Abstract
SMAD4 is the only common SMAD in TGF-β signaling that usually impedes immune cell activation in the tumor microenvironment. However, we demonstrated here that selective deletion of Smad4 in NK cells actually led to dramatically reduced tumor cell rejection and augmented tumor cell metastases, reduced murine CMV clearance, as well as impeded NK cell homeostasis and maturation. This was associated with a downregulation of granzyme B (Gzmb), Kit, and Prdm1 in Smad4-deficient NK cells. We further unveiled the mechanism by which SMAD4 promotes Gzmb expression. Gzmb was identified as a direct target of a transcriptional complex formed by SMAD4 and JUNB. A JUNB binding site distinct from that for SMAD4 in the proximal Gzmb promoter was required for transcriptional activation by the SMAD4-JUNB complex. In a Tgfbr2 and Smad4 NK cell–specific double–conditional KO model, SMAD4-mediated events were found to be independent of canonical TGF-β signaling. Our study identifies and mechanistically characterizes unusual functions and pathways for SMAD4 in governing innate immune responses to cancer and viral infection, as well as NK cell development.
Keywords: Immunology
Keywords: Innate immunity, NK cells
Introduction
Since NK cells were discovered, almost half a century has been devoted to understanding how they control the detection and spread of cancer. Studies depleting NK cells in murine tumor models, in which T and B cells were untouched, revealed significantly increased tumor outgrowth (1, 2). A recent study showed that activation of NK cells by an inhibitor of Tyro3, Axl, and Mer (TAM) markedly reduced melanoma metastases, demonstrating the critical role that NK cells have in preventing the spread of cancer (3). Elimination of tumor cells by NK cells largely relies on the constitutive expression of cytolytic molecules, including perforin and granzymes. The expression of granzyme B (Gzmb) is regulated by many factors, which include modification of the promoter (4), maturation status (5), and activation of transcription factors. A model based on CD11b and CD27 expression is well established for describing the late maturation process of NK cells, whereby CD11b+CD27+ NK cells become CD11b+CD27+ and then fully mature into CD11b+CD27+ NK cells (6). In this model, expression of Gzmb increases along this path and peaks in CD11b+CD27+ NK cells (5, 6). However, the in vivo mechanism by which Gzmb is regulated in NK cells is largely unknown.
TGF-β signaling, which usually plays a suppressive role in immune cells (7, 8), inhibits tumor growth at early stages (9, 10) and promotes tumor development or epithelial-to-mesenchymal transition (EMT) at later stages (11–16). TGF-β is considered an important negative regulator of NK cell development and function (17), and SMAD proteins are critical factors in the canonical TGF-β signaling pathway. For example, we previously found that SMAD proteins mediate TGF-β signaling to inhibit IFN-γ production by NK cells in response to proinflammatory cytokines (18, 19). The unique common SMAD (co-SMAD), SMAD4, generally acts as a central mediator of the TGF-β signaling pathway in many biological processes (20). The role of SMAD4 in cancer is complicated; it can be both a tumor promoter and a tumor suppressor, as also shown for TGF-β signaling (15, 16, 21). Patients with familial juvenile polyposis (JP) who have germline SMAD4 mutations or deletions have a higher risk of developing gastrointestinal cancer (22, 23). However, the role of Smad4 in NK cells, especially in regulating their antitumor and antiviral ability as well as NK cell homeostasis and maturation, is unknown.
In this study, we explored the role of SMAD4 in regulating NK cells and addressed whether the transcription factor acts downstream of the canonical TGF-β signaling pathway or independently from it to influence the tumor immune surveillance of NK cells. Our data demonstrate that SMAD4 is highly expressed in NK cells and that deletion of the single gene Smad4 in NK cells leads to impairment of NK cell maturation, NK cell homeostasis, and NK cell immune surveillance against melanoma metastases and cytomegalovirus. We also discovered that SMAD4 directly binds to the promoter of Gzmb and positively regulates Gzmb expression through interaction with JUNB.
Results
SMAD4 is required for antitumor and antiviral innate immunity mediated by NK cells.
SMAD4 protein was abundantly expressed in NK cells as well as in T and B cells (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI121227DS1). In NK cells, the expression of Smad4 increased as maturation proceeded (Supplemental Figure 1B). Given that homozygous Smad4 mutation leads to embryonic lethality (24), we deleted Smad4 in NK cells using an improved Cre-driven (iCre-driven) strategy. Mice with iCre under the control of the NKp46 promoter (NKp46-iCre mice) were crossed with Smad4fl/fl mice to generate NKp46iCre/+Smad4fl/fl mice (hereafter referred to as Smad4ΔNK mice) (Supplemental Figure 1C). Immunoblotting of isolated cell subsets indicated that SMAD4 expression was indeed absent from NK cells, but was present at normal levels in T and B cells from Smad4ΔNK mice (Supplemental Figure 1D). Of note, TGF-β was found to still increase phosphorylated SMAD2/3 (p-SMAD2/31) in both WT and Smad4-deficient NK cells (Supplemental Figure 1E).
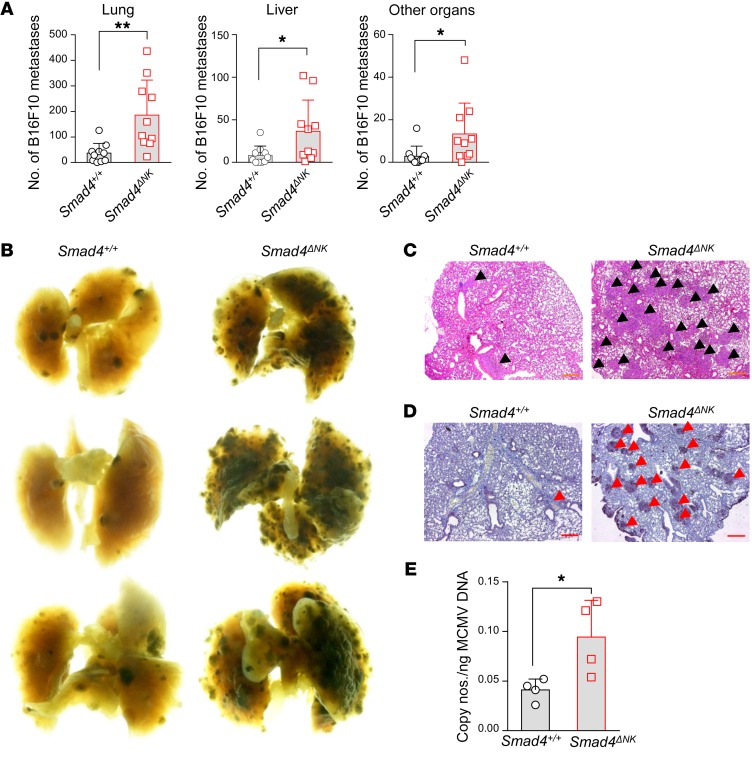
To assess the antitumor activity of NK cells in Smad4ΔNK mice, we first conducted in vivo experiments using B16F10, a melanoma cell line susceptible to NK cell killing (25) and capable of metastasizing to the lungs (26). We injected i.v. B16F10 cells into either WT mice (NKp46+/+Smad4fl/fl, hereafter referred to as Smad4+/+ mice) or Smad4ΔNK mice. Two weeks after inoculation, mice were euthanized, and metastases were quantified. Postmortem analysis revealed that the lungs of Smad4ΔNK mice were overwhelmed with melanoma metastases, while we found significantly fewer (4-fold fewer) melanoma nodules in the lungs of Smad4+/+ mice (Figure 1, A and B). We also observed more metastases in the livers, kidneys, bones, intestines, and reproductive organs of Smad4ΔNK mice compared with those of Smad4+/+ mice (Figure 1A). Histological analysis of lungs further confirmed the higher frequency of B16F10 metastases in Smad4ΔNK mice (Figure 1, C and D). Antiviral immunity is another important function of NK cells. We used a murine CMV (MCMV) model to analyze whether KO of Smad4 in NK cells affects viral clearance. We measured viral titers on day 7 after MCMV infection and detected a higher viral titer in Smad4ΔNK mice than in Smad4+/+ mice (Figure 1E), indicating impaired antiviral capacity when Smad4 is absent in NK cells. These data suggest that a single gene, Smad4, is able to control the NK cell–mediated antitumor and antiviral innate immune responses.
Figure 1. Smad4-deficient NK cells fail to control tumor metastases and MCMV infection.
(A) The frequency of metastases in lung, liver, and other organs was determined 14 days after B16F10 inoculation (n = 10). (B) Representative macroscopic lung images of tumor-bearing mice. (C and D) Lung sections from tumor-bearing mice were subjected to H&E staining (C) and IHC staining with anti-S100 mAb (D) to detect metastatic B16F10 melanoma cells. Arrowheads point to the nodules of metastatic melanoma. Scale bars: 500 μm. (E) Smad4ΔNK mice and WT mice were infected with MCMV. MCMV titers were determined by real-time RT-PCR 7 days after infection (n = 4). Data are presented as the mean ± SD. Differences between littermates were evaluated. *P < 0.05 and **P < 0.01, by 2-tailed, paired t test. The circles and squares on the bar graphs denote data for individual mice.
SMAD4 deficiency impairs the intrinsic cytotoxic activity of NK cells.
