cells.
(A) Gene expression data from EMT TF models of KrasG12D-driven lung cancer were enriched for differentially expressed genes related to c-Myc programs (data sets obtained from the Molecular Signatures Database). (B–E) Western blot of global O-GlcNAcylation levels and c-Myc protein levels in NSCLC cells treated for 48 hours with the OGT inhibitor TT04 (B) or DON (C and D) in vitro (48 hours) and A549 xenografts in vivo (3 weeks, E). Levels of c-Myc by Western blot (F) after 48-hour transient genetic knockdown of OGT, OGA, GFPT2, and UAP1 in A549 cells. (G) Western blots of SNAI1 and c-Myc levels in A549 and H358 NSCLC cell lines with SNAI1 overexpression. (H and I) c-Myc protein level in CR, CRS, and CRT mice (CR: n = 5, CRS: n = 6, CRT: n = 7 mice, same samples and blot membrane as in Figure 2, D–E and Figure 5, E–H). (J) IHC for c-Myc in CR, CRS, and CRT tumors with quantification. Positive staining was quantified in 5 different fields in each section, CR: n = 4, CRS: n = 5, CRT: n = 3. Original magnifications ×40; insets ×200. Error bars indicate mean ± SD. P values were derived from unpaired, 2-tailed Student’s t test in E; and from 1-way ANOVA and Dunnett’s multiple comparisons test in J. *P < 0.05, ****P < 0.0001.
Discussion
Alteration of cellular metabolism has been shown to be important for survival and progression of established cancer cells (42). It is likely that metabolic byproducts reinforce the cancer phenotype by modulating not just metabolic maintenance but also altering other cellular structures and functions. In particular, the importance of PTMs such as O-GlcNAcylation are becoming realized as they provide rapid, reversible adaptations to the stressors that occur during tumor progression. Importantly, targeting specific glycosylations have potential as novel cancer treatment strategies (43). The role of these metabolic adaptations for early tumorigenesis are less well appreciated. Recent evidence correlates EMT-related transcriptomic alterations with metabolic reprogramming in cancer, which include increased aerobic glycolysis and the accumulation of dihydropyrimidine (27, 44). We and others have demonstrated an important role for epithelial plasticity programs such as EMT not only during tumor progression toward metastasis and treatment resistance, but perhaps more importantly, also during the earliest stages of tumorigenesis (3–9). Herein we show that EMT and the hexosamine biosynthetic metabolic pathway form a novel axis accelerating Kras mutant lung tumorigenesis that can be targeted to produce antitumor effects.
Previous unsupervised hierarchical clustering using metabolic gene expression profiling of 978 cancer cell lines from many different representative types of cancers resulted in the clear identification of a mesenchymal subset of cancer cells. These mesenchymal cancer cell lines were composed of lines derived directly from a cancer with mesenchymal cell of origin but also epithelial cancers that were high-grade and had assumed a more plastic phenotype. When clustered together these mesenchymal cancer cell lines shared a distinct MMS that was particularly enriched for glycan biosynthesis genes, most prominently GFPT2 and UAP1 (27). A prior link between TGF-ϐ–induced EMT and increased O-GlcNAcylation was recently observed in A549 cells (45), but the functional significance for this EMT-HBP axis for tumorigenesis and tumor maintenance was not examined. Our data suggest that when epithelial plasticity programs driven by TWIST1 or SNAI1 are activated, they too metabolically reprogram epithelial lung cells and lung cancer cells by stimulating the HBP pathway, and increase global protein O-GlcNAcylation in vitro and in vivo. Due to experimental limitations of propagating and genetic manipulation of primary type II pneumocytes from mice, we were unable to directly test the senescence response to HBP inhibition in these cells. However, using human lung cells, we showed that genetic and pharmacologic manipulation of the HBP pathway resulted in changes in cellular senescence in many cell types and rescued this phenotype with the GlcN metabolite in normal human lung cells. We also directly measured the level of the UDP-GlcNAc (and downstream metabolite UDP-GalNAc) with HPLC after overexpression of EMT TFs, demonstrating that EMT TFs increased HBP metabolite flux toward total global protein O-GlcNAcylation. However, further investigation using other technologies such as DIONEX will help to determine the levels of GlcNAc more precisely in EMT TF-overexpressed cells. Furthermore, we observed that these transcriptional and metabolic changes in lung cancer cells in vivo were not necessarily associated with cells that had undergone a classical full EMT and were generally morphologically and histologically epithelial. Whether what we have observed in our mouse models could be considered a partial EMT-like state requires further study (26). Regardless, the collection of data thus far strongly suggests that when epithelial cells activate EMT programs, resulting in either partial or full EMT morphological phenotypes, there is an obligate metabolic reprogramming that favors HBP activation and increased global O-GlcNAcylation.
