Abstract
First-generation immune checkpoint inhibitors, including anti–CTLA-4 and anti–programmed death 1 (anti–PD-1) antibodies, have led to major clinical progress, yet resistance frequently leads to treatment failure. Thus, new targets acting on T cells are needed. CD33-related sialic acid–binding immunoglobulin-like lectins (Siglecs) are pattern-recognition immune receptors binding to a range of sialoglycan ligands, which appear to function as self-associated molecular patterns (SAMPs) that suppress autoimmune responses. Siglecs are expressed at very low levels on normal T cells, and these receptors were not until recently considered as interesting targets on T cells for cancer immunotherapy. Here, we show an upregulation of Siglecs, including Siglec-9, on tumor-infiltrating T cells from non–small cell lung cancer (NSCLC), colorectal, and ovarian cancer patients. Siglec-9–expressing T cells coexpressed several inhibitory receptors, including PD-1. Targeting of the sialoglycan-SAMP/Siglec pathway in vitro and in vivo resulted in increased anticancer immunity. T cell expression of Siglec-9 in NSCLC patients correlated with reduced survival, and Siglec-9 polymorphisms showed association with the risk of developing lung and colorectal cancer. Our data identify the sialoglycan-SAMP/Siglec pathway as a potential target for improving T cell activation for immunotherapy.
Keywords: Immunology, Oncology
Keywords: Cancer immunotherapy, T cells
Introduction
The landscape of oncological practice has substantially changed with the introduction of immune checkpoint inhibitors that target inhibitory receptors on the surface of CD8+ T cells (1). In particular, blocking of the inhibitory receptor programmed death 1 (PD-1) or its ligand PD-L1 has been shown to induce durable responses across many different cancer types, including non–small cell lung cancer (NSCLC), melanoma, squamous carcinoma of the head and neck, kidney cancer, and ovarian cancer (1, 2). However, most patients have primary resistance to therapeutic PD-1 or PD-L1 blockade or develop secondary resistance (2). Thus, identification of additional pathways mediating suppression of tumor-specific T cells is clearly needed to increase therapeutic efficacy of PD-1– or PD-L1–targeting strategies.
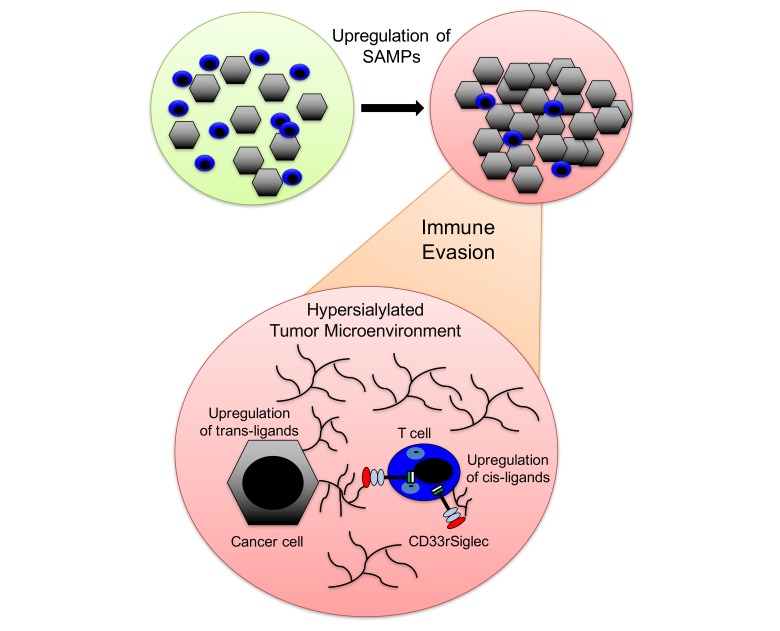
Pattern recognition receptors (PRRs), such as Toll-like receptors or nucleotide-binding oligomerization domain–like (NOD-like) receptors, detect pathogens or danger signals, and engagement of these PRRs by pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) activates the respective immune cells (3–6). Receptors that bind to PAMPs and DAMPs have also been implicated in cancer progression and anticancer immunity (6, 7). In addition to these well-described pathways, sialoglycans have been shown to serve as a self-associated molecular patterns (SAMPs) by binding to CD33-related sialic acid–binding immunoglobulin-like lectins (CD33rSiglecs) (8, 9), a family of receptors on immune cells containing intracellular immunoreceptor tyrosine-based inhibitory motifs (ITIM) (10–13). Unlike the conserved Siglecs (Siglec-1, Siglec-2, Siglec-4, and Siglec-15), CD33rSiglecs, including Siglec-3 (CD33), Siglec-5, Siglec-6, Siglec-7, Siglec-8, Siglec-9, Siglec-10, Siglec-11, Siglec-14, and Siglec-16, have evolved rapidly due to multiple selection forces, including evolutionary pressure by pathogens that can mimic sialoglycan-SAMPs (Sia-SAMPs) and bind to inhibitory CD33rSiglecs to escape innate immune control (14–18). The different Siglecs have different binding preferences for sialoglycan ligands (9). While Siglec-8 has a more defined spectrum of ligands, which include 6′-sulfo-sialyl Lewis x (19, 20), Siglec-9 has a much broader binding spectrum of sialylated ligands (9, 21). Siglec-9 is therefore a typical PRR for Sia-SAMPs.
Recent evidence suggests that the upregulation of Sia-SAMPs can facilitate evasion of immune control during cancer progression by engaging inhibitory CD33rSiglecs, such as Siglec-7 and Siglec-9 (21–26). Engagement of Siglec-7 and Siglec-9 inhibits NK cell–mediated tumor cell killing in vitro (22, 23). Siglec-9 and its murine functional paralog Siglec-E have also been implicated in myeloid cell–mediated cancer progression (21, 24). Binding of the cancer-associated sialylated glycoform of MUC1 to Siglec-9 on macrophages can induce a tumor-associated macrophage (TAM) phenotype that can promote cancer progression and immune evasion (25).
In chronic infections and cancer, expression of checkpoint molecules, including PD-1, CTLA-4, TIM-3, and LAG-3, is observed on T cells and is associated with a state of T cell dysfunction termed T cell exhaustion (27). Chronic infections, such as HIV, have also been shown to upregulate inhibitory CD33rSiglecs on peripheral CD4+ T cells, which suggests that inhibitory Siglec receptors might be involved in T cell immunoregulation (28). In contrast, previous analyses have found very low expression of CD33rSiglecs on peripheral T cells of healthy humans (29) and mice (30). We therefore sought to characterize Siglec expression on tumor-infiltrating T cells and to test the functional role of Siglecs as new potential immune checkpoints that are regulated by glycans within the tumor microenvironment.
Results
Siglec-9 is upregulated on tumor-infiltrating T cells.