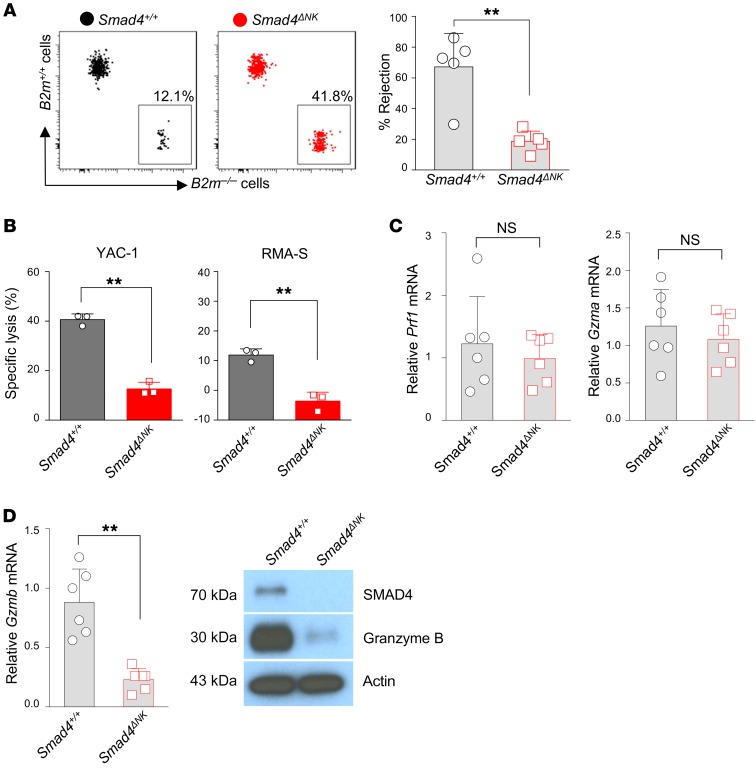
A missing-self recognition assay has previously been used to assess the “in vivo cytotoxicity” of NK cells, as described by Karo et al. and Bix et al. (27, 28). This assay is based on the understanding that through binding of NK cell inhibitory receptors to MHC class I molecules on other cells, NK cells develop tolerance to self (29), while cells lacking normal MHC class I expression are targeted and killed by NK cells (30). Here, we also used the missing-self recognition assay to assess the cytotoxicity of NK cells in Smad4ΔNK mice, as previously described (27, 28). For this purpose, splenocytes from B2m–/– mice, which lack MHC class I protein expression on the cell surface, and splenocytes from WT mice (B2m+/+) were mixed at a 1:1 ratio and i.v. injected into Smad4+/+ and Smad4ΔNK mice. One day later, we performed flow cytometry to measure donor cells in the recipient mice. We found that Smad4+/+ mice efficiently rejected B2m–/– splenocytes but that this rejection was significantly impaired in Smad4ΔNK mice (Figure 2A). Ex vivo 51Cr assays also showed that Smad4-deficient NK cells were less cytotoxic than were Smad4+/+ NK cells (Figure 2B). These data suggest that Smad4-deficient NK cells are defective in killing target cells.
Figure 2. Impaired rejection of B2m–/– target cells and decreased Gzmb in Smad4-deficient NK cells.
(A) Representative flow cytometric data showing relative B2m–/– and B2m+/+ donor splenocyte rejection by Smad4+/+ and Smad4ΔNK recipient mice. The graph shows summary data of rejection percentages of B2m–/– donor splenocytes by Smad4+/+ and Smad4ΔNK recipient mice calculated using the formula described in Methods (n = 5). Data are presented as the mean ± SD. Differences between littermates were evaluated. **P < 0.01, by 2-tailed, paired t test. The circles and squares on the bar graphs denote data for individual mice. (B) Ex vivo cytotoxicity analysis of NK cells from Smad4+/+ and Smad4ΔNK mice. Effector/target = 50:1 (n = 3). **P < 0.01, by 2-tailed, unpaired t test. (C) Real-time RT-PCR analysis of Prf1 and Gzma mRNA expression in NK cells (n = 6). Data are presented as the mean ± SD. Differences between littermates were evaluated. Statistical analysis was conducted using a 2-tailed, paired t test. The circles and squares on bar graphs denote data for individual mice. (D) Real-time RT-PCR and immunoblot analysis of Gzmb expression in NK cells. Data are presented as the mean ± SD. Differences between littermates were evaluated. **P < 0.01, by 2-tailed, paired t test. The circles and squares on the bar graphs denote data for individual mice.
As mice bearing Smad4-deficient NK cells had an increase in tumor metastasis and a decrease in the rejection of virus and target cells compared with Smad4+/+ mice, we hypothesized that the repertoire of inhibitory or activating receptors may change or that cytolysis-related genes may be downregulated in Smad4-deficient NK cells. Ly49 molecules consist of a cluster of inhibitory or activating surface receptors expressed on NK cells. After binding to MHC class I molecules, most Ly49 receptors suppress NK cell cytotoxicity (31). To determine whether any change in the expression of Ly49 molecules occurs in Smad4-deficient NK cells, we assessed the expression of Ly49A, Ly49C/I, Ly49D, or Ly49G2 by flow cytometry. We found no significant difference in the molecular profiles of Ly49s between Smad4-deficient and WT NK cells (Supplemental Figure 2). Furthermore, there was no apparent difference in the expression levels of CD94, NKG2A/C/E, NKG2D (nonclassical MHC recognition molecules), or CD69 (activation marker) between WT and Smad4-deficient NK cells (Supplemental Figure 2). Thus, the impaired rejection of target cells in Smad4-deficient NK cells does not appear to be attributable to differences in the expression of these surface receptors.
Perforin and granzymes are involved in rendering NK cells able to kill target cells effectively and rapidly (32). NK cell cytotoxicity from perforin-deficient mice is considerably impaired (33), and granzymes are required for the rapid induction of target cell DNA fragmentation (34, 35). To investigate impaired rejection of target cells in Smad4-deficient NK cells as the possible cause of increased tumor metastases, we analyzed the expression of perforin (Prf1), granzyme A (Gzma), and Gzmb in splenic NK cells isolated from Smad4+/+ and Smad4ΔNK mice by real-time RT-PCR and/or immunoblotting. We detected no significant difference in Prf1 or Gzma mRNA expression between WT and Smad4-deficient NK cells (Figure 2C). However, the expression of Gzmb was remarkably downregulated in Smad4-deficient NK cells at both the transcript and protein levels (Figure 2D), which implied that SMAD4 upregulates the expression of Gzmb in NK cells. We obtained similar data by using a siRNA approach. The expression level of Gzmb in NK cells was decreased when Smad4 was knocked down by siRNA (Supplemental Figure 3).
SMAD4 interacts with JUNB to upregulate Gzmb expression.
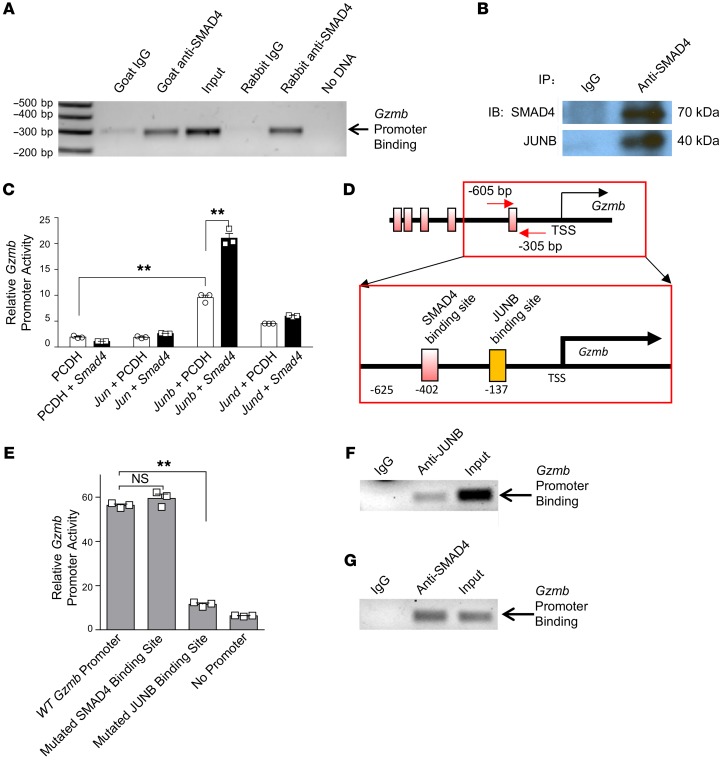
SMAD4 is a transcription factor with direct DNA-binding capability (36). The promoter region of Gzmb has 5 putative SMAD4 binding sites within 3 kb upstream of the transcription start site (TSS), including 1 proximal site around –402 bp and 4 distal sites around –1,359 bp, –2,022 bp, –2,178 bp, and –2255 bp relative to the TSS (Supplemental Figure 4, A and B). This prompted us to perform ChIP assays to investigate whether SMAD4 positively regulates the expression of Gzmb through promoter interaction (transactivation) at these sites. Chromatin from purified primary murine splenic NK cells was immunoprecipitated with 2 distinct anti-SMAD4 Abs from different species or their corresponding control IgG separately and then subjected to PCR amplification with primers specific to the fragments harboring putative SMAD4 binding sites. These assays revealed the association of SMAD4 over the –605-bp to –305-bp DNA fragment corresponding to the proximal region of the Gzmb promoter (Figure 3A and Supplemental Figure 4B). Of note, we observed similar results when the 2 anti-SMAD4 Abs from different sources (i.e., goat anti-SMAD4 and rabbit anti-SMAD4 Abs) were used (Figure 3A).
Figure 3. SMAD4 cooperates with JUNB to transactivate Gzmb.
(A) Binding of SMAD4 to the Gzmb promoter in freshly isolated NK cells as determined by ChIP assays. Two anti-SMAD4 Abs from different sources were used for validation, and the experiments were repeated at least once. (B) Co-IP was performed to assess the interaction of SMAD4 and JUNB in NK cells. (C) Luciferase reporter assays were used to assess Gzmb promoter activity. NIH-3T3 cells were cotransfected with Jun, Junb, or Jund expression plasmids and a Smad4 expression plasmid or an empty vector (n = 3). The experiment was repeated 3 times, and similar results were obtained. **P < 0.01, by 1-way ANOVA with Holm’s multiple comparisons test. (D) Scheme denoting putative SMAD4 and JUNB binding sites in the Gzmb promoter. (E) The Gzmb promoter was mutated at the SMAD4 binding site or the JUNB binding site, and the transcriptional activity was assessed after Smad4 and Junb were overexpressed. The WT Gzmb promoter construct served as a positive control. The PGL3 vector without a promoter insert served as a control for the background luciferase activity (n = 3). Results shown are representative of 3 experiments performed with similar results. Data are presented as the mean ± SD. **P < 0.01, by 1-way ANOVA with Holm’s multiple comparisons test. (F and G) ChIP assays were performed to detect the association of JUNB and SMAD4 with the Gzmb promoter. Chromatin from murine splenic NK cells was immunoprecipitated with an anti-JUNB Ab (F) or an anti-SMAD4 Ab (G), or normal IgG (F and G). PCR was used to confirm the association of JUNB or SMAD4 with the Gzmb promoter, using primers specific to the fragment harboring the putative JUNB binding site.
