Abstract
Graft-versus-host disease (GVHD) in the gastrointestinal (GI) tract remains the major cause of morbidity and nonrelapse mortality after BM transplantation (BMT). The Paneth cell protein regenerating islet-derived 3α (REG3α) is a biomarker specific for GI GVHD. REG3α serum levels rose in the systematic circulation as GVHD progressively destroyed Paneth cells and reduced GI epithelial barrier function. Paradoxically, GVHD suppressed intestinal REG3γ (the mouse homolog of human REG3α), and the absence of REG3γ in BMT recipients intensified GVHD but did not change the composition of the microbiome. IL-22 administration restored REG3γ production and prevented apoptosis of both intestinal stem cells (ISCs) and Paneth cells, but this protection was completely abrogated in Reg3g−/− mice. In vitro, addition of REG3α reduced the apoptosis of colonic cell lines. Strategies that increase intestinal REG3α/γ to promote crypt regeneration may offer a novel, nonimmunosuppressive approach for GVHD and perhaps for other diseases involving the ISC niche, such as inflammatory bowel disease.
Keywords: Immunology, Transplantation
Keywords: Adaptive immunity, Bone marrow transplantation, Innate immunity
Introduction
Acute graft-versus-host disease (GVHD) is the most serious complication of allogeneic BM transplantation (BMT) and was noted in animal models more than 6 decades ago (1, 2). Acute GVHD predominantly involves the skin, liver, and lower gastrointestinal (GI) tract (3).
Without rigorous depletion of donor T cells from the graft, this complication occurs in 30%–70% of patients after allogeneic BMT (4–6). Standard treatment of acute GVHD is the administration of systemic corticosteroids and additional immunosuppressive agents; only half of patients who develop acute GVHD will have a complete response to corticosteroid therapy, although the numbers vary widely between studies (7). The outcome for patients with severe GVHD of the lower GI tract is poor, with 25% overall survival (8). We undertook this study to understand the biologic role of regenerating islet-derived 3α/γ (REG3α/γ) in GI GVHD.
Results
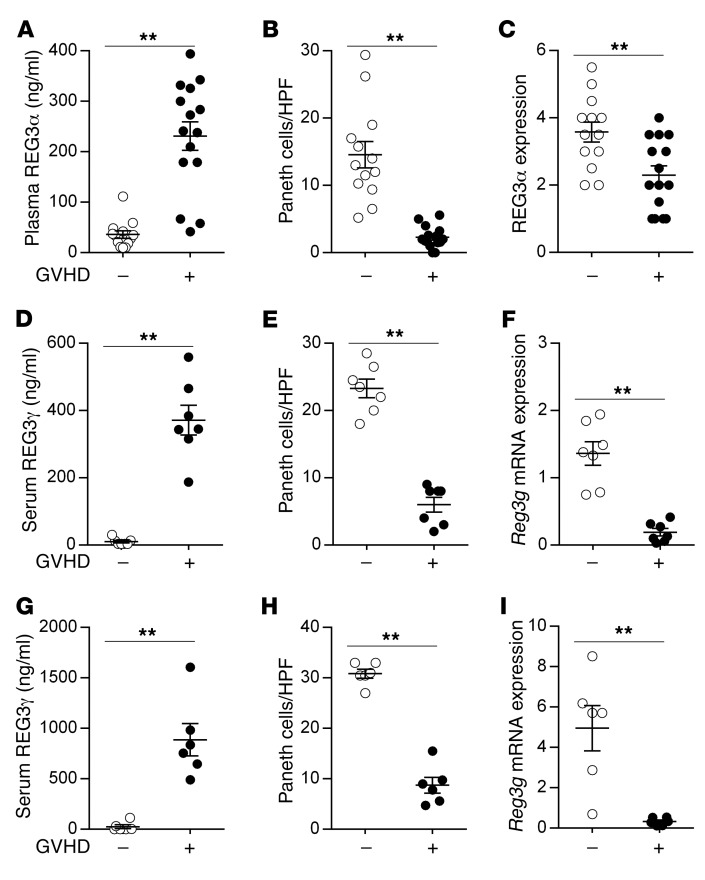
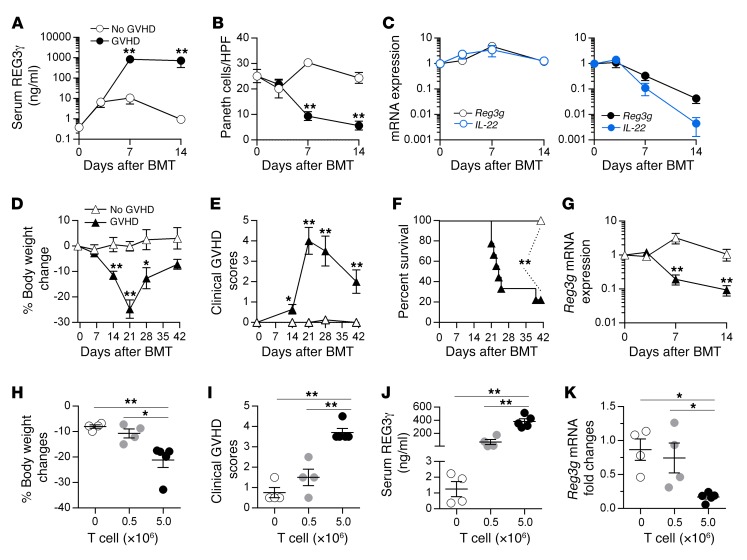
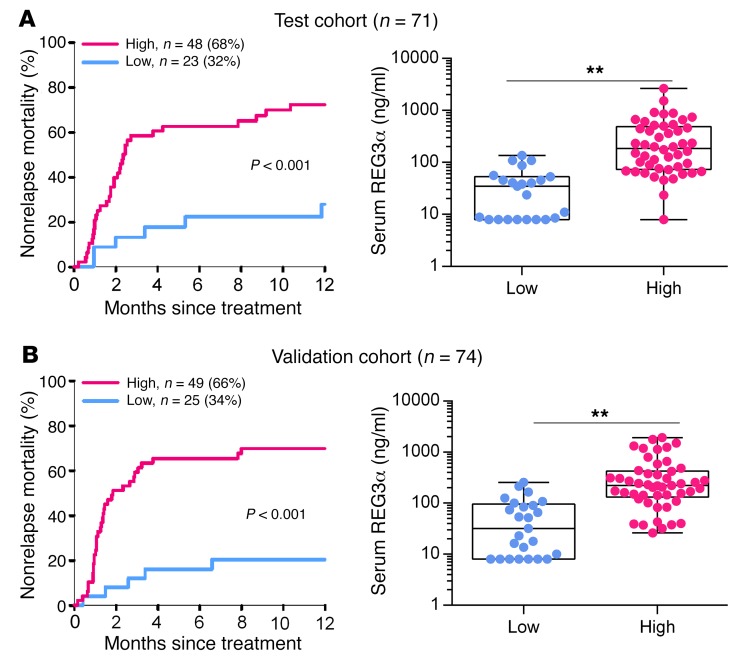
An elevated REG3α blood level is a strong indicator of GI GVHD that drives transplant reported mortality (9–12). We observed that increased blood levels of REG3α in patients with GVHD inversely correlated with the number of Paneth cells (Figure 1, A–C) and observed a similar inverse correlation in 2 mouse models of GVHD (Figure 1, D–I, and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI99261DS1). This inverse correlation continued over time and increased with GVHD severity independently of BMT conditioning (Figure 2). IL-22 induces REG3γ (13–15), and the production of IL-22 decreased very rapidly during GVHD (Figure 2). Increased Reg3g expression was not observed in any other tissue, including lung, liver, and pancreas (data not shown), suggesting that damage to the GI crypt resulted in the loss of REG3γ into the systemic circulation. REG3α serum levels remained elevated, often by an order of magnitude, after 1 week of systemic steroid therapy in patients with GI GVHD and predicted increased nonrelapse mortality, confirming the significance of continued serum elevations with respect to long-term clinical outcomes (Figure 3 and ref. 16).
