Significance
It has long been known that chromosomes, which contain life’s genetic instructions, condense into discrete bodies when cells divide. Chromosome condensation is thought to shut down gene expression and is misregulated in many diseases. The molecular details of this process are unknown because the fundamental subunits of chromosomes, called nucleosomes, are too small and fragile to be seen by conventional microscopy. We used state-of-the-art 3D cryo-electron tomography to reveal how chromosomes condense within intact cells. We found evidence that condensation is uneven, resulting in porous chromosomes that allow cells to express a limited set of genes. Furthermore, our work and those of others suggest that chromatin dynamics at the molecular scale are an underappreciated component of chromosome function.
Keywords: chromatin, condensation, cryo-ET, fission yeast
Abstract
Chromosomes condense during mitosis in most eukaryotes. This transformation involves rearrangements at the nucleosome level and has consequences for transcription. Here, we use cryo-electron tomography (cryo-ET) to determine the 3D arrangement of nuclear macromolecular complexes, including nucleosomes, in frozen-hydrated Schizosaccharomyces pombe cells. Using 3D classification analysis, we did not find evidence that nucleosomes resembling the crystal structure are abundant. This observation and those from other groups support the notion that a subset of fission yeast nucleosomes may be partially unwrapped in vivo. In both interphase and mitotic cells, there is also no evidence of monolithic structures the size of Hi-C domains. The chromatin is mingled with two features: pockets, which are positions free of macromolecular complexes; and “megacomplexes,” which are multimegadalton globular complexes like preribosomes. Mitotic chromatin is more crowded than interphase chromatin in subtle ways. Nearest-neighbor distance analyses show that mitotic chromatin is more compacted at the oligonucleosome than the dinucleosome level. Like interphase, mitotic chromosomes contain megacomplexes and pockets. This uneven chromosome condensation helps explain a longstanding enigma of mitosis: a subset of genes is up-regulated.
Chromatin structure influences key nuclear activities such as transcription, DNA repair, and replication (1). The fundamental unit of chromatin is the nucleosome, which consists of ∼147 bp of DNA wrapped around a histone octamer (2). In mammalian cells, 2–35 nucleosomes pack into irregular “clutches” (3). More than 500 sequential nucleosomes (calculated from the nucleosome repeat length) are thought to interact as topologically associating domains (4). Likewise, in the fission yeast Schizosaccharomyces pombe, some 300–7,000 nucleosomes are thought to associate as compact globular chromatin bodies called domains (5–7). Furthermore, superresolution imaging of S. pombe revealed that the chromatin is not uniformly distributed in vivo (8). These studies suggest that chromatin higher-order structure arises from physical interactions within large groups of nucleosomes. In mitotic cells, chromosomes condense into discrete structures that can be resolved in a light microscope. The factors involved in condensation have been well characterized (9), and a number of models have been considered for the large-scale organization of chromatin domains (10). However, the molecular details of chromatin reorganization are still unknown. Knowledge of how chromosomes condense in 3D at the molecular level is needed to explain the nearly global transcriptional repression that happens to the mitotic cells of most eukaryotes (11). Such a model could also explain how a subset of fission yeast genes escape this mitotic repression and get up-regulated (12–14). Some insights on in vivo chromatin organization were made possible by new methods, including chromatin-conformation capture (Hi-C), superresolution microscopy, and traditional electron microscopy (EM) of cells stained with DNA-proximal osmium (15–17). However, the resultant models are limited because these methods rely on population-averaged data, or have low resolution, or perturb the sample due to the fixation, dehydration, and staining. Cryo-EM is a label-free method to visualize cells in a life-like frozen-hydrated state (18, 19). Cryo-electron tomography (cryo-ET) goes further and can reveal the 3D positions of macromolecular complexes inside cells at ∼4-nm resolution (20). Using cryo-ET, we previously showed that nucleosomes in picoplankton and budding yeast cells pack irregularly and do not compact into discrete chromosomes in mitosis (21, 22).Yeast (budding and fission) and human chromatin have differences such as linker-DNA length, which is shorter in yeast (23), and the abundance of heterochromatin, which is rarer in yeast (24). However, fission yeast mitotic chromosome condensation resembles human chromosome condensation in both morphological and Hi-C phenotypes (6, 7, 25–27). We have now used cryo-ET to visualize the nuclear macromolecular complexes both before and after mitotic condensation in vivo in the fission yeast S. pombe. To obtain cryotomograms with sufficient contrast, we imaged cells that were thinned by cryomicrotomy. To more reproducibly characterize the heterogeneous complexes in the crowded nucleoplasm, we also took advantage of recent advances in phase-contrast hardware and image-classification software (28, 29). This approach can discriminate densities that do not have the size or shape expected of nucleosomes. Image classification failed to identify canonical nucleosomes, i.e., those resembling the crystal structure, which leads to numerous implications for the structure of fission yeast nucleosomes in vivo. Inspection of the nucleoplasm revealed the presence of multimegadalton complexes (megacomplexes) and pockets free of macromolecular complexes. In mitosis, chromosomes condense unevenly: they also have pockets and megacomplexes. These observations contribute to a model that can explain how some genes can be transcriptionally up-regulated in mitosis.