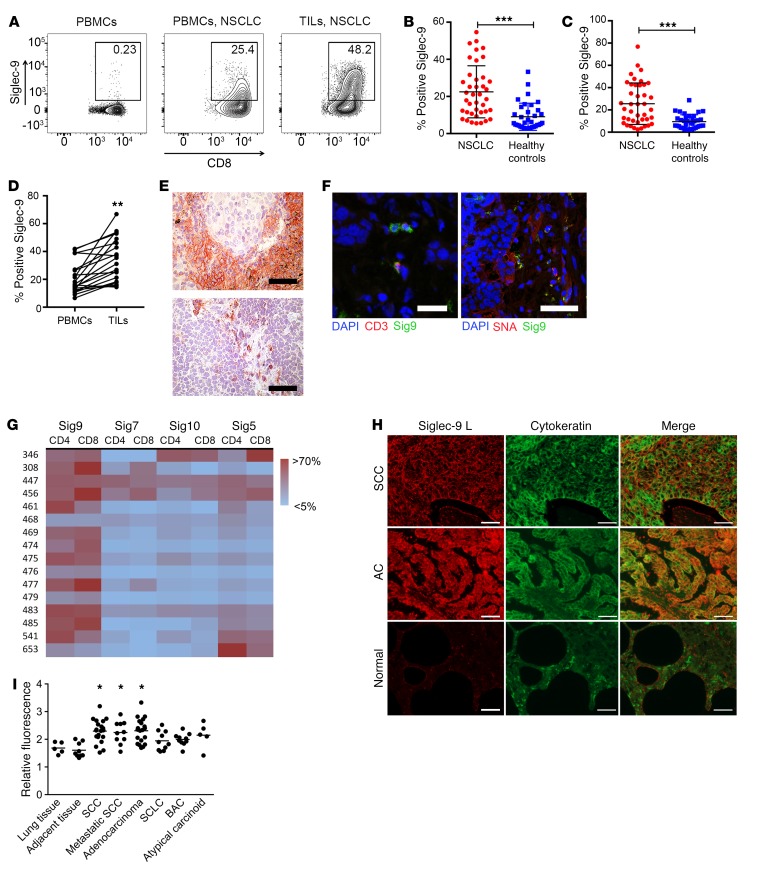
We analyzed the expression of inhibitory CD33rSiglecs on immune cells in primary NSCLC samples (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI120612DS1). As previously described, CD33rSiglecs, including Siglec-7 and Siglec-9, were expressed on NK cells and Siglec-9 was also expressed on myeloid cells (Supplemental Figure 1B). Several inhibitory CD33rSiglecs, including Siglec-3 (CD33), Siglec-5, Siglec-7, Siglec-10, and Siglec-9, were expressed on a significant proportion of tumor-infiltrating lymphocytes (TILs), including CD4+ and CD8+ T cells (Figure 1, A–G, and Supplemental Figure 1, C–F). Siglec-9 was the most prominently and consistently expressed CD33rSiglec on TILs across patients (Figure 1G). Lower levels of Siglec-9 were observed on peripheral blood T cells from healthy donors (Figure 1, A–C) or in spleens from patients undergoing splenectomy for nonmalignant disease (Supplemental Figure 1G). There were a few healthy donors that had a larger population of Siglec-9–expressing CD8+ T cells (Figure 1C). In order to further understand the heterogeneity of Siglec-9 expression, we further stained for Siglec-9 in an additional and better-characterized population of 49 healthy donors (Supplemental Figure 1H). The distribution of Siglec-9 on CD8+ T cells was similar in both cohorts, and no correlation with age could be seen (Supplemental Figure 1I). No significant difference was seen between sexes (9% in men, 7.2% in women Sig9+CD8+ T cells of total CD8+ T cells). We found an inverse correlation of Sig9+CD8+ with naive CCR7+CD45RA+ T cells (Supplemental Figure 1J). Immunostaining of intratumoral leukocytes showed that Siglec-9 was typically expressed on small lymphocytes and macrophages (Figure 1E). Double staining showed that Siglec-9–positive TILs in NSCLC samples also expressed CD3 (Figure 1F). We also found an upregulation of Siglec-9 on colorectal cancer (CRC) and epithelial ovarian cancer (EOC) TILs or pleural effusions (Supplemental Figure 1K).
Figure 1. Siglec-9 is upregulated on CD8+ TILs.
(A) Representative flow cytometry analysis of Siglec-9 expression on CD8+ T cells in PBMCs from healthy donors (left panel), PBMCs from NSCLC patients (middle panel), and TILs from a matched NSCLC patient (right panel). (B and C) Quantification of Siglec-9 expression on CD4+ (B) and CD8+ (C) TILs from NSCLC patients (PBMCs, n = 36; NSCLC, n = 41; mean ± SD). Statistical analysis by unpaired Student’s t test. (D) Paired analysis of CD8+ T cells from the peripheral blood and tumors of NSCLC patients (n = 20). Statistical analysis by paired Student’s t test. (E) Immunohistochemical staining of Siglec-9–positive cells in NSCLC sections. Scale bars: 50 μm. (F) Representative immunofluorescence analysis of CD3 and Siglec-9 double-positive cells and Siglec-9 staining or SNA staining and Siglec-9 staining (right panel). Scale bars: 30 μm (left panel); 50 μm (right panel).(G) Heatmap of expression analysis of different Siglecs in NSCLC samples on CD4+ and CD8+ TILs. (H and I) Immunofluorescence study in paraffin-embedded tissue microarrays using recombinant Siglec-9–Fc (human IgG1) fusion protein coupled to secondary PE-conjugated (Fab′)2 goat anti-human Fc antibody. Representative images (H) and Siglec ligands quantification summary (I) are shown. Original magnification, ×400. Scale bars: 50 μm. Fluorescence values were normalized against an IgG1 isotype control. Lung tissue, n = 5; adjacent lung tissue, n = 9; squamous cell carcinoma (SCC), n = 20; adenocarcinoma, n = 20; small cell lung cancer (SCLC), n = 10; broncho-alveolar carcinoma (BAC), n = 10; atypical carcinoid, n = 5. Data are shown as mean ± SEM. Statistical analysis performed by 1-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001.
There was a significant increase of Sia-SAMPs (Siglec-7 and Siglec-9 ligands) in lung carcinomas compared with healthy lung tissue as well as in acute and chronic inflammatory diseases, as assessed with Siglec-Fc proteins (Figure 1, H and I). Ligands were strongly present on cytokeratin-positive cells, suggesting they were in a trans position, and a trend toward an increase in ligands at higher stages was observed (Supplemental Figure 1L). As Siglec-9 was the most prominent and consistent inhibitory CD33rSiglec upregulated on TILs across different patients, we focused our further analysis on Siglec-9.
Characterization of Siglec-9–expressing intratumoral CD8+ T cells.
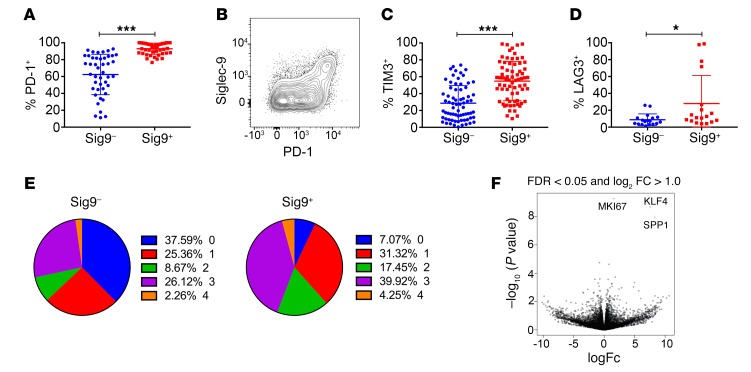
We aimed to further characterize the Sig9+CD8+ TILs in samples from NSCLC patients by multicolor flow cytometry. Sig9+CD8+ TILs coexpressed several other inhibitory receptors, including PD-1 in particular and also TIM-3 and LAG-3 (Figure 2, A–D, and Supplemental Figure 2, A–D). Most Sig9+CD8+ TILs were found within the population with the highest PD-1 expression (PD-1hi, Supplemental Figure 2A). However, not all the PD-1hiCD8+ T cells expressed Siglec-9 (Supplemental Figure 2B), suggesting that Sig9+CD8+ TILs are a subpopulation of tumor-specific PD-1hi TILs. Sig9+CD8+ T cells also expressed high levels of the transcription factor Eomesodermin (Eomeshi) and low levels of T-bet (T-betlo, Supplemental Figure 2E). In general, Sig9+CD8+ TILs had more inhibitory receptors upregulated and coexpressed than Sig9–CD8+ TILs from NSCLC patients (Figure 2E). Several costimulatory receptors were also enriched on Sig9+CD8+ TILs as compared with Sig9–CD8+ TILs (Supplemental Figure 2, F–J). RNA sequencing (RNA-seq) revealed that several genes were differentially regulated between Sig9+CD8+ TILs and Sig9–CD8+ TILs (Supplemental Figure 2K). The 3 main genes upregulated in Sig9+CD8+ TILs were SPP1 (osteopontin), Ki67, and KLF4 (Figure 2F). We also looked for the expression of genes involved in the generation of Sia-SAMPs. Expression of the rate-limiting enzyme for sialic acid biosynthesis, UDP-N-acetylglucosamine-2-epimerase (GNE), was higher in Sig9+CD8+ TILs than in Sig9–CD8+ TILs (Supplemental Figure 2L). Consequently, an upregulation of sialoglycans, as shown by increased staining with Sambucus nigra lectin (SNA) (Supplemental Figure 2M), and an upregulation of cis ligands on TILs, as shown by increased staining with Siglec-9-Fc protein, occurred (Supplemental Figure 2N). Protein levels of osteopontin (SPP1) ( Supplemental Figure 2, O and P) and Ki-67 (Supplemental Figure 2Q) were also significantly higher in Sig9+CD8+ TILs than in Sig9–CD8+ TILs (Supplemental Figure 3 shows the gating strategy for sorting of primary T cells from tumor samples).
Figure 2. Sig9+CD8+ TILs coexpress inhibitory receptors.
(A and B) Expression of PD-1 in primary NSCLC samples on Sig9–CD8+ (Sig9–) or Sig9+CD8+ (Sig9+) TILs (A, n = 44) and representative FACS analysis (B). Statistical analysis by paired Student’s t test. (C and D) Expression of TIM-3 (C, n = 71) and LAG-3 (D, n = 18) on Sig9–CD8+ or Sig9+CD8+ TILs from NSCLC samples. Statistical analysis by paired Student’s t test. (E) Analysis of the number of coexpressed inhibitory receptors on Sig9–CD8+ or Sig9+CD8+ TILs. (F) Volcano plot of RNA-seq on sorted TILs according to their Siglec-9 expression. The 3 significantly differentially expressed genes were MIK67 (Ki67), KLF4, and SPP1. *P < 0.05; ***P < 0.001. Data are presented as mean ± SD.