OIS and OIA elicited by oncoproteins such as KrasG12D represent early tumor suppressive barriers that must be overcome for premalignant cells to ultimately emerge as neoplastic (46). We and others have reported previously that EMT TFs could suppress OIS in vitro and in vivo, resulting in acceleration of tumorigenesis and tumor progression (3–9). Mechanisms implicated for EMT TF suppression of OIS/OIA involved inhibition of p53 (reviewed in ref. 42) and direct transcriptional regulation of BMI1 (47). Now, in this report, we have also implicated the HBP in the suppression of cellular senescence and OIS. The HBP results in altered protein glycosylation and is well positioned to mediate many cellular changes associated with EMT, including cell-cell adhesion, responsiveness to growth factors, immune system evasion, and prooncogenic signal transduction programs (38). One intriguing factor that can be O-GlcNAcylated and has been shown to suppress KrasG12D-induced OIS, resulting in accelerated tumorigenesis in vivo, is c-Myc (38–40, 48). HBP O-GlcNAcylation has been shown to stabilize c-Myc levels (41), which we confirmed in human and mouse lung cancer cells can be driven by EMT TFs. Thus, we have nominated c-Myc as a likely O-GlcNAcylated candidate that suppresses KrasG12D-induced OIS. Myc has been shown previously to suppress KrasG12D-induced OIS and contribute to increased tumorigenesis (38–40, 48). Future studies will be directed at confirming the causal role of O-GlcNAcylated c-Myc and isolating any other proteins mediating KrasG12D-induced OIS suppression.
The HBP utilizes glycolytic intermediates to generate the metabolite UDP-GlcNAc. This and other charged nucleotide sugars serve as the basis for biosynthesis of glycoproteins and other glycoconjugates (18–21). KrasG12D has been previously shown to drive increased flux through the HBP in an inducible mouse model of pancreatic ductal adenocarcinoma (24). The requirement for increased HBP flux for KrasG12D-induced tumorigenesis and tumor maintenance was not explored in this model. Using complimentary genetic and pharmacologic methods in vitro and in vivo, we showed for the first time that the HBP was required for KrasG12D-induced lung tumorigenesis and lung tumor maintenance. As the HBP is required for various glycosylation processes, such as protein N-linked glycosylation, O-GlcNAcylation, and glycolipid synthesis (31, 49), it was important for us to examine whether this HBP requirement constituted N-linked downstream events or O-GlcNAcylation. Using more specific genetic (OGT and OGA manipulation) and pharmacologic (TT04) tools, we established that O-GlcNAcylation was not only required for KrasG12D-induced lung tumorigenesis and tumor maintenance in vitro and in vivo, but was also sufficient to accelerate KrasG12D-induced lung tumorigenesis. Although our data showed that no significant change in the level of N-linked glycosylation was detected with overexpression of SNAI1 or TWIST1 (Supplemental Figure 8) with a concanavalin A (ConA) blot, this technique only recognizes the core of the N-linked glycans. However, the HBP can also impact N-linked glycan branching and many other glycosylation pathways, and this aspect of the EMT-HBP axis will need to be more fully explored in future studies.
The involvement of EMT-mediated HBP metabolic reprogramming may extend beyond KRAS-mutant lung cancers, as our prior work suggests that TWIST1 knockdown can result in activation of latent OIS-OIA in other oncogene-driven lung cancer cells where KRAS is WT (4). As HBP flux is central to many glycosylation events, EMT-driven changes in the HBP can potentially impact other glycan classes, such as the O-linked glycoproteins. Many O-linked glycoproteins are large molecules, such as mucin, that are involved in supporting cell-cell and cell–extracellular matrix (ECM) interactions, and can also promote cancer cell growth and survival. This EMT-HBP axis may also be broadly important in other types of lung cancer, as we have shown previously that TWIST1 was found to be overexpressed in a majority of human lung cancer samples including not only adenocarcinoma but also other major lung cancer histologies such as squamous cell carcinoma (3). This hypothesis can be further explored in vivo through introduction of our inducible SNAI1 and Twist1 mouse models into other relevant transgenic oncogene-driven models of lung tumorigenesis.
Importantly, regardless of whether there is an exclusive association between KRAS mutation and EMT TF overexpression in human lung cancer cells, the data presented strongly support that EMT TF expression in KRAS mutant lung cancers represents a novel and particularly promising therapeutic target. Evidence suggests that EMT plays important roles in therapeutic resistance, tumor recurrence, and metastatic progression, therefore, targeting EMT is an attractive therapeutic approach to inhibit cancer progression (50). However, similar to mutant KRAS, due to the complexity of the signaling networks of different EMT TFs and that all the master regulators are transcription factors in the nucleus, the EMT remains difficult to pharmacologically target. Fortunately, our data demonstrate O-GlcNAcylation is a product of EMT-driven metabolic reprogramming that contributes to KrasG12D-induced lung tumorigenesis and tumor progression. Thus, inhibition of the HBP and O-GlcNAcylation can impact lung cancer cell death and senescence, suggesting the HBP is a novel targetable pathway to indirectly target EMT- and KRAS-driven lung cancers.
Methods
Cell lines and drug treatments.