Results
S. pombe Subcellular Structures Are Revealed at Molecular Resolution by Cryo-ET.
To study how native chromatin organization differs between interphase and mitosis, we imaged S. pombe cells by cryo-ET of frozen-hydrated sections (cryosections, ∼70 or 100 nm nominal thickness). To ensure that the imaged cells had a known cell-cycle state, we arrested temperature-sensitive cdc25-22 cells in G2 phase (30) and cold-sensitive nda3-KM311 cells in prometaphase (25). For brevity, we refer to these as G2-phase and prometaphase cells. Fluorescence microscopy confirmed that prometaphase cells have the well-known mitotic chromosome-condensation phenotype (SI Appendix, Fig. S1 A and B). In a typical cryotomogram of an S. pombe cell, we could recognize organelles such as the endoplasmic reticulum, vesicles, and the nucleus due to their location and membrane morphology (SI Appendix, Fig. S1 C and D). To assess the quality of the cryosections and data, we performed subtomogram averaging of cytoplasmic ribosomes (SI Appendix, Fig. S2 A and B). Ribosomes were manually picked and then subjected to reference-free 3D classification. All particles converged to a single class and were then 3D-refined using the class average as a reference. The resulting average resembles a low-pass–filtered budding yeast ribosome crystal structure (SI Appendix, Fig. S2C) (31). Therefore, the conformations of large complexes are preserved at the molecular level.
Interphase Chromatin Is Loosely Packed Among Much Larger Complexes.
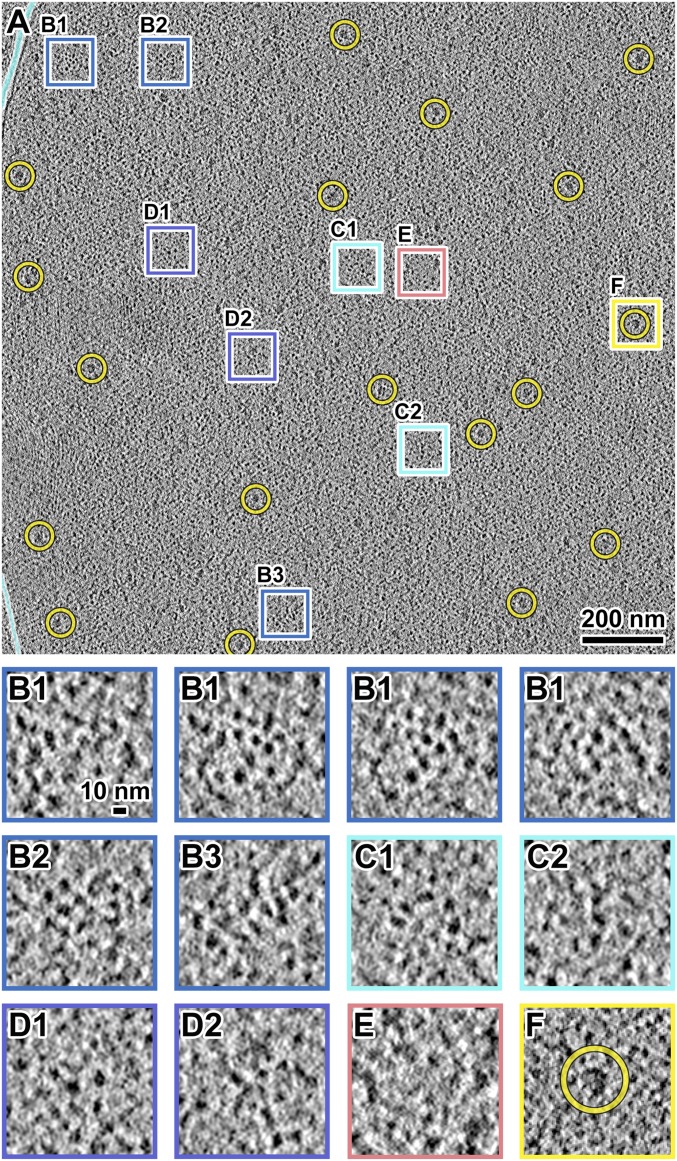
G2-phase cell nuclei are populated with many nucleosome-like particles, which we define as globular densities ∼10 nm wide along at least one axis (Fig. 1A and Movie S1). Groups of nucleosome-like particles appear as irregular clusters or irregular linear arrangements (Fig. 1 B and C). The nucleosome-like particles within these motifs are packed ∼10–12 nm center-to-center. We also observed positions where the nucleosome-like particles are more dispersed (Fig. 1D) and positions that are free of macromolecular complexes, which we call “pockets” (<50 nm diameter) (Fig. 1E; see also template-matching and classification analysis below). The boundaries of these pockets are also heterogeneous due to the irregular distribution of macromolecular complexes. Globular particles the size of ribosomes (>20 nm) are spread throughout the nucleus (Fig. 1F). These particle dimensions are consistent with multimegadalton nuclear assemblies such as preribosomes, spliceosomes, and transcription-preinitiation complexes (32–34); we call these “megacomplexes” for brevity. As expected, megacomplexes were more abundant in the nucleolus (SI Appendix, Fig. S3), which occupies a hemispherical volume in the fission yeast nucleus and is the key site of preribosome assembly. While most of the nucleoplasmic megacomplexes are likely to be preribosomes at various stages of maturation, the identities of individual megacomplexes are presently unknown. In summary, the G2-phase S. pombe nucleoplasm is rich in irregular clusters of nucleosome-like particles that commingle with megacomplexes and pockets.
