Abstract
Renin cells are crucial for survival — they control fluid-electrolyte and blood pressure homeostasis, vascular development, regeneration, and oxygen delivery to tissues. During embryonic development, renin cells are progenitors for multiple cell types that retain the memory of the renin phenotype. When there is a threat to survival, those descendants are transformed and reenact the renin phenotype to restore homeostasis. We tested the hypothesis that the molecular memory of the renin phenotype resides in unique regions and states of these cells’ chromatin. Using renin cells at various stages of stimulation, we identified regions in the genome where the chromatin is open for transcription, mapped histone modifications characteristic of active enhancers such as H3K27ac, and tracked deposition of transcriptional activators such as Med1, whose deletion results in ablation of renin expression and low blood pressure. Using the rank ordering of super-enhancers, epigenetic rewriting, and enhancer deletion analysis, we found that renin cells harbor a unique set of super-enhancers that determine their identity. The most prominent renin super-enhancer may act as a chromatin sensor of signals that convey the physiologic status of the organism, and is responsible for the transformation of renin cell descendants to the renin phenotype, a fundamental process to ensure homeostasis.
Keywords: Endocrinology, Nephrology
Keywords: Epigenetics, Homeostasis, Hypertension
Introduction
Renin-expressing cells appeared more than 400 million years ago. Throughout evolution, they acquired numerous defensive functions that rendered them as perfect machines to maintain homeostasis: they control blood pressure, fluid-electrolyte balance, vascular development, and glomerular regeneration (1). In addition, renin-expressing cells share a lineage relationship with erythropoietin-producing pericytes and participate in the regulation of oxygen delivery to tissues (1).
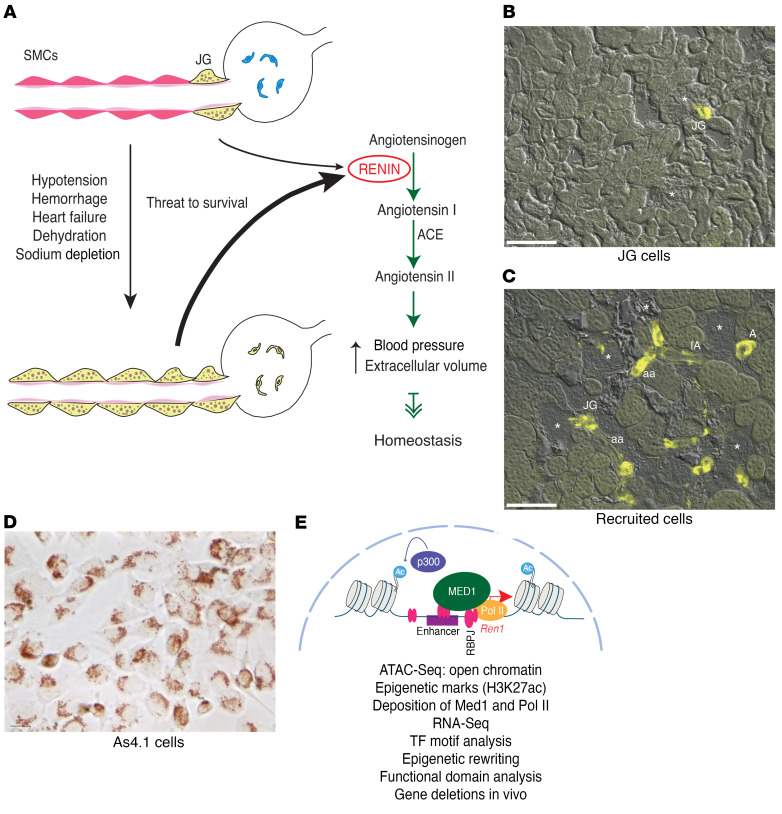
In adult unstressed mammals, renin-expressing cells are located in the walls of the renal arterioles at the entrance to the glomeruli and are termed juxtaglomerular (JG) cells (Figure 1) (2). JG cells are sensors that synthesize and release the hormone and enzyme RENIN in response to changes in blood pressure and extracellular fluid volume (3). Upon reaching the circulation, RENIN initiates an enzymatic cascade that culminates in the production of angiotensin II, a potent vasoconstrictor that elevates blood pressure and extracellular fluid volume (Figure 1). Although the number of JG cells is quite small, constituting 0.01% of the total kidney cell mass, under normal circumstances rapid and precise secretion of RENIN by those few cells usually suffices to maintain blood pressure and fluid-electrolyte homeostasis. However, if an adult animal is subjected to a more serious threat such as dehydration, hemorrhage, heart failure, sodium depletion, or hypovolemia, smooth muscle cells along the kidney arterioles, mesangial cells within glomeruli, and pericytes within the kidney interstitium are transformed to synthesize RENIN until homeostasis is regained (Figure 1A) (4). This process has been termed “recruitment” to indicate that cells along the renal arteriole adopt the renin phenotype when more RENIN is needed to overcome a major threat to survival (5) (Figure 1). Once the crisis passes, smooth muscle cells (SMCs) switch off the renin gene and regain their smooth muscle phenotype.
Figure 1. Identity of the renin cells and transformation of renin cell descendants to the renin phenotype when homeostasis is threatened.
The 3 renin cell models used in the present study. (A) Schematic showing juxtaglomerular (JG) cells, the classical renin-expressing cells, located at the entrance to the glomerulus (yellow) in the adult unstressed animal and along the afferent arteriole in response to a threat to survival. These cells secrete the hormone RENIN, a key enzyme of the renin angiotensin system that culminates in the generation of angiotensin II, a powerful vasoconstrictor that regulates blood pressure and fluid electrolyte homeostasis. (B) Kidney section from a Ren1c-YFP WT mouse. In this mouse YFP expression is driven by the renin promoter thus marking the JG renin-expressing cells. The YFP signal is restricted to the afferent arterioles at the entrance to the glomerulus. Scale bar: 100 μM. (C) In renin-deficient mice, SMCs that descended from renin precursors are chronically stimulated to adopt the renin phenotype. Kidney section from a Ren1c–/– Ren1c-YFP mouse. YFP+ cells reporting the activity of the renin promoter are distributed along the afferent arterioles (aa), interlobular arteries (IA), larger intrarenal arteries (A), at the entrance to the glomerulus (JG), and in the mesangium. Scale bar: 100 μM. (D) As4.1 cells, mouse kidney tumoral cells that constitutively produce renin. Numerous renin granules detected with the vital marker neutral red are present in the cytoplasm of these cells. Scale bar: 10 μM. (E) Schematic depicting our working hypotheses and the methods used to test the renin cells using as an example the chromatin at the renin locus. Briefly, we hypothesize that renin cells possess unique sets of renin cell–specific genomic elements such as super-enhancers that determine renin cell identity and the molecular memory of the renin phenotype, allowing renin cell descendants to switch on the renin program when homeostasis is threatened.
The ability to switch on and off the renin phenotype depends on the developmental history of the transformed cells (5). Thus, renin cell descendants (such as arteriolar SMCs) retain the memory to synthesize RENIN when a physiological or pathological circumstance requires that more renin is produced to regain homeostasis. Preservation of such molecular memory is a fundamental mechanism to respond to life threats. However, the mechanisms that control the identity and fate of renin cells are unknown. We hypothesize that the molecular memory of the renin phenotype resides in the chromatin state of renin cell descendants. Using a genome-wide assessment of the chromatin status of WT unstressed JG cells, native renin cells chronically and acutely stimulated to produce RENIN, and tumoral As4.1 cells, which produce RENIN constitutively, we uncovered a unique set of super-enhancers that determine the identity of renin cells. Of those, the most prominent renin super-enhancer harbors the molecular memory of the renin phenotype and is ultimately responsible for the transformation of renin cell descendants to the renin phenotype, a fundamental mechanism to preserve homeostasis.
Results
Accessible chromatin and enhancer elements characteristic of renin cells.
The identity of a cell is ultimately determined by its unique and specific pattern of gene expression. In turn, gene expression is facilitated by active regulatory elements in the genome that display open, accessible chromatin configuration. To define regulatory distal enhancers and promoter regions along the genome of renin-producing cells where the chromatin is open, we performed the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-Seq) (6). We compared several types of renin cells. JG cells are the classical renin-expressing cells found in the adult unstressed animal (Figure 1B). To isolate JG cells, we generated mice bearing the Ren1c promoter fused to yellow fluorescent protein (YFP), thus identifying cells in which the renin promoter is actively transcribing renin and YFP (Figure 1B). JG cells were freshly dispersed and purified using fluorescence-activated cell sorting. We also looked at “recruited” cells along the afferent arterioles chronically stimulated to adopt the renin phenotype (Figure 1C). They are an invaluable model to understand the developmental memory of renin cell identity. Recruited cells were sorted from the kidneys of Ren1c–/– Ren1c-YFP+ mice (7). These mice contain intact 5′ and 3′ regulatory regions of the renin locus, but the gene itself is inactivated by gene targeting, therefore providing a unique tractable model system to study the regulatory activity of the renin locus (8). These animals are hypotensive and fluid depleted, 2 conditions that constantly stimulate the recruitment of cells attempting to produce RENIN along the kidney arterioles (Figure 1C). They harbor the same Ren1c-YFP transgene described above, reporting the activity of the renin promoter via YFP (9), which marks the recruited renin cells along the arterioles (Figure 1C). These mice have 5 to 10 times more YFP+ cells (range: 1,770–80,369; median: 16,175; mean: 18,334) along the arterioles than JG cells from WT mice (range: 841–6,917; median: 3,771; mean: 3,863; P < 0.001), and although these native renin cells do not produce RENIN, they maintain a super-activated renin promoter and conserve the molecular program of the renin cell phenotype (7). The results from those in vivo–isolated cells were compared with a third cellular model of renin-producing cells, the As4.1 kidney tumoral cells, derived from transgenic mice that harbor a transgene containing the Ren2 5′ regulatory noncoding region fused to the SV40 T antigen gene (Figure 1D). These cells express authentic renin from the Ren1c gene — similarly to the freshly dispersed native cells described above — and secrete large amounts of RENIN constitutively in long-term culture (10). As4.1 cells contain all the necessary machinery for the correct tissue- and cell-specific expression of renin (11). Figure 1D shows the morphology of As4.1 cells and the presence of RENIN granules stained with the vital dye neutral red, resembling the appearance of JG precursor cells in the mammalian kidney (12). Figure 1E illustrates our hypotheses and the methods used to test the renin cells using as an example the chromatin at the renin locus. We hypothesize that renin cells possess unique sets of renin cell–specific genomic elements such as enhancers and super-enhancers characterized by open chromatin configuration, deposition of H3K27ac, mediator subunit 1 (Med1), polymerase II (Pol II), and high density of transcription factor binding. We suggest that those genomic regions are functionally responsible for the unique transcription profile that determines the identity of the renin cell. We further propose that the memory of the renin phenotype resides in and is controlled by a dynamic super-enhancer at the renin locus, which can be reactivated in response to external signals and/or epigenetic rewriting.