The human NSCLC cell lines (H460 and A549) and embryonic kidney cell lines (HEK-293T and Gryphon) were obtained from ATCC and grown in media as recommended. The NSCLC cell lines (H358 and H23) and human bronchial epithelial cell (HBEC-3KT) line were gifts from Steve Baylin (Johns Hopkins University School of Medicine, Baltimore, MD, USA). OgtY/fl MEFs were generated and maintained as previously described. Inducible SNAI1 MEFs were isolated from E13.5 embryos and propagated as previously described (3). MEFs were grown for 2 population doublings and then frozen for future experiments. MEFs were grown in DMEM supplemented with 10% fetal calf serum. All cell lines were checked by short tandem repeat profiling and mycoplasma testing services of the Johns Hopkins Medicine Genetic Resources Core Facility. Cells were treated with the appropriate inhibitor for 48 hours prior to analysis using the following compounds: 6-Diazo-5-oxo-L-norleucine (D2141, Sigma), TT04 (ST045849, TimTec), TMG (4390, Bio-Techne), tunicamycin (T7765, Sigma), and azaserine (A4142, Sigma).
PCR genotyping.
DNA was isolated from mouse tails by boiling in 100 μl Solution A (0.5M EDTA pH 8 and 10M NaOH in diH2O) for 1 hour and then adding Solution B (Tris HCl pH 8 in diH2O). The CCSP-rtTA (C), tetO7-KrasG12D (R), Twist1-tetO7-luc (T), and LSL-KrasG12D (LSL) transgenic lines were screened as previously described. The SNAI1-tetO7-luc line was detected with the following primers: SNAI1-Luc.S2 59-CCTTATGCAGTTGCTCTCCAG-39 and SNAI1-Luc.AS2 59-GCTTGCCTATGTTCTTTTGGA-39. DNA was amplified using PCR, and PCR products were resolved on a 2% agarose gel.
Transgenic mice.
The human SNAI1-6SA cDNA was PCR cloned into the bidirectional tetO7 vector S2f-IMCg at EcoRI and NotI sites, replacing the eGFP ORF. The resultant construct, SNAI1-6SA-tetO7-luc, was sequence confirmed, digested with KpnI and XmnI to release the bidirectional transgene, and then used for injection of FVB/N pronuclei by the Stanford Transgenic Facility. We obtained 1 founder after screening by tail genotyping using PCR as described below. This founder was mated to CCSP-rtTA mice to screen for inducible SNAI1-tetO7-luc expression and was used for all the experiments in this study.
We use the β-actin-rtTA, CCSP-rtTA, tetO7-KrasG12D, Twist1-tetO7-luc, and LSL-KrasG12D single transgenic lines and bred these mouse lines to generate the appropriate double and triple transgenic mouse lines. SNAI1, Twist1, and/or KrasG12D expression was activated in the CS, CT, CR, CRS, CRT, CS-LSL, and CT-LSL lung lines by administering DOX (Sigma) to the drinking water (2 mg/ml) starting at the age of 4–5 weeks and was changed with fresh DOX weekly. The conditional LSL-KrasG12D lines were activated by intranasal delivery of Ad-CMV-iCre (catalog 1045, Vector Biolabs). DON (D2141, Sigma) was administered i.p. (20 mg/kg) to mice BID and TMG (4390, Bio-Techne; 1.5 mg/ml) was administered in mouse drinking water changed every 3 days.
LSL-KrasG12D Ogt knockout mice.
The Cre-conditional X-linked OGTfl/fl mouse line was genotyped and mouse lines were maintained as previously described (37). Male LSL-KrasG12D mice with WT OGTY/+ were crossed with female homozygous OGTfl/fl mice, resulting in all homozygous OGTY/fl males. Only male LSL OGTY/fl mice were used in experiments, as males were homozygous for the floxed allele compared with hemizygous females. Intranasal administration of Ad-CMV-iCre (1045, Vector Biolabs) was used to infect lung epithelial cells, simultaneously inducing KrasG12D expression while knocking out OGTY/–. OGT knockout was validated by qPCR from frozen lung tissue samples.
Small animal imaging and quantification.
CBCT was performed on an in-house standalone system. Each animal was placed in a rotatable animal stage situated between the stationary and parallel opposed x-ray source and the detector panel. Mice were screened serially every 1–5 weeks following DOX activation and/or intranasal adenoviral Ad-CMV-iCre (1045, Vector Biolabs) and images were reviewed by a board-certified radiation oncologist (PTT). No attenuation correction or partial volume corrections was applied. CBCT images were reviewed by a board-certified radiation oncologist (PTT) for quantification at 6 months post-DOX.
Mouse xenograft model.
Female athymic nude mice 4 to 5 weeks old were purchased from Harlan Laboratories. Mice were maintained under pathogen-free conditions and given food and water ad libitum in accordance with guidelines from the Johns Hopkins Animal Care and Use Committee. A549 cells (1 × 1010) or H358 cells (8 × 1010) in 100 ml RPMI and Matrigel (BD Biosciences) mixed 1:1 were injected subcutaneously in right and left flanks. Mice were split into DON-treated and untreated groups (n ≥ 16 tumors) and tumor measurements were taken every 3 days for 3–5 weeks. DON (D2141, Sigma) was dissolved in sterile diH2O and was administered once a week i.p. within 1 week posttumor injection (or when small tumors were first visible, ~50 mm3) for 3–5 weeks. Mice were euthanized by CO2 when tumors reached the volume of 500 mm3 (1,000 mm3 per mouse).