Fig. 1.
S. pombe G2-phase chromatin consists of irregularly packed nucleosome-like particles and dispersed megacomplexes. (A) Cryotomographic slice (10 nm) of an interphase nucleus. Yellow circles, a subset of megacomplexes. The cyan lines follow the outer-nuclear membrane. The boxes are enlarged fourfold in B–F. (B1–B3) Three examples of clusters of nucleosome-like particles. The four B1 panels show left-to-right a series of cryotomographic slices (10 nm) in a step size of 4 nm. (C1 and C2) Two examples of chain-like arrangements of nucleosome-like particles. It is not possible to determine if any of these densities are nucleosomes on the basis of linker DNAs, which generally cannot be seen. (D1 and D2) Two examples of loosely packed nucleosome-like particles. (E) An example position that has few nucleosome-like particles in the center. (F) An example megacomplex (circled). Note that SI Appendix, Fig. S5G shows the 2D-classified nucleosome-like particles reproduce the packing phenomena seen in B–E. See Movie S1 for more tomographic slices.
Condensed Mitotic Chromosomes Contain Fewer Megacomplexes.
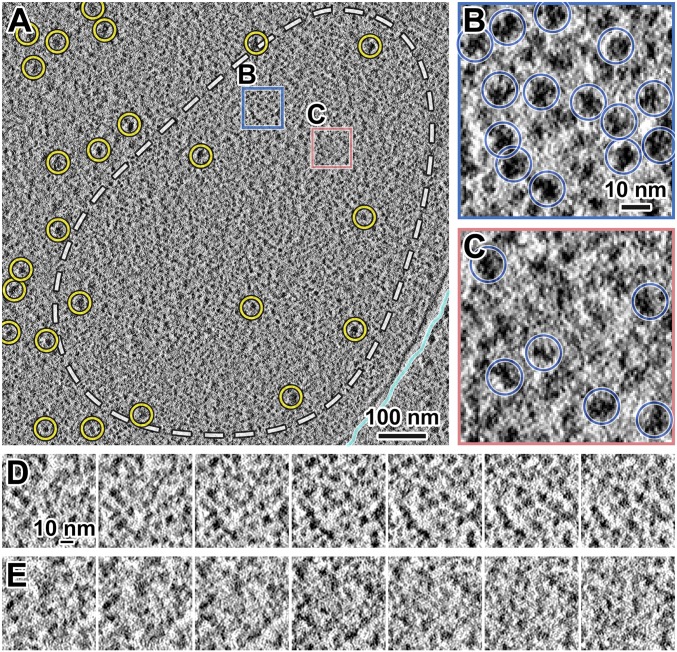
Unlike in G2-phase cells, in each cryotomogram of a prometaphase cell we found a single position that has features consistent with its being a section through a condensed chromosome (Fig. 2A and Movie S2). For example, nucleosome-like particles are abundant within these positions (Fig. 2 B and D) and rarer outside (Fig. 2 C and E). Compared with G2-phase cells, in which megacomplexes are distributed throughout the nucleoplasm (Fig. 1A), megacomplexes are mostly absent from these large contiguous regions in prometaphase cells. These positions are hundreds of nanometers wide, matching the size of condensed chromosomes seen by fluorescence microscopy (SI Appendix, Fig. S1). If these positions are indeed chromosomes, which are thicker than cryosections, the megacomplex-depleted positions should also span multiple cryosections. To test this hypothesis, we sampled a larger nuclear volume by cryo-ET of five nearly sequential cryosections, covering ∼700 nm in the z axis within the same cell (SI Appendix, Fig. S4A). There is a single megacomplex-poor region in each of these cryosections, which is consistent with these positions being sections through at least one of the three fission yeast chromosomes (SI Appendix, Fig. S4 B–F). Taken together, we located mitotic chromosomes and found that they have fewer megacomplexes within.
Fig. 2.