Sig9+ TILs are a distinct subset within the CD8+ T cell population.
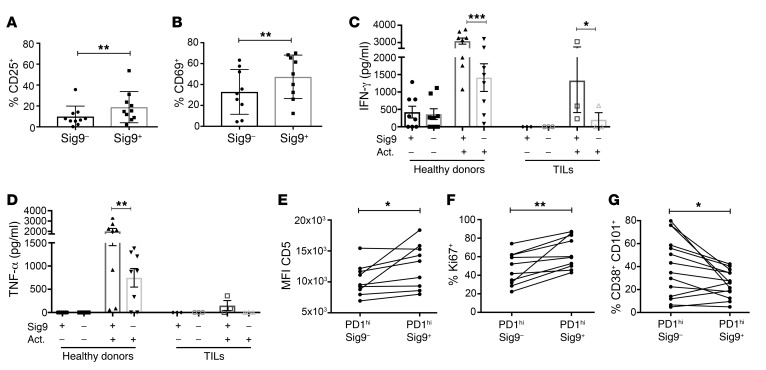
We further tested to determine whether Siglecs are upregulated by activation of T cells. While Siglec-5 expression increased significantly on polyclonally activated CD8+ T cells from healthy donors, Siglec-9 was only slightly upregulated and Siglec-7 expression was unchanged (Supplemental Figure 4A). Antigen-specific stimulation of T cell clones with reactivity against peptides from influenza antigens, CMV antigens, and melanA led to a slight upregulation of Siglec-9 (Supplemental Figure 4B). Sig9+CD8+ TILs from NSCLC samples activated with anti-CD3/anti-CD28 antibodies expressed significantly higher surface levels of CD25 (Figure 3A), CD69 (Figure 3B), and the newly identified activation marker Siglec-5 (Supplemental Figure 4C) compared with similarly activated Sig9–CD8+ TILs. Similar results were seen for polyclonally activated peripheral Sig9+CD8+ cells from healthy donors (Supplemental Figure 4, D–G). CD8+ T cells from NSCLC samples were clearly dysfunctional or exhausted, as demonstrated by reduced production of cytokines such as IFN-γ and TNF-α compared with that of CD8+ T cells from healthy donor peripheral blood mononuclear cells (PBMCs) (Figure 3, C and D). However, restimulation of TILs from NSCLC patients with anti-CD3/anti-CD28 antibodies showed that Sig9+CD8+ TILs were much easier to restimulate and secrete at higher levels of multiple cytokines, including IFN-γ, than Sig9–CD8+ TILs (Figure 3, C and D, Supplemental Figure 5, A–N). In addition, Sig9+CD8+ cells expressed a distinct pattern of chemokine receptors, including CXCR3, CXCR5, CCR4, CCR6, and CX3CR1 (Supplemental Figure 5, O–T). A recent study also found a subpopulation of PD-1hiCD8+ TILs with a distinct, higher functionality (31); this subpopulation was associated with an increased expression of CD5 and a lower expression of CD38 and CD101 (31). We therefore stained for these markers and found an enrichment of CD5 and Ki67 expression (Figure 3, E and F) on Sig9+ cells in the PD-1hi gate as well as a reduced presence of CD38+ and CD101+ (Figure 3G, Supplemental Figure 6). Our data provide evidence that intratumoral Sig9+CD8+ cells are a distinct subpopulation of tumor-specific CD8+ TILs.
Figure 3. Sig9+CD8+ TILs are a distinct subset within intratumoral CD8+ T cells.
(A and B) Upregulation of the activation markers CD25 (A) and CD69 (B) on Sig9–CD8+ or Sig9+ TILs sorted from primary NSCLC samples and activated with anti-CD3/28 antibodies for 48 hours (n = 9). Statistical analysis by paired Student’s t test. (C) ELISA analysis of IFN-γ in the supernatant of sorted Sig9–CD8+ T cells or Sig9+CD8+ T cells (n = 3–7, independent patients). Cells were sorted from PBMCs of healthy donors or primary NSCLC samples (TILs). Act, activated. Supernatants from 50,000 cells were analyzed. (D) Analysis of TNF-α in the supernatant of sorted Sig9–CD8+ or Sig9+CD8+ cells from healthy donors or NSCLC patient samples (n = 3–7, independent donors/patients). Statistical analysis performed by 1-way ANOVA. (E) Expression level of CD5 in the CD8+PD-1hi population on Sig9– TILs and Sig9+ TILs (n = 9). (F) Percentage of Sig9–CD8+ TILs or Sig9+CD8+ TILs in primary NSCLC samples that express Ki67 within the PD-1hi population (n = 9). (G) Frequency of CD38hiCD101hi cells on Sig9– and Sig9+CD8+PD-1hi TILs determined by flow cytometric analysis (n = 13). Statistical analysis by paired Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001. Data are presented as mean ± SD.
Sia-SAMP/Siglec-9 interaction is a target for improving T cell activation.
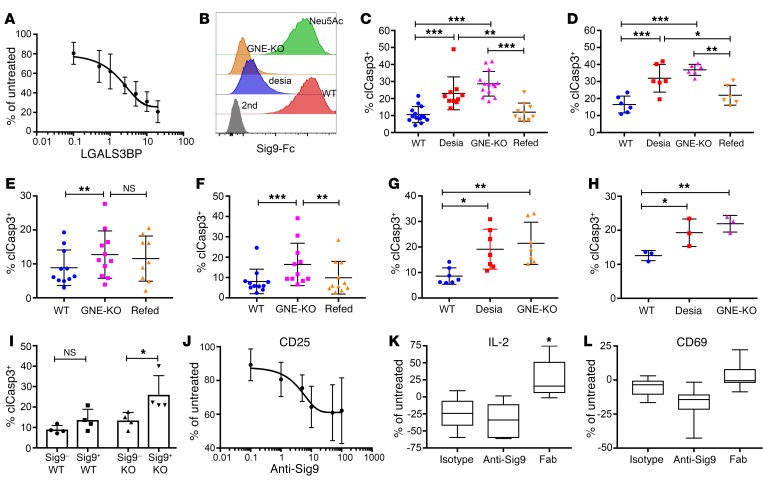
Next, we wanted to explore the functional implication of Siglec expression on T cells. We used the high-affinity CD33rSiglec ligand LGALS3BP, which is upregulated in NSCLC (24). LGALS3BP inhibited CD8+ T cell activation in a sialic acid–dependent fashion (Figure 4A, Supplemental Figure 7, A and B). We further generated cell lines that expressed lower surface levels of Sia-SAMPs by knocking out GNE. GNE-deficient A549 lung adenocarcinoma cells showed lower binding to Siglec-9 compared with WT A549 cells (Figure 4B). Feeding GNE-KO A549 cells with N-acetylneuraminic acid (Neu5Ac) to metabolically bypass the mutation led to a recovery of ligands on the cell surface and binding to Siglec-9 (Figure 4B). CD8+ T cell–mediated tumor cell killing was tested by coincubation of T cells from healthy donors and different A549 tumor cell variants in the presence of a CD3/EpCAM T cell–bispecific (TCB) antibody (catumaxomab). T cell–mediated killing of GNE-KO A549 cells and desialylated A549 cells was higher compared with that of WT A549 cells and GNE-KO A549 cells fed with Neu5Ac (Figure 4C). Similar results were obtained with catumaxomab when using HT29 tumor cells (Supplemental Figure 7C and Figure 4D) and TILs obtained from primary NSCLC samples together with A549 and HT29 cells (Figure 4, E and F). Experiments with the anti-CD3/CD19 TCB antibody blinatumomab and CD19+ Ramos cells (Figure 4G and Supplemental Figure 7D) or primary CLL samples (Figure 4H) showed similar results. Sig9+CD8+ T cells from NSCLC TILs were significantly more reactive toward GNE-KO A549 cells than Sig9– TILs, with the difference being sialic acid dependent (Figure 4I). Similarly, Sig9+CD8+ T cells induced more apoptosis in HT-29 and Ramos cells, respectively, than Sig9–CD8+ T cells (Supplemental Figure 7, E and F).
Figure 4. Sia-SAMPs inhibit T cell–mediated tumor cell killing in vitro.