SYBR-green quantitative RT-PCR.
Total RNA was isolated from tissue using TRIzol RNA Isolation Reagent (Life Technologies) according to the manufacturer’s protocol. cDNA was generated from 1 μg total RNA using the iScript cDNA synthesis kit (Bio-Rad). cDNA was amplified with iTaq Universal SYBR Green Supermix (Bio-Rad) using the CFX384 Touch Real-Time PCR Detection System (Bio-Rad) with the C1000 TouchThermal Cycler (Bio-Rad) for up to 40 cycles. PCR reactions were performed at least in triplicate in a final volume of 10 μl.
For every sample, the difference in cycle threshold (ΔCt) values were determined as the difference between the Ct value of a specific transcript and the Ct value of 18s rRNA, serving as the housekeeping control messenger RNA. Relative transcript levels (for example, treated versus untreated) were calculated based on 2(–ΔΔCt), with ΔΔCt = (ΔCt treated) – (ΔCt untreated). Primers for qPCR are listed in Supplemental Table 4.
Immunoblot, immunofluorescence, and lectin blot analysis.
Cells were lysed on ice for 60 minutes in RIPA lysis buffer (R0278, Sigma) supplemented with Protease Inhibitor Cocktail (P8340, Sigma), phenylmethylsulfonyl fluoride (PMSF; 10837091001, Sigma), β-hexosaminidase inhibitor (376820, Millipore), and TMG (4390, Bio-Techne) inhibitors and clarified by centrifugation. Protein concentrations were determined by Pierce660nm Protein Assay (Bio-Rad). Equal aliquots corresponding to 20–60 μg protein were resolved on 6%–12% SDS-polyacrylamide gels and transferred to PVDF or nitrocellulose membranes. Antigen detection was carried out with antibodies against the following proteins: Actin (C-11, Santa-Cruz Biotechnology, 1:5,000), Twist1 (TWIST2C1a, Santa-Cruz Biotechnology, 1:100), SNAI1 (NBP1-80022, Novus, 1:2,000; H-130, Santa-Cruz Biotechnology, 1:500), GFPT2 (EPR19095, Abcam, 1:500), UAP1 (EPR10259, Abcam, 1:500), c-Myc (Y69, Abcam/Epitomics, 1:10,000), p21 (12D1, Cell Signaling Technology, 1:1,000), p53 (9282, Cell Signaling Technology, 1:1,000), OGT (DM-17, Sigma-Aldrich, 1:2,000), O-GlcNAc (CTD110.6, 1:2,000, Cell Signaling Technology), and cleaved caspase 3 (9664S, Cell Signaling Technology). After washing, corresponding anti-mouse IgG (GE Healthcare, 1:2,500), anti-rabbit IgG (GE Healthcare, 1:2,500), and anti-rabbit IgM (MilliporeSigma, 1:2,500) peroxidase-conjugated secondary antibodies were used. Antigen-antibody complexes were visualized by Amersham electrochemiluminescence (ECL detection system; GE Healthcare). CTD110.6 specificity for O-GlcNAc was tested by blotting with or without 200 mM N-Acetylglucosamine (GlcNAc; MilliporeSigma). Relative O-GlcNAcylated protein levels were measured by analyzing the band density using ImageJ (http://imagej.nih.gov/ij/download.html) then normalized to the density of actin. All experiments were performed in triplicate and average relative fold changes were calculated.
For immunofluorescence, SNAI1 (H-130, Santa-Cruz Biotechnology, 1:50) and O-GlcNAc (RL2, Novus, 1:200) were incubated at 4°C overnight. After washing, Alexa 488–conjugated anti-mouse (green) and Alexa 594–conjugated anti-rabbit (red) (Invitrogen, 1:300) were used as secondary antibodies and incubated at room temperature for 30 minutes. DAPI (blue) was used as a nuclear stain and slides were mounted in aqueous mounting media (Vector Laboratories).
For N-glycosylation analysis, membrane was blocked in 3% BSA in TBS, washed in TBS, and incubated at 4°C overnight with Concanavalin A-HRP lectin (ConA; L6397, Sigma, 0.25 μg/ml) in high-salt TBST (HS-TBST; 1M NaCl, TBST). The blot was washed extensively in HS-TBST and developed using ECL detection system (GE Healthcare). ConA specificity was tested by blotting with or without 200 mM mannose (MilliporeSigma).
Histology and immunohistochemistry.