Condensed S. pombe mitotic chromosomes contain few megacomplexes. (A) Cryotomographic slice (11 nm) of a prometaphase cell. Cyan line, nuclear envelope; yellow circles, megacomplexes. The dashed white line roughly outlines a condensed chromosome. Rectangular boxes are enlarged sixfold in B and C. (B) An example position with many nucleosome-like particles. (C) An example position with a megacomplex and fewer nucleosome-like particles. Yellow circle, a megacomplex. (D and E) Consecutive cryotomographic slices (10 nm) in a 2-nm step size of positions shown in B and C, respectively. See Movie S2 for more tomographic slices.
No Evidence of Abundant Canonical Nucleosome Conformations in Vivo.
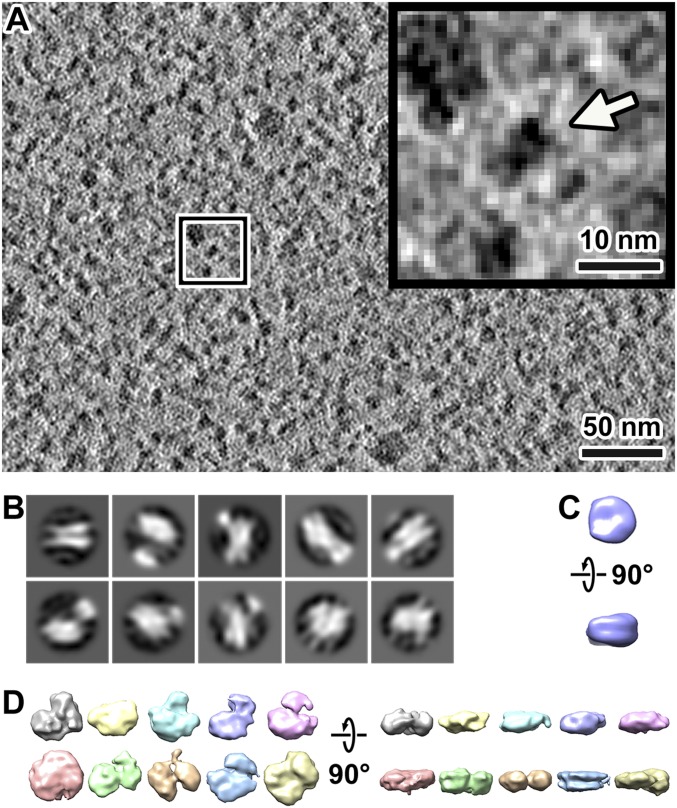
We imaged a few cryosection positions of G2-phase S. pombe cells on an electron cryomicroscope equipped with a Volta phase plate, a direct-detection camera operated in counting mode, and an imaging filter operated in zero-loss mode (Fig. 3A). Notably, Volta cryo-EM data have much more contrast, making it possible to locate and determine the orientations of smaller protein complexes and to resolve protein complexes packed to near-crystalline density inside of cells (29, 35). We template-matched candidate nucleosomes by using a cylinder as a reference, and then we filtered the candidates using a low cross-correlation cutoff (SI Appendix, Fig. S5). This low cutoff decreases the number of false negatives and increases the number of false positives. We then attempted to “purify” the nucleosomes in silico by classification (SI Appendix, Fig. S5) (36). As shown in SI Appendix, Fig. S5D, 2D classification allows obvious false positives to be identified and excluded from analysis. Examples of false positives are megacomplexes, particles with the wrong shape, and globular particles that are too small. Owing to the crowdedness of nucleoplasm, adjacent densities influenced the 2D classification (Fig. 3B), thereby reducing the resolution of some class averages. Some 2D class averages had pairs of linear densities that resemble the DNA gyres seen in the “side” views of the canonical nucleosome (Fig. 3B). For 3D classification, canonical nucleosomes are expected to have the characteristic DNA gyres and the linker-DNA density stumps. However, 3D classification of these 2D-classified nucleosome-like particles did not reveal any class averages that met these criteria (Fig. 3 C and D).
Fig. 3.
Subtomogram averages of nucleosome-like particles do not resemble the canonical nucleosomes. (A) Cryotomographic slice (10 nm) of a G2-phase cell nucleoplasm. (Inset) A nucleosome-like particle, indicated by an arrow, enlarged fivefold from the position boxed in white. (B) The 2D classification of nucleosome template-matching hits from four Volta cryotomograms of G2-phase chromatin. (C) A simulated density map of the canonical nucleosome [PDB 1ID3 (65)], low-pass–filtered to 40-Å resolution. (D) Isosurface renderings of representative 3D class averages of template-matching hits in four Volta cryotomograms of G2-phase chromatin. The particles contributing to each class average are likely to be heterogeneous.