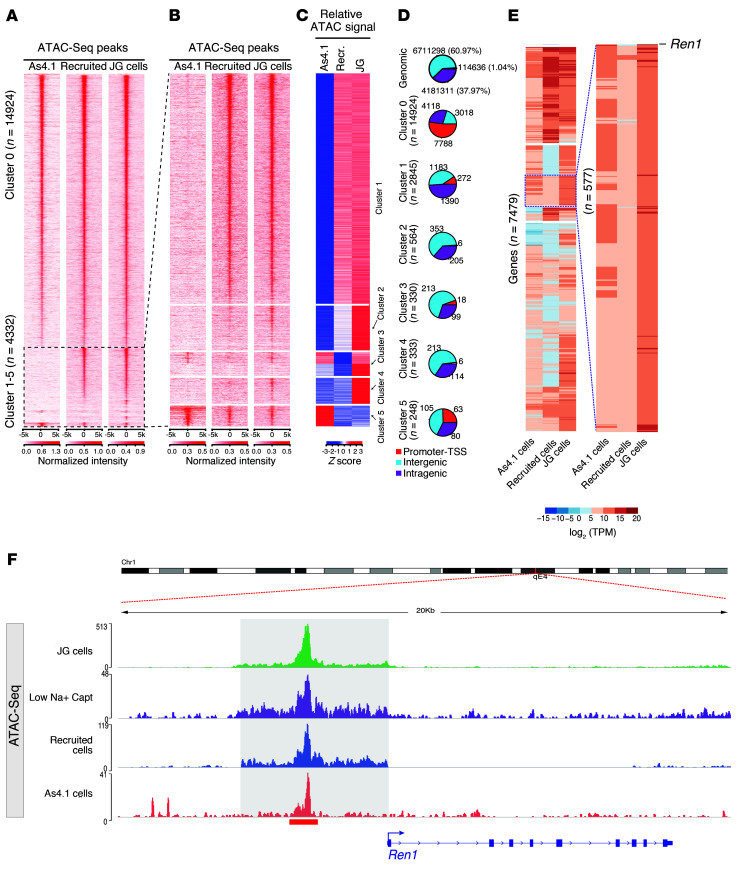
Figure 2 shows the DNA accessibility profiles found in each cell type. ATAC-Seq performed in JG cells identified 16,905 open chromatin regions, whereas recruited renin cells and As4.1 cells contained 37,802 and 49,501 accessible chromatin regions, respectively. These 3 cell types shared 19,256 consensus ATAC-Seq peaks genome-wide (Figure 2A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI121361DS1). In the heatmaps, each row represents an ATAC-Seq region with significant signal intensity in any of the 3 renin cell types that were clustered according to their chromatin state. Cluster 0 illustrates the ATAC-Seq peaks shared among the 3 cell types, indicating regulatory elements that may characterize the renin cells (Figure 2A). Clusters 1–5 identified regions of open chromatin differentially distributed among cell types (Figure 2B). We observed that clusters 1 and 2 shared ATAC-Seq peaks between JG and recruited cells, cluster 3 shows shared peaks between JG and As4.1 cells, cluster 4 shows specific peaks in JG cells, and cluster 5 shows specific peaks for the As4.1 cell line. The intensities of the ATAC-Seq peaks were normalized (Z scores) for these clusters to highlight the relative cell type–specific regions for each of them (Figure 2C). Figure 2D illustrates the proportions of ATAC-Seq peaks in each cluster according to the genomic compartments. For instance, cluster 0 shows the greater enrichment for ATAC-Seq peaks at the promoter regions (–1 kb to +1 kb of the transcription start site [TSS]), suggesting that these are active regions in the genome.
Figure 2. Comparative analysis of chromatin states across renin cell types.
(A and B) Heatmaps depict the sequencing read coverage intensity at 19,256 consensus ATAC-Seq peaks. Each row represents the DNA accessibility profile of a regulatory element demonstrating significant ATAC-Seq signal intensity. While cluster 0 shows ATAC-Seq and H3K27ac ChIP-Seq (Supplemental Figure 1A) peaks that are shared by the 3 cell types, clusters 1–5 demonstrate cell type–specific differential distributions and ATAC-Seq reads (B) at 5 kb upstream and downstream of each of the ATAC-Seq peaks. (C) Read coverage intensities at cell type–specific ATAC-Seq peaks: cluster 1 and 2 show ATAC-Seq peaks observed in mouse isolated cell types; cluster 3 shows ATAC-Seq peaks shared by As4.1 cell line and in JG cells; cluster 4 shows specific peaks in JG cells; cluster 5 shows specific ATAC-Seq peaks in As4.1 cells. Normalized ATAC-Seq signal intensities (Z scores) at peaks in clusters 1–5 to highlight the relative cell type–specific ATAC-Seq signal. (D) Pie charts show proportions of ATAC-Seq peaks in each cluster in the indicated genomic compartments. Promoter regions are defined as 1,000 bp upstream to 100 bp downstream of the transcription start site (TSS), intragenic regions are defined as introns, exons, 5′ UTR, 3′ UTR, and TTS, and the rest of the genome is defined as the intergenic regions. Genomic control sites are defined as all the sites in the genome with bins equivalent to the median ATAC peak size and a sliding window of half that size. (E) Expression levels for genes associated to ATAC-Seq peaks in cluster 0 shown in B. The inset to the right shows the renin gene with notable differences in its expression between the 3 cell types. (F) Genome browser image at the renin locus shows a remarkably similar pattern of ATAC-Seq among the renin cell types.
To correlate the activity at these regulatory elements with their transcriptional state, we acquired RNA-Seq gene expression profiles in these 3 cell types. Using the Genomic Regions Enrichment of Annotations Tool (GREAT) (13), we annotated genes near these overlap regions, using the GRCh38/mm10 assembly under default settings, thereby assigning each gene to a regulatory region nearby and either upstream or downstream to the TSS. As a result, we obtained 7,479 genes corresponding to cluster 0. Comparative transcriptome analysis among JG cells, recruited cells, and As4.1 cells are shown in Figure 2E and Supplemental Table 2. The inset in Figure 2E shows 577 genes clustered with a similar expression pattern. We highlighted the presence of the renin gene within this group, indicating that although the gene is not expressed in recruited cells, the intact regulatory region is important to the identity of the cells.
To define whether similar patterns of chromatin accessibility were present in renin cells from animals subjected to more acute hemodynamic stress, we studied cells obtained from WT (Ren1c+/+) mice harboring the same Ren1c-YFP transgene described above, subjected to sodium depletion plus captopril for 7 days as we previously described (14, 15). As expected, with this physiological challenge, YFP+ cells increase in number and are distributed along the afferent arterioles in a pattern similar to the one found in chronically recruited animals (Supplemental Figure 1A, right). The recruitment of renin-expressing cells was accompanied by a significant increase in circulating RENIN (328,689.78 ± 104,567 vs. 81,353.52 ± 44,573 pg/ml; P < 0.01; Supplemental Figure 1B). Underlying these morphological and functional changes, ATAC signals along the regulatory regions of the renin gene indicated that the chromatin was open in a pattern remarkably similar to the patterns observed in JG cells, chronically recruited cells, and As4.1 cells (Figure 2F, Supplemental Figure 1C). Further comparisons of chromatin accessibility across the whole genome of these 4 cell types indicated that although the overall chromatin accessibility was quite similar among the 4 cells studied, the pattern of chromatin accessibility in acutely recruited (low sodium plus captopril) cells was more similar to the pattern observed in JG and chronically recruited cells than to the pattern observed in As4.1 cells (Supplemental Figure 1D). These results suggest that under short-term physiological stimulation, newly recruited cells may possess a chromatin configuration that resembles the chromatin of JG cells. On the other hand, chronic protracted stimulation, such as occurs in Ren1c knockout animals or in tumoral As4.1 cells, may result in additional chromatin changes unrelated to the core chromatin features that determine the renin phenotype but rather to concomitant pathological changes that may occur in these 2 situations (7, 10).
To further narrow the active distal and proximal regulatory elements, we also acquired epigenome maps of regions occupied by histone 3 lysine 27 acetylation (H3K27ac), which marks active enhancers and promoters (16). We performed chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) in biological replicates of native recruited renin cells and As4.1 cells. Using the model-based analysis of ChIP-Seq (MACS) (17), we identified 10,056 and 36,568 significantly enriched regions throughout the genome of recruited and As4.1 cells, respectively. Supplemental Figure 2A shows a heatmap with the enrichment of H3K27ac for the same 19,256 ATAC-Seq regions found among the 3 cell types (shown in Figure 2A). Supplemental Figure 2B shows an average plot of H3K27ac signal distribution in recruited renin cells across gene bodies, adjacent to TSSs throughout the genome. The average plot for promoter regions shows enrichment in both upstream and downstream regions of the TSS where enhancers are usually located. Supplemental Figure 2B shows the same distribution pattern of H3K27ac signal in As4.1 cells. The peak locations relative to genomic annotations in both samples indicate that H3K27ac is equally distributed between introns and intergenic regions in native recruited renin cells as well as in As4.1 cells (Supplemental Figure 2C). The pattern of H3K27ac enrichment along all chromosomes is remarkably similar in both cell types, including the notoriously sparse signals in the sexual pair of chromosomes (Supplemental Figure 2D). Given that As4.1 cells continuously transcribe the renin gene, we investigated 2 other marks strongly associated with active enhancers and super-enhancers. We performed ChIP-Seq for the mediator subunit 1 (Med1) of the mediator complex, a coactivator mark associated with the preinitiation complex and which plays a crucial role in gene transcription (18). Using the MACS tool, we found 10,633 enhancers marked by MED1 deposition along the genome, with a strong enrichment –1.5 kb upstream as well as downstream of the TSSs (Supplemental Figure 2E). In addition, Chip-Seq for RNA Pol II was performed to identify genomic areas undergoing transcription in As4.1 cells. Using spatial clustering for identification of ChIP-enriched regions (SICERs) (19), we identified 40,897 peaks for Pol II (Supplemental Figure 2F). The most enriched binding for Pol II occurred at proximity to the TSS, indicative of the formation of the preinitiation complex or a paused state. The distribution of Pol II found along gene bodies is suggestive of genes undergoing active transcription, while the occupancy of Pol II at the 3′ end of genes indicates relative pausing as transcription is terminated.
Overall, in addition to the similarities identified in the renin cell types, the results also indicate that each cell type has its own exclusive set of open chromatin regions reflecting their particular physiological state.
Renin cells possess a unique group of super-enhancers that differentiate them from other cell types.
Unlike typical enhancers, which are characterized by sharp and narrow (2–5 kb) peaks of H3K27ac, super-enhancers contain larger domains of H3K27ac (20, 21) and are densely loaded with the mediator complex and transcription and chromatin regulators. Super-enhancers are associated with genes that function as master regulators of lineage-controlling transcription factors and oncogenes (20–25).