Figure 1. REG3α/γ levels in the plasma/serum and intestinal mucosa during GVHD.
(A–C) Samples were collected from 28 allogeneic BMT patients without GVHD (white circles, n = 13) and with GVHD (black circles, n = 15). (A) Plasma concentrations of REG3α measured by ELISA. (B) Average Paneth cell numbers per high-power field (HPF) in the same biopsies. (C) Semiquantitative REG3α expression in the duodenum. (D–F) B6 mice underwent BMT from syngeneic B6-Ly5.1 donors (GVHD –, white circles, n = 7) or allogeneic C3H.SW donors (GVHD +, black circles, n = 7), and samples were analyzed on day +7 after BMT. (D) Serum REG3γ levels measured by ELISA. (E) Average Paneth cell numbers per HPF in ileal tissue from the same mice. (F) Ileal tissue Reg3g mRNA expression measured by quantitative PCR (qPCR). (G–I) B6D2F1 mice underwent BMT from syngeneic B6D2F1 donors (GVHD –, white circles, n = 6) or allogeneic B6 donors (GVHD +, black circles, n = 6), and samples were analyzed as before on day +7 after BMT. (G) Serum REG3γ levels. (H) Average Paneth cell numbers per HPF in ileal tissue. (I) Ileal tissue Reg3g mRNA expression. **P < 0.01, unpaired 2-tailed t test. Data are expressed as mean ± SEM.
Figure 2. Time course of REG3γ and IL-22 expression during GVHD.
(A–C) B6D2F1 mice underwent BMT from syngeneic B6D2F1 donors (no GVHD, white circles) or allogeneic B6 donors (GVHD, black or blue circles), and samples were analyzed on days +3, +7, and +14 after BMT (n = 5 for each group on each day). (A) Serial REG3γ levels in the serum as measured by ELISA. (B) Serial average Paneth cell numbers per HPF in ileal tissue. (C) Serial Reg3g and IL-22 mRNA expression in the ileum was measured by qPCR. (D–G) B6D2F1 mice underwent BMT, without irradiation conditioning, from syngeneic B6D2F1 donors (no GVHD, white triangles) or allogeneic B6 donors (GVHD, black triangles). (D–F) Serial body weight measurement (D), clinical GVHD score (E), and survival (F) of mice after BMT. (G) Serial Reg3g mRNA expression in the ileum on days +3, +7, and +14 after BMT (n = 6 for each group on each day). *P < 0.05, **P < 0.01, unpaired 2-tailed t test (A–E and G); **P < 0.01, log-rank test (F). (H–K) Following irradiation, B6 mice received 5.0 × 106 BM cells plus 0 × 106 to 5.0 × 106 T cells, as indicated, from C3H.SW donors. Samples were analyzed on day +7 after BMT (n = 4 for each group). (H) Body weight measurement. (I) Clinical GVHD score. (J) Serum REG3γ levels. (K) Reg3g mRNA expression in the ileum. *P < 0.05, **P < 0.01, 1-way ANOVA. Data are expressed as mean ± SEM.
Figure 3. Serum REG3α levels and correlates with nonrelapse mortality in patients with GI GVHD.
Patients with GI GVHD (n = 145) who were treated with systemic steroids for at least 7 days provided blood samples for biomarker analysis and were divided according to biomarker probabilities into low (blue) and high (pink) groups as previously published (16). Twelve-month nonrelapse mortality and REG3α serum levels are shown for test (n = 71) (A) and validation (n = 74) (B) cohorts. **P < 0.01, unpaired 2-tailed t test. Data are expressed as box-and-whisker plots.
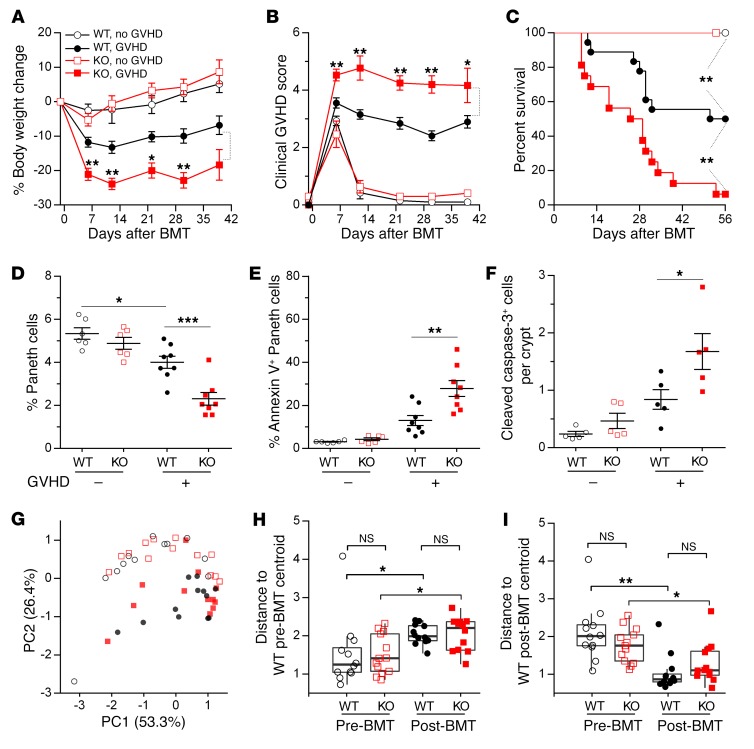
We next investigated the role of REG3γ in GVHD using REG3γ-deficient (Reg3g−/−) mice as BMT recipients. Reg3g−/− mice experienced more severe clinical GVHD and greater mortality than WT mice in 2 GVHD models (Figure 4, A–C). The histologic severity of GVHD, as measured by Paneth cell apoptosis and loss, also increased in Reg3g−/− BMT recipients; so did the number of crypt cells expressing cleaved caspase-3, a hallmark of apoptosis (Figure 4, D–F, and Supplemental Figures 2 and 3). Reg3g−/− allogeneic BMT recipients also exhibited significantly more CD8+ IFN-γ+ effector T cells in the lamina propria as well as increased hepatic infiltrates, confirming greater histologic damage in the absence of recipient REG3γ (Supplemental Figures 3 and 4). The composition of fecal microbial communities did not differ significantly, however, between WT and Reg3g−/− BMT recipients either before transplant or during GVHD (Figure 4, G–I, and Supplemental Figure 5).