Formalin-fixed, paraffin-embedded (FFPE) tissues were processed and stained with H&E according to common procedures. Tissues were prepared on plus slide antigens, and antigens were retrieved with Target Retrieval Solution (S1699, Dako) and probed for antigen with the following antibodies: Ki-67 (NCL-Ki67p, Leica, 1:1,000), p16 (F-12, Santa-Cruz Biotechnology, 1:200), pERK (4370, Cell Signaling Technology, 1:400), cleaved caspase-3 (9664, Cell Signaling Technology, 1:2,000), SNAI1 (NBP-80022, Novus, 1:200), c-Myc (Y69, Abcam/Epitomics, 1:500), and O-GlcNAc (RL2, Novus, 1:200), as previously described. For pathological analysis, a certified veterinarian pathologist (BWS) reviewed the disease burden for each mouse. For immunohistochemistry analysis, immunopositive cells were counted and analyzed from at least 5 different fields of view per section from more than 3 different animals in total. For SA-βGal stain, 10-μm sections were cut from Tissue-Tek cryo-OCT–infused (14-373-65, Thermo Fisher Scientific) frozen lung tissue, and staining was carried out as previously described (3). H&E, tissue processing, and c-Myc IHC were performed using the Johns Hopkins Oncology Tissue Services Core Facility.
Viral transduction and siRNA transfection.
Lentivirus was generated by transfecting HEK-293T cells with SNAI1 shRNA constructs obtained from the Broad RNAi Consortium using pLKO.1-shRNA scramble vector (MilliporeSigma) as a negative control. H358 NSCLC cell lines were then transduced with virus as previously described. Retrovirus was produced using the Gryphon Amphotropic Packaging Cell Line (Allele Biotechnology), per the company’s recommendation. Early passage MEFs, noncancer HBEC3-KT cells, and NSCLC cell lines (A549, H460, and H23) were transfected with pWZL-Hygro vector (Ador pWZL-Hygro, Addgene) SNAI1 constructs twice in succession over a 36-hour period, followed by selection, as previously described for Twist1 cell lines. HBEC3-KT cells were infected with OGA or GFP control adenovirus at a MOI of 50 for 6 hours, and cell lysate was collected 48 hours after transduction. For siRNA transfection, cells were transfected with ON-TARGETplus nontargeting siRNA 1, human GFPT2 SMARTpool siRNA (L-010390-00-0005, Dharmacon), human UAP1 SMARTpool siRNA (L-017160-01-0005, Dharmacon), or human OGT SMARTpool siRNA (L-019111-00-0005, Dharmacon), for a final concentration of 50 nM. Samples were analyzed 48 hours after transfection.
Colony formation and proliferation assays.
Cells were transiently transfected with OGT siRNA and plated at 500 cells per 10-cm dish. After 2 weeks, colonies were fixed and stained with crystal violet (0.5% in 95% methanol, MilliporeSigma) and stained colonies (defined as ≥50 cells) were counted. Quantification of MEFs was performed as previously described by using crystal violet staining and quantification by spectrophotometer. For all other cell proliferation experiments, CyQUANT NF Cell Proliferation assay kit (Molecular Probes, Invitrogen) was used to minimize influence of mitochondrial metabolism, according to the manufacturer’s protocol.
SA-βGal staining.
After genetic or pharmacological treatment, noncancer and cancer cells were washed twice with PBS and then fixed with PBS containing 2% formaldehyde and 0.2% glutaraldehyde for 5 minutes. The cells were then incubated at 37°C overnight with staining solution (catalog 9860, Cell Signaling Technology). After incubation, cells were washed twice with PBS and viewed with bright-field microscopy. Cell number was measured by using the cell count feature of ImageJ (http://imagej.nih.gov/ij/download.html). All experiments were performed in triplicate and average relative fold changes were calculated.
Microarray data processing and analysis.
Total RNA was isolated and purified from cells using TRIzol RNA Isolation Reagent (Life Technologies) as detailed by the manufacturer’s recommendation, with tissue homogenization. Labeled RNA was hybridized to the Illumina Mouse WG-6 Expression Array and scanned as previously described. Gene expression data were normalized using the Robust Multichip Average in the oligo Bioconductor package at the transcript level. Genes and gene sets with Benjamini-Hochberg P values below 0.05 were considered statistically significant. Gene set enrichment analysis was performed using the C2 Curated Gene Sets collection from the Molecular Signature Database 3.0 and statistical comparisons by Fisher’s exact test. All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE117519).
Determining differentially expressed genes, enriched gene sets, and gene ontology.
Nonspecific filtering was used to remove genes with an interquartile range less than 0.05. To find differentially expressed genes between EMT/KrasG12D-driven tumors versus KrasG12D alone, Significance Analysis of Microarray (samr R package version 2.0) was applied using a 2-class unpaired comparison, minimal Z-score fold change of 1.2, and median false discovery rate of 0.05. Unannotated transcripts were not considered. To test whether gene sets were enriched in response to different conditions, we used gene set analysis as implemented in the GSA R package version 1.0329. The maximum mean test statistic was used to test enrichment using a 2-class comparison. All P values and false discovery rates were based on 500 to 1,000 permutations. For restandardization, a method that combines randomization and permutation to correct permutation values of the test statistic and to take into account the overall distribution of individual test statistics, the entire data set was used rather than only the genes in the gene sets tested. Gene ontology for comprehensive protein evolutionary and functional classification was performed using Protein Analysis THrough Evolutionary Relationships (PANTHER; http://pantherdb.org).