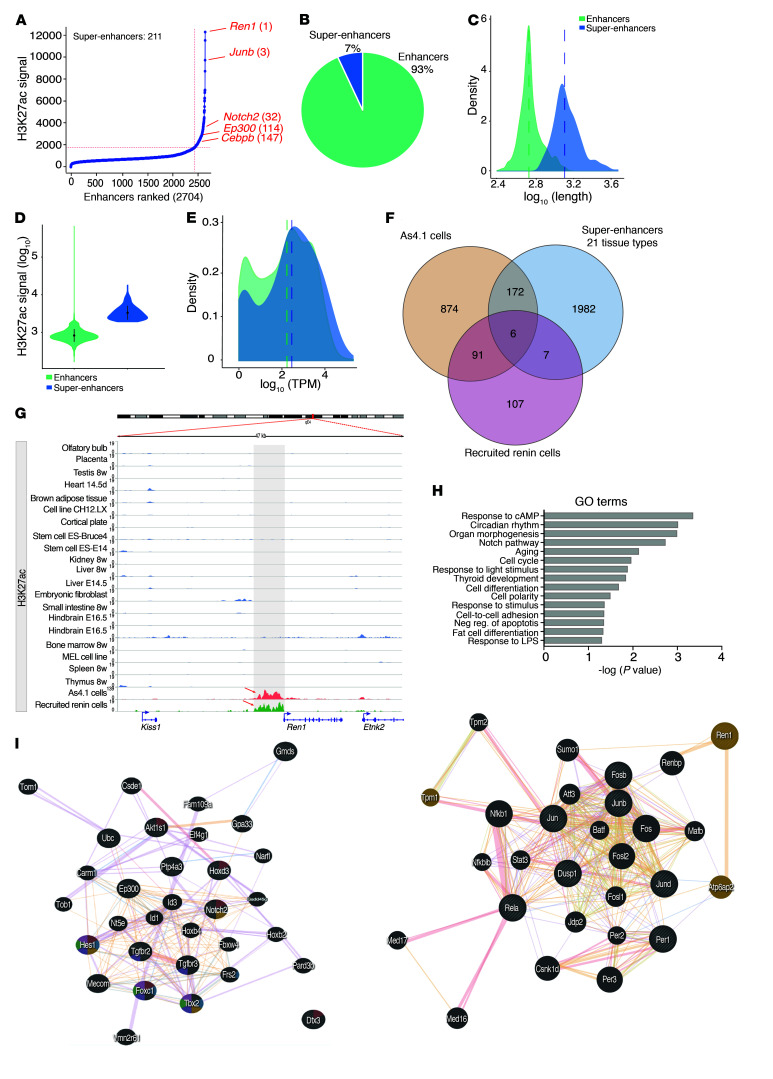
To determine whether super-enhancers occur in renin cells, we used the ranking ordering of super enhancer (ROSE) software (21, 22). We identified 211 super-enhancers out of 2,704 total enhancers, based on the signal strength for H3K27ac in recruited native renin cells. Figure 3A shows a hockey plot depicting the super-enhancers found in those cells. Overall, in recruited native renin cells, 7% of the total H3K27ac peaks corresponded to super-enhancers (Figure 3B). The strong enrichment of H3K27ac signals found at super-enhancers was characterized by their occupancy of broader areas in the genome in comparison with typical enhancers (the median length in logarithmic scale was 3.150 bp as compared with 2.771 bp for typical enhancers; Figure 3C). While typical enhancers covered regions shorter than 1 kb, super-enhancers are gathered into clusters over 12.5 kb, the optimal distance found for stitching together the closely spaced enriched regions with very high signals (20). As expected, super-enhancers displayed higher signal intensity than typical enhancers (median intensity in log scale 3.482 and 2.904 for super-enhancers and typical enhancers, respectively; Figure 3D).
Figure 3. The renin cells possess a unique set of super-enhancers that differentiate them from other cell types.
(A) There were 211 super-enhancers identified in native recruited cells, with the renin gene exhibiting the highest enrichment of H3K27ac signal, according to the ROSE software. (B) Pie chart illustrating the percentage of super-enhancers identified by H3K27ac ChIP-Seq. (C and D) Super-enhancer regions are larger in length and had more cumulative enrichment of H3K27ac signal than typical enhancers. (E) Super-enhancer–associated genes (blue) have higher expression levels than enhancer-associated genes (green). The histogram shows the density (number of counts in a bin, y axis) of genes as a function of the expression range in a logarithm scale (x axis). The dashed lines show the mean for each group. (F) Ninety-one super-enhancers, including the renin super-enhancer, are shared by the renin cells and absent in the other 21 tissue types. (G) Genome browser image showing deposition of H3K27ac signal upstream of the coding region of the renin gene only in renin cells. (H) GO terms significantly enriched using the genes associated with the 91 super-enhancers in renin cells. The GO terms exhibited a fold enrichment greater than 2 and a P < 0.05. (I) Left. Genes involved in Notch signaling (striped circles) and predicted associations returned by GeneMANIA. Right. Gene network illustrating predicted associations between genes involved in the cAMP signaling pathway as determined by GeneMANIA. Highlighted are genes related to the regulation of systemic blood pressure. Each gene is represented by a node (circles). The node size is proportional to the degree of connectivity and the node colors represent the functions in which genes are involved. Lines represent the relationship between genes, and the line width is proportional to the confidence of the connection. Wilcoxon signed rank sum test was performed for C, D, and E, P < 0.0001.
To investigate whether genes associated with super-enhancers had higher expression levels than those possessing typical enhancers, we used GeneMargin, utilizing arbitrary margins of 10 kb upstream of the transcription start site to 10 kb downstream from the 3′UTR. We assigned 563 genes to 211 super-enhancers (Supplemental Table 3). Figure 3E illustrates the distribution of genes with and without super-enhancers as a function of their level of expression. Whereas a large group of genes was associated with traditional enhancers, a greater density of highly expressed genes possessed super-enhancers (Figure 3E, right; Kruskal-Wallis, P = 0.0007447; Wilcoxon’s rank sum test, P = 9.383 × 10–5), resulting in a shift of the curve to the right. These results suggest that gene transcription is increased in genes driven by super-enhancers.
Our analysis of enhancers in As4.1 cells revealed 14,871 and 8,165 enhancers identified using the H3K27ac and Med1 marks, respectively. Of these, 888 and 477 super-enhancers were associated with either H3K27ac or Med1 enrichment, respectively (Supplemental Figure 3A), representing 6% of the peaks identified (Supplemental Figure 3B). Similarly, and as shown in native renin cells, super-enhancers in As4.1 cells were larger than typical enhancers (Supplemental Figure 3C), and they displayed more intense signals for H3K27ac and MED1 than traditional enhancers (Supplemental Figure 3D). Further, genes associated with super-enhancers had higher levels of expression than those associated with traditional enhancers (Supplemental Figure 3E; P < 2.2 × 10–16). Interestingly, genes associated with super-enhancers containing both H3K27ac and MED1 peaks had higher levels of expression than those containing one (Supplemental Figure 3E; P < 0.005633). However, a signal for H3K27ac had a higher impact on gene expression levels than a Med1 signal (P < 2.2 × 10–16) (Supplemental Figure 3E).
To investigate whether the super-enhancers found in renin cells were exclusive to them, we compared H3K27ac ChIP-Seq data from native recruited cells and As4.1 cells with H3K27ac ChIP-Seq data from 21 different tissues available in the Super-Enhancer Archive database (SEA) (26). The Venn diagram in Figure 3F shows that the recruited native renin cells possess an exclusive set of 107 super-enhancers that were not present in either the 21 tissue types from the SEA or the As4.1 cells. Equally importantly, a group of 91 super-enhancers, including the renin super-enhancer, was common to native renin cells and As4.1 cells, but was absent in available databases (Supplemental Table 4). Only 6 super-enhancers were shared among the native renin cells, As4.1 cells, and the super-enhancer database (Figure 3F). Figure 3G indicates that the enrichment of H3K27ac at the renin locus is unique to native renin cells and As4.1 cells, and it is not found in any other cell types from the ENCODE database (27). Given their specificity to renin-expressing cells, it is likely that this core group of super-enhancers regulates renin cell identity (Supplemental Table 4). The heatmap in Supplemental Figure 4A shows the expression pattern of the super-enhancer–associated genes in renin cells with the renin gene exhibiting the highest level of expression. The distribution of these super-enhancer–associated genes along the mouse chromosomes is not homogenous. Several chromosomes possess clustered super-enhancer genes (Chr: 11, 15, 17) whereas others (Chr: 5, 6, 13, 14, 18, X, Y) do not have any (Supplemental Figure 4B). Taken together, these results indicate that native renin cells can be recognized and discriminated from other cell types by their super-enhancer repertoire and their gene expression profile. This finding led us to investigate promising super-enhancer–associated candidate genes that are shared between native and As4.1 renin-producing cells. In order to identify the gene ontology (GO) terms significantly overrepresented in the given set of super-enhancer–associated genes, and to investigate which pathways could be enriched in recruited renin cells and As4.1, we utilized the KEGG (Kyoto Encyclopedia of Genes and Genomes) database and the DAVID annotation tool (28) for genes that are expressed in either one or both cell types. GO terms and KEGG pathway analysis taking into account only the 91 super-enhancers common to As4.1 and native recruited renin cells indicated the importance of the Notch and cAMP pathways and their associated genes in renin cell identity (Supplemental Table 5). Figure 3H shows the GO terms with fold enrichment greater than 2 when compared with the mouse genomic background (Benjamini P < 0.05). Interestingly, the Notch pathway was one of the most prominently enriched, suggesting that this pathway is important to maintain the identity of renin-expressing cells. In fact, genomic deletion or pharmacological inhibition of the Notch pathway in renin cells results in depletion of renin-positive cells (14). Figure 3I shows a predicted gene interaction, generated using GeneMANIA software (29), that includes genomic, proteomic, and molecular network interactions from different online sources. Genes included under Notch signaling that also possess associated super-enhancers include Ncor (nuclear receptor corepressor 2), Dtx3 (deltex 3, E3 ubiquitin ligase), Dvl3 (disheveled segment polarity protein 3), Notch2 (notch 2), Ep300 (E1A binding protein p300), and Hes1 (hairy and enhancer of split 1). Supplemental Table 6 lists all genes and their functions as returned by GeneMANIA. In addition, components of the cyclic AMP signaling pathway were also significantly enriched against the mouse genomic background. These results agree with our previous demonstration that the cAMP pathway is crucial for the development of renin cells, expression of the renin gene, and RENIN release (9). Genes annotated with this GO term include Jun (jun proto-oncogene), Per1 (period circadian clock 1), Fosb (FBJ osteosarcoma oncogene B), Dusp1 (dual specificity phosphatase 1), Jund (jun D proto-oncogene), Ren1 (renin 1), Rela (v-rel reticuloendotheliosis viral oncogene homolog A [avian]), and Junb (jun B proto-oncogene). These findings support previous work demonstrating the importance of these 2 signaling pathways in the maintenance of the renin cell phenotype (9, 14, 30).