(A) Inhibition of T cell activation by LGALS3BP in a dose-dependent manner measured by intracellular IFN-γ by flow cytometry. CD8+ T cells from healthy donors were activated with anti-CD3 and anti-CD28 antibodies in the presence of increasing amounts of LGALS3BP (μg/ml, n = 3). (B) Representative histograms of binding of Sig9-Fc to A549 WT cells, enzymatically desialylated A549 cells (desia), GNE-deficient A549 cells (GNE-KO), and GNE-KO A549 cells refed with 10 mM Neu5Ac. (C) Percentage of cleaved caspase-3–positive (clCasp3+) WT A549 cells, desialylated A549 cells, GNE-KO A549 cells, or GNE-KO A549 cells fed with Neu5Ac (refed) after incubation with CD8+ T cells and catumaxomab (n = 10). (D) Apoptosis of WT, desialylated, GNE-KO, and refed GNE-KO HT-29 cells measured by upregulation of cleaved caspase-3 in tumor cells (n = 6). (E and F) clCaps3+ A549 (E, n = 11) or HT-29 (F, n = 11) tumor cells after coincubation with TILs from NSCLC or CRC samples. (G) CD8+ T cells were sorted according to their Siglec-9 expression and incubated with either WT or GNE-KO A549 cells (n = 4). (H) CD19+ RAMOS cells were incubated with CD8+ T cells from healthy donors in the presence of CD3 and CD19 bispecific antibody blinatumomab (n = 7). (I) GNE-KO RAMOS cells incubated with CD8+ T cells from patients with chronic lymphocytic leukemia (n = 3). (J) Activation measured by CD25 on CD8+ T cells treated with anti-CD3 and anti-CD28 antibodies in the presence of anti–Siglec-9 antibody (clone 191240, g/ml, n = 4). (K) Relative IL-2 production of NSCLC primary tumor samples incubated with SEB and Siglec-9–blocking antibody and the Fab fragments (clone 191240, n = 5). (L) Measurement of CD69 upregulation on CD8+ TILs from NSCLC patients upon incubation with SEB in the presence of antibodies or Fab fragments (n = 5). Statistical analyses in this figure were performed by 1-way ANOVA. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
To test the effect of Siglec-9 blockade on T cell activation in vitro, we used a Siglec-9 antibody and the previously described staphylococcal enterotoxin B (SEB) test (32). Two full IgG Siglec-9 antibodies (191240 and E10-286) inhibited T cell activation and therefore were agonistic (Supplemental Figure 7, G and H). Moreover, Siglec-9 antibodies led to a dose-dependent inhibition of T cell activation tested by upregulation of the activation markers CD25 (Figure 4J) and CD69 (Supplemental Figure 7I). Addition of the Siglec-9 antibody (clone 191240) inhibited IL-2 production and CD69 expression on the surface of TILs upon SEB stimulation of primary NSCLC samples (Figure 4, K and L). We hypothesized that bivalent binding and subsequent dimerization of Siglec-9 might be responsible for the agonistic effect on Siglec-9 by the clone 191240. Indeed, monovalent carbohydrate-recognition domain–blocking (CRD-blocking) Fab fragments from clone 191240 (Supplemental Figure 7J) significantly increased IL-2 (Figure 4K), but not CD69 (Figure 4L), production upon SEB stimulation. These findings demonstrate that T cell activation in NSCLC tumors can be increased by targeting Sia-SAMPs/Siglec-9 interactions.
Siglec-E is expressed on TILs in mice.
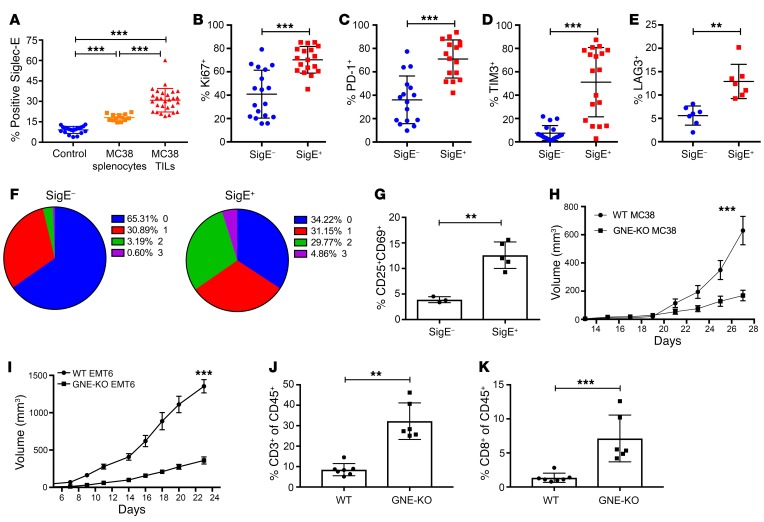
We further investigated whether the murine functionally equivalent paralog (Siglec-E) was upregulated on TILs in mouse models. We found a significant upregulation of Siglec-E on murine CD8+ TILs when compared with splenocytes from control mice and tumor-bearing mice in the MC38 subcutaneous tumor model (Figure 5A) and other models (LLC, B16, and EMT6 subcutaneous tumor models; Supplemental Figure 8A). While other inhibitory CD33rSiglecs, including Siglec-F and Siglec-G, were also upregulated on the surface of murine TILs, Siglec-E was the most prominently upregulated inhibitory Siglec tested (Supplemental Figure 8B). We further analyzed the phenotype of SigE+CD8+ murine TILs. The proliferation marker Ki67, the activation markers granzyme B and CD69, and the activating receptor CD27 (Figure 5B, and Supplemental Figure 8, C–E) were all significantly increased on murine SigE+CD8+ TILs compared with SigE–CD8+ TILs, similarly to what is seen in human TIL samples. The expression and number of murine inhibitory receptors, including PD-1, were also higher on SigE+CD8+ TILs than on SigE–CD8+ TILs (Figure 5, C–F, and Supplemental Figure 8, F and G). SigE+CD8+ TILs were predominantly terminally differentiated Eomeshi, Tbetlo, and CD127+ T cells (Supplemental Figure 8H). Sorted murine SigE+CD8+ TILs again showed a stronger reactivation than SigE–CD8+ TILs, as shown by upregulation of the activation markers CD25 and CD69 (Figure 5G). Taken together, these findings suggest that SigE+CD8+ TILs in mice are phenotypically similar to Sig9+CD8+ TILs in humans.
Figure 5. Sialylated SAMPs enhance immune escape and tumor growth in vivo.
(A) Siglec-E expression was determined by flow cytometry on control splenocytes, splenocytes from tumor-bearing mice, and CD8+ TILs from subcutaneous MC38 tumors (n = 25–28). Statistical analysis performed by 1-way ANOVA. (B) Expression of intracellular Ki67 was examined by flow cytometry on SigE–CD8+ and SigE+CD8+ TILs (n = 18). Statistical analysis by paired Student’s t test. (C–E) Frequencies of inhibitory immune receptor expression on SigE–CD8+ and SigE+CD8+ TILs from MC38 tumors, as studied by flow cytometry. PD-1 (C, n = 16), TIM-3 (D, n = 18), and LAG-3 (E, n = 7) were analyzed. Statistical analysis by paired Student’s t test. (F) Number of coexpressed inhibitory receptors on SigE–CD8+ or SigE+CD8+ TILs. (G) Upregulation of CD25+CD69+ upon restimulation of sorted SigE–CD8+ and SigE+CD8+ TILs. Statistical analysis by paired Student’s t test. (H) Growth curves of subcutaneous WT or GNE-KO MC38 tumors (n = 8–9). (I) Growth curves of subcutaneous WT and GNE-KO EMT6 tumors (n = 13-14). Experiments were replicated 2 to 3 times. Statistical analysis by 2-way ANOVA. (J and K) Frequencies of CD3+ and CD8+ cells in the tumor (n = 7). Statistical analysis by unpaired Student’s t test. **P < 0.01; ***P < 0.001. Data are presented as mean ± SD.
Sia-SAMPs mediate immune escape in vivo.