On the basis of the ChIP assay data, we cloned the Gzmb promoter ranging from –625 bp to +93 bp into a luciferase reporter plasmid to determine whether SMAD4 could transactivate the Gzmb promoter. This reporter construct was cotransfected into NIH-3T3 mouse fibroblast cells with either a Smad4 expression plasmid or an empty vector, and the transactivation of the Gzmb promoter was evaluated by a luciferase assay. After confirming the overexpression of SMAD4 by immunoblot analysis (Supplemental Figure 4C), we found that cotransfection of Smad4 did not significantly improve the relative luciferase activity of the Gzmb promoter (Supplemental Figure 4D). This suggested that SMAD4 alone cannot activate the Gzmb promoter and that one or more coactivators are required.
In fact, it has been reported that under physiological conditions, the affinity between SMAD4 and the DNA sequence is too weak to support unassisted binding (20, 37). Coactivators or cosuppressors are therefore considered necessary for SMAD4 to transregulate target genes. Thus, we aimed to identify a coactivator that cooperated with SMAD4 to activate the Gzmb promoter in NK cells. After a pilot screen using IP, we identified JUNB as a candidate cofactor. As shown in Figure 3B, JUNB was immunoprecipitated with anti-SMAD4 Abs, implying that SMAD4 interacts with JUNB in NK cells.
To evaluate the functional significance of the interaction between SMAD4 and JUNB, we measured the transcriptional activity of the Gzmb promoter by luciferase assays when Smad4 was overexpressed together with Junb in NIH-3T3 cells. We observed modest or no enhanced transactivation when the Junb or Smad4 expression plasmids were transfected alone, respectively. However, we found that cotransfection of Smad4 with Junb plasmids, but not with Jun or Jund plasmids, significantly enhanced the activity of the Gzmb promoter (Figure 3C). Together, these data suggested that SMAD4 interacts with JUNB and that JUNB is required for transactivation of the Gzmb promoter by SMAD4.
Both SMAD4 and JUNB have DNA binding activity, and a SMAD4 binding site and a JUNB binding site are closely located in the proximal region of the Gzmb promoter, with an interval of approximately 260 bp (Figure 3D). Thus, there are at least 3 potential mechanisms by which the SMAD4-JUNB complex transactivates the Gzmb promoter (Supplemental Figure 5A): (a) the SMAD4-JUNB complex binds to the SMAD4 binding site alone; (b) the SMAD4-JUNB complex binds to the JUNB binding site alone; or (c) SMAD4 and JUNB each bind to their respective sites but interact with each other, as the distance between the SMAD4 and JUNB binding sites is short. To investigate this, we performed site-directed mutagenesis of either the SMAD4 binding site or the JUNB binding site (Supplemental Figure 5B) and analyzed promoter activity with cotransfection of SMAD4 and JUNB. Our data showed that mutating the SMAD4 binding site did not decrease the Gzmb promoter reporter activity; however, mutating the JUNB binding site resulted in a significant decrease in promoter activity when Smad4 and Junb were cotransfected (Figure 3E). Besides, ChIP assays indicated the binding of JUNB to its binding site in the proximal Gzmb promoter (Figure 3F), and SMAD4 was also observed to be associated with this promoter region (Figure 3G), consistent with our IP results showing that these 2 proteins interact with each other (Figure 3B). These data revealed that the JUNB binding site, but not the SMAD4 binding site, is required for the transcriptional activation of Gzmb by the SMAD4-JUNB complex and support the hypothesis that the SMAD4-JUNB complex binds to the JUNB binding site and then activates the Gzmb promoter (Supplemental Figure 5A).
Since SMAD4 was previously found to be associated with the Junb promoter (38) and Junb is also required for Gzmb expression (39, 40), SMAD4 may upregulate Gzmb through the induction of Junb expression. To test whether Junb is a target gene of SMAD4 in NK cells, we compared the expression levels of Junb in NK cells from Smad4+/+ and Smad4ΔNK mice at different stages of maturation. We detected similar expression levels of Junb in WT and Smad4-deficient NK cells at each stage analyzed (Supplemental Figure 5C), suggesting that SMAD4 requires JUNB as a cofactor, but does not modulate its expression to control the expression of Gzmb in NK cells.
SMAD4 positively controls NK cell homeostasis.
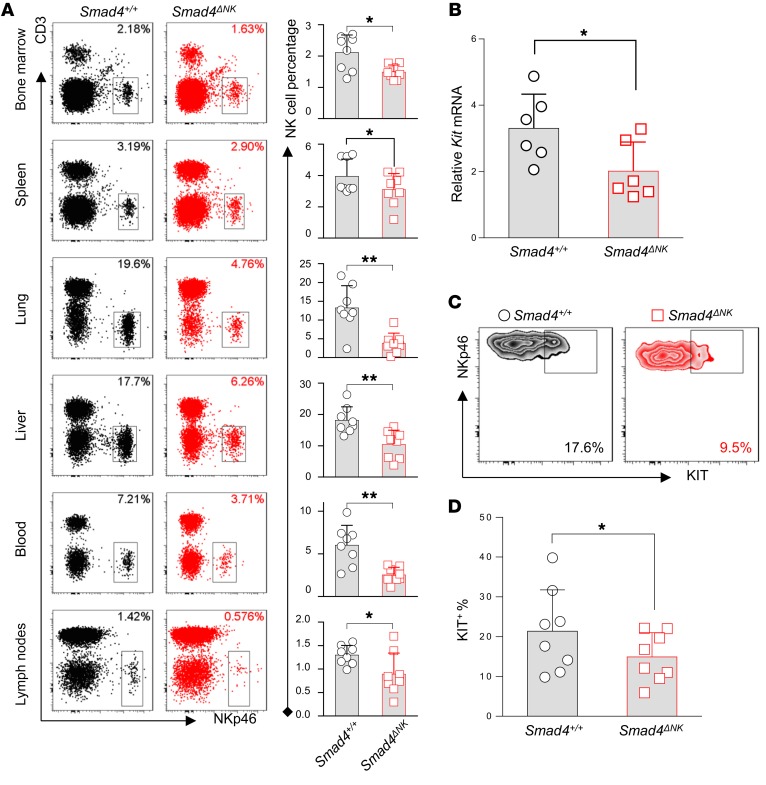
We also measured the quantity of NK cells in Smad4ΔNK mice. We analyzed the proportion of NK cells in bone marrow, spleen, lung, liver, blood, and lymph nodes by flow cytometry. Compared with Smad4+/+ mice, the percentage of NK cells among lymphocytes in Smad4ΔNK mice was significantly decreased in all of the tissues and organs analyzed (Figure 4A). However, consistent with a recent report (41), we observed that the NKp46+CD49a+ cell population that is similar to group 1 innate lymphoid cells (ILC1-like) significantly increased when Smad4 was absent in NKp46+ cells (Supplemental Figure 6). We continued to explore potential mechanisms responsible for the decrease of NK cells in Smad4ΔNK mice.
Figure 4. SMAD4 is required to maintain NK cell homeostasis.
(A) The proportion of NK cells among lymphocytes in bone marrow, spleen, lung, liver, blood, and lymph nodes was analyzed by flow cytometry (n = 8). (B) Kit mRNA expression in CD11b–CD27+ NK cells was assessed by real-time RT-PCR (n = 6). (C) Representative flow cytometric analysis of KIT expression. NKp46+CD3– splenocytes were first gated, and then expression of KIT was determined in CD11b–CD27+ cells. (D) Cumulative data for KIT expression analyzed by flow cytometry (n = 8). Data are presented as the mean ± SD. Differences were evaluated between littermates. *P < 0.05 and **P < 0.01, by 2-tailed, paired t test. The circles and squares on the bar graphs denote data for individual mice.
Myc has been previously identified as a target gene of SMAD4 in both tumor cells (42) and T cells (43), where SMAD4 binds to the promoter of Myc to activate its transcription and drive cell proliferation. However, our data showed that Myc expression was not significantly different in NK cells from Smad4ΔNK versus those from Smad4+/+ mice, suggesting that Myc is unlikely to be a target gene of SMAD4 in NK cells (Supplemental Figure 7) and that different mechanisms may be involved in regulating the aforementioned NK cell homeostasis.
KIT is expressed in immature NK cells (44) and functions as an essential regulator of NK cell homeostasis (45). Previous studies have clearly demonstrated that the quantity of NK cells in mice is reduced when KIT is absent (45). We found that at both the mRNA and protein levels, KIT expression levels were highest in CD11b–CD27+ immature NK cells (Supplemental Figure 8). The expression of Kit was dramatically decreased when NK cells mature into CD11b+CD27+ and then CD11b+CD27– cells. Therefore, we compared the expression levels of KIT in CD11b–CD27+ NK cells between Smad4ΔNK and Smad4+/+ mice. We found that KIT was downregulated in Smad4-deficient NK cells at both the mRNA and protein levels (Figure 4, B–D), coincident with the reduced quantity of NK cells in Smad4ΔNK mice and suggesting that KIT may mediate SMAD4 to control NK cell homeostasis.
SMAD4 promotes NK cell terminal maturation.