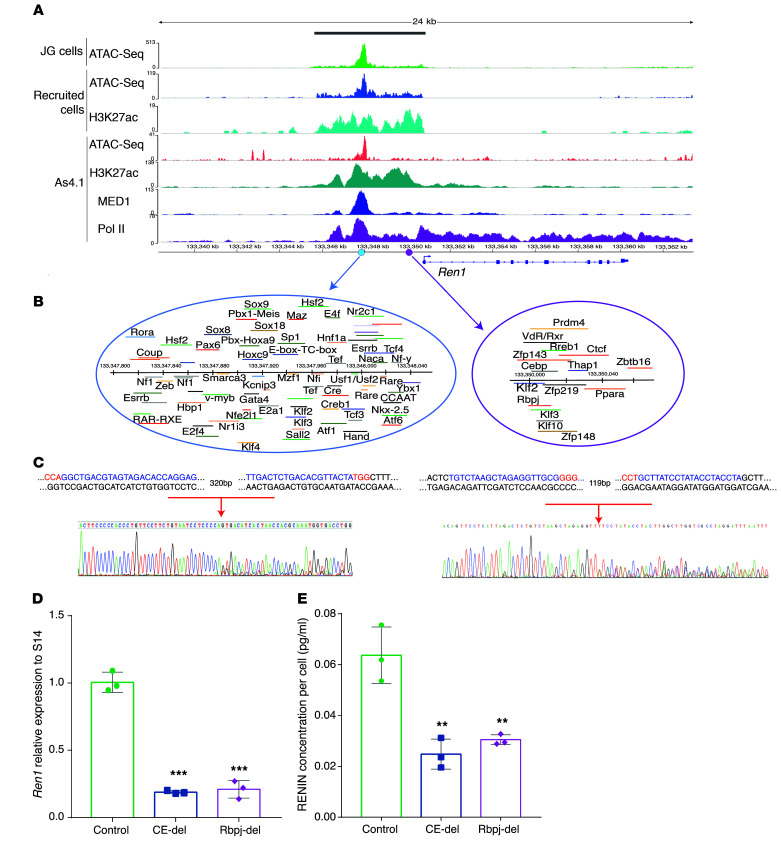
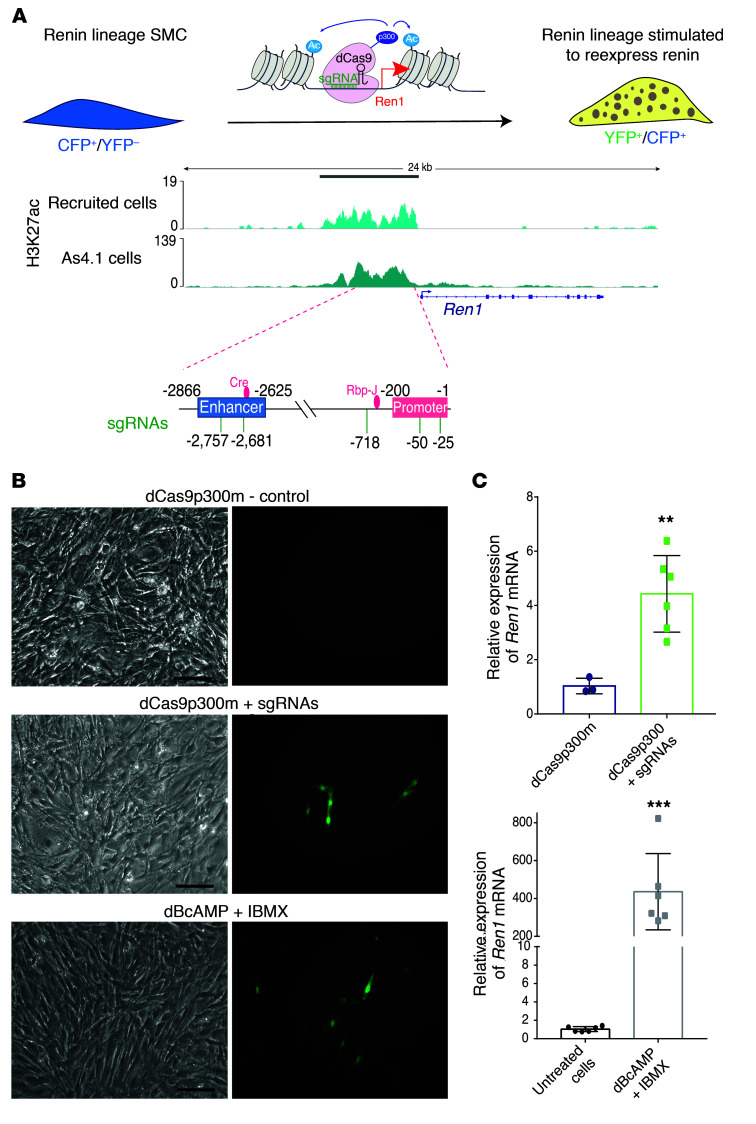
Figure 4. A super-enhancer at the renin locus contributes to renin cell identity.
(A) The renin super-enhancer is indicated by the grey bar at the top of the figure. The y axis shows the scales used for the signal enrichment for each assay. The x axis shows the coordinates across the genome. The ATAC-Seq profile shows overlap of the signals among the JG cells (green), recruited cells (blue), and As4.1 cells (pink) upstream of the coding region of the renin gene. Recruited cells and As4.1 cells share a similar distribution of H3K27ac signals. Also shown are the ChIP-Seq signals corresponding to MED1 (dark blue) and Pol II (purple). Pol II shows a strong signal all along the renin gene, suggesting that the gene is actively transcribed. All the ChIP-Seq signals overlapped with the accessible chromatin shown for the JG cells and recruited native renin cells. (B) The classic enhancer (CE, 241 bp) is located between –2866 to –2625 (light blue circle). It contains a bona fide CRE element where Creb binds. The purple circle marks the Rbpj-Enh. (C) Targeted deletion of the CE and the Rbpj-Enh using the CRISPR-Cas9 system. Shown are the genomic sequences of the targeted areas with the PAM sequences indicated in red and the Sanger sequencing chromatograms confirming the deletions. (D) Renin mRNA expression levels were significantly decreased in the targeted populations of cells in comparison with control samples for both the CE and the Rbpj-Enh. (E) Measurement of renin protein in the supernatants of targeted populations after 48 hours of transfection shows a significant decrease in secreted renin levels. These data indicate that these regions within the renin SE are involved in the regulation of renin synthesis and release. Data are represented as mean ± SD (n = 3 in each group). **P < 0.01, ***P < 0.001, by unpaired, 2-sided Student’s t test.
The renin locus possesses a super-enhancer that contributes to renin cell identity.
Expression of renin is the most important distinctive feature of the renin cell. Thus, it was important to define the chromatin status at the renin locus. We found that the renin gene is associated with a super-enhancer (Chr1: 133345631–133350823) ranking at the first position for H3K27ac enrichment in native renin cells as shown in the hockey plot of Figure 3A. Figure 4A provides an overview of the renin super-enhancer identified in recruited native renin cells and As4.1 cells. H3K27ac ChIP-Seq and ATAC-Seq results revealed that in the recruited native renin cells there is an enrichment of the active enhancer mark and that the chromatin is accessible upstream of the coding region of the renin gene. These results confirm that under conditions of severe and unremitting physiological challenge such as the inability to produce renin, resulting in hypotension, dehydration, lack of angiotensin, and inhibition of renin synthesis, the recruited cells are constantly attempting to produce RENIN and reenact the renin phenotype. Thus, the conserved regulatory regions upstream and downstream of the gene respond appropriately to the physiological challenge and maintain the memory of the renin phenotype via the establishment of the renin super-enhancer. Supporting this idea, Figure 4A shows binding of MED1, a coactivator usually found in super-enhancer regions required to bind Pol II and initiate transcription. MED1 enrichment is at the same location as the H3K27ac signal in As4.1 cells. ROSE analysis of the MED1 signal also indicated that this region of the renin locus can be classified as a super-enhancer in strict correspondence with the H3K27ac findings (Figure 4 and Supplemental Figure 3A, right). Also, ChIP-Seq for Pol II revealed a strong signal upstream and all along the renin gene, suggesting active renin transcription in As4.1 cells. Therefore, in all the renin cells studied, the epigenetic marks show a similar spatial distribution upstream of the renin gene (Figure 4A). It should be noted that this super-enhancer region contains the classical renin enhancer known to induce high levels of transcription (11) and the Rbpj enhancer (Rbpj-Enh) described below.
Identification of transcription factor binding motifs at the renin super-enhancer in native renin cells.
Because super-enhancers are characterized by high enrichment of transcription factor binding, we sought to identify transcription factors that could potentially bind to the renin super-enhancer and possibly affect renin transcription. Using MatInspector tool (31) from Genomatix software v3.9, we identified 1,448 predicted binding sites within a region comprising –5000 bp to the ATG site of the renin gene. A complete list of transcription factors and their predicted binding sites is shown in Supplemental Table 7. To narrow down our search for transcription factors that are most likely to bind the renin super-enhancer, we focused on those that were expressed in native renin cells as determined by RNA-Seq. Accordingly, we found 171 transcription factors (Supplemental Table 8) that are expressed in either JG cells or recruited renin cells and that putatively bind to the renin super-enhancer. From those, 125 transcription factors, expressed at a level higher than 1 TPM in JG cells, could be involved in the regulation of renin expression. To continue further filtering the number of transcription factors potentially relevant for the renin phenotype, we searched for those that contained enhancer or super-enhancers associated to them using H3K27ac as the relevant discriminating signal and the GeneMargin settings as described above. We found 12 transcription factors that possessed super-enhancers in recruited native renin cells (Supplemental Table 9). The top expressed transcription factor containing a super-enhancer at its loci was kruppel-like factor 2 (lung) (Kfl2), a member of a zinc finger family, which is involved in cell differentiation, angiogenesis, erythropoiesis, and immune regulation (32, 33). Moreover, Klf2 has significant antiangiogenic effects preventing vascular diseases (34), and is essential for the correct development of mouse embryonic vasculature (35), including maintenance of endothelial integrity in adult mice (36). Notably, 2 members of the HES family, which are Notch target genes, are included in the list of transcription factors: hairy and enhancer of split 1 and 7 (Drosophila) (Hes1 and Hes7). Hes1 regulates the timing of biological events in many cell types and Hes7 functions as a segmentation clock in mouse presomitic mesoderm, its wave-like expressions leading to the generation of a pair of somites (37). It remains to be determined what roles Hes1 and Hes7 have in native renin cells and whether they affect other genes related to renin expression.