To further investigate the role of Sia-SAMPs in immune evasion, we generated GNE-deficient MC38 cells and compared their subcutaneous growth with that of WT MC38 tumor cells. GNE-KO MC38 tumors showed a clear delay in tumor growth compared with WT MC38 tumors injected subcutaneously in C57BL/6 mice (Figure 5H). Similar observations were made for GNE-KO and WT EMT6 tumors injected subcutaneously into BALB/c mice (Figure 5I). The GNE-KO cell lines showed no growth differences or survival differences in vitro (Supplemental Figure 8I). The frequencies of CD3+, CD8+, and CD4+ T cells in leukocyte infiltrates were significantly higher in GNE-KO MC38 tumors compared with WT MC38 tumors (Figure 5, J and K, and Supplemental Figure 8J), whereas there was no difference in the infiltration of regulatory T cells (Supplemental Figure 8J). GNE-KO tumor infiltrates also contained more classical dendritic cells (CD11c+MHCII+ cells within CD45+ cells) than WT MC38 tumors, whereas other myeloid cell types did not differ significantly (Supplemental Figure 8J). In order to analyze the role of adaptive immunity, we repeated the experiment in mice deficient for the adaptive immune system (NOD scid gamma [NSG] mice). Both MC38 and EMT6 GNE-KO cell lines showed no growth difference when compared with WT cell lines in NSG mice (Supplemental Figure 8K). From these findings, we concluded the sialylation directly affects tumor growth and also the antitumor immune response by enhancing the adaptive immune response to cancer (Supplemental Figure 9 shows the gating strategy for determining Siglec-E expression on tumor-infiltrating T cells).
Expression of Siglec receptors on T cells directly modulate antitumor immunity.
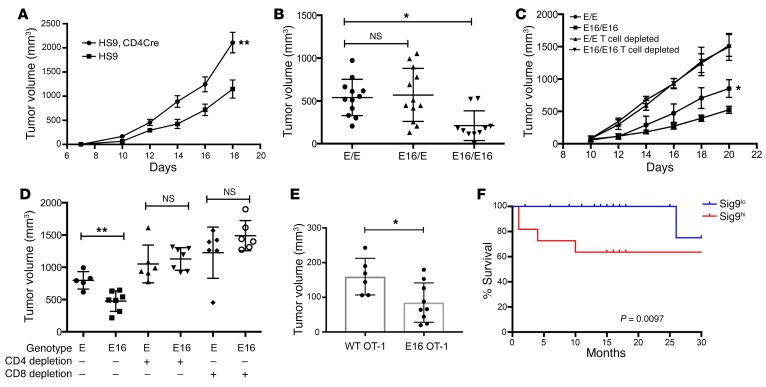
To further investigate the role of Sia-SAMPs and their interactions with CD33rSiglecs, we used a human Siglec-9 transgenic mouse (HS9) that allows selective expression of Siglec-9 in cells producing Cre recombinase (21). Previous analysis demonstrated that human Siglec-9 binds to ligands on murine tumor cells (21). HS9 mice were crossed with CD4-Cre mice to express human Siglec-9 in T cells. These mice expressed Siglec-9 in both CD4+ and CD8+ T cells (Supplemental Figure 10A). The growth of syngeneic MC38 tumor cells was accelerated in mice expressing Siglec-9 in T cells, and the tumors grew larger compared with those in littermate control mice (Figure 6A). This finding supports the hypothesis that inhibitory Siglecs on T cells can mediate immune evasion. We also used the recently developed Siglec-E16 (SigE16) transgenic mice (18). These mice express a chimeric Siglec receptor with the extracellular domain of Siglec-E and the transmembrane and intracellular domain of the activating human Siglec-16 receptor under the Siglec-E promoter (18). No major differences were observed between the frequency of naive and memory T cells in different genetic animal models (Supplemental Figure 10B). Siglec-E expression in WT and SigE16 mice was similar (Supplemental Figure 10C). Compared with those in WT littermate control mice, subcutaneous syngeneic MC38 tumors grew more slowly and remained smaller in homozygous SigE16 mice (Figure 6B). MC38 tumors had an increased induction of antitumor antibodies (Supplemental Figure 10D). Depletion experiments demonstrated the dependence of the effect on both CD4+ and CD8+ T cells and directly showed that the genetic reversal of an inhibitory into an activating Siglec receptor can also influence T cell activation (Figure 6, C and D). We also used an adoptive transfer model to study the role of Siglec-E on T cells. We transferred OT-I T cells expressing either WT Siglec-E or activating SigE16 into WT mice with OVA-expressing MC38-OVA tumors and measured the growth of tumors. MC38-OVA tumors in mice transferred with OT-I from SigE16 mice had significantly reduced tumor growth (Figure 6E). In addition, OT-I T cells expressing SigE16 had increased proliferation (Supplemental Figure 10E). In addition, a trend toward increased infiltration was also seen in mice adoptively transferred with SigE16-expressing OT-I T cells (Supplemental Figure 10F). To determine the effect of sialoglycans in trans position on the growth difference in SigE16 mice, we used MC38 WT and GNE-KO cells. The previous growth difference could no longer be seen in the SigE16 background (Supplemental Figure 10G). This finding suggests that the effect seen in SigE16 mice is mediated by interactions of trans ligands with activating SigE16 receptor.
Figure 6. Engagement of inhibitory Siglecs on T cells mediates immune escape.
(A) Subcutaneous growth curves of MC38 tumors in littermate Siglec-9 transgenic control mice (HS9) or Siglec-9 transgenic mice crossed to CD4-Cre mice (HS9, CD4Cre) (n = 8–10). The experiment was repeated 3 times. Statistical analysis by 2-way ANOVA. (B) Tumor volumes after 21 days of subcutaneous MC38 tumors in littermate control mice (E/E) or homozygous (E16/E16) mice that express the chimeric receptor SigE16 (n = 7). Statistical analysis performed by 1-way ANOVA. (C) Subcutaneous growth curves of MC38 tumors in E16 mice or littermate control mice after CD4 and CD8 cell depletion by antibodies (n = 7–8). The experiment was repeated 2 times. Statistical analysis by 2-way ANOVA. (D) MC38 tumor volumes after 21 days in E16 mice and littermate control mice and independent depletion of CD4+ and CD8+ T cells. (E) Tumor volume of MC38-OVA tumors after adoptive transfer of OVA-specific OT-I CD8+ T cells from WT or SigE16 (E16) mice. Statistical analysis performed by 1-way ANOVA. *P < 0.05; **P < 0.01. Data are presented as mean ± SD. (F) Survival analysis of NSCLC patients with low (n = 18) and high percentage (above 30%, n = 11) of Siglec-9 expression on their CD8+ T cells. Differences were analyzed by Wilcoxon’s test. A multivariate analysis of the 2 groups for age and stage showed slightly reduced significance, with a P value of 0.0669 (multivariate, univariate analysis P = 0.0097) and a hazard ratio of 14.6.
Sig9+ TILs and Siglec-9 polymorphisms are associated with clinical end points.
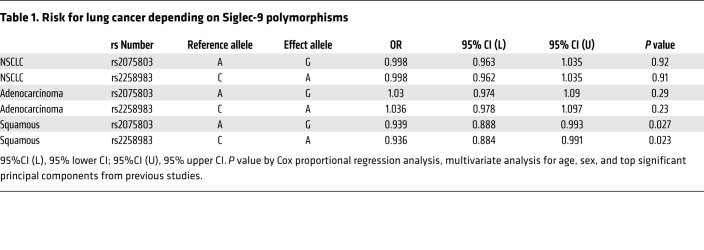
We further analyzed the correlation of Siglec expression on T cells with clinical parameters. Patients with a high frequency of Sig9+ TILs (>30% of CD8+ T cells) had significantly worse overall survival (Figure 6F). The significance was slightly reduced in a multivariate analysis taking age and also stage into account (P = 0.0668 with a hazard ratio of 14.6 by a Cox proportional hazard regression analysis). Recently, polymorphisms of Siglec-9 were associated with pulmonary diseases (33). We studied to determine whether the risk of developing lung cancer was associated with the rs2075803 and rs2258983 polymorphisms in the TRICL4 cohort (34). The risk of developing squamous cell lung cancer was significantly reduced in the presence of these polymorphisms (Table 1). We also examined the polymorphisms rs2075803 and rs2258983 in 3,795 cases of CRC and 3,044 controls to estimate the relative risk of developing CRC. Individuals who were homozygous A for the rs2075803 SNP and homozygous C for the rs2258983 SNP had a significantly reduced risk of developing CRC (Supplemental Table 1).
Table 1. Risk for lung cancer depending on Siglec-9 polymorphisms.
Discussion
Recent studies have shown that inhibitory CD33rSiglecs can modulate the interactions of immune cells with tumor cells by sialic acid–dependent mechanisms (21–23, 25, 26), raising the possibility that these Siglecs could be used for therapeutic checkpoint inhibition, analogous to the remarkable recent therapeutics directed against PD-1 or CTLA-4 (2). We and others have shown that T cells from healthy humans express low levels of CD33-related Siglecs (29, 35). However, we show here that expression of the inhibitory Siglec-9 is clearly increased on CD4+ and CD8+ TILs compared with peripheral T cells from healthy donors. We also observed the presence of Siglec ligands in both cis and trans positions. In addition, we confirmed previous observations that have shown a small population of peripheral T cells with Siglec-7 and Siglec-9 expression (35). Properly glycosylated CD52 has previously been identified as a ligand for Siglec-10 on human T cells (36). Furthermore, inhibitory mouse Siglec-G (often referred to as the murine paralog of human Siglec-10) was found to inhibit DAMP-associated T cell activation (37). These investigations demonstrate that Siglecs can dampen T cell responses in the context of general inflammation.