Figure 4. REG3γ absence increases GVHD severity.
WT B6 and B6-Reg3g–/– (KO) mice underwent BMT from B6-Ly5.1 donors (GVHD –: WT, white circles; KO, white squares) or C3H.SW donors (GVHD +: WT, black circles; KO, red squares). (A–C) Serial body weight measurement (A), clinical GVHD score (B), and survival (C) of mice after BMT (WT, white circles, n = 7; KO, white squares, n = 7; WT, black circles, n = 18; KO, red squares, n = 16). (D–F) On day +7 after BMT, WT or KO recipients (Lgr5-EGFP+) were euthanized. Small intestine crypt cells were isolated and analyzed by flow cytometry. Small intestine sections were stained by immunochemistry. (D and E) Quantification of Paneth cells (D) and quantification of Paneth cells undergoing early apoptosis by annexin V+ staining (E) (WT, white circles, n = 6; KO, white squares, n = 6; WT, black circles, n = 8; KO, red squares, n = 8). (F) Quantification of cleaved caspase-3+ cells per crypt of ileum sections in recipients (each group, n = 5). *P < 0.05, **P < 0.01, ***P < 0.001, unpaired 2-tailed t test (A, B, and D–F); **P < 0.01, log-rank test (C). Data are expressed as mean ± SEM. (G–I) Microbiome composition of fecal samples from WT and KO mice 1 day before BMT (Pre-BMT) (WT, white circles, n = 12; KO, white squares, n = 13) or day +7 after BMT (Post-BMT) (WT, black circles, n = 12; KO, red squares, n = 13). (G) Principal component analysis was computed from the Bray-Curtis β-diversity matrix among all samples. (H and I) Bray-Curtis compositional distances were measured from the centroid of WT pre-BMT samples (H) or post-BMT samples (I). NS, P > 0.05; *P < 0.05, **P < 0.01, Wilcox test.
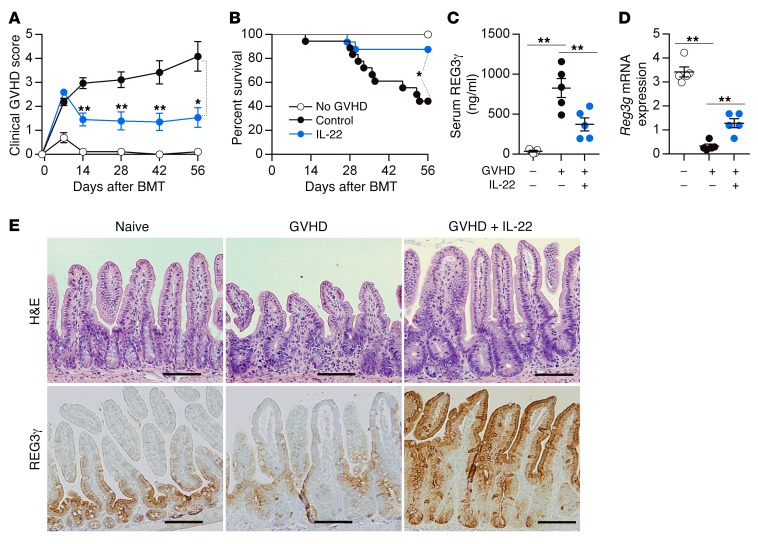
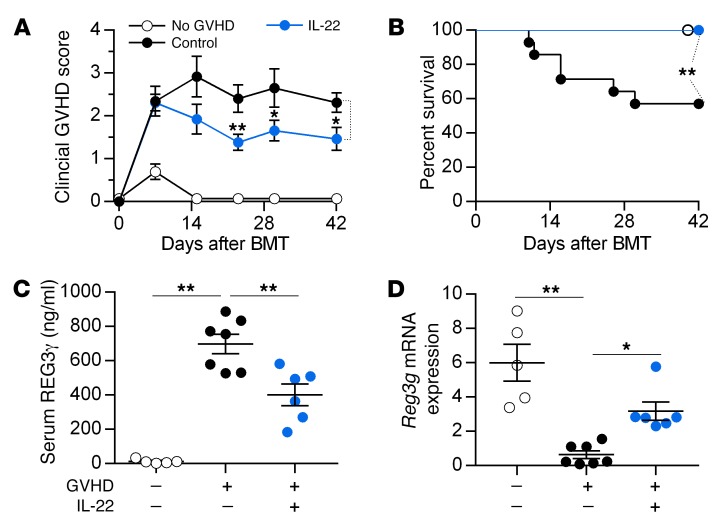
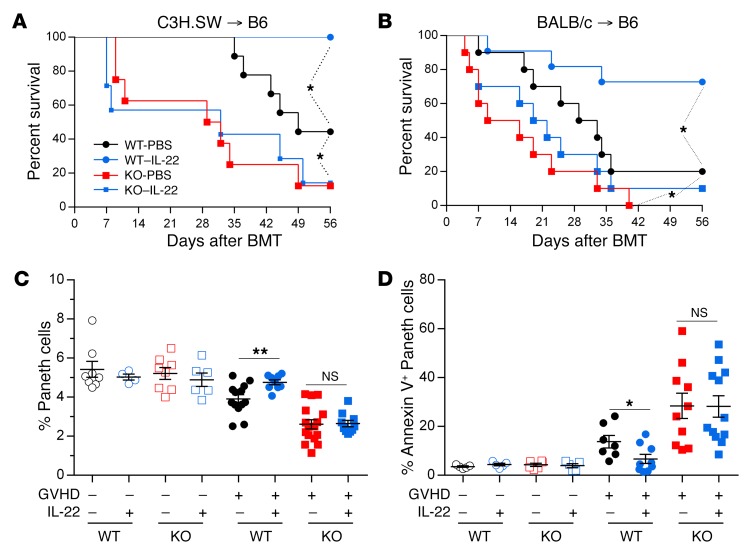
Although IL-22 has pleiotropic effects and can increase GVHD severity in some models (17, 18), prophylactic administration of IL-22 prevented GVHD and increased REG3γ production in the ileum as previously reported (15) while simultaneously decreasing serum REG3γ levels (Figure 5 and Supplemental Figure 6). IL-22 treatment of mice also reversed established GVHD in 2 separate models, and decreased release of REG3γ into serum (Figure 6 and Supplemental Figure 7). IL-22 was completely unable, however, to prevent clinical and histologic GVHD and Paneth cell loss in Reg3g−/− recipients in 2 separate models, confirming the need for REG3γ to repair the integrity of GI epithelium (Figure 7 and Supplemental Figures 8–10). mRNA expression of other antimicrobial Paneth cell proteins that are disrupted during GVHD (19), such as Defa1, Defa2, S100a8, and S100a9, did not consistently change in Reg3g−/− recipients (Supplemental Figure 11).