The absence of canonical nucleosomes in our 3D class averages could be due to insufficient resolution, even though recent studies (including our own) done with similar hardware and imaging parameters suggest otherwise (37, 38). Alternatively, if S. pombe nucleosomes cannot form canonical nucleosomes at all, none of the class averages would resemble canonical nucleosomes. To test this possibility, we imaged the chromatin released from cellular lysates (SI Appendix, Fig. S6 A–C). The 3D classification shows that one of the class averages (the most abundantly populated one) has features expected of the canonical nucleosome (SI Appendix, Fig. S6 D and E). This result suggests that S. pombe nucleosomes with the canonical conformation can be observed in some conditions. One potential explanation is that nucleosomes are conformationally heterogeneous, such as being partially unwrapped to varying degrees. Another explanation is that nucleosomes are constitutionally heterogeneous; i.e., the types of proteins bound are variable (SI Appendix). At present, we cannot distinguish between these two possibilities. Because we cannot identify nucleosomes conclusively, as a compromise we combined template-matching and 2D classification (SI Appendix, Fig. S5) to remove from analysis particles that are too small or too large. This approach also filters out nucleosomes that have different structures relative to canonical nucleosomes and nucleosomes that are tightly associated with many proteins; the distribution of filtered hits is nevertheless consistent with the crowding seen by eye (SI Appendix, Fig. S5G). Our 3D packing analysis below takes into account the 2D-classified template-matched particles, which we will continue to refer to as nucleosome-like particles.
Mitotic Chromosomes Condense Unevenly and Support Transcription.
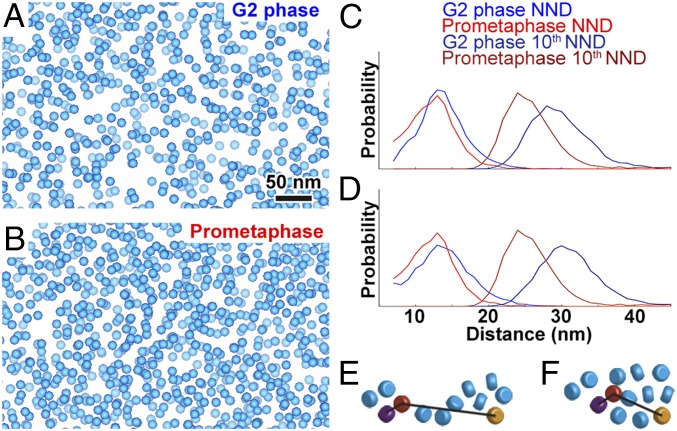
Volta cryotomograms of prometaphase nuclei also revealed that nucleosome-like particles are packed irregularly in megacomplex-poor chromosome positions (Fig. 4A and Movie S3). Importantly, we confirmed that some positions (Fig. 4 B and D) have more nucleosome-like particles than others (Fig. 4 C and E), meaning that condensation is uneven. The pockets are also irregular (Fig. 4C), as in G2-phase cells. These phenotypes also hold for chromatin in lysates (SI Appendix, Fig. S7). To detect changes in chromosome packing, we analyzed the positions of 2D-classified nucleosome-like particles in Volta cryotomograms of G2-phase and prometaphase cells (Fig. 5 A and B and SI Appendix, Fig. S8). If chromosomes condense via uniform accretion, the average nearest-neighbor distance (NND) distribution of the nucleosome-like particles should shorten. The NNDs of nucleosome-like particles in G2-phase were longer than in prometaphase, but not by much (two-tailed t test, P < 0.01, difference of means: 1.6 nm) (Fig. 5C). We previously used simulated distributions of particles to show that the 10th NND is sensitive to changes in particle cluster size and separation (36). We found that the 10th NNDs shifted to shorter distances in prometaphase cells (two-tailed t test, P < 0.001, difference of means: 4.1 nm), which explains the more crowded appearance of prometaphase chromosomes. To test if the cryomicrotomy compression artifact changed the apparent packing, we transformed the distributions of nucleosome-like particles, assuming that compression moves nucleosomes uniformly closer along with cutting direction and further apart perpendicular to the section surface. The NND (two-tailed t test, P < 0.01, difference of means: 1.6 nm) and 10th NND distributions (two-tailed t test, P < 0.01, difference of means: 5.3 nm) of the transformed data yield the same conclusion (Fig. 5D). Given the irregular spacing and packing of nucleosome-like particles, there are multiple ways that chromatin could rearrange to produce a small NND change and a larger 10th NND change. The simplest model is that nucleosome-like–particle clusters enlarge in mitosis (Fig. 5 E and F).
Fig. 4.
The macromolecular complexes are unevenly packed within S. pombe mitotic condensed chromosomes. (A) Cryotomographic slice (11 nm) of a nucleus in a prometaphase cell, imaged with Volta phase contrast. White dashed line, approximate chromosome boundary. Yellow circles, megacomplexes. The positions in the blue and salmon boxes are enlarged sixfold in B and C, respectively. (B) A position within the mitotic chromosome that contains many closely packed nucleosome-like particles. (C) A position within the mitotic chromosome that contains fewer, loosely packed nucleosome-like particles. Blue circles, 2D-classified nucleosome-like particles. Notice that the centers of mass of some particles are “above” or “below” the tomographic slice shown. These particles therefore appear either smaller or less dense than the other nucleosome-like particles. (D and E) Consecutive cryotomographic slices (10 nm) in a 2-nm step size of the positions shown in B and C, respectively. See Movie S3 for more tomographic slices.