How Siglec-9 is upregulated on TILs and how Siglecs are regulated in T cells, including intracellular signaling, remain unclear. We observed only a slight upregulation of Siglec-9 upon polyclonal and antigen-specific stimulation of healthy T cells. Other immune checkpoint receptors are upregulated through repetitive antigenic stimulation or the tumor microenvironment, including inhibitory cytokines (27). We have seen a similar effect in several mouse models, indicating that the mechanism for Siglec upregulation in T cells appears to be conserved across species, even though Siglec receptors have diverged quite a lot between mice and humans. We observed a heterogeneity of the frequency in Sig9+CD8+ T cells in the peripheral blood of healthy donors and an inverse correlation with the frequency of naive CD8+ T cells. This finding suggests that healthy individuals with more circulating peripheral memory T cells also have a higher number of Sig9+CD8+ T cells. Until now, mainly Siglec-9–expressing NK cells and myelomonocytic cells have been associated with immune modulation in cancer. Myeloid cells were more reactive in a mouse model of Siglec-E deficiency, and increased immunosurveillance was observed in these mice (21). Engagement of Siglec-9 on monocytes/macrophages by a cancer-associated, sialylated glycoform of MUC1 showed polarization toward immune-suppressive TAMs with upregulation of PD-L1 (25). Two analyses examined the function of inhibitory Siglec-7 and Siglec-9 in NK cell–mediated tumor cell killing (22, 23). Other immune receptors, including PD-1, are also expressed on myeloid cells and NK cells within the tumor (38, 39). Further experiments are needed to understand the functional relevance of these immune-modulatory receptors, including Siglec-9 or PD-1, on cell types other than T cells. However, it is likely that CD8+ T cells continue to play central roles in cancer immunotherapy, and we define Siglec-9 on CD8+ TILs as a potential therapeutic target for cancer immunotherapy.
Further characterization of Sig9+CD8+ TILs revealed that these cells coexpress several inhibitory receptors, including PD-1, TIM-3, Lag3, and others. Our analysis suggests that Sig9+CD8+ TILs are a subpopulation of CD8+PD-1hi TILs, which are often tumor-specific (40). Coexpression of multiple inhibitory receptors on T cells and also PD-1hi TILs was associated with an exhaustion phenotype in cancer, including in patients with NSCLC (27, 41). Indeed, functional analysis of TILs from NSCLC patients showed a significantly reduced capacity to produce cytokines and chemokines (Figure 3). However, Sig9+CD8+ T cells from NSCLC patients were easier to reactivate than Sig9–CD8+ T cells despite the increased expression of PD-1 and other inhibitory immune receptors on Sig9+ TILs. This finding suggests that Sig9+CD8+ TILs belong to a specific subtype within PD-1hi TILs. A recent analysis of chromatin states in murine and human TILs from melanoma and NSCLC patients demonstrated a subpopulation within PD-1hi TILs that had a relatively higher expression of CD5 and lower expression of CD38 and CD101 and that showed a greater potential for reprogrammability and reactivation (31). Our findings showed an enrichment of high CD5 expression and low CD38 and CD101 expression on Sig9+CD8+ TILs in the PD-1hi gate. This finding could be interpreted as an overlap of Sig9+CD8+ TILs with this previously described tumor-specific TIL subpopulation that could be potentially reactivated by immunotherapy. Other studies have also shown that, not only exhausted TILs coexpress a high frequency of inhibitory receptors, but also activated TILs (42). This could mean that the Sig9+CD8+ TILs are in a higher activation state that could potentially be uncovered by blocking these different inhibitory receptors (43).
Our analysis also includes functional studies both in vitro with human tissue and in vivo in different mouse models to demonstrate the relevance of inhibitory Siglecs on TILs and their interactions with cancer-associated Sia-SAMPs. The previous studies have provided evidence that Sia-SAMP/Siglec interactions can inhibit immune cell activation in in vitro models (21–23, 25). The use of bivalent anti–Siglec-9 antibodies (clones 191240 and E10-286) led to an inhibition of T cell activation, which might be due to the activation of Siglec-9 signaling by dimerization. In contrast, the use of monovalent Fab fragments of the antibody clone 191240, which inhibits Sia-SAMP/Siglec-9 interactions by hampering ligand binding to the CRD, led to increased T cell activation. This result suggests that the Sia-SAMP/Siglec-9 interaction can be targeted for the enhancement of TIL activation in lung tumors. Functional relevance is further supported by the analysis of a Siglec-9 polymorphism in cancer patients. This shows an association of hypomorphic alleles with the risk of developing squamous NSCLC. In addition, the same alleles were also associated with the risk for CRC. These polymorphisms were previously associated with an increased frequency of pulmonary emphysema, exacerbations of chronic obstructive pulmonary disease, and a hyperactive immune response of myeloid cells (33). While the function of these polymorphisms needs to be tested in T cells, their linkage to disease outcome suggests a role for Siglec-9 in the development of lung and CRC.
Our studies also show that the inhibitory Siglec-E receptor, the functional paralog of Siglec-9 and Sia-SAMPs in the tumor, influences the T cell–mediated immune response to tumors in mouse models. Reduction of Sia-SAMPs by using GNE-KO tumor cell lines led to a reduced growth of tumors. In particular, the use of the more antigenic cell line EMT6-HER2 showed rejection of the tumors in some mice when hyposialylated GNE-KO tumor cells were used. The KO of GNE in those cell lines most likely induces additional changes in glycosylation besides reducing the level of sialylation. An upregulation of polylactosamine could change the interaction with other immune-modulatory lectins, such as galectins (44). Also, during the growth of tumors in mice, the cells could potentially acquire sialic acids from the host. Nevertheless, we observed a relevant growth difference in 2 different models. Previous experiments with B16 melanoma cell lines have also shown similar results, although different approaches were used for the reduction of tumor sialylation (45). Another analysis with hyposialylated methylcholanthrene-induced (MCA-induced) tumors has shown growth inhibition of subcutaneous tumors (46). Overexpression of inhibitory Siglec-9 on CD4+ and CD8+ T cells led to increased tumor growth, which indicates a functional role of inhibitory Siglecs on T cells. Exchange of inhibitory Siglec-E with activating SigE16 (extracellular Siglec-E domain with transcellular activating human Siglec-16) showed T cell–dependent growth inhibition. In an adoptive transfer model, we demonstrated that Siglec-E on T cells is functionally involved, although the model we used, with OVA-specific OT-I T cells, has certain limitations and OVA is not directly comparable to naturally occurring tumor antigens.
Cancer immunotherapy and, in particular, checkpoint blockade with inhibitors of the PD-1/PD-L1/L2 axis and CTLA-4 are now routinely used in daily oncological practice (1, 47). However, only a minority of cancer patients show objective responses under checkpoint blockade and only a few develop long-term remissions. Thus, combination therapies are a promising approach to improving response rates of immunotherapies and outcomes for patients (48, 49). Our data suggest that targeting the Sia-SAMP/Siglec-9 pathway could improve antitumor immunity and define this pathway as an inhibitory immune checkpoint for T cell activation.
Methods
Patients and sample preparation.
Surgical specimens were mechanically dissociated, digested with accutase (PAA Laboratories), collagenase IV (Worthington), hyaluronidase (MilliporeSigma), and DNAse type I (MilliporeSigma), filtered, washed, and frozen for future use. Single-cell suspensions were prepared. Human PBMCs were isolated by density gradient centrifugation using Histopaque-1077 (MilliporeSigma) from buffy coats obtained from healthy blood donors (Blood Bank, University Hospital Basel). Single-cell suspensions and PBMCs were frozen for later use in liquid nitrogen.
Multicolor flow cytometry.
For multicolor flow cytometry, dead cells were stained using LIVE/DEAD Fixable Blue Dye (Invitrogen) and various panels of antibodies. Doublets were excluded in all analyses. Corresponding isotype antibodies or fluorescence-minus-one (FMO) samples were used as a control, in particular for the Siglec staining. All tumor samples were analyzed with a Fortessa LSR II Flow Cytometer (BD Biosciences). For infiltration analysis, mice were euthanized and tumors were mechanically dissociated and digested as described for the human sample preparation.
Staining for Siglec-7 and Siglec-9 ligands on tumor microarray.