Figure 5. Prophylactic administration of IL-22 reduces GVHD.
B6D2F1 mice received PBS or IL-22 injections from day +1 after BMT from B6 donors (GVHD, +) or B6D2F1 donors (No GVHD, –). (A and B) Clinical GVHD score (A) and survival (B) of mice after BMT (GVHD +: PBS, black circles, n = 18; IL-22, blue circles, n = 16; GVHD –, white circles, n = 7). (C−E) Samples were analyzed on day +7 after BMT. (C) Serum REG3γ levels. (D) Ileal tissue Reg3g mRNA expression. *P < 0.05, **P < 0.01, unpaired 2-tailed t test (A), 1-way ANOVA (C and D); *P < 0.05, log-rank test (B). (E) H&E (top) staining and REG3γ (bottom) staining by immunohistochemistry in ileum from individual mice with or without IL-22 treatment. Naive B6D2F1 mice were used as controls. Images were taken with an Olympus BX51 with a ×20 objective. Scale bars: 100 μm. Data are expressed as mean ± SEM.
Figure 6. IL-22 treatment reverses GVHD.
B6 mice received BM and T cells from C3H.SW donors (GVHD +) treated with PBS (black circles) or IL-22 (blue circles) from day +7. As a control, mice received BM only (No GVHD, –, white circles). (A and B) Clinical GVHD score (A) and survival (B) of mice after BMT (PBS, n = 14; IL-22, n = 13; no GVHD, n = 5). Samples were analyzed on day +10. (C) Serum REG3γ protein. (D) Small intestine tissue Reg3g mRNA. *P < 0.05, **P < 0.01, unpaired 2-tailed t test (A), 1-way ANOVA (C and D); **P < 0.01, log-rank test (B). Data are expressed as mean ± SEM.
Figure 7. IL-22 requires REG3γ to reduce GVHD.
(A) Survival of WT B6 and B6-Reg3g–/– (KO) mice received PBS or IL-22 injections from day +1 after BMT from C3H.SW donors (WT-PBS, black circles, n = 9; WT–IL-22, blue circles, n = 8; KO-PBS, red squares, n = 8; KO–IL-22, blue squares, n = 7). (B) Survival of WT B6 and B6-Reg3g–/– (KO) mice received PBS or IL-22 injections from day +1 after BMT from BALB/c donors (WT-PBS, black circles, n = 10; WT–IL-22, blue circles, n = 11; KO-PBS, red squares, n = 10; KO–IL-22, blue squares, n = 10). *P < 0.05, log-rank test. (C and D) WT B6 and B6-Reg3g−/− (KO) mice (Lgr5-EGFP+) received PBS or IL-22 injections from day +1 after BMT from B6-Ly5.1 donors (GVHD –) or C3H.SW donors (GVHD +). On day +7 after BMT, small intestine crypt cells were analyzed by flow cytometry. (C) Quantification of Paneth cells (GVHD –: WT-PBS, empty black circles, n = 8; WT–IL-22, empty blue circles, n = 4; KO-PBS, empty red squares, n = 8; KO–IL-22, empty blue squares, n = 6; GVHD +: WT-PBS, filled black circles, n = 13; WT–IL-22, filled blue circles, n = 9; KO-PBS, filled red squares, n = 16; KO–IL-22, filled blue squares, n = 10). (D) Quantification of Paneth cells undergoing early apoptosis by annexin V+ staining (GVHD –: WT-PBS, empty black circles, n = 5; WT–IL-22, empty blue circles, n = 5; KO-PBS, empty red squares, n = 6; KO–IL-22, empty blue squares, n = 5; GVHD +: WT-PBS, filled black circles, n = 7; WT–IL-22, filled blue circles, n = 9; KO-PBS, filled red squares, n = 10; KO–IL-22, filled blue squares, n = 12). NS, P > 0.05; *P < 0.05, **P < 0.01, unpaired 2-tailed t test. Data are expressed as mean ± SEM.
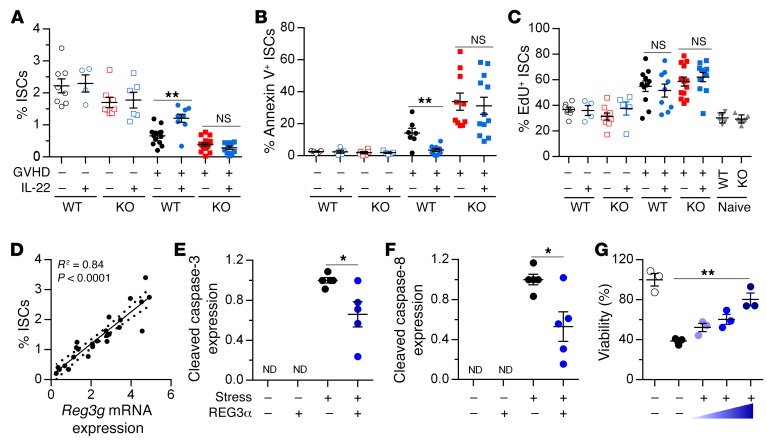
To investigate the effect of REG3γ on intestinal stem cells (ISCs), we bred Reg3g−/− mice with mice heterozygous for green fluorescent protein (GFP) in Lgr5+ enterocytes (20) to use as BMT recipients. We confirmed that the loss of ISCs during GVHD (21) was not reversed by IL-22 treatment in Reg3g−/− mice (Figure 8A and Supplemental Figure 12). We next quantified ISC apoptosis and proliferation using established flow cytometry protocols (22). Surprisingly, IL-22 did not affect ISC proliferation under any conditions, but reversed apoptosis only in WT and not in Reg3g−/− recipients; the number of ISCs correlated directly with Reg3g mRNA expression (Figure 8, B–D, and Supplemental Figures 13 and 14). We then directly evaluated the antiapoptotic properties of REG3α in intestinal epithelium in vitro. Addition of REG3α in vitro to colonic epithelial cell lines undergoing apoptotic stress reduced the cleavage of caspase-3, caspase-8, and PARP and increased cell viability in a dose-dependent manner, confirming its role as a survival factor for epithelial cells (Figure 8, E–G, and Supplemental Figure 15).
Figure 8. REG3γ protects ISCs from apoptosis.