Fig. 5.
Comparison of macromolecular complex packing in G2-phase and prometaphase S. pombe cells. (A and B) Template-matched, 2D-classified filtered hits of nucleosome-like particles rendered as blue spheres in Volta cryotomograms of a (A) G2-phase and (B) prometaphase cell. (C) Nearest-neighbor distance (NND) and 10th NND analysis of template-matching hits. x axis, NND or 10th NND, 1-nm bins; y axis, normalized probability. (D) NND and 10th NND of compression-compensated template-matching hits. (E and F) Cartoons showing how (E) two small or (F) one large cluster of nucleosomes (rounded cylinders) could have similar NND, but different 10th NND. In each panel, the purple and orange nucleosomes are the respective nearest and 10th-nearest neighbors of the red nucleosome.
The existence of loosely packed nucleosome-like particles in prometaphase cells suggests that mitotic chromatin is permissive to transcriptional machinery, which is consistent with the observation that some genes are up-regulated during mitosis (12–14). However, these earlier studies could not determine if S. pombe mitotic transcripts are from active mitotic RNA polymerase II transcription or are stable mRNAs left over from G2 phase. Phosphorylation of RNA polymerase II at Serine 2 of the carboxyl-terminal domain (phospho-Ser2 CTD) heptamer repeat is a conserved marker of transcription elongation (39, 40). Using immunofluorescence, we confirmed the existence of RNA polymerase II with phospho-Ser2 CTD, and therefore active transcription, in prometaphase chromosomes (SI Appendix, Fig. S9). To rule out the possibility of abnormal transcriptional activity in the mutants, we imaged asynchronous wild-type cultures, in which a small minority of cells was undergoing mitosis. We also detected phospho-Ser2 CTD signal in early mitotic cells. Therefore, we confirmed that transcription is not shut down globally in mitotic S. pombe cells.
Dinucleosome-Like Particles Are Irregular in Vivo.
Nucleosome-like particles are so crowded that it is infeasible to manually search for higher-order motifs. Our previous study of natural Saccharomyces cerevisiae chromatin in vitro showed that if a higher-order motif is abundant, it can be found automatically by reference-free 2D classification (36). One functionally important motif is the face-to-face–stacked dinucleosome-like particle, which if present in vivo would limit access to regulatory motifs like the acidic patch (2). We therefore performed 2D classification analysis on dinucleosome-like particles extracted from our cryotomograms. Our 2D classification of S. pombe chromatin in vivo did not reveal any classes containing face-to-face–stacked dinucleosome-like particles (SI Appendix, Fig. S10), meaning that such interactions must be rare in vivo. We did not find any other classes that convincingly resemble pairs of nucleosomes in any relative orientation. G2-phase chromatin (SI Appendix, Fig. S10A) was more irregularly packed in terms of the separation between nucleosome-like particles than in prometaphase cells (SI Appendix, Fig. S10B). Taken together, our 2D classification analyses did not produce evidence of ordered chromatin in vivo.
Discussion
We have directly visualized the nuclei in both interphase and mitotic S. pombe cells by cryo-ET. The chromosome-condensation phenotypes are subtle. In both interphase and mitotic cells, most of the nucleosome-like particles are packed irregularly, sometimes into cluster-like arrangements. The nearest-neighbor distance between these particles changes little in mitotic cells. In other words, there is no wholesale accretion as commonly depicted in textbooks. The most notable—yet still subtle—changes are that mitotic chromatin contains fewer megacomplexes in vivo and that this chromatin remains more compact than interphase chromatin after being released from lysed cells.
Cryo-ET Reveals Insights into Higher-Order Chromatin Organization.
If the majority of nucleosomes are partially unwrapped in vivo, then the unwrapped states would be a large source of conformational variability. Recent studies showed that isolated nucleosomes can adapt to a range of unwrapped and wrapped-but-mildly distorted conformations in vitro (41, 42). In higher-order chromatin, linker-DNA variability alone can give rise to irregular oligonucleosome folding (43). These sources of conformational distortions can explain how our 2D classification did not uncover any evidence of abundant dinucleosome-like motifs or any other higher-order motifs. Irregular chromatin is therefore a conserved feature in single-celled eukaryotes (21, 22). A study using a DNA negative-staining method concluded that chromatin structure is also irregular in mammalian cells (17), which is consistent with earlier studies of other mammalian cells and chromosomes (18, 19, 44–48).
Cryo-ET, Hi-C, and Live-Cell Imaging Paint a Complex Picture of Chromatin.