Tissue microarrays from US Biomax were used for screening of Siglec-9 ligand expression in lung cancer samples. For immunofluorescence staining, recombinant human Siglec-hFcs (R&D Systems) were mixed with PE-conjugated anti-human IgG (Jackson ImmunoResearch Laboratories) for 1 hour at 4°C prior to use. For the specific visualization of cancer cells, Anti–Multi-Cytokeratin (Leica Biosystems) staining was performed and visualized with a goat anti-mouse IgG coupled to Alexa Fluor 488 (Life Technologies). The general fluorescence per sample was measured in a ScanRI Microarray Scanner (PerkinElmer) and processed with ImageJ software, version 1.48 (NIH). Pictures were taken using a fluorescence microscope with original magnification of ×400 (Zeiss). Fluorescence was quantified using ImageJ software and normalized to background staining (secondary antibody only).
In vitro T cell activation and inhibition.
Murine and human T cells were polyclonally stimulated with anti-CD3 and anti-CD28 antibodies (BioLegend). Anti-CD3 antibodies (0.5 μg/ml) were either coated overnight at 4°C for human T cells or added for mouse T cells. Anti-CD28 was added at 1 μg/ml. For inhibition assays with sialylated LGALS3BP, LGALS3BP was coated overnight as well. Activation of T cells was determined by staining for activation and proliferation markers by flow cytometry or measuring cytokines in supernatants by ELISA. For the SEB assay, primary NSCLC samples were incubated at 200,000 cells/well with SEB (10 ng/ml, MilliporeSigma) for 48 hours in the presence of full-length or digested Fab fragments of the 191240 anti–Siglec-9 antibody (R&D Systems). T cell activation was assessed by FACS staining, and IL-2 secretion was measured in the supernatant by ELISA (BioLegend).
RNA-seq and analysis.
CD8+ TILs were sorted from frozen NSCLC samples by FACS according to their Siglec-9 expression. Gates used were as follows: lymphocytes, singlets, DAPI–CD3+, and CD8+CD4–. RNA was isolated and the library prepared by Microsynth AG. Next-generation sequencing of the library and data analysis were performed by Microsynth AG. Count data from RNA-seq samples were analyzed using the edgeR Bioconductor package in R. Filtered genes, expressed at more than 1 count per million (cpm) in at least 3 samples, were analyzed using the QLF functions with batch correction for patients. All genes were ranked according to their F statistics comparing Siglec-9–positive and –negative samples. A weighted gene set enrichment analysis (GSEA) was performed using this preranked list using the GSEA java application (http://www.broad.mit.edu/gsea/). Box plots comparing expression levels between Siglec-9–positive and –negative cells were performed in R using logarithmically transformed cpm values. All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE115305).
GNE-deficient cells and in vitro tumor cell killing with CD3/EpCAM TCB or CD3/CD19 TCB.
Human and murine GNE-deficient tumor cell lines were created using CRISPR/CAS9. Guide RNAs were designed online using e-crisp.org, synthesized by Microsynth AG, and cloned into the pX458 vector (Addgene). A549, HT-29, and RAMOS cell lines were bought from ATCC. After transient transfection in tumor cells, single-cell sorting and subsequent screening for cell-surface sialylation were performed. Multiple GNE-deficient clones were pooled in order to avoid clonal selection when comparing with WT cell lines.
GNE-deficient cell lines and cell lines were refed with 5 mM Neu5Ac for 24 hours. Coincubation with magnetically isolated CD8+ T cells from healthy donors or tumor samples in the presence of catumaxomab (anti-CD3/anti-EpCAM; Fresnius) or blinatumomab, (anti-CD3/anti-CD19; Amgen) was performed at a concentration of 10 ng/ml. Alternatively, the respective cell lines were stained with carboxyfluorescein succinimidyl ester (CSFE) (Sigma-Aldrich) and spiked into full-tumor digests obtained from primary NSCLC or CRC samples. Tumor cell killing was analyzed by FACS staining for cleaved caspase-3 and by staining with the LIVE/DEAD Viability/Cytotoxicity Kit (Thermo Fisher Scientific). Tumor cells were gated by size and expression of EpCAM or CD19, respectively, as well as by negative gating using CD3, CD8, and CD4.
Genetic mouse models for in vivo analysis of Siglec function on T cells.
Siglec-9 transgenic mice were described previously (21). Siglec-9 transgenic mice (B6.Cg-Tg[CAG-Siglec-9]1Avrk) were crossed with CD4-Cre mice (B6Tg[CD4-cre]1Cwi) to obtain a T cell–specific overexpression of inhibitory human Siglec-9 in the C57BL/6 background. Transgenic mice expressing SigE16 chimeric Siglec-E with transmembrane and intracellular domain of activating human Siglec-16 (B6.Cg-Siglece < tm4E16Avrk>) were also described previously (18). Tumor cell lines were injected into 8- to 12-week-old female mice, and tumor growth was monitored as described above.
SigE16 was crossed into OT-I transgenic mice. For the adoptive transfer of OVA-specific OT-I cells from WT or SigE16 mice, T cells were harvested from splenocytes and injected into WT C57BL/6 mice bearing subcutaneous MC38-OVA tumors. T cells were followed for proliferation by labeling with Cell Trace Violet (CTV) (Thermo Fisher Scientific). Tumor growth was measured, and cell numbers were determined by counting beads and flow cytometry.
Association study of Siglec-9 polymorphism.
The association analysis of Siglec-9 polymorphisms in lung cancer was studied on the TRILC cohort (34). In total, 11,348 patients with lung cancer and 15,861 controls were studied for the rs2075803 and the rs2258983 SNPs. An association test was performed after adjusting for age, sex, and top principal components. Then we conducted a fixed effects meta-analysis with an inverse variance-weighted average method to combine the summary data from each association test. Meta-analysis was performed using SAS version 9.4 (SAS Institute Inc.). For the analysis of an association of Siglec-9 polymorphisms with the risk of CRC, SNP genotypes for rs2075803 and rs2258983 among cases and controls from imputed genotype data derived from samples genotyped on the Oncoarray platform and USC Norris Cancer Center were used. Using contingency table analysis (Fisher’s test), we calculated odds ratios (OR), 95% CIs, and P values using an additive model and genotype-specific OR.
For additional information, see Supplemental Methods.
Statistics.
Statistical significance between 2 groups was determined using 2-tailed Student’s t tests. Significance among more than 2 conditions was assessed using 1-way ANOVAs with multiple comparisons. For survival curve analysis, 2-way ANOVA was used. Survival analysis was performed by the Gehan-Breslow-Wilcoxon test, and multivariate analysis was done by Cox proportional hazard analysis. P < 0.05 was considered statistically significant.
Study approval.
The local ethical committee in Basel, Switzerland, approved the sample collection and the use of clinical data (Ethikkomission Nordwest- und Zentralschweiz, EK321/10 and UBE 15–106). Informed consent was obtained from all patients prior to the sample collection. Mouse experiments were approved by the local committee of Basel Stadt (approval number 2747).
Author contributions
MAS and SSS planned and conducted the experiments and analyzed the data. HL conceived the study and planned the project. MAS, DES, AV, AZ, and HL wrote the manuscript. MPT, DST, KFB, SVG, AT, and FS performed experiments. LT, DL, VHS, MVBB, AE, and MS provided important material for the research. WZ, HJL, YH, and CIA analyzed the Siglec polymorphisms.
Supplementary Material
Acknowledgments
This work was supported by funding from the Goldschmidt-Jacobson Foundation (to HL), the Promedica Foundation (to MAS and AZ), Krebsliga Beider Basel (KLBB) (to HL), the Lichtenstein Foundation (to HL), Schoenmakers Foundation (to HL), the Huggenberger Foundation (to HL), and the NIH (P01 HL107150 to AV). We thank Patrick Secrest, Petra Herzig, and Béatrice Dolder-Schlienger for their technical assistance. We thank Pedro Romero, Camilla Jandus, and Marc Donath for critically reading the manuscript. We also thank all the patients that allowed us to use their material and made this work possible.
Version 1. 08/21/2018
In-Press Preview
Version 2. 09/24/2018
Electronic publication
Version 3. 10/22/2018
Corrected spelling of Michael von Bergwelt-Baildon in author list.
Version 4. 11/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(11):4912–4923.https://doi.org/10.1172/JCI120612.
Contributor Information
Michal A. Stanczak, Email: michal.stanczak@unibas.ch.
Shoib S. Siddiqui, Email: shsiddiqui@ucsd.edu.
Marcel P. Trefny, Email: marcel.trefny@unibas.ch.
Daniela S. Thommen, Email: d.thommen@nki.nl.
Kayluz Frias Boligan, Email: kayluz.frias@pki.unibe.ch.
Stephan von Gunten, Email: stephan.vongunten@pki.unibe.ch.