(A–D) WT B6 and B6-Reg3g–/– (KO) mice (Lgr5-EGFP+) received PBS or IL-22 injections from day +1 after BMT from B6-Ly5.1 donors (GVHD –) or C3H.SW donors (GVHD +). On day +7 after BMT, small intestine crypt cells were analyzed by flow cytometry, and Reg3g mRNA was measured by qPCR. (A) Quantification of ISCs from mice in Figure 7. (B) Quantification of ISCs undergoing early apoptosis by annexin V+ staining in the mice from Figure 7. Data are combined from 3 separate experiments. (C) Quantification of EdU+ ISCs in syngeneic recipients (GVHD –: WT-PBS, empty black circles, n = 7; WT–IL-22, empty blue circles, n = 4; KO-PBS, empty red squares, n = 9; KO–IL-22, empty blue squares, n = 5), allogeneic recipients (GVHD +: WT-PBS, filled black circles, n = 8; WT–IL-22, filled blue circles, n = 9; KO-PBS, filled red squares, n = 13; KO–IL-22, filled blue squares, n = 11), and naive WT (downward triangles, n = 5) and KO (upward triangles, n = 5) controls. (D) Correlation of ISCs and Reg3g mRNA expression in WT mice from 2 of the 3 experiments shown in A (n = 25), as calculated by the Pearson correlation coefficient. (E–G) HT-29 cells were cultured with recombinant REG3α protein and apoptotic stimuli as described in Methods. Data are representative of 3 independent experiments. (E and F) Cell lysates were collected after 16-hour cell culture for Western blot analysis. Quantification of cleaved caspase-3 (E) and cleaved caspase-8 (F) expression. (G) Quantification of cell viability by CellTiter-Glo 2.0 assay after 24-hour cell culture. *P < 0.05, **P < 0.01, unpaired 2-tailed t test. Data are expressed as mean ± SEM. ND, not detectable.
Discussion
We report here that the production of REG3α/γ in the GI tract falls as acute GVHD progresses and inversely correlates with its concentration in the serum, supporting the concept of this biomarker as a “liquid biopsy” that quantifies crypt damage and the loss of ISCs and Paneth cells. Administration of IL-22 restores GI epithelial integrity and reduces GVHD because REG3α/γ acts as a survival signal for ISCs and Paneth cells, preventing their apoptosis both in vitro and in vivo. The requirement of REG3α/γ to repair damaged GI epithelium treatment indicates a previously unknown function for this protein in GI crypt homeostasis.
These results suggest the possibility of a more physiologic approach to the prevention and treatment of GI GVHD through restoration of the GI mucosal barrier rather than intensified systemic immunosuppression. Paneth cells, the “guardians of the crypt,” secrete multiple antimicrobial peptides, including REG proteins, that help to generate the chemical/physical barrier of the GI mucosa (23). A disrupted microbiome constitutes a significant risk factor for both experimental and clinical GVHD (24, 25) and might amplify GVHD by exposing enterocytes to Gram-positive pathogens (26). But analysis of the fecal microbiome demonstrated that the absence of REG3γ produced no significant difference in the microbial community architecture either before or during GVHD even though it caused greater crypt destruction and significantly increased the loss of ISCs and Paneth cells. This discrepancy suggested that the antimicrobial activity of REG3γ was not primarily responsible for its protective properties. Absence of host REG3γ thus amplified GVHD by accelerating apoptosis of the crypts, allowing microbes to break the mucosal epithelial barrier and activate antigen-presenting cells that attracted and stimulated alloreactive donor T cells, furthering crypt damage (27).
Previous experiments with cultured pancreatic acinar cells and hepatocytes suggested that expression of REG3β could reduce the apoptosis mediated by TNF-α, an important mediator of GVHD (28, 29). Our results directly demonstrate a survival function for REG3α/γ that is key to restoration of epithelial integrity during GVHD. A recently published approach to GVHD prevention is the promotion of Paneth cell development from ISCs through the administration of R-spondin-1, a Wnt agonist (30). It is unknown whether R-spondin-1 requires REG3γ to increase Paneth cell number as does IL-22. In this regard it is interesting to note that although IL-22 increases ISC number in vivo and stimulates organoid growth in vitro, it does not act as a mitogen as was initially thought; rather it prevents the apoptosis (or necroptosis) caused by the inflammation of GVHD through the induction of REG3γ. Thus, in addition to its well-defined antimicrobial properties, REG3γ provides critical survival signals to cells in the stem cell niche.
The potential limitations of this study should be noted. First, the mechanistic insights derive from mouse models and cell lines that cannot reproduce the complexity of human disease. Despite many tight correlations between human and murine data, some experiments do not and cannot have precise human equivalents, e.g., quantification of ISCs through a green fluorescent protein. Another important limitation relates to differences along the axial geography of the bowel. Both the large and the small intestine contain Lgr5+ ISCs, but Paneth cells are largely absent in the colon, where the stem cell niche is supported by deep secretory cells that possess some similarities to Paneth cells (31). Experiments are currently in progress to determine whether REG3γ also acts as a survival signal for colonic ISCs in vivo. If restoration of the survival signal provided by REG3γ can repair damaged intestinal epithelium of both small and large intestines, this approach may offer a novel, nonimmunosuppressive strategy not only for GVHD but for other immunologically mediated diseases of the bowel.
Methods
Patients.
Patients were studied with the approval of the Institutional Review Boards at the University of Michigan, and all patients gave written informed consent in accordance with the Declaration of Helsinki. Twenty-eight patients, who consented to a prospective observational trial of allogeneic BMT complications and underwent allogeneic BMT from 2002 to 2007, were available for analysis; samples were obtained from patients who developed GI symptoms and had a duodenal biopsy and plasma collection at the time of onset of symptoms. All patients without GVHD had a histologic grade of 0, and the median time of biopsy was day +37 (range 15–78); all those with GVHD had a histologic grade of 3, and the median time of biopsy was day +65 (range 13–138).
Mice.
C57BL/6 (B6; H-2b, CD45.2+), B6-Ly5.1 (H-2b, CD45.1+), B6 × DBA2 F1 (B6D2F1; H-2b/d), and BALB/c (H-2d, CD90.2+) mice were purchased from Charles River Laboratories. C3H.SW (H-2b, CD45.2+) mice were purchased from The Jackson Laboratory. Reg3g−/− (H-2b) mice were obtained from Lora V. Hooper (26). B6-Lgr5-EGFP-IRES-creERT2 (Lgr5-EGFP+) mice were a gift from Linda C. Samuelson (University of Michigan, Ann Arbor, Michigan, USA). Reg3g−/− mice were bred with WT Lgr5-EGFP+ mice to generate dual Reg3g−/−/Lgr5-EGFP+ mice (B6 background). For BMT experiments, WT and Reg3g−/− recipient mice were cohoused for a minimum of 2 weeks, in order to reduce heterogeneity in the microbiota between the strains. All animals were cared for under the regulations reviewed and approved by the University Committee on the Use and Care of Animals, per University Laboratory Animal Medicine guidelines at the University of Michigan or the Icahn School of Medicine at Mount Sinai.
BMT.