Isolation of type II pneumocyte from mouse lung.
Type 2 pneumocytes were isolated from mouse lung following an adaptation of a previously described protocol (51). Briefly, a euthanized mouse was dissected, renal arteries sectioned, and lungs perfused with PBS to remove blood. Then, intratracheal lung perfusion of 1 ml dispase was applied for 10 minutes for predigestion. Lungs were isolated, sliced, and incubated for 30 minutes in dissociation buffer (1 ml collagenase, 3 ml dispase, 1–50 μl DNAse, 6 ml DMEM medium). Then, cells were washed in DMEM and negatively selected with CD45/CD31 antibodies and magnetic microbeads (MACS system, Miltenyi Biotec; LS columns catalog 130-042-401, CD45 mouse microbeads catalog 130-052-301, CD31 mouse microbeads catalog 130-097-418) to remove immune and endothelial cells. The CD45/CD31 negative fraction, composed of type 2 pneumocytes, red blood cells, and fibroblast, was collected and subjected to EpCAM-positive selection using EpCAM-FITC antibody (MACS system, Miltenyi Biotec; CD326 EpCAM-FITC; mouse, clone caa7-9G8, catalog 130-102-214) and anti-FITC microbeads (MACS system, Miltenyi Biotec; anti-FITC microbeads catalog 130-048-701) to isolate the type 2 pneumocytes. Once isolated, type 2 pneumocytes were maintained in culture in keratinocyte media.
Nucleotide sugar analysis.
Nucleotide sugars were rapidly extracted from adherent cells using ice-cold acetonitrile as previously reported (52). Briefly, cells were washed twice with ice-cold PBS. Ice-cold acetonitrile spiked with the internal standard BRDu (10 μM; 1 ml) was added to cells prior to 300 μl water. Cellular debris were scraped and transferred to an Eppendorf tube. Proteins were pelleted at 14,000g (10 minutes at 4°C) and extracted in 0.5% (wt/vol) SDS in 50 mM Tris-HCl pH 8.0 prior to a BSA protein estimation. The supernatant was lyophilized in a Speedvac and stored until analysis at –80°C.
Nucleotide sugars were separated and quantified using an approach previously reported (53). Briefly, nucleotides and nucleotide sugars were resuspended in ultrapure water and filtered through a 0.22-μm filter. Samples were analyzed on an Amersham Pharmacia SMART uHPLC and detected at 214, 254, and 280 nm using a diode array detector. The HPLC was equipped with a SUPELCOSIL LC-18-T column (15 cm × 4.6 mm, 3 μm particle size), which features an octadecylsilane bonded phase. The column was equilibrated in 100 mM phosphate buffer containing 8 mM tetrabutylammonium bisulfate at pH 5.5 (250 μl/minute). Nucleotides and nucleotide sugars (10%) were eluted from the column using a linear triphasic gradient: 0–5 minutes 0% (vol/vol) B; 5–15 minutes 0%–40% (vol/vol) B, 15–52.5 minutes 40%–85% (vol/vol) B. In addition to the samples, a standard curve was generated using fixed concentrations of BRDu (1,000 pmol) and decreasing concentrations of UDP-GlcNAc (1,000, 500, 250, 125 pmol). We noted that baseline separation of UDP-GlcNAc and GalNAc was not achieved, and the area under the curve (A280) for UDP-HexNAc (GlcNAc/GalNAc) and BRDu was calculated. BRDu is an internal standard spiked into the extraction buffer, and it was used to normalize each sample. The concentration of nucleotides is reported as picomoles of UDP-HexNAc per mg of protein.
Measurement of oxygen consumption rate (OCR) and proton production rate (PPR).
Assessment of cellular respiration and aerobic glycolysis was performed by measuring OCR and PPR with a Seahorse Bioscience XF24 Extracellular Flux Analyzer (Seahorse Bioscience) as previously described. Briefly, cell lines were plated in standard growth media at a density of 4 × 104 cells per well 24 hours prior to metabolic analysis. To measure OCR, 1 hour prior to metabolic analysis the normal growth media was replaced with XF minimal basal medium (Seahorse Bioscience) supplemented with 11 mM glucose, 1 mM sodium pyruvate, and 1X GlutaMax (Life Technologies). To measure PPR, 1 hour prior to metabolic analysis the normal growth media was replaced with XF minimal basal medium (Seahorse Bioscience) supplemented with 11 mM glucose and 1X GlutaMax (Life Technologies). After changing to respective XF media, cells were incubated at 37°C in a humidified atmosphere without CO2. Oligomycin (1 μM), FCCP (400 nM), rotenone (2 μM), antimycin A (2 μM), and 2-deoxy-D-glucose (100 mM) were added serially to determine mitochondrial respiration measurements. Oligomycin (1 μM) and 2-deoxy-D-glucose (100 mM) were added serially to determine glycolytic capacity measurements. Data presented are normalized per cell number per well.
Statistics.