Another gene highly expressed in renin cells and associated to a super-enhancer is Junb, a subunit of the activated protein-1 (AP-1) family that plays a crucial role in angiogenesis activating Vegfa (38) and facilitating cell proliferation. Mice lacking Junb die at early embryonic stages due to severe vasculogenic defects (39). Given the fundamental role of renin cells in vascular development, it would be important to define whether Junb is involved in the angiogenic properties of renin cells in early development.
The thyroid hormone receptor alpha (Thra) is the primary thyroid receptor expressed in endothelial and SMCs. These hormone receptors have a significant function in regulating vascular tone, but also enhance renal sodium reabsorption, stimulate secretion of erythropoietin, and increase blood volume, affecting renin release (40). The effects of thyroid hormone receptors in renin cells needs to be investigated.
The CCAAT enhancer–binding protein beta (Cebpb) is a member of a family of transcription factors containing a leucine zipper motif required for the interaction with regulatory domains of enhancers and promoters of target genes (41). Cebpb has critical roles in many tissues, affecting adipocyte differentiation and inflammatory responses (42), both characteristics found in our Ren1c–/– Ren1c-YFP mice. Although Cebpb regulation and function in vascular cells is still unknown, mice lacking Cebpb show glomerular enlargement and an increase in mesangial cells. With age, these mice develop splenomegaly and lymphadenopathy (43). Together, the data suggest that Cebpb could be part of the machinery involved in renin transcription.
Hoxa9 and Hoxa10 genes are located at chromosome 6 in the mouse genome adjacent to each other. These genes play a pivotal role in body patterning and tissue identity during development; both genes are involved in hematopoietic development. Hoxa9 plays a central role in myeloid leukemia where it has been shown to bind MEIS1 and PBX1 proteins (44). High levels of Hoxa9 and Meis1 results in increased p300/CBP recruitment at enhancer regions to upregulate leukemic-associated target genes (45).
We next sought to map transcription factor domains within the renin super-enhancer. Figure 4A shows the chromatin landscape at the renin locus and 2 major transcription factor binding (TFB) clusters (Figure 4B) that could be functionally relevant for the operation of the renin super-enhancer. These clusters include (a) the classic renin enhancer (CE; 242 bp, –2866 to –2625), which contains a cAMP-responsive element to which CREB binds, and 72 other putative TFB sites, and (b) the Rbpj-Enh (119 bp, –728 to –609), characterized by the binding of Rbpj and other major transcriptional regulators such as Vdr, Ctcf, Zfp143, Cebpb, and Klf2.
Major domains within the renin super-enhancer control the renin phenotype.
To define whether the Renin super-enhancer functionally controls the renin phenotype, we used the CRISPR/Cas9 system (46) to delete the CE and the Rbpj-Enh in cultured As4.1 cells. We constructed multiplex vectors designed to simultaneously express the Streptococcus pyogenes Cas9 protein (Cas9), 2 single-guide RNAs (sgRNAs), and a green fluorescent protein (eGFP), allowing the isolation of positively transfected cells (Supplemental Figure 5). The targeting guides to the CE and the Rbpj-Enh are shown in Supplemental Table 10. Deletion of the 2 regions within the larger renin super-enhancer (Figure 4C) had a significant influence on renin expression. Renin mRNA expression levels were significantly diminished in the 2 GFP+ populations of transfected cells when compared with cells transfected with a control plasmid lacking sgRNAs (CE-del 0.19 ± 0.03, Rbpj-del 0.21 ± 0.07 vs. control 1.01 ± 0.11; P < 0.001) (Figure 4D). Moreover, ELISA measurements conducted in culture supernatants of As4.1 cells showed a significant decrease in RENIN released compared with the control cell population (Figure 4E). These data indicate that these regions within the renin super-enhancer are functionally involved in the regulation of RENIN synthesis and release.
The molecular memory of the renin phenotype resides within the renin super-enhancer.
We have previously shown that renin cells are progenitors for arteriolar SMCs, mesangial cells, and interstitial pericytes, and that renin cell descendants retain the plasticity to regain the renin phenotype upon threats to survival (5).
We hypothesized that the molecular memory of the renin phenotype resides within the renin super-enhancer and could be elicited by H3K27 acetylation mediated by the histone acetyltransferase p300. To test this hypothesis, we used our immortalized CFP/YFP cell line derived from the kidney arterioles of mice generated by breeding Ren1d-cre mice with ROSAloxp (CFP) reporter mice also harboring the Ren1c-YFP transgene (9). These immortalized arteriolar SMCs express CFP constitutively, indicating that they previously expressed renin and are therefore derived from renin cell precursors. A well-known characteristic of renin cells is that when placed in culture, they stop expressing renin within 48 hours. Similarly, our arteriolar CFP/YFP cells do not express renin (or YFP) until they are stimulated to do so. When the cells are treated with cAMP analogs the renin gene is activated, the cells become YFP+, and they synthesize renin mRNA. Thus, the cells are able to switch the renin gene on and off as they do in vivo in response to a physiological challenge. We therefore targeted CFP/YFP cells with p300 to specific loci within the renin super-enhancer using a CRISPR/Cas9-based system consisting of the nuclease-null dCas9 protein fused to the catalytic core of the acetyltransferase p300 (dCas9p300) and sgRNAs directed to the renin super-enhancer (Figure 5A, and Supplemental Table 10). Upon treatment with dCas9p300+sgRNAs or with di-Butyryl cAMP, a subset of cells became YFP+ (Figure 5B) and synthesized renin mRNA (Figure 5C, 4.37-fold and 477.36-fold, respectively). These results indicate that the switch for expression of the renin gene and the memory of the renin phenotype resides within the renin super-enhancer and is at least in part governed by the activity of p300 and H3K27 acetylation at this locus of the renin gene.
Figure 5. The molecular memory of the renin phenotype resides within the renin super-enhancer.
(A) In vitro model to study the molecular memory of the renin cell phenotype. The immortalized SMCCFP/YFP cell line derived from the mouse kidney constitutively expresses the renin lineage marker CFP and can be induced to express renin (and YFP) upon stimulation. The nuclease-null dCas9 protein is fused to the catalytic core of p300 and catalyzes the acetylation of H3K27. The renin locus showing the renin super-enhancer region in renin cells (indicated by the grey bar on top) previously characterized H3K27ac signal in renin cells. The schematic shows the locations of the sgRNAs used to target the SE region of renin. (B) Bright field images (left panels) of corresponding fluorescent images (right panels). Top, no YFP+ cells were observed in the controls where cells were transfected with a plasmid containing dCas9 bearing mutated p300. Middle, SMCCFP/YFP expressing YFP 72 hours after transfection with a plasmid containing the sequence for the dCas9p300 core and 5 sgRNAs targeted to the renin super-enhancer. Bottom, YFP expression in SMCCFP/YFP treated with 1 mM dB-cAMP and 0.1 mM IBMX for 72 hours to stimulate renin expression. Scale bars: 10 μm. (C) Relative mRNA expression of Ren1, determined by qRT-PCR. The levels of renin expression increased 4.37 to 477 times in response to dCas9p300 + sgRNAs (n = 6) or to exogenous cAMP (n = 6), respectively, versus control samples (dCas9p300-mutated and untreated cells, n = 3 and n = 6, respectively). Data are mean ± SD. **P < 0.01; ***P < 0. 001, by unpaired, 2-sided Student’s t test.
Med1, a component of the renin super-enhancer, is essential for renin expression and circulating RENIN in vivo.
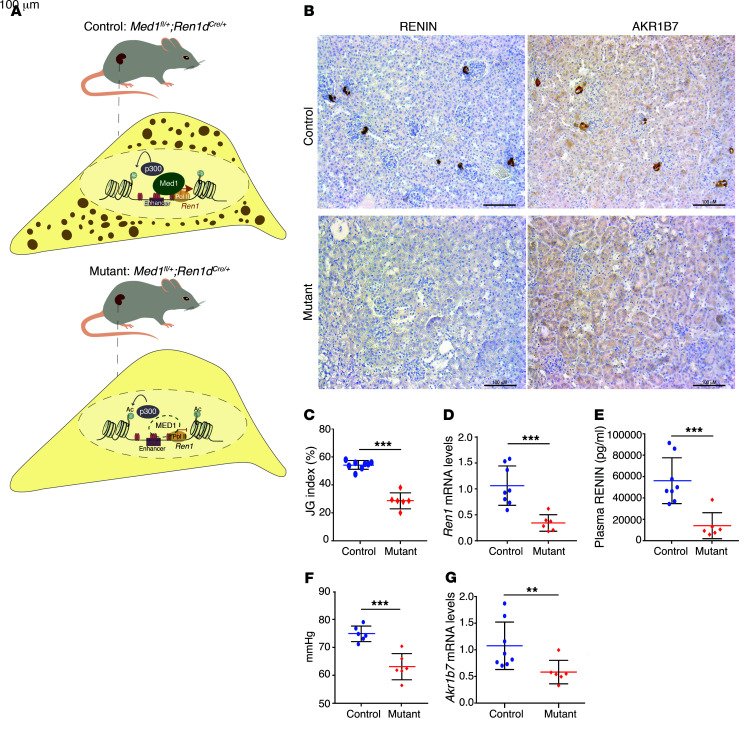
MED1 integrates a multiprotein complex that interacts with RNA Pol II to initiate gene transcription (47, 48). MED1 is involved in determining chromatin architecture, and it has been proposed as a characteristic feature of super-enhancers (21). Because renin-synthesizing cells possess a super-enhancer characterized by MED1 enrichment, we sought to examine the role of MED1 in renin-expressing cells in vivo. We generated mice with conditional deletion of Med1 in renin cells (see Methods). Figure 6A shows the study mice and a scheme of their renin cells. Kidneys from adult mutant mice were practically devoid of renin-positive cells as shown by RENIN immunostaining (Figure 6B, bottom left). Because we have previously identified that aldo-keto reductase family 1, subfamily B member 7 (Akr1b7) is an independent marker of cells programmed for the renin phenotype (49), we performed immunostaining for AKR1B7 to investigate whether deletion of Med1 is also affecting Akr1b7 expression. Figure 6B shows a markedly decreased signal for AKR1B7 in renin cells of Med1 mutant mice when compared with controls, indicating that Med1 affects AKR1B7 protein in a similar manner as it affects renin. Figure 6C shows a reduced number of renin-positive JG areas in Med1 mutant mice when compared with control mice (28.75% ± 5.71 vs. 54.11% ± 3.15; P < 0.0001). Relative renin mRNA levels measured by q-PCR from kidney cortices (Figure 6D) were also decreased in Med1 mutant mice when compared with the control group (0.389 ± 0.173 vs. 1.050 ± 0.357; P < 0.001). As a consequence, plasma RENIN concentration was reduced in Med1 mutant mice (14,437.42 ± 11,948.89 pg/ml vs. 56,070.33 ± 21,093.8 pg/ml; P < 0.001; Figure 6E). Consistent with those results, Med1 mutant mice exhibited a significantly lower mean arterial pressure than control mice (63.98 ± 1.975 mmHg vs. 75.32 ± 1.048 mmHg; P < 0.001; Figure 6F). Relative Akr1b7 mRNA levels were also decreased when compared with controls (0.708 ± 0.260 vs. 1.050 ± 0.401; P < 0.01; Figure 6G). Further characterization of this new mouse model showed no macroscopic or microscopic differences between the kidneys of mutant and control mice (Supplemental Figure 6A). Similarly, immunostaining for myosin heavy chain and α-smooth muscle actin (α-SMA) did not reveal differences between mutant and control animals (Supplemental Figure 6B, top, middle and low panel, respectively), indicating that lack of Med1 does not affect the overall development or morphology of the kidney. Med1 mutant mice did not show any differences in body weight, kidney weight, kidney/body weight ratio, liver or spleen weight (Supplemental Figure 6, C–G) when compared with control animals. Plasma sodium and chloride concentrations were not different between groups (Supplemental Figure 6, H and I); however, a slightly elevated blood urea nitrogen was found in mutant mice when compared with control mice (Supplemental Figure 6J). As expected, Med1 mRNA levels were decreased in mutant mice compared with the control group (0.362 ± 0.145 vs. 1.077 ± 0.478; P < 0.001; Supplemental Figure 6K). Overall, the results indicate that Med1 plays an important role in the maintenance of the renin phenotype, which is physiologically reflected in the availability of circulating RENIN.