BMT was performed as previously described (32, 33). Briefly, BM cells were collected from the femurs and tibiae of euthanized donor mice. Donor T cells were obtained from the spleens of donor mice by positive selection using CD90.2 magnetic beads (Miltenyi Biotec), according to the manufacturer’s instructions. Unless otherwise stated, B6D2F1 recipients were given a total of 1,250 cGy total-body irradiation, split into 2 doses separated by 4 hours on day –1, and then injected with 2 × 106 to 4 × 106 splenic T cells and 5 × 106 BM cells from allogeneic B6 donors on day 0. B6 recipients were irradiated with 1,100 cGy total-body irradiation, as a single dose on day –1, and injected on day 0 with 50 × 106 whole splenic cells and 5 × 106 BM cells from MHC-matched, minor antigen–disparate C3H.SW donors or 5 × 106 whole splenic cells and 5 × 106 BM cells from MHC-mismatched BALB/c donors.
In all experiments, transplantation with cells from the appropriate syngeneic donor or BM only was used as a control. Donor and recipient mice were sex-matched and were 8–16 weeks of age at the time of transplantation. Survival was monitored daily, and clinical GVHD was assessed weekly by a previously described scoring system (34).
Administration of IL-22 in vivo.
Recombinant mouse IL-22 (carrier-free) was purchased from BioLegend. For prophylaxis experiments, mice received i.p. injections of 10 μg of IL-22 in PBS, or PBS as a control, on alternate days from day +1 after BMT. Mice were given either 3 doses (when GI tissues were analyzed on day +7) or 6 doses (for longer-term assessment of GVHD scores and survival).
To investigate the effect of treatment with IL-22 after development of GVHD, mice received i.p. injections of 10 μg of IL-22 in PBS, or PBS as a control, starting on day +7 after BMT. Mice were given either 3 daily doses (when GI tissues were analyzed on day +10) or a total of 7 total doses on days 7, 8, 9, 11, 13, 15, and 17 (for longer-term GVHD studies).
All animals were individually ear-tagged before BMT (4 animals per cage). At the time of randomization to receive IL-22 or PBS alone (day 0 for prophylaxis experiments and day 7 for treatment experiments), the first 2 mice were assigned to the IL-22 group and the second 2 mice were assigned to the PBS group. For experiments evaluating treatment beginning on day 7, animals with severe GVHD (weight loss of more than 25%) were excluded before randomization. This criterion was established before BMT and the subsequent development of disease.
5-Ethynyl-2′-deoxyuridine incorporation in vivo assay.
ISC proliferation was determined using 5-ethynyl-2′-deoxyuridine (EdU) incorporation into DNA. Lgr5-EGFP+ mice were injected i.p. with 200 μl of 5 mM EdU solution (Invitrogen) in PBS 2 hours before small-intestinal samples were collected. EdU was detected with the Click-iT Plus EdU Flow Cytometry Assay kit according to the manufacturer’s instructions (Invitrogen).
Histopathology and immunohistochemistry.
For pathologic analyses, samples from patients (duodenal biopsies) and mice (distal ilea and livers) were fixed in 10% neutral-buffered formalin, embedded in paraffin, sectioned, slide-mounted, and stained with H&E (12). Paneth cells were identified by their eosinophilic granules by an experienced observer blinded to the treatment groups, in at least 3 high-power fields (HPFs) with an Olympus BX51 microscope. Using a highly reproducible technique (12), HPF was defined as a ×40 objective with a field of 0.345 mm2. The counts from each HPF were averaged to give the number of Paneth cells per HPF.
Immunohistochemistry was performed with polyclonal rabbit anti-REG3α (Abcam, ab134309) at a 1:200 dilution for human samples and rabbit anti-REG3γ from the Lora V. Hooper laboratory (26) at a 1:8,000 dilution or rabbit anti–cleaved caspase-3 (clone 5A1E, Cell Signaling) at a 1:400 dilution for murine samples using a DAKO AutoStainer Link; slides were subsequently coated with a goat anti-rabbit IgG HRP conjugate (DAKO) at a 1:200 dilution and finally a diaminobenzidine (DAB) dilution to generate brown-colored signals. Slides were counterstained with Harris hematoxylin. Semiquantitative REG3α expression from duodenum biopsies was scored on a 0–2 scale for 3 parameters: Paneth cells, epithelial cytoplasm, and stromal cells. Pictures from tissue sections were taken using a digital camera (DP70, Olympus) mounted on a microscope (BX51, Olympus) or an EVOS XL Core Imaging System from Life Technologies.
Quantitative PCR.
Total RNA was isolated from the small intestine using the RNeasy RNA Isolation Kit (Qiagen) and was used to synthesize cDNA with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR analysis was performed using the SYBR Green Master Mix (Invitrogen) and specific primers. Signals were normalized to Gapdh levels within each sample, and normalized data were used to calculate relative levels of gene expression using ΔΔCt analysis. Details of all primers used can be found in Supplemental Table 1.
ELISA.
REG3α in human plasma was quantified with the REG3α ELISA kit (MBL International, Japan) as described previously (11). REG3γ in mouse serum was measured by a REG3γ ELISA Kit (Cloud-Clone Corp.) according to the manufacturer’s protocol. Absorbance was measured with a SpectraMax M2 (Molecular Devices), and results were calculated with SoftMax Pro version 5.4 software (Molecular Devices).
Western blot.
HT-29 cells were treated with human recombinant REG3α protein (5940-RG-050, R&D Systems) (1,000 ng/ml) followed by treatment with birinapant (10 nM) and recombinant human TNF-α (rhTNF-α; 10 ng/ml) for various time periods (4, 8, and 16 hours). Cells were lysed in RIPA buffer containing protease and phosphatase inhibitors and sonicated with Bioruptor (Diagenode). Analysis was performed on total cell lysate. Protein was quantified using Pierce Rapid Gold BCA protein assay kit (Thermo Fisher Scientific). Thirty micrograms of lysate was loaded onto NuPAGE 4%–12% Bis-Tris protein gel or 12% Bis-Tris protein gel, and protein was separated by electrophoresis followed by transfer into nitrocellulose membrane. Membranes were blocked for 1 hour at room temperature with tris-buffered saline with tween (TBST) and 5% nonfat milk and incubated overnight at 4°C with the following primary antibodies (Cell Signaling): PARP (9532S), cleaved PARP (5625S), caspase-8 (9746S), caspase-3 (29629S), cleaved caspase-3 (9661S), and β-actin (5125S). Proteins were detected with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) or Lumigen ECL Ultra (Lumigen) following 1 hour of incubation at room temperature with anti-rabbit–HRP (7074S; Cell Signaling) or anti-mouse–HRP (7076S; Cell Signaling) antibody.
Apoptosis-induced colorectal epithelial cell viability measurement.