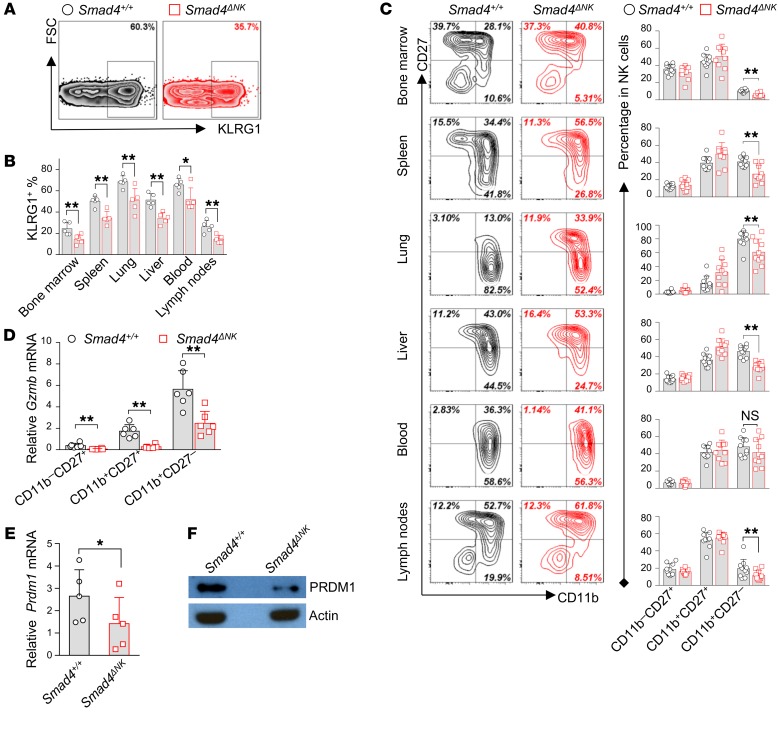
NK cells are derived from hematopoietic stem cells, and NK progenitor cells traffic to different tissues that provide tissue-specific microenvironments for NK cells to achieve terminal maturation (46, 47). Optimum effector functions and the maximum expression levels of cytotoxicity-related factors, including Prf1 and Gzmb, culminate in mature NK cells (5), while delayed maturation of NK cells leads to impaired tumor surveillance and increased melanoma metastasis (48). We compared NK cell maturation in different tissues from Smad4ΔNK and Smad4+/+ mice using the final-stage NK cell maturation marker KLRG1 (49). Our data showed that blood and lung had the highest proportion of KLRG1+ NK cells, followed by liver and spleen (Figure 5, A and B). Lymph nodes and bone marrow showed the lowest percentage of KLRG1+ NK cells. While the distribution pattern of KLRG1+ NK cells among different tissues and organs was similar between Smad4+/+ and Smad4ΔNK mice, the percentage of KLRG1+ cells in all of the tissues and organs analyzed was significantly lower in Smad4ΔNK mice (Figure 5, A and B, and Supplemental Figure 9).
Figure 5. Smad4 deficiency impairs the maturation of NK cells.
(A) Representative flow cytometric analysis of KLRG1 in lung NK cells. Gating was performed on the CD3–NKp46+ lymphocyte population in both WT and Smad4ΔNK mice. Results from 1 of 5 pairs of littermate mice with similar data are presented. (B) Cumulative data for KLRG1 expression in NKp46+ cells in the bone marrow, spleen, lungs, liver, blood, and lymph nodes (n = 5). Differences were evaluated between littermates. (C) Representative flow cytometric analysis and cumulative data showing the distribution of CD27 versus CD11b in NK cells from bone marrow, spleen, lungs, liver, blood, and lymph nodes (n = 10). Differences were evaluated between littermates. P values were calculated using a 2-tailed, paired t test. (D) Expression of Gzmb in NK cells at different stages of maturation was analyzed by real-time RT-PCR (n = 6). (E and F) Expression of Prdm1 in NK cells from Smad4+/+ and Smad4ΔNK mice (n = 5) was assessed by real-time RT-PCR (E) and immunoblotting (F). Data are presented as the mean ± SD. The circles and squares on the bar graphs denote data for individual mice. Differences were evaluated between littermates. *P < 0.05 and **P < 0.01, by 2-tailed, paired t test.
We next assessed NK cell maturation in Smad4+/+ and Smad4ΔNK mice on the basis of CD11b and CD27 surface expression. In Smad4ΔNK mice, the proportion of the most mature (CD11b+CD27–) NK cell subset was significantly decreased in bone marrow, spleen, lung, liver, and lymph nodes, while in blood, this decrease was moderate and did not reach statistical significance (Figure 5C). Together, these data indicate that NK cell maturation is impaired in Smad4ΔNK mice and that Smad4 is required for the terminal maturation of NK cells.
Given that terminally mature NK cells were decreased in Smad4ΔNK mice (Figure 5) and the expression of Gzmb is upregulated in mature NK cells (5), the decreased Gzmb expression we detected in NK cells from Smad4ΔNK mice could simply be due to a smaller number of mature cells. To test this, we sorted NK cells from Smad4+/+ and Smad4ΔNK mice at different stages of maturation (CD11b–CD27+, CD11b+CD27+, CD11b+CD27–) and assessed Gzmb levels. In NK cells from both Smad4+/+ and Smad4ΔNK mice, the expression of Gzmb increased as NK cells proceeded toward terminal maturation (Figure 5D). We reasoned that if decreased Gzmb is a secondary effect of impaired maturation, Smad4-sufficient and Smad4-deficient NK cells should have similar Gzmb expression at a specific stage of maturation. However, we found that the expression of Gzmb in Smad4-deficient NK cells was much lower than that in Smad4-sufficent NK cells at every maturation stage analyzed (Figure 5D). These data indicate that SMAD4 intrinsically regulates the expression of Gzmb and that decreased Gzmb expression in Smad4-deficient NK cells is therefore a bona fide change that is not solely due to the reduced number of terminal mature NK cells in Smad4ΔNK mice.
Because our data indicated a blockage at the transition from KLRG1– to KLRG1+ NK cells and from CD11b+CD27+ to CD11b+CD27– NK cells in Smad4ΔNK mice, we hypothesized that SMAD4 may control other transcription factors that regulate NK cell maturation. To test this, we analyzed expression levels of the transcription factors known to regulate NK cell maturation. We detected a significant difference in the expression of Foxo1 (50), Tbx21 (51), Gata3 (52), Id2 (53), Elf4 (54), Nifl3 (55), or Eomes (56) in NK cells between Smad4+/+ and Smad4ΔNK mice (Supplemental Figure 10). However, we observed a striking decrease in Prdm1 expression at both the transcript and protein levels in Smad4-deficient NK cells (Figure 5, E and F). We further assessed mRNA expression of Prdm1 at different stages of NK cell maturity. In both Smad4+/+ and Smad4ΔNK mice, Prdm1 transcript levels were gradually increased along with the maturation of NK cells and achieved maximal levels in the CD11b+CD27– cell population (Supplemental Figure 11), consistent with findings from a previous report (57). Importantly, we found that at each of the 3 stages of maturity, the expression of Prdm1 in Smad4-deficient NK cells was significantly lower than that in Smad4-sufficient NK cells (Supplemental Figure 11). These data suggest that PRDM1 may mediate SMAD4 to control NK cell maturation.
TGF-β–independent and –dependent SMAD4 signaling pathways in NK cells.
SMAD4 is the unique co-SMAD in the canonical TGF-β signaling pathway (36). Previous studies found that deletion of Smad4 disrupted TGF-β signaling (15, 58, 59). We then asked whether impaired homeostasis, maturation, and cytotoxic capability in Smad4-deficient NK cells is caused by disruption of the TGF-β signaling pathway. For this purpose, we undertook a genetic approach using TGF-β receptor type 2 (Tgfbr2) single–conditional KO mice and Tgfbr2/Smad4 double–conditional KO mice.
We compared NK cell homeostasis, development, Gzmb expression, and tumor metastases between Tgfbr2ΔNK (NKp46iCre/+Tgfbr2fl/fl) and Tgfbr2+/+ (NKp46+/+Tgfbr2fl/fl) mice. Unlike Smad4ΔNK mice (Figure 4), the Tgfbr2+/+ and Tgfbr2ΔNK mice did not show significant differences in the proportion of NK cells among lymphocytes (i.e., NK cell homeostasis) (Supplemental Figure 12A). We observed increased terminally mature NK cells in the bone marrow, spleens, and livers of Tgfbr2ΔNK mice (Supplemental Figure 12B), in contrast to the diminished maturation of NK cells in Smad4ΔNK mice (Figure 5). These results support a role for SMAD4 that is separate from TGF-β signaling.