Figure 6. The coactivator MED1 is essential for renin expression in vivo.
(A) Control mouse where one allele of Med1 gene is floxed and the other is WT. The cell below indicates that in the presence of Med1 the renin gene is transcribed and there is granule formation and renin storage. Mutated mouse where one allele of Med1 is floxed and the other one is deleted; the Cre gene driven by the renin promoter deletes the floxed allele, generating a Med1 homozygous deletion in renin lineage cells. In the cell corresponding to the mutant mouse, absence of MED1 (green dashed line) indicates that there is no renin transcription and no granule formation in the renin cells. (B) Immunostaining for RENIN and AKR1B7 shows a decrease in signal for both proteins in JG cells of mutant mice (n = 6) compared with control mice (signal is indicated by the brown color; n = 8). (C) The JG index, measured as the percentage of RENIN-positive areas over the total number of glomeruli, indicates that mutant mice (n = 6) have fewer renin-positive JG areas when compared with control mice (n = 8). (D) Relative expression of renin mRNA levels measured by qPCR. Mutant mice (n = 6) have significantly lower expression than the controls (n = 8). (E) Plasma RENIN levels are lower in mutant mice (n = 6) than in controls (n = 8). (F) Med1 mutant mice have lower blood pressures than control mice (n = 6 per group). (G) Akr1b7 mRNA levels are decreased in mutant mice (n = 6). Data are mean ± SD. **P < 0.01; ***P < 0. 001, by unpaired, 2-sided Student’s t test.
Discussion
In this study, we have acquired and comparatively analyzed the transcriptomes and epigenomes of several renin cells at different states of stimulation to understand their chromatin profiles. Our results show that renin cells possess a unique set of super-enhancers and traditional enhancers that define their identity. Thus, all cells programmed for the renin phenotype can be easily discriminated from other cell types not only by the expression of renin or their overall gene expression profile but also by their epigenetic landscape. In fact, renin cells share a core set of 91 super-enhancers that are only found in cells programmed for the renin phenotype. These findings led us to investigate promising super-enhancer–associated candidate genes that are shared between native and As4.1 renin-producing cells. GO terms and KEGG pathway analysis, taking into account only those 91 super-enhancers common to these cell types, indicated that 2 pathways, the Notch and cAMP pathways, and their associated genes were likely to be crucial in determining renin cell identity (Figure 3H). In agreement with these findings, we have shown that the Notch pathway controls the myo-epithelioid phenotype of renin-expressing cells. The deletion of Rbpj in renin lineage cells of mice results in a remarkable switch in the fate of renin-expressing cells: they stop synthesizing renin, Akr1b7, and miR330, they lose their renin-containing granules, and they stop making contractile proteins (14, 15). Strikingly, they express transcripts characteristic of hematopoietic or immune cells (15). These findings indicate that in mice, the Notch pathway via Rbpj controls the dual endocrine-contractile phenotype of the renin cell while suppressing the expression of genes from unwanted ectopic lineages. Also, SMCs along the renal arterioles lose the plasticity to regain the renin phenotype when confronted with an external threat to homeostasis such as salt restriction and pharmacologically induced hypotension with captopril (15). The loss of plasticity generally maintained by the Notch pathway has devastating consequences for the health of the animal who is now unable to maintain homeostasis when confronted with challenges that renin cells are typically designed to overcome (14). Interestingly, the actions of the Notch pathway in controlling renin cell identity are highly conserved. Using zebrafish harboring transgenes that reported the activity of the renin promoter, Rider and colleagues demonstrated that mind bomb, a ubiquitin ligase essential for activation of Notch signaling, is crucial for the development of renin expression (50). In complete agreement with the current, unbiased analysis, these results indicate that the Notch pathway is essential in the determination of the renin phenotype.
As mentioned above, it has been suggested that the cAMP pathway is a convergence point for a variety of stimuli that control RENIN synthesis and release (9), and several studies have hypothesized that the cAMP pathway may be involved in the differentiation and maintenance of renin-expressing cells. Conditional deletion of membrane-bound beta-adrenergic receptors in renin cells results in a significant decrease in renin release (51). Further, in vivo deletion of Gsa (a G protein coupled to the beta-adrenergic receptor) in renin cell precursors results in a massive decrease in the number of renin-expressing cells accompanied by defects in the branching of terminal arterioles in the kidneys of neonatal mice (52). More importantly, conditional homozygous deletion of both histone acetyltransferases (CBP and p300, which acetylate lysine 27 of histone H3, H3K27ac) leads to significant structural alterations in the kidney including lack of renin cells, vascular abnormalities, and focal fibrosis (53). It is likely that deletion of both histone acetyltransferases leads to a dissipation of the renin super-enhancer and a change in renin cell fate. This hypothesis, however, remains to be tested. Overall, those studies show that the cAMP pathway is crucially involved in the control of the renin phenotype. Furthermore, the studies suggested that a major control point in the determination of the renin phenotype is exerted by H3K27ac, a modification characteristic of active enhancers and super-enhancers.
To define whether local deposition of H3K27ac via p300 results in activation of the renin gene and acquisition of renin phenotype, we performed additional experiments in CFP/YFP SMCs. These cells are arteriolar SMCs that descended from renin precursors; their lineage relationship with renin progenitors is marked by their inerasable constitutive expression of CFP. Equally important, these SMCs also express YFP under the control of the renin super-enhancer, which is silent until it is activated by an external stimulus (9). In this study, exposure of these cells to exogenous cAMP results in a robust reexpression of Renin. The cells become YFP+ and increase renin mRNA almost 500-fold, indicating active transcription of the renin gene. Targeting of dCas9p300 to the renin super-enhancer elicited a qualitatively similar response, indicating that acetylation of H3K27 is essential for assembling/activation of the renin super-enhancer and control of the renin phenotype. Overall, these results indicate that the molecular memory of the renin phenotype is a cell-autonomous, intrinsic property of the renin cell descendants. This memory resides primarily at the renin super-enhancer and can be elicited by external stimuli such as cAMP and by specific targeting of the acetyltransferase p300 to the renin super-enhancer.
The present study also allowed us to compare cells programmed for the renin phenotype at different physiological states: the normal unstressed JG cells, the chronically recruited renin-null cells, the acutely recruited cells from mice subjected to sodium depletion plus captopril, and the constitutively active As4.1 cells. ATAC-Seq revealed that all those cells share a notably reproducible pattern of open chromatin along the genome. It should be noted, however, that whereas those cells share many features that define them as renin cells, such as areas of open chromatin, super-enhancers, and core gene expression profiles, they also display recognizable differences (Figure 2, A–E), likely a reflection of their varying biological/physiological states within the spectrum of the renin phenotype. Equally important, those states can be traced for each cell type and can be identified and discriminated from one another by their chromatin state.
In addition to the repertoire of 91 super-enhancers, we found that all cells driven to express Renin possess a unique super-enhancer at the renin locus. Not only is the renin super-enhancer unique to renin-expressing cells not found in other cell types, but the fact that it scored at the highest level (number 1 in the ROSE analysis) suggested that this super-enhancer controls the expression of renin, the defining feature of the renin cell. To test the functional relevance of the super-enhancer in the determination of this cell’s identity, we performed a series of regulatory DNA island deletions. Our deletion studies indicate that the renin super-enhancer (at least through 2 major islands, the classical renin enhancer, and the Rbp-Enh) controls the expression of the renin gene. Interestingly, the classical enhancer contains a binding site for CREB and its coactivators CBP/p300, constituting the site of convergence for the cAMP pathway. Deletion of members of this pathway results in the disappearance of renin cells, decreased circulating renin, and low blood pressure (53). Furthermore, smooth muscle cell descendants from renin cell progenitors are unable to reexpress renin in response to sodium depletion and captopril, 2 manipulations leading to fluid volume depletion and hypotension, major challenges to homeostasis. Similar results were obtained when we deleted the Rbpj-Enh. Thus, our data indicate that the renin super-enhancer, through at least these 2 major regulatory islands, is needed to activate renin transcription in a diverse set of circumstances, including volume depletion, hypotension, and lack of angiotensin II generation. Finally, our results indicate that Med1 has an essential role in renin activation. MED1 has been reported as a bridging factor between promoters and enhancers, interacting preferentially with transcription activators localized at the enhancers and interacting transiently with Pol II during transcription initiation (18). In addition, it has been shown that reduction of Med1 levels affects the expression of super-enhancer–associated genes in embryonic stem cells (21) and loss of mediator at enhancer and promoter regions changes their gene expression program (54). Because Med1-null mice die at embryonic day 11.5 attributed to defects in the development of the placenta vasculature (55), we generated a conditional knockout mouse for Med1 in cells of the renin lineage to test its role in renin expression. Our results showed that Med1 is required for the maintenance of renin cells. The absence of Med1 in cells of the renin lineage resulted in decreased renin expression, reduced number of renin-positive cells and decreased circulating renin. Ultimately this results in decreased arterial blood pressure, emphasizing the relevance of Med1 function in renin cells for blood pressure control even in the basal state. Given that absence of Med1 also affects the expression of Akr1b7, a renin-independent marker of cells programmed for the renin phenotype, the fate of the cells that stopped making renin remains to be determined by carefully designed lineage tracing experiments (7). We anticipate that lack of Med1 in renin cells leads to a dissipation of the renin super-enhancer, making it impossible for the transcription machinery to activate the renin gene. This possibility remains to be investigated.