Statistical analysis was carried out using GraphPad Prism v5.04 (GraphPad Software). Based on previous experience with our KrasG12D transgenic lung cancer mouse models, the sample sizes used typically reflect 3 or more individual primary tumors, each from different mice (that is, biological, not technical, replicates). The survival and tumor latency of mouse cohorts were calculated using Kaplan-Meier estimation and compared with the log-rank (Mantel-Cox) test. Statistical analysis of pharmacological treatments (TMG and DON) and contingency tables were calculated using Fisher’s exact test. Statistical analysis of the data was performed using 2-tailed, unpaired Student’s t test or 1-way ANOVA with Dunnet post hoc test, or 2-way ANOVA with Sidak post hoc test. Values are mean ± SD, where appropriate. Significance is noted as P < 0.05 unless stated.
Study approval.
All animal experiments were performed in accordance with Stanford Administrative Panel on Lab Animal Care (APLAC) and Johns Hopkins Animal Care and Use Committee (ACUC) protocols. Animals were housed in a pathogen-free environment.
Author contributions
KT, HW, and PTT conceived the project. KT, HW, RM, and PTT designed and performed all the experimental work unless specified below, and wrote the manuscript. RDW, KT, and HW performed microarray experiment and data analysis. BWS served as the veterinarian pathologist and reviewed all mouse tissues and staining. KN, MB, HW, KT, and RM performed all animal husbandry and tumor tissue IHC, and provided technical assistance with the bulk of the experiments. TS and JV performed the Seahorse experiment. KT, HW, AL, MAB, GY, RH, JG, and XW performed the glycosylation-related experiments. KN, MB, BZ, and KKHW performed CT imaging. JW, BDN, KJP, DF, and NEZ provided critical intellectual contributions throughout, and reagents. Members of the Tran lab (KT, HW, RM, MAB, MB, KN, RDW, and PTT) provided critical intellectual contributions throughout. NEZ and PTT designed and directed the project, as well as contributed to the manuscript production.
Supplementary Material
Acknowledgments
NZ acknowledges support from the National Institutes of Health (NIH) (NHLBI P01HL107153, NCI CA199806) and the Mizutani Foundations for the Glycosciences. KT is funded by the NIH (F31CA189588). HW is funded by a Uniting Against Lung Cancer Young Investigator Award and Johns Hopkins and Allegheny Health Network Award. RM is funded by the Prostate Cancer Foundation (PCF). PT is funded by the Nesbitt-McMaster Foundation, Ronald Rose, Joan Lazar, the Department of Defense (W81XWH-13-1-0182), a Kimmel Translational Science Award (SKF-13-021), an ACS Scholar award (122688-RSG-12-196-01-TBG), the American Lung Association (LCD-339465), Movember-PCF, and the NIH (R01CA166348).
Version 1. 08/21/2018
In-Press Preview
Version 2. 09/24/2018
Electronic publication
Version 3. 11/01/2018
Print issue publication
Funding Statement
Natasha Zachara
Natasha Zachara
Natasha Zachara
Kekoa Taparra
Hailun Wang
Reem Malek
Phuoc Tran
Phuoc Tran
Phuoc Tran
Phuoc Tran
Phuoc Tran
Phuoc Tran
Phuoc Tran
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(11):4924–4937.https://doi.org/10.1172/JCI94844.
Contributor Information
Kekoa Taparra, Email: taparra.kekoa@mayo.edu.
Hailun Wang, Email: hwang110@jhmi.edu.
Reem Malek, Email: rymalek@gmail.com.
Audrey Lafargue, Email: alafarg1@jhmi.edu.
Mustafa A. Barbhuiya, Email: mbarbhu1@jhmi.edu.
Xing Wang, Email: angwxx@gmail.com.
Brian W. Simons, Email: bsimons3@jhmi.edu.
Matthew Ballew, Email: mjballew2@gmail.com.
Katriana Nugent, Email: katriana.nugent@gmail.com.
Jennifer Groves, Email: jengroves89@gmail.com.
Russell D. Williams, Email: russell.d.williams@gmail.com.
Takumi Shiraishi, Email: takumi14@koto.kpu-m.ac.jp.
James Verdone, Email: James.verdone58@gmail.com.
Gokben Yildirir, Email: gyildirir@hotmail.com.
Roger Henry, Email: rogerhnr@gmail.com.
Bin Zhang, Email: binzhang@dlut.edu.cn.
John Wong, Email: jwong35@jhmi.edu.
Ken Kang-Hsin Wang, Email: kwang27@jhmi.edu.
Barry D. Nelkin, Email: bnelkin@jhmi.edu.
Kenneth J. Pienta, Email: kpienta1@jhmi.edu.
Dean Felsher, Email: dfelsher@stanford.edu.
Natasha E. Zachara, Email: nzachara@jhmi.edu.
Phuoc T. Tran, Email: tranp@jhmi.edu.