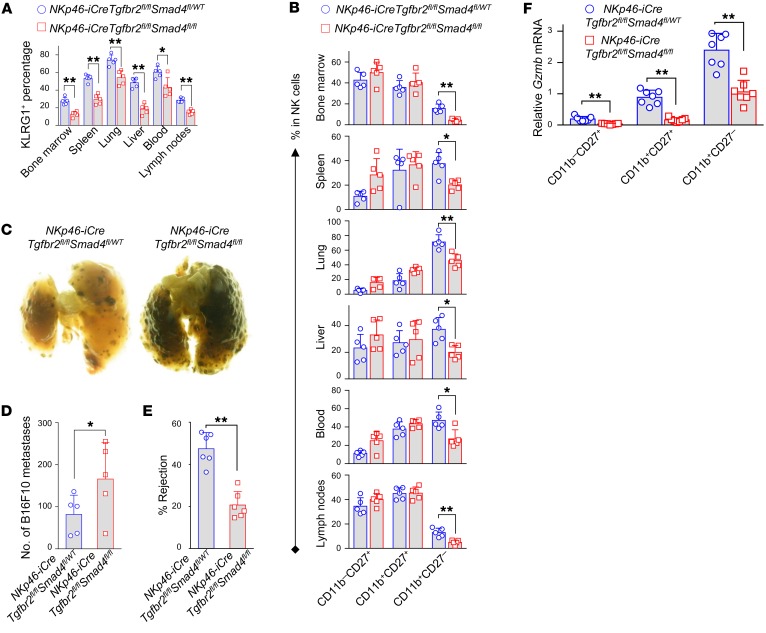
To more rigorously validate the dispensability of TGF-β signaling in the SMAD4 signaling pathway that we characterized in NK cells, we adopted an additional genetic approach built on the model used in Supplemental Figure 1C. Tgfbr2 and Smad4 double–conditional KO mice were generated by crossing NKp46-iCre mice with mice that had loxP sites flanking both Tgfbr2 and Smad4 (Tgfbr2fl/flSmad4fl/fl). We hypothesized that if the canonical TGF-β signaling is dispensable for SMAD4-mediated positive regulation, the impaired NK cell homeostasis, maturation, and cytotoxicity that we observed in Smad4 NK–conditional KO mice would be recapitulated in the absence of TGF-β signaling, i.e., under a Tgfbr2-deficient background. Indeed, we observed decreased percentages of NK cells among lymphocytes in the tested organs and tissues of NKp46-iCreTgfbr2fl/flSmad4fl/fl mice (absence of both Tgfbr2 and Smad4) compared with those of their NKp46-iCreTgfbr2fl/flSmad4fl/WT littermates (absence of Tgfbr2 but presence of 1 allele of Smad4) (Supplemental Figure 13). Significant downregulation of KLRG1 due to the deletion of Smad4 in the presence of TGF-β signaling was also recapitulated in NK cells from bone marrow and other tissues in NKp46-iCreTgfbr2fl/flSmad4fl/fl mice compared with NKp46-iCreTgfbr2fl/flSmad4fl/WT mice (Figure 6A and Supplemental Figure 14A). Likewise, NKp46-iCreTgfbr2fl/flSmad4fl/fl mice had a lower proportion of CD11b+CD27– NK cells and a higher proportion of CD11b+CD27+ NK cells in bone marrow and other tissues compared with their NKp46-iCreTgfbr2fl/flSmad4fl/WT littermates (Figure 6B and Supplemental Figure 14B). Fourteen days after inoculation with B16F10 cells, NKp46-iCreTgfbr2fl/flSmad4fl/fl mice showed more metastatic melanoma nodules than did their NKp46-iCreTgfbr2fl/flSmad4fl/WT littermates (Figure 6, C and D). We also observed impaired NK cell–mediated rejection of MHC class I–deficient cells in NKp46-iCreTgfbr2fl/flSmad4fl/fl mice compared with NKp46-iCreTgfbr2fl/flSmad4fl/WT mice, although only 1 allele difference in Smad4 exists between the 2 mouse strains (Figure 6E). Not surprisingly, we also observed decreased expression levels of Kit, Prdm1, and Gzmb in NKp46-iCreTgfbr2fl/flSmad4fl/fl mice compared with levels in their NKp46-iCreTgfbr2fl/flSmad4fl/WT littermates (Supplemental Figure 15, A and B, and Figure 6F). These data support our hypothesis that, independent of the canonical TGF-β/SMAD signaling pathway, SMAD4 promotes homeostasis, maturation, cytotoxicity, and the expression of Gzmb in NK cells.
Figure 6. TGF-β signaling is not required for SMAD4 to positively regulate NK cell maturation and effector functions.
Summary data comparing NK cell maturation between NKp46-iCreTgfbr2fl/flSmad4fl/fl mice and NKp46-iCreTgfbr2fl/flSmad4fl/WT mice, based on the expression of KLRG1 (A) and the distribution of CD11b and CD27 (B) (n = 5). Representative images (C) and cumulative data (D) for B16F10 melanoma metastases (n = 5). (E) Rejection of B2m–/– donor cells by NKp46-iCreTgfbr2fl/flSmad4fl/fl and NKp46-iCreTgfbr2fl/flSmad4fl/WT mice (n = 6). (F) Expression of Gzmb in NK cell subsets was analyzed by real-time RT-PCR (n = 7). Data are presented as the mean ± SD. Differences were evaluated between littermates. *P < 0.05 and **P < 0.01, by 2-tailed, paired t test. The circles and squares on the bar graphs denote data for individual mice.
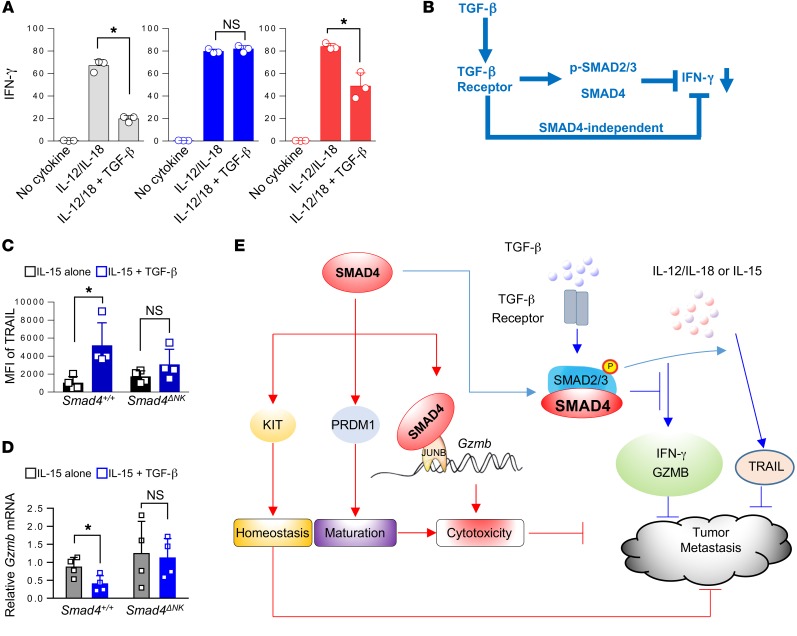
Our previous studies have shown that TGF-β represses IFN-γ in NK cells through a SMAD-dependent pathway (18, 19). We therefore investigated whether SMAD4 participates in TGF-β signaling to inhibit IFN-γ secretion in ex vivo–stimulated NK cells. Splenocytes from Tgfbr2ΔNK (NKp46iCre/+Tgfbr2fl/fl) or Smad4ΔNK mice were costimulated with IL-12 and IL-18 in the presence or absence of TGF-β, and NK cell IFN-γ secretion was assayed by flow cytometry. In the absence of TGF-β, costimulation with IL-12 and IL-18 initiated a robust increase in IFN-γ secretion by NK cells from all mice including WT, Tgfbr2ΔNK, and Smad4ΔNK mice (Figure 7A), and the increase was similar between Smad4+/+ and Smad4ΔNK mice (Supplemental Figure 16). TGF-β treatment markedly abrogated the IFN-γ production in IL-12– and IL-18–stimulated NK cells from WT mice, as evidenced by an appreciable decrease in the proportion of IFN-γ–producing NK cells (Figure 7A). In stark contrast, we found that the inhibitory effect of TGF-β on IFN-γ production was significantly blunted in IL-12– and IL-18–stimulated NK cells from Tgfbr2ΔNK mice and was only partially blunted in IL-12– and IL-18–stimulated NK cells from Smad4ΔNK mice (Figure 7A). These data provide ex vivo evidence that TGF-β utilizes TGFBR2 and SMAD4 to inhibit IFN-γ production in NK cells, which is consistent with our previous studies (18, 19). However, the partial blunting of IFN-γ production by TGF-β in NK cells from Smad4ΔNK mice implies the existence of a noncanonical or SMAD4-independent TGF-β pathway, by which TGF-β represses IFN-γ expression in NK cells (Figure 7B).
Figure 7. TGF-β–dependent and –independent SMAD4 signaling pathways in NK cells.
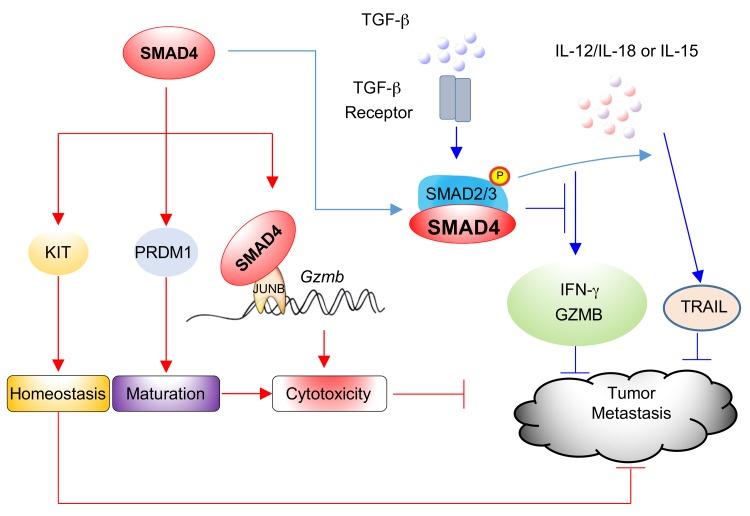
(A) Splenocytes isolated from WT, Tgfbr2ΔNK, and Smad4ΔNK mice were costimulated overnight with IL-12 (10 ng/ml) and IL-18 (10 ng/ml) in the presence or absence of 10 ng/ml TGF-β. GolgiStop was added 6 hours before intracellular staining of IFN-γ (n = 3). Data are presented as the mean ± SD. The circles and squares on the bar graphs denote data for individual mice. Differences were evaluated between littermates by 1-way ANOVA with Holm’s multiple comparisons test; *P < 0.05. (B) Model of the inhibition of IFN-γ production through both the canonical TGF-β signaling pathway and the SMAD4-independent TGF-β signaling pathway. (C) Splenic NK cells were isolated from WT and Smad4ΔNK mice and stimulated with the indicated cytokines for 3 days. TRAIL expression was analyzed by flow cytometry (n = 4). Data are presented as the mean ± SD. *P < 0.05, by 2-tailed, paired t test. The circles and squares on the bar graphs denote data for individual mice. (D) NK cells from the spleens of WT and Smad4ΔNK mice were isolated and enriched. Enriched NK cells were cultured with IL-15 in the presence or absence of TGF-β. After 3 days in culture, Gzmb mRNA expression levels were analyzed by real-time RT-PCR (n = 4). Data are presented as the mean ± SD. *P < 0.05, by 2-tailed, paired t test. The circles and squares on the bar graphs denote data for individual mice. (E) Model of SMAD4 signaling pathways.
Two more TGF-β target genes, Trail and Gzmb, were assessed in WT and Smad4-deficient NK cells in the presence of IL-15, a critical cytokine that regulates NK cells. We found that TGF-β induced TRAIL but inhibited Gzmb in WT NK cells (Figure 7, C and D), which is consistent with the results of a previous study (60). However, in Smad4-deficient NK cells, we observed that TGF-β failed to upregulate TRAIL or downregulate Gzmb (Figures 7, C and D). These data indicate that SMAD4 is also required for TGF-β to downregulate Trail and Gzmb in NK cells.