Our S. pombe cryo-ET data allow for a comparison with previous Hi-C studies (SI Appendix). Two recent studies reported that short-range Hi-C contacts are less probable in mitotic S. pombe cells than in interphase cells (6, 7), echoing a study of mitotic human cells (27). This counter intuitive phenomenon was also observed in quiescent B cells, which have condensed chromosomes and lower short-range Hi-C contact probabilities than active B cells, which have open chromatin (49). If Hi-C detections arise solely from nucleosome–nucleosome proximity, then nucleosomes should move further apart in mitosis; we did not observe evidence of this. What else, then, could explain the lower short-range Hi-C contact frequency in mitotic cells? This conundrum might be reconciled if the dynamics of groups of nucleosomes are taken into consideration (6, 50, 51). A slowdown of individual nucleosome dynamics within a group could explain the lower frequency that cross-linkable groups can interact with each other in mitotic cells. We propose a model of mitotic condensation that incorporates our structural data and published dynamic principles from recent studies (Fig. 6). In this model, the average distance between nearest-neighbor nucleosomes and other similarly sized macromolecular complexes does not decrease much. Instead, small nucleosome clusters enlarge and become less dynamic. This mechanism could be similar to local condensation in human interphase chromatin, which is proposed to be organized as “nanodomains” that cluster together (52).
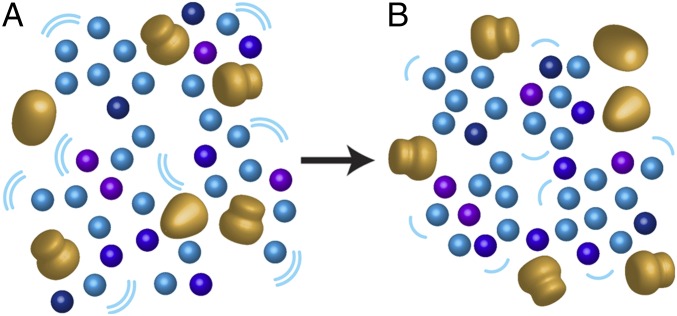
Fig. 6.
A molecular model of interphase and mitotic chromatin. (A) Interphase chromatin is loosely packed, with many megacomplexes and pockets. Blue, purple, and dark blue, nucleosome-like particles, with heterogeneity denoted by the variability in colors; large gold bodies, megacomplexes. Megacomplexes (preribosomes, spliceosomes, transcription-preinitiation complexes, etc.) are interspersed between the nucleosome-like particles and macromolecular-complex–free pockets. (B) During mitotic chromosome condensation, most nucleosome-like particles form large clusters, thereby excluding or inhibiting the assembly of megacomplexes. Furthermore, chromatin dynamics (magnitude proportional to the size and number of the curved lines) also decrease. In both interphase and mitotic chromatin, ordered motifs are largely absent.
Role of Uneven Condensation in Transcriptional Regulation.
The strong correlation between mitotic chromosome condensation and transcriptional repression is well established (53, 54). Mechanistic evidence of mitotic repression came from the observation that mitotic condensation is coincident with transcription-factor displacement (55). Even though transcription is largely down-regulated in mitotic S. pombe cells, many cell-cycle–regulated genes actually get expressed more (12–14). In agreement, our immunofluorescence experiments detected elongating RNA polymerase II in mitotic S. pombe chromosomes. We propose that chromatin structure at multiple size scales permits access to regulatory sequences, somewhat independently of dynamics. At the mononucleosome level, many chromatin-regulatory complexes recognize and bind to the nucleosome’s face (56). Because face-to-face nucleosome stacking is also exceptionally rare in mitotic cells, this critical surface remains accessible for interactions. Partial nucleosome unwrapping could also expose more DNA. At the level of the chromosome, fewer megacomplexes are interspersed within the more densely packed chromatin. However, uneven condensation leaves open pockets that could remain permissive to the occasional passage or assembly of megacomplexes such as spliceosomes and transcription-preinitiation complexes. Cryo-fluorescence microscopy has been used in combination with cryo-EM of vitreous sections to detect subcellular structures (57, 58). This form of correlative imaging could potentially facilitate the identification of some megacomplexes. If uneven chromosome condensation is conserved, the mechanisms proposed here could explain the recent observations that mitotic chromatin remains accessible and somewhat transcriptionally active in mammalian cells (59–61).