2.5 × 105 cells of the colorectal cell line HT-29 were cultured in serum-deprived DMEM overnight. Human recombinant REG3α protein (5940-RG-050, R&D Systems) (10, 100, and 1,000 ng/ml) was added to culture for 2 hours, and then apoptosis was induced by addition of birinapant (10 nM) and rhTNF-α (10 ng/ml) to the culture for 24 hours. Cell viability was measured by CellTiter-Glo 2.0 assay kit (Promega) according to the manufacturer’s protocol. Luminescence was measured with a SpectraMax M2 (Molecular Devices).
Lamina propria lymphocyte isolation.
Lamina propria lymphocytes (LPLs) from small intestines were isolated as described previously (26). Briefly, 5 cm distal ileum was opened longitudinally and rinsed in cold PBS, cut into 5-mm pieces, and washed twice with 2 mM EDTA (Thermo Fisher Scientific) and 1 mM DTT (Thermo Fisher Scientific) in serum-free RPMI (MilliporeSigma) for 15 minutes at 37°C in a shaker at 150 rpm. Tissues were then washed with PBS supplemented with 3% FBS (MilliporeSigma) in a 100-μm cell strainer. Tissue was then cut into 1- to 3-mm pieces and incubated in a solution containing RPMI, 2% FBS, 3.7 U Liberase (Roche Diagnostics), and 400 U DNase I (MilliporeSigma) for 15 minutes in an orbital shaker under the same conditions as above. Tissues were subsequently serially filtered through a 70-μm and a 40-μm cell strainer using ice-cold PBS, and LPLs were isolated after centrifugation on a Percoll gradient (GE Healthcare).
Intestinal crypt isolation and single-cell dissociation.
Isolation of mouse intestinal crypts and dissociation of cells for analysis by flow cytometry were performed as previously described (35). Briefly, 5 cm proximal duodenum was opened longitudinally, rinsed in cold PBS, and cut into 5-mm pieces. To detach the crypts, small intestine pieces were incubated on ice in 10 mM EDTA (Thermo Fisher Scientific) for 30 minutes. Intestine was then resuspended vigorously in PBS, leading to supernatant enriched in crypts, which was subsequently filtered through a 70-μm cell strainer. Following centrifugation of isolated crypts, the pellet was disaggregated into single cells by incubation for 5 minutes with prewarmed (37°C) 1× TrypLE Express (Gibco, Life Technologies) supplemented with 10 μM ROCK inhibitor Y-27632 dihydrochloride (Thermo Fisher Scientific) and 0.5 mM N-acetylcysteine (MilliporeSigma).
Flow cytometry.
After isolation, LPLs were further incubated with Cell Stimulation Cocktail (eBioscience) for 4 hours, and 1 × 106 cells were subsequently surface-stained with CD4, CD8, and TCRβ antibodies. For detection of polarized T cell subsets, cells were incubated overnight at 4°C in 400 μl of Fix/Perm solution (BD Biosciences), and then stained in Perm/Wash buffer (BD Biosciences) with IFN-γ, IL-10, and IL-17A antibodies. For detection of Tregs, surface-stained LPLs were stained with FoxP3 antibody using the FoxP3/Transcription Factor Staining Buffer Set (eBioscience). Isolated, dissociated intestinal crypt cells were surface-stained with EpCAM, CD45, CD31, TER-119, and CD24 antibodies. Apoptotic intestinal crypt cells were detected using the Annexin V Apoptosis Detection Kit (eBioscience). DAPI, 7-aminoactinomycin D, and Fixable Live/Dead Cell Stain Kits (Invitrogen) were used for viability staining. All flow cytometry was performed on a FACSCanto or LSR Fortessa, using FACSDiva software (all BD Biosciences). Data were analyzed using FlowJo software (Tree Star). Full details of all antibodies used are in Supplemental Table 1.
16S sequencing.
DNA from fecal pellets of individual mice were frozen until DNA was extracted by mechanical disruption (bead-beating) in phenol-chloroform. The V4–V5 region of the ribosomal RNA gene was amplified, sequenced, and computationally analyzed as described (36, 37).
Statistics.
Statistical analysis was performed and graphs were generated using Prism software (GraphPad). Survival curves were plotted using Kaplan-Meier estimates and compared using the log-rank test. For survival experiments, a sample size of 7 per group provided 80% power to detect survival benefit of at least 40% with an α of 0.05. For comparison of 2 groups, an unpaired 2-tailed t test for parametric data was used. In the case of multiple comparisons, 1-way ANOVA with Bonferroni correction was used. Principal component analysis was performed using the R package, and Bray-Curtis compositional distances were compared using a Wilcoxon test. All data were tested for normality through application of the F test with Prism software, and all tests were 2-sided. Differences were considered significant when the P value was less than 0.05. Data are always presented as mean and SEM for the different groups. Unless otherwise specified, all studies for which data are presented are representative of at least 2 independent experiments.
Study approval.
Study of human blood samples was approved by the Icahn School of Medicine at Mount Sinai (IRB 15-0093). The mouse research project was approved by the Icahn School of Medicine at Mount Sinai (IACUC 2014-0202).
Author contributions
DZ, YHK, and JLMF designed the study. DZ, YHK, SJ, JKG, MSC, MH, ERA, MRMVDB, JUP, ALCG, AES, MJD, ACH, JEL, UO, TSS, AVH, TCL, and PR performed the experiments and analysis. LVH provided key reagents. DZ, MSC, and JLMF prepared the manuscript with input from all authors.
Supplementary Material
Acknowledgments
The authors thank the physicians and data management teams of the BMT programs at the University of Michigan and the University of Regensburg. This work was supported by NIH grants P01-CA039542 (to JLMF), 5T32AI078892 (to ERA), and R01-CA228358 and R01-CA228308 (to MRMVDB), the American Cancer Society (CRP-13-306-06-COUN to JLMF), and Deutscher Akademischer Austauschdienst (to MH).
Version 1. 08/14/2018
In-Press Preview
Version 2. 09/24/2018
Electronic publication
Version 3. 11/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(11):4970–4979.https://doi.org/10.1172/JCI99261.
Contributor Information
Dongchang Zhao, Email: dongchang.zhao@mssm.edu.
Yeung-Hyen Kim, Email: yhkim61@gmail.com.
Seihwan Jeong, Email: seihwan.jeong@mssm.edu.
Joel K. Greenson, Email: facjkgmd@med.umich.edu.
Mohammed S. Chaudhry, Email: mohammed.chaudhry@mssm.edu.
Matthias Hoepting, Email: Matthias.Hoepting@klinik.uni-regensburg.de.
Erik R. Anderson, Email: adahn1986@gmail.com.
Marcel R.M. van den Brink, Email: vandenbm@mskcc.org.
Jonathan U. Peled, Email: peledj@mskcc.org.
Antonio L.C. Gomes, Email: gomesa@mskcc.org.
Ann E. Slingerland, Email: slingera@mskcc.org.
Michael J. Donovan, Email: michael.donovan@mssm.edu.
Andrew C. Harris, Email: Andrew.Harris@hsc.utah.edu.
John E. Levine, Email: john.levine@mssm.edu.
Lora V. Hooper, Email: lora.hooper@utsouthwestern.edu.