Our studies reveal two different SMAD4 pathways in NK cells (Figure 7E). The first pathway is a TGF-β–independent one by which SMAD4 positively regulates NK cell homeostasis and maturation, promotes elimination of target cells by NK cells, upregulates the expression of Gzmb, and inhibits tumor metastasis. The second one is the TGF-β–dependent pathway, by which SMAD4 contributes to repression of IFN-γ and Gzmb as in the presence of TGF-β, which may promote tumor metastasis (61). However, the TGF-β–dependent pathway also upregulates the expression of TRAIL, which is toxic to tumor cells. Additional studies will be required to dissect the relative importance of the two SMAD4 signaling pathways under normal and pathological conditions.
Discussion
NK cells are important antitumor, large granular effector lymphocytes. Accumulating clinical evidence supports the notion that NK cells play a critical role in tumor surveillance, and NK cell intratumoral infiltration positively correlates with favorable prognosis in cancer patients (62, 63). Our recent data on enhanced antitumor activity of NK cells armed with a chimeric antigen receptor (CAR) indicate that CAR NK cells could be an excellent alternative to CAR T cells for clinical application (64–66). However, the detailed mechanism(s) underlying the antitumor immunity of NK cells remain to be addressed.
In this study, we described functions of SMAD4 in NK cells. We showed impaired homeostasis, maturation, and antitumor immunity in Smad4-deficient NK cells. A recent study by the Colonna group supports these findings (41). Different from the study by the Colonna group, we also characterized mechanisms. We identified 3 important SMAD4 target genes, Kit, Prdm1, and Gzmb, which positively regulate homeostasis, maturation, and cytotoxicity, respectively, and the expression levels of all of these genes were decreased in NK cells from Smad4ΔNK mice. We showed that SMAD4 and JUNB interact with each other and bind to the Gzmb promoter in order to cooperatively and positively regulate the expression of Gzmb. Additional work will be required to determine the origin of the extracellular signals that initiate or induce the positive role of SMAD4. In the TGF-β superfamily, BMPs provide another important signal upstream of SMAD4. However, we found that expression levels of the BMP receptor BMPR1a on the surface of murine NK cells were very low or undetectable (Supplemental Figure 17).
The development and maturation of NK cells are governed by many transcription factors (67). After analyzing the expression of well-characterized transcription factors that regulate NK cell maturation, we found that only Prdm1 was significantly decreased in Smad4-deficient NK cells (Figure 5E). Expression levels of both Prdm1 and Smad4 increased as NK cells matured, implying the important roles of these genes in promoting NK cell maturation. Our data show that some phenotypes of Smad4-deficient NK cells closely resemble Prdm1-deficient NK cells, in that both NK cell populations have impaired maturation blocked in the CD11b+CD27+ or KLRG1– state, and their expression of Gzmb is decreased (57). The significant correlation between Prdm1 and Smad4 implies the close link of the 2 genes in regulating NK cell maturation.
JUNB, a member of the activating protein 1 (AP-1) family, regulates multiple target genes and physiological functions (68–70). Although a JUNB binding site has been identified on the Gzmb promoter (39), detailed mechanisms of regulation involving JUNB transactivation have been lacking. For example, to our knowledge, no cofactors had previously been identified for JUNB in the transcriptional activation of Gzmb. GZMB is a key cytolytic molecule in both innate and adaptive immune functions. Thus, its optimal expression should be critical for effective immune responses against malignant cells. Our data not only show that SMAD4 cooperates with JUNB to achieve an optimal expression of Gzmb, but also support a model whereby JUNB directly interacts with the Gzmb proximal promoter that carries both SMAD4 and JUNB binding sites, with an interval of approximately 260 bp; SMAD4 is able to be associated with this promoter region by interacting with JUNB. However, despite being unable to transactivate the Gzmb proximal promoter without JUNB, it remains to be determined whether SMAD4 has direct DNA binding activity in this promoter region (ChIP assays were unable to distinguish direct DNA binding or an indirect association with a promoter). In addition, as NK cells are activated by a variety of stimuli, especially proinflammatory cytokines (18, 46), whether and how the SMAD4-JUNB complex mediates the signals from these stimuli and cross talks with other signaling pathways to quickly shape NK cell immune responses requires further exploration.
The continuous interactions of tumor cells and immune effector cells, including NK cells, in the tumor microenvironment can control tumorigenesis and metastasis (71). Tumor cells or other cells in the microenvironment can secrete TGF-β to suppress the normal function of immune effector cells and enhance immune evasion (72, 73); however, here we show that effector cells can also evolve mechanisms, e.g., the expression of Smad4 shown in this report, to circumvent the immune suppression caused by TGF-β. Indeed, our data show that in NK cells, SMAD4 plays a positive role in controlling tumor metastasis and maintaining one arm of antitumor immunity while TGF-β can mediate the suppression of IFN-γ production. Our data also suggest that IFN-γ production by NK cells alone may not be effective in controlling tumor formation, as Smad4ΔNK mice, deficient in Gzmb expression but not in IFN-γ production, are more susceptible to B16F10 tumor dissemination when compared with Smad4+/+ mice. In fact, the role of IFN-γ in antitumor activity can be a double-edged sword. IFN-γ can activate macrophage and T cell polarization (74, 75), while it can also induce PD-L1 expression on tumor cells, leading to tumor immune evasion (76).
Our data support the concept that, unlike somatic mutation(s) affecting only tumor cells or only immune cells, germline mutation(s) of a gene such as Smad4 that has a tumor-suppressive role in tumor cells and an antitumor role in immune cells could have a greater contribution to malignant transformation. In the instance of Smad4, the germline mutation would not only alter TGF-β signaling in tumor cells and other immune cells in the tumor microenvironment, but would also disarm the capacity of NK cells to lyse tumor cells. Of note, patients with JP, who carry a heterozygous germline mutation of SMAD4, are more susceptible to developing gastric cancer (22, 23, 77). Although many approaches, such as chemotherapy, directly target tumor cells but can also suppress the immune response (78), an approach to systemically enhance SMAD4 expression, at least in certain cancer patients, such as those with colon or pancreatic cancer, may have dual positive effects. In the clinical setting, systemic upregulation of SMAD4 could suppress tumor cells as well as enhance the cellular innate immune responses, both of which could theoretically contribute to control tumor development and progression.
Like many other signaling pathways, the SMAD4 signaling pathway is complex. The positive role of SMAD4 should be independent of TGF-β or p-SMAD2/3 signaling, at least when taking into account the following 2 pieces of evidence: (a) our data showed no significant difference in p-SMAD2/3 levels and Smad2/3 gene expression between Smad4-sufficient and -deficient NK cells under physiological conditions (Supplemental Figure 1E and Supplemental Figure 18), and (b) when we compared WT and Smad4-KO mice in the absence of SMAD2/3 signaling (i.e., under a TGFRII-KO background), we found the same phenotypes as those in the presence of SMAD2/3 signaling (i.e., on a TGFRII WT background). However, the positive role of SMAD4 elucidated in the current study does not exclude its TGF-β–dependent negative role, whereby SMAD4 cooperates with p-SMAD2 and p-SMAD3 to modulate the expression of IFN-γ, TRAIL, and even Gzmb in the presence of TGF-β. Whether the positive or negative SMAD4 pathway plays a predominant role should be context specific. For example, at early stages of primary tumor development or at remote sites of early tumor metastasis, the antitumor role of SMAD4 in NK cells may dominate over the immunosuppressive role, leading to effective immune surveillance. However, in the later stages of cancer development, especially within the tumor microenvironment, abundant TGF-β produced by tumor and/or stromal cells will likely dominate over the antitumor role of SMAD4, leading to immunosuppression of NK cells. Under such circumstances, it would be ideal to therapeutically enhance SMAD4 signaling at the early stages of tumor growth but inhibit its signaling at the latter stages of tumor growth. Thus, SMAD4 may be a critical regulator in the switch from effective antitumor immune surveillance to immunosuppression and immune evasion. Of note, recent studies reveal that the expression of TGF-β can determine the efficacy of PD-1 and PD-L1 immune checkpoint inhibitors in certain settings (79, 80).
In conclusion, we discovered a TGF-β–independent role of SMAD4 in the positive regulation of NK cell antitumor activity. These findings provide insight into how innate immune cells effectively survey against tumor metastases, which may allow us to better harness NK cells for cancer immunotherapy.
Methods
Mice.
WT C57BL/6, Smad4fl/fl, B2m–/–, and Tgfbr2fl/fl mice were purchased from The Jackson Laboratory. NKp46-iCre mice were a gift of Eric Vivier (Centre d’Immunologie de Marseille-Luminy, Marseille, France). All mice were on a C57BL/6 background. Six- to twelve-week-old male and female mice were used for the experiments.
Metastatic melanoma experiment and MCMV challenge.
B16F10 cells (0.25 × 106) were injected i.v. into mice. Fourteen days after injection, the mice were euthanized for postmortem analysis. Melanoma nodules in lung and extrapulmonary metastases were analyzed by macroscopic examination of organs. Lungs were fixed for H&E staining and IHC analysis. For IHC analysis, anti-S100 (Dako, Z0311) mAbs were used to identify B16F10 metastasis. The B16F10 cell line was provided by Gregory B. Lesinski (Emory University School of Medicine, Atlanta, Georgia, USA).
Smad4ΔNK and Smda4+/+ mice were infected with i.p. injections of Smith strain MCMV (1 × 105 PFU), which was provided by Lewis Lanier (UCSF, San Francisco, California, USA). Peripheral blood samples were obtained through submandibular puncture 7 days after infection. To measure viral loads in the peripheral blood, DNA was isolated using a QIAGEN DNeasy Blood and Tissue Kit for quantitative PCR analysis. The following primers were used: MCMV-IE1, 5′-AGCCACCAACATTGACCACGCAC-3′ (forward) and MCMV-IE1, 5′-GCCCCAACCAGGACACACAACTC-3′ (reverse).