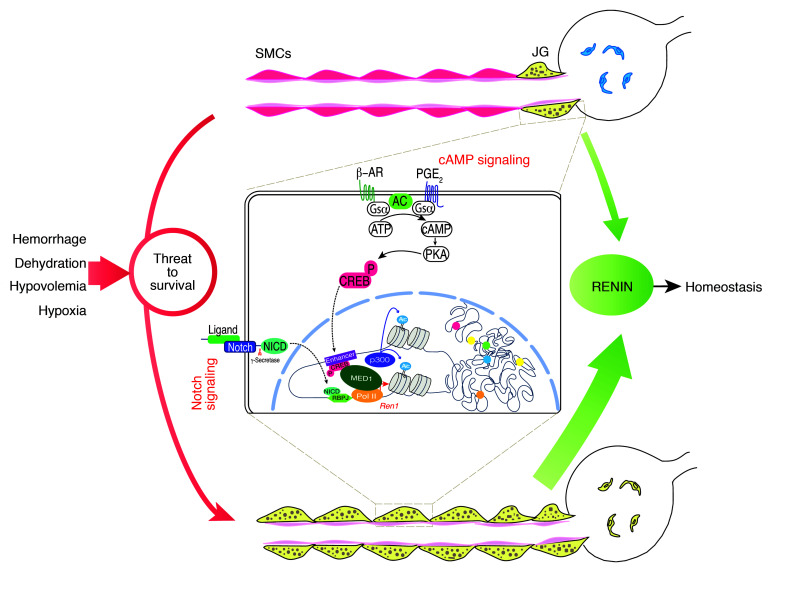
Based on the results of the present studies, we propose a model linking renin cell identity to the control of homeostasis (Figure 7). Briefly, renin-expressing cells possess a unique set of super-enhancers that determine renin cell identity. These super-enhancers are likely to operate together to determine the full phenotype of the renin cell. The most prominent super-enhancer, located at the renin locus, constitutes the point of convergence of extracellular signals that convey the physiologic status of the organism, such as volume and composition of the extracellular fluid, perfusion pressure, and sympathetic activation. In response to a threat to survival (dehydration, hemorrhage, sodium depletion, or hypoxia) this super-enhancer acts as a chromatin sensor of the external milieu and is responsible for the transformation of renin cell descendants to the renin phenotype. Because it can be activated in response to stimuli, the renin super-enhancer is at the core of the molecular memory of renin cell function, a fundamental mechanism to preserve homeostasis.
Figure 7. Super-enhancers acting as chromatin sensors of the extracellular milieu control the identity and memory of renin cells to maintain homeostasis.
See main text for details. The inset illustrates events that occur in renin cells in order to maintain or reenact the renin phenotype. The cAMP pathway is activated through the β-adrenergic receptor or the prostaglandin E2 receptor. Phosphorylated CREB enters the nucleus and binds the CRE element at the enhancer of the renin gene. As the Notch receptor is activated by cell-cell interaction, its intracellular domain (NICD) enters the nucleus and binds the transcription factor RBPJ. As these events occur, the chromatin is made accessible at the renin locus, which is characterized by the deposition of H3K27ac due to p300 histone acetyl transferase activity; MED1 establishes the bridge between the renin enhancer and Pol II. These key components act together to activate the transcriptional machinery. Moreover, other super-enhancers (color dots) may cooperate to fully establish the renin phenotype. β-AR, beta adrenergic receptor; PGE2, prostaglandin E2; AC, adenylate cyclase; Gsα, activating G protein–coupled subunit; PKA, protein kinase A; CREBP, phosphorylated cAMP responsive element binding protein.
Methods
Cell culture.
The renin-expressing As4.1 cell line (ATCC, CRL-2193) (10) was cultured to perform ChIP-Seq, RNA-Seq, and transient transfections for CRISPR-Cas9 experiments.
We used cultured arteriolar SMCs of the renin lineage that express CFP, a renin lineage marker, and YFP when cells are induced to produce renin (9) in CRISPR-dCas9 experiments.
Mice.
Ren1c-YFP and Ren1c–/– Ren1c-YFP mice were used to isolate native renin cells (8, 9). Med1fl/fl mice were obtained from Reddy and Jia’s lab at Northwestern University (56). We generated Med1Δ/fl Ren1dCre/+ mice after several crosses using Med1fl/fl (56) with a mouse expressing Sox2Cre and Ren1dCre/+ (5) to generate the conditional knockout mice. Mice were studied at 4–5 months. There were 12 mutants (6 females and 6 males) and 14 controls (4 females and 10 males). Of these, 6 mutants and 6 controls were used to measure blood pressure.
Neutral Red staining.
As4.1 cells were stained with the vital dye Neutral Red to visualize RENIN granules.
Renin cell isolation.
Native renin cells were isolated from the kidney cortex of Ren1c-YFP and Ren1c–/– Ren1c-YFP adult reporter mice by FACS as we previously described (7, 9).
Chromatin immunoprecipitation followed by sequencing.
As4.1 cells were grown to a final count of 1.5 × 107 cells per experiment and fixed according to the manufacturer’s protocol. From Ren1c–/– Ren1c-YFP mice, 1 × 105 YFP+ cells were isolated and snap-frozen unfixed in liquid nitrogen. Samples were sent to Active Motif for ChIP, library preparation, sequencing, and initial data analysis.
Identification of super-enhancers.
We used the rank ordering of super-enhancers software (ROSE) (21, 22). The MACS peaks identified were used as input for the algorithm, which allows regions within 12.5 kb of each other to be stitched together. Enhancers were ranked (x axis) according to their total reads per million per base pair (y axis) (Figure 3A). An inflection point in the distribution of the occupancy of the factor was used to establish the cutoff for super-enhancers. ROSE was run with a stitching distance of 12.5 kb by default and without promoter exclusion.
Assay for transposase accessible chromatin with high-throughput sequencing.
ATAC-Seq was performed to identify open chromatin regions in JG cells, cells from acutely and chronically recruited mice, and in As4.1 cells. We collected 5 × 104 cells for As4.1 and from chronically recruited mice, and 17 × 104 JG cells and cells from acutely recruited mice, as described elsewhere (6, 57). Paired-end ATAC-Seq libraries were sequenced by the HudsonAlpha Institute for Biotechnology.
RNA-Seq.
As4.1 cells were grown to a final count of 1.5 × 107 cells and fixed according to the manufacturer’s protocol. The sample was sent to Active Motif to perform RNA isolation, library preparation, sequencing, and initial data analysis. For renin cells isolated from Ren1c-YFP mice, the Clontech SMARTer-Seq Lysis, RT, and PCR solutions (catalog 634833) were used to generate bulk cDNA samples.
The data discussed in this publication have been deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE117725).
CRISPR/Cas9 system.
Cells were transiently transfected with a multiplex plasmid (Addgene 1000000054) generated according to Sakuma et al. (58) to delete the classical renin enhancer and the Rbpj-Enh. We transfected the cells using the 4D-Nucleofector System (Lonza). After 48 hours, the supernatant was collected to measure renin by ELISA, and cells were trypsinized and resuspended to be sorted for eGFP expression. For CRISPR/dCas9p300 experiments, 5 sgRNAs targeting the super-enhancer region of Renin were cloned first in a multiplex plasmid. Subsequently, the Cas9 sequence was replaced by the dCas9p300 sequence from plasmid pcDNA-dCas9p300 Core (Addgene 61357; deposited by Charles Gersbach, Duke University, Durham, NC). The sgRNAs were designed using the CRISPR design tool (Fen Zhang, Massachusetts Institute of Technology). Cells were analyzed for YFP and renin expression 72 hours after transfection.
Immunohistochemistry.
Mice were anesthetized with tribromoethanol. Immunostaining was performed on 5-μm-thick paraffin sections using primary antibodies against renin, α-SMA, Akr1b7, and myosin heavy chain, and the appropriate Vectastain ABC kit (Vector Laboratories) as previously described (12, 59).
RNA extraction and quantitative real-time PCR.
Total RNA was extracted as we previously described (60). Renin, Med1, and Akr1b7 mRNA levels were normalized to S14 expression, and the changes in expression were determined by the ΔΔCt method.
Blood pressure measurement.
Mice were anesthetized with 1.5% isoflurane and placed on a thermostatically controlled heating table at 37.5°C. Measurements of mean arterial pressure, systolic, and diastolic blood pressure were taken over a 10-minute period (61, 62) utilizing an RX104A transducer coupled to a data acquisition system and AcqKnowledge software (Biopac Systems, Inc.).
Acute recruitment study.
Ren1cYFP mice at 2 months of age were treated with a low sodium diet (0.1%, 7034, ENVIGO) plus captopril added to the drinking water (0.5 g/l) for 7 days (14, 15). At the end of the treatment period, YFP+ cells were isolated by FACS and cells were used to perform ATAC-Seq.
Blood chemistry.
Blood was collected by cardiac puncture or through the carotid catheter. After blood was collected for renin measurements, blood samples were collected into heparinized and EDTA plasma separator tubes (Microtainer). Complete blood count and the basic metabolic panels were performed by the University of Virginia Hospital clinical laboratory (7).
Plasma renin.
Plasma RENIN concentration was determined using ELISA following the manufacturer’s instructions (RayBiotech) (63).
Statistics.
Statistical analysis was performed using GraphPad Prism 7. Two-tailed Student’s t test or Wilcoxon signed rank sum test was used when comparing 2 related samples not normally distributed. A P value less than 0.05 was considered significant.
Study approval.
Animal experiments were handled in accordance with the National Institutes of Health guidelines for the care and use of experimental animals, and the study was approved by the IACUC of the University of Virginia. Additional details for all methods can be found in the Supplemental Material.
Authors contributions
RAG conceived the project. MFM, SM, RAG, MLSSL, and MA conceived the experiment design. MFM, RIB, SM, OG, NB, and TT performed experiments. MFM, RIB, OG, SS, and SM analyzed and interpreted data. RAG, MLSSL, and MA provided resources. MFM and RAG wrote the manuscript with the contribution of all the authors. RAG, BCB, and MLSSL supervised the research. RAG provided the financial support for the project.
Supplementary Material
Acknowledgments
Studies were funded by National Institutes of Health grants DK-096373 and HL-096735 to RAG, and DK-091330 and DK-096373 to MLSSL. We thank Yang Jiekun for help with the bioinformatics analysis, and Masafumi Oka and Hirofumi Watanabe for cell isolation and other support. We thank Robert Paxton, Tiffany Southard, Fang Xu, Minghong Li, and Xiuyin Liang for technical assistance, and Ellen S. Pentz for initial help in the pursuit of this work.
Version 1. 08/21/2018
In-Press Preview
Version 2. 10/02/2018
Electronic publication
Version 3. 11/01/2018
Print issue publication
Version 4. 02/02/2024
Robin Isadora Brown's name updated in author list (ORCID: 0000-0002-1398-5721)
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.Submitted: March 29, 2018; Accepted: August 14, 2018.
Reference information: J Clin Invest. 2018;128(11):4787–4803.https://doi.org/10.1172/JCI121361.
See the related Commentary at ATAC-ing the Mechanisms of Renin Regulation.
Contributor Information
Maria Florencia Martinez, Email: MFM6B@hscmail.mcc.virginia.edu.