References
- 1.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 2.Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28(2):99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran PT, et al. Twist1 suppresses senescence programs and thereby accelerates and maintains mutant Kras-induced lung tumorigenesis. PLoS Genet. 2012;8(5):e1002650. doi: 10.1371/journal.pgen.1002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns TF, et al. Inhibition of TWIST1 leads to activation of oncogene-induced senescence in oncogene-driven non-small cell lung cancer. Mol Cancer Res. 2013;11(4):329–338. doi: 10.1158/1541-7786.MCR-12-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansieau S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14(1):79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 7.Morel AP, et al. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS Genet. 2012;8(5):e1002723. doi: 10.1371/journal.pgen.1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni T, et al. Snail1-dependent p53 repression regulates expansion and activity of tumour-initiating cells in breast cancer. Nat Cell Biol. 2016;18(11):1221–1232. doi: 10.1038/ncb3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krebs AM, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19(5):518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 10.Ingthorsson S, Andersen K, Hilmarsdottir B, Maelandsmo GM, Magnusson MK, Gudjonsson T. HER2 induced EMT and tumorigenicity in breast epithelial progenitor cells is inhibited by coexpression of EGFR. Oncogene. 2016;35(32):4244–4255. doi: 10.1038/onc.2015.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 12.Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158(1):171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janda E, et al. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol. 2002;156(2):299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer KR, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng X, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabbah M, et al. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Updat. 2008;11(4-5):123–151. doi: 10.1016/j.drup.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2010;1800(2):80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau KS, Dennis JW. N-Glycans in cancer progression. Glycobiology. 2008;18(10):750–760. doi: 10.1093/glycob/cwn071. [DOI] [PubMed] [Google Scholar]
- 19.Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010;1800(2):96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21(5):576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005(312):re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 22.Hanover JA, Krause MW, Love DC. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol. 2012;13(5):312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- 23.Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol. 2015;208(7):869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis FM, Stewart TA, Thompson EW, Monteith GR. Targeting EMT in cancer: opportunities for pharmacological intervention. Trends Pharmacol Sci. 2014;35(9):479–488. doi: 10.1016/j.tips.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 27.Shaul YD, et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158(5):1094–1109. doi: 10.1016/j.cell.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato M, et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol Cancer Res. 2013;11(6):638–650. doi: 10.1158/1541-7786.MCR-12-0634-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez RD, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64(24):9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 30.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5(4):375–387. doi: 10.1016/S1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 31.Taparra K, Tran PT, Zachara NE. Hijacking the hexosamine biosynthetic pathway to promote EMT-mediated neoplastic phenotypes. Front Oncol. 2016;6:85. doi: 10.3389/fonc.2016.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itkonen HM, et al. UAP1 is overexpressed in prostate cancer and is protective against inhibitors of N-linked glycosylation. Oncogene. 2015;34(28):3744–3750. doi: 10.1038/onc.2014.307. [DOI] [PubMed] [Google Scholar]
- 33.Caldwell SA, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29(19):2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- 34.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011;11(9):678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SY, et al. Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J. 2010;29(22):3787–3796. doi: 10.1038/emboj.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259(5):3308–3317. [PubMed] [Google Scholar]
- 37.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97(11):5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran PT, et al. Combined inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PLoS ONE. 2008;3(5):e2125. doi: 10.1371/journal.pone.0002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105(13):5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell. 1987;49(4):465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 41.Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270(32):18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 42.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi W, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337(6097):975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong C, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23(3):316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucena MC, et al. Epithelial mesenchymal transition induces aberrant glycosylation through hexosamine biosynthetic pathway activation. J Biol Chem. 2016;291(25):12917–12929. doi: 10.1074/jbc.M116.729236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muñoz-Espín D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15(7):482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 47.Yang MH, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12(10):982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 48.Hydbring P, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci U S A. 2010;107(1):58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Traxinger RR, Marshall S. Coordinated regulation of glutamine:fructose-6-phosphate amidotransferase activity by insulin, glucose, and glutamine. Role of hexosamine biosynthesis in enzyme regulation. J Biol Chem. 1991;266(16):10148–10154. [PubMed] [Google Scholar]
- 50.Malek R, Wang H, Taparra K, Tran PT. Therapeutic targeting of epithelial plasticity programs: focus on the epithelial-mesenchymal transition. Cells Tissues Organs (Print) 2017;203(2):114–127. doi: 10.1159/000447238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bantikassegn A, Song X, Politi K. Isolation of epithelial, endothelial, and immune cells from lungs of transgenic mice with oncogene-induced lung adenocarcinomas. Am J Respir Cell Mol Biol. 2015;52(4):409–417. doi: 10.1165/rcmb.2014-0312MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kultti A, et al. Methyl-beta-cyclodextrin suppresses hyaluronan synthesis by down-regulation of hyaluronan synthase 2 through inhibition of Akt. J Biol Chem. 2010;285(30):22901–22910. doi: 10.1074/jbc.M109.088435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kochanowski N, et al. Intracellular nucleotide and nucleotide sugar contents of cultured CHO cells determined by a fast, sensitive, and high-resolution ion-pair RP-HPLC. Anal Biochem. 2006;348(2):243–251. doi: 10.1016/j.ab.2005.10.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.