In vivo and ex vivo cytotoxicity assays.
Splenocytes were isolated from B2m-deficient (B2m–/–) and B2m-sufficient (B2m+/+) mice. Splenocytes from B2m–/– mice were labeled with eF670. Splenocytes from B2m+/+ mice were labeled with CFDA-SE. Labeled splenocytes from B2m–/– mice and B2m+/+ mice were mixed together at a ratio of 1:1 and i.v. injected into Smad4+/+ and Smad4ΔNK mice. One day after injection, spleens were harvested from the recipient mice, and the percentage of labeled donor cells among recipient splenocytes was analyzed by flow cytometry. The percentage of rejection was then calculated using the following formula: 100 – ([(% B2m–/–/% B2m+/+) output/(% B2m–/–/% B2m+/+) input] ×100). Ex vivo cytotoxicity of NK cells was evaluated by standard 51Cr release assays. YAC-1 cells, purchased from the American Type Culture Collection (ATCC), and RMA-S cells, a gift of André Veillette (McGill University, Montréal, Canada), were used as target cells. Fresh NK cells were cocultured with targets cells at a ratio of 50:1.
Flow cytometry.
LSRII and FACSAria II cytometers (BD Biosciences) were used for flow cytometry and cell sorting. FlowJo software was used to analyze the data. The following Abs were obtained from BD Biosciences: CD335-BV421 (catalog 562850, clone 29A1.4); IFN-γ–PE (catalog 554412, clone XMG1.2); NKG2D-PE (catalog 558403, clone CX5); NKG2A/C/E-FITC (catalog 550520, clone 20d5); CD27-PE (catalog 558754, clone LG.3A10); CD27-PE-Cy7 (catalog 563604, clone LG.3A10); CD11b-V450 (catalog 560455, clone M1/70); CD11b-FITC (catalog 557396, clone M1/70); CD69-PE (catalog 553237, clone H1.2F3); Ly49G2-FITC (catalog 555315, clone 4D11); Ly49D-FITC (catalog 555313, clone 4E5); Ly49C/I-FITC (catalog 553276, clone 5E6); Ly49A-FITC (catalog 553677, clone A1); CD3-V450 (catalog 560801, clone 500A2); CD3-FITC (catalog 553062, clone 145-2C11); CD45-FITC (catalog 553772, clone 104); and NKp46-AF647 (catalog 560755, clone 29A1.4). CD94-FITC (catalog 11-0941-81, clone 18d3) was purchased from eBioscience.
Real-time RT-PCR.
RNA was isolated using a Total RNA Purification Kit (Norgen, 17200)and then reverse transcribed to cDNA with SuperScript VILO Master Mix (Life Technologies, Thermo Fisher Scientific, 11755050) following the instructions provided by the manufacturer. mRNA expression levels were analyzed using SYBR Green PCR Master Mix (catalog 4334973) and a Vii 7 Real-Time RT-PCR System (both from Thermo Fisher Scientific). Primer sequences are listed in Supplemental Table 1.
Immunoblotting, co-IP, and ChIP assays.
Immunoblotting and co-IP were performed according to standard procedures. Anti-SMAD4 mAbs were purchased from MilliporeSigma (catalog 04-1033/clone EP618Y) or from Abcam (catalog ab40759, clone EP618Y). Anti-SMAD4 polyclonal Abs were purchased from Cell Signaling Technology (catalog 9515) or from Santa Cruz Biotechnology (sc-1909). The anti-GZMB polyclonal Ab (catalog 4275) and anti-JUNB mAb (catalog 3753, clone C37F9) were purchased from Cell Signaling Technology. ChIP assays were carried out using a Magna A/G Chromatin Immunoprecipitation Kit (Merck Millipore) or a Pierce Magnetic ChIP Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, an equal amount of an anti-SMAD4 or anti-JUNB Ab and the corresponding control normal IgG were separately used to precipitate the cross-linked DNA-protein complexes derived from 5 × 106 purified mouse primary NK cells, respectively. Following reversal of cross-linking, the DNA immunoprecipitated by the indicated Ab was tested by PCR. The primer pair used for detecting the Gzmb promoter region spanning from –605 bp to –305 bp relative to the TSS, where the putative SMAD4 binding site was harbored, was 5′-TCGGACAAATATCATCTTTCCC-3′ (forward) and 5′-AAGCATGACCATCTAGTGAG-3′ (reverse). The primer pair used for detecting the Gzmb promoter region spanning from –150 bp to +3 bp relative to the TSS, where the putative JUNB binding site was harbored, was 5′-CTGTAAGTGGTGGTTCTC-3′ (forward) and 5′-TCTCTGCTTTTATGATGCT-3′ (reverse).
Luciferase reporter assay.
The Gzmb promoter region ranging from –625 bp to +93 bp of the TSS was amplified from murine NK cells and cloned into a pGL3-basic luciferase reporter vector (Promega) to generate a pGL3-Gzmb reporter plasmid. Murine NIH-3T3 cells purchased from ATCC were cotransfected with the pGL3-Gzmb reporter plasmid as well as Smad4 and/or Junb overexpression plasmids or an empty vector, together with a pRL-TK Renilla reporter plasmid (Promega) for normalization of transfection efficiency. The cells were harvested for lysis 48 hours after transfection, and luciferase activity was quantified fluorimetrically with the Dual Luciferase System (Promega). The QuikChange II Site-Directed Mutagen Kit (200523) from Agilent Technologies was used for mutating the SMAD4 or JUNB binding site in the proximal Gzmb promoter. The SMAD4 binding site “TGTCTGT” was mutated to “TAAAAAT,” and the JUNB binding site “TTCTCTGAGTCA” was mutated to “TTCTCAAAAACA”.
Statistics.
For continuous and normally distributed measurements, 2-sample t tests were used to compare the group mean of 2 independent groups such as Smad4ΔNK versus Smad4+/+. Paired t tests were used to compare 2 paired groups such as littermates split into 2 groups. ANOVA models were used to compare multiple groups. P values were adjusted for multiple comparisons by Holm’s procedure. A P value of 0.05 or less was considered significant. Data are presented as the mean ± SD.
Study approval.
All mice were bred and housed in the animal facility at The Ohio State University. All animal experiments were approved by the IACUC of The Ohio State University (protocol 201200000090) and performed in accordance with institutional animal care and use guidelines. No human subjects were used in this study.
Author contributions
Youwei Wang designed experiments, performed experiments, and wrote the manuscript. JC, Yufeng Wang, PY, WD, HW, JS, SS, and YD performed experiments. JZ analyzed the data. LCW, XH, BMB, AGF, and LSW interpreted results and reviewed the manuscript. MAC designed experiments and reviewed and edited the manuscript. JY conceived and directed the project, designed the research, interpreted the results, acquired funding, and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank Eric Vivier (Centre d’Immunologie de Marseille-Luminy) for providing the NKp46-iCre mice. This project was supported in part by grants from the NIH (AI129582, NS106170, CA185301, CA068458, CA095426, CA210087, and CA163205). This project was also supported in part by an American Cancer Society Research Scholar Grant (RSG-14-243-01-LIB); 2 grants from The Leukemia and Lymphoma Society; and a grant from Gabrielle’s Angel Foundation for Cancer Research. Youwei Wang was supported by the Pelotonia Postdoctoral Fellowship from The Ohio State University Comprehensive Cancer Center. The authors would like to thank David M. Lucas and Tiffany Hughes at The Ohio State University for their critical reading of the manuscript.
Version 1. 09/05/2018
In-Press Preview
Version 2. 10/15/2018
Electronic publication
Version 3. 11/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(11):5123–5136.https://doi.org/10.1172/JCI121227.
Contributor Information
Youwei Wang, Email: youwei.wang@osumc.edu.
Jianhong Chu, Email: chujianhong@hotmail.com.
Ping Yi, Email: yp_yiping@foxmail.com.
Yufeng Wang, Email: wangyufengilu@163.com.
Hongwei Wang, Email: whw6626@sina.com.
Jianying Zhang, Email: jianying.zhang@osumc.edu.
Lai-Chu Wu, Email: Laichu.Wu@osumc.edu.
Youcai Deng, Email: dengyc125@hotmail.com.
Xiaoming He, Email: shawnhe@umd.edu.
Aharon G. Freud, Email: aharon.freud@osumc.edu.
Li-Shu Wang, Email: liswang@mcw.edu.
Michael A. Caligiuri, Email: mcaligiuri@coh.org.
Jianhua Yu, Email: jianhua.yu@osumc.edu.