Thaddeus S. Stappenbeck, Email: stappenb@pathology.wustl.edu.
Ta-Chiang Liu, Email: tliu@path.wustl.edu.
Pavan Reddy, Email: reddypr@umich.edu.
James L.M. Ferrara, Email: james.ferrara@mssm.edu.
References
- 1.Billingham RE. The biology of graft-versus-host reactions. Harvey Lect. 1966;62:21–78. [PubMed] [Google Scholar]
- 2.Gatti RA, Kersey JH, Yunis EJ, Good RA. Graft-versus-host disease. Prog Clin Pathol. 1973;5:1–18. [PubMed] [Google Scholar]
- 3.Weisdorf DJ, et al. Acute upper gastrointestinal graft-versus-host disease: clinical significance and response to immunosuppressive therapy. Blood. 1990;76(3):624–629. [PubMed] [Google Scholar]
- 4.Przepiorka D, et al. Risk factors for acute graft-versus-host disease after allogeneic blood stem cell transplantation. Blood. 1999;94(4):1465–1470. [PubMed] [Google Scholar]
- 5.Verneris MR, et al. HLA mismatch is associated with worse outcomes after unrelated donor reduced-intensity conditioning hematopoietic cell transplantation: an analysis from the Center for International Blood and Marrow Transplant Research. Biol Blood Marrow Transplant. 2015;21(10):1783–1789. doi: 10.1016/j.bbmt.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pidala J, et al. Amino acid substitution at peptide-binding pockets of HLA class I molecules increases risk of severe acute GVHD and mortality. Blood. 2013;122(22):3651–3658. doi: 10.1182/blood-2013-05-501510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westin JR, et al. Steroid-refractory acute GVHD: predictors and outcomes. Adv Hematol. 2011;2011:601953. doi: 10.1155/2011/601953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castilla-Llorente C, et al. Prognostic factors and outcomes of severe gastrointestinal GVHD after allogeneic hematopoietic cell transplantation. Bone Marrow Transplant. 2014;49(7):966–971. doi: 10.1038/bmt.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill GR, Ferrara JL. The primacy of the gastrointestinal tract as a target organ of acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood. 2000;95(9):2754–2759. [PubMed] [Google Scholar]
- 10.MacMillan ML, DeFor TE, Weisdorf DJ. What predicts high risk acute graft-versus-host disease (GVHD) at onset?: identification of those at highest risk by a novel acute GVHD risk score. Br J Haematol. 2012;157(6):732–741. doi: 10.1111/j.1365-2141.2012.09114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrara JL, et al. Regenerating islet-derived 3-α is a biomarker of gastrointestinal graft-versus-host disease. Blood. 2011;118(25):6702–6708. doi: 10.1182/blood-2011-08-375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine JE, et al. Low Paneth cell numbers at onset of gastrointestinal graft-versus-host disease identify patients at high risk for nonrelapse mortality. Blood. 2013;122(8):1505–1509. doi: 10.1182/blood-2013-02-485813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14(3):282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 14.Sanos SL, et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10(1):83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanash AM, et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 2012;37(2):339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Major-Monfried H, et al. MAGIC biomarkers predict long-term outcomes for steroid-resistant acute GVHD. Blood. 2018;131(25):2846–2855. doi: 10.1182/blood-2018-01-822957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couturier M, et al. IL-22 deficiency in donor T cells attenuates murine acute graft-versus-host disease mortality while sparing the graft-versus-leukemia effect. Leukemia. 2013;27(7):1527–1537. doi: 10.1038/leu.2013.39. [DOI] [PubMed] [Google Scholar]
- 18.Zhao K, et al. Interleukin-22 aggravates murine acute graft-versus-host disease by expanding effector T cell and reducing regulatory T cell. J Interferon Cytokine Res. 2014;34(9):707–715. doi: 10.1089/jir.2013.0099. [DOI] [PubMed] [Google Scholar]
- 19.Eriguchi Y, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of α-defensins. Blood. 2012;120(1):223–231. doi: 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 20.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 21.Lindemans CA, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528(7583):560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184(1):39–51. doi: 10.1016/0022-1759(95)00072-I. [DOI] [PubMed] [Google Scholar]
- 23.Ganz T. Paneth cells — guardians of the gut cell hatchery. Nat Immunol. 2000;1(2):99–100. doi: 10.1038/77884. [DOI] [PubMed] [Google Scholar]
- 24.Jenq RR, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209(5):903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber D, et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood. 2015;126(14):1723–1728. doi: 10.1182/blood-2015-04-638858. [DOI] [PubMed] [Google Scholar]
- 26.Vaishnava S, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeiser R, Blazar BR. Acute graft-versus-host disease — biologic process, prevention, and therapy. N Engl J Med. 2017;377(22):2167–2179. doi: 10.1056/NEJMra1609337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malka D, et al. Tumor necrosis factor α triggers antiapoptotic mechanisms in rat pancreatic cells through pancreatitis-associated protein I activation. Gastroenterology. 2000;119(3):816–828. doi: 10.1053/gast.2000.16491. [DOI] [PubMed] [Google Scholar]
- 29.Simon MT, et al. HIP/PAP stimulates liver regeneration after partial hepatectomy and combines mitogenic and anti-apoptotic functions through the PKA signaling pathway. FASEB J. 2003;17(11):1441–1450. doi: 10.1096/fj.02-1013com. [DOI] [PubMed] [Google Scholar]
- 30.Hayase E, et al. R-Spondin1 expands Paneth cells and prevents dysbiosis induced by graft-versus-host disease. J Exp Med. 2017;214(12):3507–3518. doi: 10.1084/jem.20170418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki N, et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc Natl Acad Sci U S A. 2016;113(37):E5399–E5407. doi: 10.1073/pnas.1607327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teshima T, et al. Acute graft-versus-host disease does not require alloantigen expression on host epithelium. Nat Med. 2002;8(6):575–581. doi: 10.1038/nm0602-575. [DOI] [PubMed] [Google Scholar]
- 33.Reddy P, Maeda Y, Liu C, Krijanovski OI, Korngold R, Ferrara JL. A crucial role for antigen-presenting cells and alloantigen expression in graft-versus-leukemia responses. Nat Med. 2005;11(11):1244–1249. doi: 10.1038/nm1309. [DOI] [PubMed] [Google Scholar]
- 34.Cooke KR, et al. LPS antagonism reduces graft-versus-host disease and preserves graft-versus-leukemia activity after experimental bone marrow transplantation. J Clin Invest. 2001;107(12):1581–1589. doi: 10.1172/JCI12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staffas A, et al. Nutritional support from the intestinal microbiota improves hematopoietic reconstitution after bone marrow transplantation in mice. Cell Host Microbe. 2018;23(4):447–457.e4. doi: 10.1016/j.chom.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shono Y, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med. 2016;8(339):339ra71. doi: 10.1126/scitranslmed.aaf2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.