Silvia Medrano, Email: sm8z@virginia.edu.
Robin Isadora Brown, Email: eab2dh@virginia.edu.
Turan Tufan, Email: tt7z@virginia.edu.
Stephen Shang, Email: ss9af@virginia.edu.
Nadia Bertoncello, Email: nsb6qb@hscmail.mcc.virginia.edu.
Omar Guessoum, Email: oeg5qc@virginia.edu.
Mazhar Adli, Email: ma9fe@eservices.virginia.edu.
Brian C. Belyea, Email: bcb9e@hscmail.mcc.virginia.edu.
R. Ariel Gomez, Email: rg@virginia.edu.
References
- 1.Gomez RA, Sequeira-Lopez MLS. Renin cells in homeostasis, regeneration and immune defence mechanisms. Nat Rev Nephrol. 2018;14(4):231–245. doi: 10.1038/nrneph.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez RA, et al. Renin and angiotensinogen gene expression and intrarenal renin distribution during ACE inhibition. Am J Physiol. 1988;254(6 Pt 2):F900–F906. doi: 10.1152/ajprenal.1988.254.6.F900. [DOI] [PubMed] [Google Scholar]
- 3.Keeton TK, Campbell WB. The pharmacologic alteration of renin release. Pharmacol Rev. 1980;32(2):81–227. [PubMed] [Google Scholar]
- 4.Gomez RA, Sequeira-Lopez ML. Novel functions of renin precursors in homeostasis and disease. Physiology (Bethesda) 2016;31(1):25–33. doi: 10.1152/physiol.00039.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sequeira López ML, Pentz ES, Nomasa T, Smithies O, Gomez RA. Renin cells are precursors for multiple cell types that switch to the renin phenotype when homeostasis is threatened. Dev Cell. 2004;6(5):719–728. doi: 10.1016/S1534-5807(04)00134-0. [DOI] [PubMed] [Google Scholar]
- 6.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oka M, Medrano S, Sequeira-Lόpez MLS, Gómez RA. Chronic stimulation of renin cells leads to vascular pathology. Hypertension. 2017;70(1):119–128. doi: 10.1161/HYPERTENSIONAHA.117.09283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi N, et al. Ren1c homozygous null mice are hypotensive and polyuric, but heterozygotes are indistinguishable from wild-type. J Am Soc Nephrol. 2005;16(1):125–132. doi: 10.1681/ASN.2004060490. [DOI] [PubMed] [Google Scholar]
- 9.Pentz ES, Lopez ML, Cordaillat M, Gomez RA. Identity of the renin cell is mediated by cAMP and chromatin remodeling: an in vitro model for studying cell recruitment and plasticity. Am J Physiol Heart Circ Physiol. 2008;294(2):H699–H707. doi: 10.1152/ajpheart.01152.2007. [DOI] [PubMed] [Google Scholar]
- 10.Sigmund CD, et al. Isolation and characterization of renin-expressing cell lines from transgenic mice containing a renin-promoter viral oncogene fusion construct. J Biol Chem. 1990;265(32):19916–19922. [PubMed] [Google Scholar]
- 11.Petrovic N, et al. Role of proximal promoter elements in regulation of renin gene transcription. J Biol Chem. 1996;271(37):22499–22505. doi: 10.1074/jbc.271.37.22499. [DOI] [PubMed] [Google Scholar]
- 12.Sequeira Lopez ML, Pentz ES, Robert B, Abrahamson DR, Gomez RA. Embryonic origin and lineage of juxtaglomerular cells. Am J Physiol Renal Physiol. 2001;281(2):F345–F356. doi: 10.1152/ajprenal.2001.281.2.F345. [DOI] [PubMed] [Google Scholar]
- 13.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castellanos Rivera RM, et al. Transcriptional regulator RBP-J regulates the number and plasticity of renin cells. Physiol Genomics. 2011;43(17):1021–1028. doi: 10.1152/physiolgenomics.00061.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellanos-Rivera RM, et al. Recombination signal binding protein for Ig-κJ region regulates juxtaglomerular cell phenotype by activating the myo-endocrine program and suppressing ectopic gene expression. J Am Soc Nephrol. 2015;26(1):67–80. doi: 10.1681/ASN.2013101045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuras L, Borggrefe T, Kornberg RD. Association of the mediator complex with enhancers of active genes. Proc Natl Acad Sci U S A. 2003;100(24):13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25(15):1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hnisz D, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58(2):362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herranz D, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med. 2014;20(10):1130–1137. doi: 10.1038/nm.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei Y, et al. SEA: a super-enhancer archive. Nucleic Acids Res. 2016;44(D1):D172–D179. doi: 10.1093/nar/gkv1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dennis G, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 29.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9 Suppl 1:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everett AD, Carey RM, Chevalier RL, Peach MJ, Gomez RA. Renin release and gene expression in intact rat kidney microvessels and single cells. J Clin Invest. 1990;86(1):169–175. doi: 10.1172/JCI114680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cartharius K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 32.Feinberg MW, Lin Z, Fisch S, Jain MK. An emerging role for Krüppel-like factors in vascular biology. Trends Cardiovasc Med. 2004;14(6):241–246. doi: 10.1016/j.tcm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Krüppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40(10):1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Atkins GB, Jain MK. Role of Krüppel-like transcription factors in endothelial biology. Circ Res. 2007;100(12):1686–1695. doi: 10.1161/01.RES.0000267856.00713.0a. [DOI] [PubMed] [Google Scholar]
- 35.Chiplunkar AR, et al. The Krüppel-like factor 2 and Krüppel-like factor 4 genes interact to maintain endothelial integrity in mouse embryonic vasculogenesis. BMC Dev Biol. 2013;13:40. doi: 10.1186/1471-213X-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Z, et al. Kruppel-like factor 2 regulates endothelial barrier function. Arterioscler Thromb Vasc Biol. 2010;30(10):1952–1959. doi: 10.1161/ATVBAHA.110.211474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134(7):1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt D, et al. Critical role for NF-kappaB-induced JunB in VEGF regulation and tumor angiogenesis. EMBO J. 2007;26(3):710–719. doi: 10.1038/sj.emboj.7601539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schorpp-Kistner M, Wang ZQ, Angel P, Wagner EF. JunB is essential for mammalian placentation. EMBO J. 1999;18(4):934–948. doi: 10.1093/emboj/18.4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein I, Ojamaa K. Thyroid hormone and the cardiovascular system. N Engl J Med. 2001;344(7):501–509. doi: 10.1056/NEJM200102153440707. [DOI] [PubMed] [Google Scholar]
- 41.Chinery R, Brockman JA, Dransfield DT, Coffey RJ. Antioxidant-induced nuclear translocation of CCAAT/enhancer-binding protein beta. A critical role for protein kinase A-mediated phosphorylation of Ser299. J Biol Chem. 1997;272(48):30356–30361. doi: 10.1074/jbc.272.48.30356. [DOI] [PubMed] [Google Scholar]
- 42.Akira S, Kishimoto T. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol Rev. 1992;127:25–50. doi: 10.1111/j.1600-065X.1992.tb01407.x. [DOI] [PubMed] [Google Scholar]
- 43.Screpanti I, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J. 1995;14(9):1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shestakova EA, Boutin M, Bourassa S, Bonneil E, Bijl JJ. [Identification of proteins associated with transcription factors HOXA9 and E2A-PBX1 by tandem affinity purification] Mol Biol (Mosk) 2017;51(3):490–501. doi: 10.7868/S0026898417030132. [DOI] [PubMed] [Google Scholar]
- 45.Huang Y, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119(2):388–398. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conaway RC, Conaway JW. Origins and activity of the mediator complex. Semin Cell Dev Biol. 2011;22(7):729–734. doi: 10.1016/j.semcdb.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malik S, Roeder RG. The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11(11):761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin EE, Pentz ES, Sequeira-Lopez ML, Gomez RA. Aldo-keto reductase 1b7, a novel marker for renin cells, is regulated by cyclic AMP signaling. Am J Physiol Regul Integr Comp Physiol. 2015;309(5):R576–R584. doi: 10.1152/ajpregu.00222.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rider SA, Mullins LJ, Verdon RF, MacRae CA, Mullins JJ. Renin expression in developing zebrafish is associated with angiogenesis and requires the Notch pathway and endothelium. Am J Physiol Renal Physiol. 2015;309(6):F531–F539. doi: 10.1152/ajprenal.00247.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neubauer B, et al. Development of vascular renin expression in the kidney critically depends on the cyclic AMP pathway. Am J Physiol Renal Physiol. 2009;296(5):F1006–F1012. doi: 10.1152/ajprenal.90448.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L, et al. Regulation of renin in mice with Cre recombinase-mediated deletion of G protein Gsalpha in juxtaglomerular cells. Am J Physiol Renal Physiol. 2007;292(1):F27–F37. doi: 10.1152/ajprenal.00193.2006. [DOI] [PubMed] [Google Scholar]
- 53.Gomez RA, Pentz ES, Jin X, Cordaillat M, Sequeira Lopez ML. CBP and p300 are essential for renin cell identity and morphological integrity of the kidney. Am J Physiol Heart Circ Physiol. 2009;296(5):H1255–H1262. doi: 10.1152/ajpheart.01266.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Y, et al. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J Biol Chem. 2000;275(18):13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- 56.Jia Y, et al. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J Biol Chem. 2004;279(23):24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 57.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep. 2014;4:5400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin EE, Sequeira-Lopez ML, Gomez RA. RBP-J in FOXD1+ renal stromal progenitors is crucial for the proper development and assembly of the kidney vasculature and glomerular mesangial cells. Am J Physiol Renal Physiol. 2014;306(2):F249–F258. doi: 10.1152/ajprenal.00313.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belyea BC, Xu F, Sequeira-Lopez ML, Ariel Gomez R. Loss of Jagged1 in renin progenitors leads to focal kidney fibrosis. Physiol Rep. 2015;3(11):e12544. doi: 10.14814/phy2.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pentz ES, Moyano MA, Thornhill BA, Sequeira Lopez ML, Gomez RA. Ablation of renin-expressing juxtaglomerular cells results in a distinct kidney phenotype. Am J Physiol Regul Integr Comp Physiol. 2004;286(3):R474–R483. doi: 10.1152/ajpregu.00426.2003. [DOI] [PubMed] [Google Scholar]
- 62.Pentz ES, Lopez ML, Kim HS, Carretero O, Smithies O, Gomez RA. Ren1d and Ren2 cooperate to preserve homeostasis: evidence from mice expressing GFP in place of Ren1d. Physiol Genomics. 2001;6(1):45–55. doi: 10.1152/physiolgenomics.2001.6.1.45. [DOI] [PubMed] [Google Scholar]
- 63.Belyea BC, et al. Identification of renin progenitors in the mouse bone marrow that give rise to B-cell leukaemia. Nat Commun. 2014;5:3273. doi: 10.1038/ncomms4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.