References
- 1.Kim S, Iizuka K, Aguila HL, Weissman IL, Yokoyama WM. In vivo natural killer cell activities revealed by natural killer cell-deficient mice. Proc Natl Acad Sci U S A. 2000;97(6):2731–2736. doi: 10.1073/pnas.050588297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sathe P, et al. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nat Commun. 2014;5:4539. doi: 10.1038/ncomms5539. [DOI] [PubMed] [Google Scholar]
- 3.Paolino M, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. 2014;507(7493):508–512. doi: 10.1038/nature12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juelich T, et al. Interplay between chromatin remodeling and epigenetic changes during lineage-specific commitment to granzyme B expression. J Immunol. 2009;183(11):7063–7072. doi: 10.4049/jimmunol.0901522. [DOI] [PubMed] [Google Scholar]
- 5.Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113(22):5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 6.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176(3):1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 7.Zhang N, Bevan MJ. TGF-β signaling to T cells inhibits autoimmunity during lymphopenia-driven proliferation. Nat Immunol. 2012;13(7):667–673. doi: 10.1038/ni.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas DA, Massagué J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8(5):369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Forrester E, et al. Effect of conditional knockout of the type II TGF-beta receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res. 2005;65(6):2296–2302. doi: 10.1158/0008-5472.CAN-04-3272. [DOI] [PubMed] [Google Scholar]
- 10.Siegel PM, Shu W, Cardiff RD, Muller WJ, Massagué J. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc Natl Acad Sci U S A. 2003;100(14):8430–8435. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han J, Alvarez-Breckenridge CA, Wang QE, Yu J. TGF-β signaling and its targeting for glioma treatment. Am J Cancer Res. 2015;5(3):945–955. [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Pang Y, Moses HL. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31(6):220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui W, et al. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86(4):531–542. doi: 10.1016/S0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 14.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David CJ, et al. TGF-β Tumor Suppression through a Lethal EMT. Cell. 2016;164(5):1015–1030. doi: 10.1016/j.cell.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bardeesy N, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20(22):3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcoe JP, et al. TGF-β is responsible for NK cell immaturity during ontogeny and increased susceptibility to infection during mouse infancy. Nat Immunol. 2012;13(9):843–850. doi: 10.1038/ni.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, et al. Pro- and antiinflammatory cytokine signaling: reciprocal antagonism regulates interferon-gamma production by human natural killer cells. Immunity. 2006;24(5):575–590. doi: 10.1016/j.immuni.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Trotta R, et al. TGF-beta utilizes SMAD3 to inhibit CD16-mediated IFN-gamma production and antibody-dependent cellular cytotoxicity in human NK cells. J Immunol. 2008;181(6):3784–3792. doi: 10.4049/jimmunol.181.6.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massagué J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1(3):169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 21.Malkoski SP, Wang XJ. Two sides of the story? Smad4 loss in pancreatic cancer versus head-and-neck cancer. FEBS Lett. 2012;586(14):1984–1992. doi: 10.1016/j.febslet.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aytac E, et al. Genotype-defined cancer risk in juvenile polyposis syndrome. Br J Surg. 2015;102(1):114–118. doi: 10.1002/bjs.9693. [DOI] [PubMed] [Google Scholar]
- 23.Howe JR, et al. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280(5366):1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 24.Sirard C, et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12(1):107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viant C, Fenis A, Chicanne G, Payrastre B, Ugolini S, Vivier E. SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nat Commun. 2014;5:5108. doi: 10.1038/ncomms6108. [DOI] [PubMed] [Google Scholar]
- 26.Cillo C, Dick JE, Ling V, Hill RP. Generation of drug-resistant variants in metastatic B16 mouse melanoma cell lines. Cancer Res. 1987;47(10):2604–2608. [PubMed] [Google Scholar]
- 27.Karo JM, Schatz DG, Sun JC. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell. 2014;159(1):94–107. doi: 10.1016/j.cell.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bix M, Liao NS, Zijlstra M, Loring J, Jaenisch R, Raulet D. Rejection of class I MHC-deficient haemopoietic cells by irradiated MHC-matched mice. Nature. 1991;349(6307):329–331. doi: 10.1038/349329a0. [DOI] [PubMed] [Google Scholar]
- 29.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319(6055):675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 30.Ljunggren HG, Kärre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11(7):237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 31.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358(6381):66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 32.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kägi D, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature. 1994;369(6475):31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 34.Heusel JW, Wesselschmidt RL, Shresta S, Russell JH, Ley TJ. Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells. Cell. 1994;76(6):977–987. doi: 10.1016/0092-8674(94)90376-X. [DOI] [PubMed] [Google Scholar]
- 35.Pardo J, Balkow S, Anel A, Simon MM. Granzymes are essential for natural killer cell-mediated and perf-facilitated tumor control. Eur J Immunol. 2002;32(10):2881–2887. doi: 10.1002/1521-4141(2002010)32:10<2881::AID-IMMU2881>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 36.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94(5):585–594. doi: 10.1016/S0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 38.Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem. 1998;273(33):21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 39.Babichuk CK, Bleackley RC. Mutational analysis of the murine granzyme B gene promoter in primary T cells and a T cell clone. J Biol Chem. 1997;272(30):18564–18571. doi: 10.1074/jbc.272.30.18564. [DOI] [PubMed] [Google Scholar]
- 40.Babichuk CK, Duggan BL, Bleackley RC. In vivo regulation of murine granzyme B gene transcription in activated primary T cells. J Biol Chem. 1996;271(28):16485–16493. doi: 10.1074/jbc.271.28.16485. [DOI] [PubMed] [Google Scholar]
- 41.Cortez VS, et al. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat Immunol. 2017;18(9):995–1003. doi: 10.1038/ni.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim SK, Hoffmann FM. Smad4 cooperates with lymphoid enhancer-binding factor 1/T cell-specific factor to increase c-myc expression in the absence of TGF-beta signaling. Proc Natl Acad Sci U S A. 2006;103(49):18580–18585. doi: 10.1073/pnas.0604773103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu AD, Zhang S, Wang Y, Xiong H, Curtis TA, Wan YY. A critical role for transcription factor Smad4 in T cell function that is independent of transforming growth factor β receptor signaling. Immunity. 2015;42(1):68–79. doi: 10.1016/j.immuni.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narni-Mancinelli E, et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc Natl Acad Sci U S A. 2011;108(45):18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colucci F, Di Santo JP. The receptor tyrosine kinase c-kit provides a critical signal for survival, expansion, and maturation of mouse natural killer cells. Blood. 2000;95(3):984–991. [PubMed] [Google Scholar]
- 46.Yu J, Freud AG, Caligiuri MA. Location and cellular stages of natural killer cell development. Trends Immunol. 2013;34(12):573–582. doi: 10.1016/j.it.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity. 2017;47(5):820–833. doi: 10.1016/j.immuni.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Helden MJ, et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med. 2015;212(12):2015–2025. doi: 10.1084/jem.20150809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huntington ND, et al. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J Immunol. 2007;178(8):4764–4770. doi: 10.4049/jimmunol.178.8.4764. [DOI] [PubMed] [Google Scholar]
- 50.Deng Y, et al. Transcription factor Foxo1 is a negative regulator of natural killer cell maturation and function. Immunity. 2015;42(3):457–470. doi: 10.1016/j.immuni.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20(4):477–494. doi: 10.1016/S1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 52.Samson SI, et al. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity. 2003;19(5):701–711. doi: 10.1016/S1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 53.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204(5):1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lacorazza HD, et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17(4):437–449. doi: 10.1016/S1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 55.Kamizono S, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. 2009;206(13):2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gordon SM, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36(1):55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kallies A, et al. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood. 2011;117(6):1869–1879. doi: 10.1182/blood-2010-08-303123. [DOI] [PubMed] [Google Scholar]
- 58.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.da Silva S, et al. Proper formation of whisker barrelettes requires periphery-derived Smad4-dependent TGF-beta signaling. Proc Natl Acad Sci U S A. 2011;108(8):3395–3400. doi: 10.1073/pnas.1014411108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Viel S, et al. TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci Signal. 2016;9(415):ra19. doi: 10.1126/scisignal.aad1884. [DOI] [PubMed] [Google Scholar]
- 61.Takeda K, et al. IFN-γ production by lung NK cells is critical for the natural resistance to pulmonary metastasis of B16 melanoma in mice. J Leukoc Biol. 2011;90(4):777–785. doi: 10.1189/jlb.0411208. [DOI] [PubMed] [Google Scholar]
- 62.Ishigami S, et al. Prognostic value of intratumoral natural killer cells in gastric carcinoma. Cancer. 2000;88(3):577–583. doi: 10.1002/(SICI)1097-0142(20000201)88:3<577::AID-CNCR13>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 63.Coca S, et al. The prognostic significance of intratumoral natural killer cells in patients with colorectal carcinoma. Cancer. 1997;79(12):2320–2328. doi: 10.1002/(SICI)1097-0142(19970615)79:12<2320::AID-CNCR5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 64.Chen X, et al. A combinational therapy of EGFR-CAR NK cells and oncolytic herpes simplex virus 1 for breast cancer brain metastases. Oncotarget. 2016;7(19):27764–27777. doi: 10.18632/oncotarget.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu J, et al. CS1-specific chimeric antigen receptor (CAR)-engineered natural killer cells enhance in vitro and in vivo antitumor activity against human multiple myeloma. Leukemia. 2014;28(4):917–927. doi: 10.1038/leu.2013.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han J, et al. CAR-engineered NK cells targeting wild-type EGFR and EGFRvIII enhance killing of glioblastoma and patient-derived glioblastoma stem cells. Sci Rep. 2015;5:11483. doi: 10.1038/srep11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun JC. Transcriptional Control of NK Cells. Curr Top Microbiol Immunol. 2016;395:1–36. doi: 10.1007/82_2015_452. [DOI] [PubMed] [Google Scholar]
- 68.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 69.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 70.Piechaczyk M, Farràs R. Regulation and function of JunB in cell proliferation. Biochem Soc Trans. 2008;36(Pt 5):864–867. doi: 10.1042/BST0360864. [DOI] [PubMed] [Google Scholar]
- 71.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ghiringhelli F, et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-beta-dependent manner. J Exp Med. 2005;202(8):1075–1085. doi: 10.1084/jem.20051511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 74.Varesio L, Blasi E, Thurman GB, Talmadge JE, Wiltrout RH, Herberman RB. Potent activation of mouse macrophages by recombinant interferon-gamma. Cancer Res. 1984;44(10):4465–4469. [PubMed] [Google Scholar]
- 75.Morandi B, Bougras G, Muller WA, Ferlazzo G, Münz C. NK cells of human secondary lymphoid tissues enhance T cell polarization via IFN-gamma secretion. Eur J Immunol. 2006;36(9):2394–2400. doi: 10.1002/eji.200636290. [DOI] [PubMed] [Google Scholar]
- 76.Loke P, Allison JP. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc Natl Acad Sci U S A. 2003;100(9):5336–5341. doi: 10.1073/pnas.0931259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woodford-Richens KL, et al. Comprehensive analysis of SMAD4 mutations and protein expression in juvenile polyposis: evidence for a distinct genetic pathway and polyp morphology in SMAD4 mutation carriers. Am J Pathol. 2001;159(4):1293–1300. doi: 10.1016/S0002-9440(10)62516-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zitvogel L, Apetoh L, Ghiringhelli F, Kroemer G. Immunological aspects of cancer chemotherapy. Nat Rev Immunol. 2008;8(1):59–73. doi: 10.1038/nri2216. [DOI] [PubMed] [Google Scholar]
- 79.Mariathasan S, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tauriello DVF, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–543. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.