Alexandar Tzankov, Email: alexandar.tzankov@usb.ch.
Lothar Tietze, Email: Lothar.Tietze@le.ortenau-klinikum.de.
Didier Lardinois, Email: didier.lardinois@usb.ch.
Viola Heinzelmann-Schwarz, Email: viola.heinzelmann@usb.ch.
Wu Zhang, Email: Wu.Zhang@med.usc.edu.
Heinz-Josef Lenz, Email: LENZ@med.usc.edu.
Christopher I. Amos, Email: Christopher.I.Amos@Dartmouth.edu.
Mohammedyaseen Syedbasha, Email: m.syedbasha@unibas.ch.
Adrian Egli, Email: adrian.egli@usb.ch.
Frank Stenner, Email: Frank.Stenner@usb.ch.
Daniel E. Speiser, Email: doc@dspeiser.ch.
Ajit Varki, Email: a1varki@ucsd.edu.
Alfred Zippelius, Email: alfred.zippelius@usb.ch.
Heinz Läubli, Email: heinz.laeubli@unibas.ch.
References
- 1.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 3.Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat Rev Immunol. 2016;16(1):35–50. doi: 10.1038/nri.2015.8. [DOI] [PubMed] [Google Scholar]
- 4.Schaefer L. Complexity of danger: the diverse nature of damage-associated molecular patterns. J Biol Chem. 2014;289(51):35237–35245. doi: 10.1074/jbc.R114.619304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22(2):240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen GY, Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10(12):826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang RF, Miyahara Y, Wang HY. Toll-like receptors and immune regulation: implications for cancer therapy. Oncogene. 2008;27(2):181–189. doi: 10.1038/sj.onc.1210906. [DOI] [PubMed] [Google Scholar]
- 8.Varki A. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology. 2011;21(9):1121–1124. doi: 10.1093/glycob/cwr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padler-Karavani V, et al. Rapid evolution of binding specificities and expression patterns of inhibitory CD33-related Siglecs in primates. FASEB J. 2014;28(3):1280–1293. doi: 10.1096/fj.13-241497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 11.Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 2014;14(10):653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearce OM, Läubli H. Sialic acids in cancer biology and immunity. Glycobiology. 2016;26(2):111–128. doi: 10.1093/glycob/cwv097. [DOI] [PubMed] [Google Scholar]
- 13.Nitschke L. CD22 and Siglec-G regulate inhibition of B-cell signaling by sialic acid ligand binding and control B-cell tolerance. Glycobiology. 2014;24(9):807–817. doi: 10.1093/glycob/cwu066. [DOI] [PubMed] [Google Scholar]
- 14.Chang YC, Nizet V. The interplay between Siglecs and sialylated pathogens. Glycobiology. 2014;24(9):818–825. doi: 10.1093/glycob/cwu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang YC, et al. Group B Streptococcus engages an inhibitory Siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog. 2014;10(1):e1003846. doi: 10.1371/journal.ppat.1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlin AF, et al. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med. 2009;206(8):1691–1699. doi: 10.1084/jem.20090691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113(14):3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz F, et al. Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen. EMBO J. 2017;36(6):751–760. doi: 10.15252/embj.201695581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pröpster JM, Yang F, Rabbani S, Ernst B, Allain FH, Schubert M. Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8. Proc Natl Acad Sci U S A. 2016;113(29):E4170–E4179. doi: 10.1073/pnas.1602214113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, et al. Siglec-8 and Siglec-9 binding specificities and endogenous airway ligand distributions and properties. Glycobiology. 2017;27(7):657–668. doi: 10.1093/glycob/cwx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Läubli H, et al. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proc Natl Acad Sci U S A. 2014;111(39):14211–14216. doi: 10.1073/pnas.1409580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jandus C, et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J Clin Invest. 2014;124(4):1810–1820. doi: 10.1172/JCI65899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudak JE, Canham SM, Bertozzi CR. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat Chem Biol. 2014;10(1):69–75. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Läubli H, et al. Lectin galactoside-binding soluble 3 binding protein (LGALS3BP) is a tumor-associated immunomodulatory ligand for CD33-related Siglecs. J Biol Chem. 2014;289(48):33481–33491. doi: 10.1074/jbc.M114.593129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beatson R, et al. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat Immunol. 2016;17(11):1273–1281. doi: 10.1038/ni.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.RodrÍguez E, Schetters STT, van Kooyk Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat Rev Immunol. 2018;18(3):204–211. doi: 10.1038/nri.2018.3. [DOI] [PubMed] [Google Scholar]
- 27.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soto PC, Karris MY, Spina CA, Richman DD, Varki A. Cell-intrinsic mechanism involving Siglec-5 associated with divergent outcomes of HIV-1 infection in human and chimpanzee CD4 T cells. J Mol Med. 2013;91(2):261–270. doi: 10.1007/s00109-012-0951-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen DH, Hurtado-Ziola N, Gagneux P, Varki A. Loss of Siglec expression on T lymphocytes during human evolution. Proc Natl Acad Sci U S A. 2006;103(20):7765–7770. doi: 10.1073/pnas.0510484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crocker PR, McMillan SJ, Richards HE. CD33-related siglecs as potential modulators of inflammatory responses. Ann N Y Acad Sci. 2012;1253:102–111. doi: 10.1111/j.1749-6632.2011.06449.x. [DOI] [PubMed] [Google Scholar]
- 31.Philip M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545(7655):452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C, et al. In vitro characterization of the anti-PD-1 antibody nivolumab, BMS-936558, and in vivo toxicology in non-human primates. Cancer Immunol Res. 2014;2(9):846–856. doi: 10.1158/2326-6066.CIR-14-0040. [DOI] [PubMed] [Google Scholar]
- 33.Ishii T, et al. Influence of SIGLEC9 polymorphisms on COPD phenotypes including exacerbation frequency. Respirology. 2017;22(4):684–690. doi: 10.1111/resp.12952. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46(7):736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikehara Y, Ikehara SK, Paulson JC. Negative regulation of T cell receptor signaling by Siglec-7 (p70/AIRM) and Siglec-9. J Biol Chem. 2004;279(41):43117–43125. doi: 10.1074/jbc.M403538200. [DOI] [PubMed] [Google Scholar]
- 36.Bandala-Sanchez E, et al. T cell regulation mediated by interaction of soluble CD52 with the inhibitory receptor Siglec-10. Nat Immunol. 2013;14(7):741–748. doi: 10.1038/ni.2610. [DOI] [PubMed] [Google Scholar]
- 37.Toubai T, et al. Siglec-G represses DAMP-mediated effects on T cells. JCI Insight. 2017;2(14):e92293. doi: 10.1172/jci.insight.92293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gordon SR, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545(7655):495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, et al. Increased expression of programmed cell death protein 1 on NK cells inhibits NK-cell-mediated anti-tumor function and indicates poor prognosis in digestive cancers. Oncogene. 2017;36(44):6143–6153. doi: 10.1038/onc.2017.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gros A, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124(5):2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thommen DS, et al. Progression of lung cancer is associated with increased dysfunction of T cells defined by coexpression of multiple inhibitory receptors. Cancer Immunol Res. 2015;3(12):1344–1355. doi: 10.1158/2326-6066.CIR-15-0097. [DOI] [PubMed] [Google Scholar]
- 42.Legat A, Speiser DE, Pircher H, Zehn D, Fuertes Marraco SA. Inhibitory receptor expression depends more dominantly on differentiation and activation than “exhaustion” of human CD8 T cells. Front Immunol. 2013;4:455. doi: 10.3389/fimmu.2013.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol. 2016;16(10):599–611. doi: 10.1038/nri.2016.80. [DOI] [PubMed] [Google Scholar]
- 44.Rabinovich GA, Croci DO. Regulatory circuits mediated by lectin-glycan interactions in autoimmunity and cancer. Immunity. 2012;36(3):322–335. doi: 10.1016/j.immuni.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Perdicchio M, et al. Tumor sialylation impedes T cell mediated anti-tumor responses while promoting tumor associated-regulatory T cells. Oncotarget. 2016;7(8):8771–8782. doi: 10.18632/oncotarget.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen M, et al. Sialylation of 3-methylcholanthrene-induced fibrosarcoma determines antitumor immune responses during immunoediting. J Immunol. 2010;185(10):5869–5878. doi: 10.4049/jimmunol.1001635. [DOI] [PubMed] [Google Scholar]
- 47.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 48.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161(2):205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melero I, Berman DM, Aznar MA, Korman AJ, Pérez Gracia JL, Haanen J. Evolving synergistic combinations of targeted immunotherapies to combat cancer. Nat Rev Cancer. 2015;15(8):457–472. doi: 10.1038/nrc3973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.