Materials and Methods
S. pombe nda3-KM311 and cdc25-22 cells were arrested at their restrictive temperatures and checked by fluorescence light microscopy. Immunofluorescence of spheroplasted cells was done with antibodies against RNA polymerase II CTD-repeat YSPTSPS (phospho S2) or fibrillarin. For cryo-EM, cells were self-pressurized frozen, cryosectioned, then attached to either a perforated- or continuous-carbon grid, which was precoated with 10-nm gold colloids as fiducials for cryo-ET alignment. To minimize occlusion by grid bars at high tilt during cryo-ET imaging, the grid was aligned so that the ribbon was in between and parallel with the grid bars. For lysates, log-phase cells were spheroplasted, lysed on ice, applied to a freshly glow-discharged perforated-carbon grid, blotted with filter paper, then plunged into liquid ethane. Cryo-ET data were collected on the National University of Singapore (NUS) Titan Krios cryo-TEM or on the Netherlands Centre for Electron Nanoscopy (NeCEN) Titan Krios-1. Imaging details are shown in SI Appendix, Table S1. Tomograms were reconstructed using the IMOD software package (62) and template-matched with PEET (63). Subtomogram classification and 3D averaging of nucleosomes were done in RELION (28, 64). Nearest-neighbor distance analysis of template-matching hits were performed on coordinates that were “compensated” for compression. Tomography data were deposited in the EMDataBank (EMD-6846) and Electron Microscopy Public Image Archive (EMPIAR-10125). All custom scripts are available at https://github.com/anaphaze/ot-tools.
Supplementary Material
Acknowledgments
We thank the Centre for BioImaging Sciences microscopy staff for support and training; Snezhka Oliferenko and Mohan Balasubramanian for feedback, sharing yeast strains, and cell-culture advice; and Kazuhiro Maeshima for feedback. High-resolution EM data were collected at NeCEN with assistance from scientific operator Christoph Diebolder. S.C., C.C., Z.Y.T., and L.G. were supported by NUS startup Grants R-154-000-515-133, R-154-000-524-651, D-E12-303-154-217, R-154-000-558-133, MOE T2 R-154-000-624-112, and MOE T1 R-154-000-A49-114.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720476115/-/DCSupplemental.
References
- 1.Dixon JR, Gorkin DU, Ren B. Chromatin domains: The unit of chromosome organization. Mol Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 4.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizuguchi T, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 2014;516:432–435. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kakui Y, Rabinowitz A, Barry DJ, Uhlmann F. Condensin-mediated remodeling of the mitotic chromatin landscape in fission yeast. Nat Genet. 2017;49:1553–1557. doi: 10.1038/ng.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanizawa H, Kim KD, Iwasaki O, Noma KI. Architectural alterations of the fission yeast genome during the cell cycle. Nat Struct Mol Biol. 2017;24:965–976. doi: 10.1038/nsmb.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuda A, et al. Highly condensed chromatins are formed adjacent to subtelomeric and decondensed silent chromatin in fission yeast. Nat Commun. 2015;6:7753. doi: 10.1038/ncomms8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano T. Condensin-based chromosome organization from bacteria to vertebrates. Cell. 2016;164:847–857. doi: 10.1016/j.cell.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 10.Maeshima K, Eltsov M. Packaging the genome: The structure of mitotic chromosomes. J Biochem. 2008;143:145–153. doi: 10.1093/jb/mvm214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 12.Rustici G, et al. Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 13.Oliva A, et al. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng X, et al. Identification of cell cycle-regulated genes in fission yeast. Mol Biol Cell. 2005;16:1026–1042. doi: 10.1091/mbc.E04-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beliveau BJ, et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun. 2015;6:7147. doi: 10.1038/ncomms8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pombo A, Dillon N. Three-dimensional genome architecture: Players and mechanisms. Nat Rev Mol Cell Biol. 2015;16:245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 17.Ou HD, et al. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357:eaag0025. doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDowall AW, Smith JM, Dubochet J. Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J. 1986;5:1395–1402. doi: 10.1002/j.1460-2075.1986.tb04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan L, Jensen GJ. Electron tomography of cells. Q Rev Biophys. 2012;45:27–56. doi: 10.1017/S0033583511000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gan L, Ladinsky MS, Jensen GJ. Chromatin in a marine picoeukaryote is a disordered assemblage of nucleosomes. Chromosoma. 2013;122:377–386. doi: 10.1007/s00412-013-0423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, et al. Budding yeast chromatin is dispersed in a crowded nucleoplasm in vivo. Mol Biol Cell. 2016;27:3357–3368. doi: 10.1091/mbc.E16-07-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lantermann AB, et al. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- 24.Bühler M, Gasser SM. Silent chromatin at the middle and ends: Lessons from yeasts. EMBO J. 2009;28:2149–2161. doi: 10.1038/emboj.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: A cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 26.Yam C, Gu Y, Oliferenko S. Partitioning and remodeling of the Schizosaccharomyces japonicus mitotic nucleus require chromosome tethers. Curr Biol. 2013;23:2303–2310. doi: 10.1016/j.cub.2013.09.057. [DOI] [PubMed] [Google Scholar]
- 27.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bharat TA, Scheres SH. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc. 2016;11:2054–2065. doi: 10.1038/nprot.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khoshouei M, Radjainia M, Baumeister W, Danev R. Cryo-EM structure of haemoglobin at 3.2 Å determined with the Volta phase plate. Nat Commun. 2017;8:16099. doi: 10.1038/ncomms16099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fantes P. Epistatic gene interactions in the control of division in fission yeast. Nature. 1979;279:428–430. doi: 10.1038/279428a0. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 32.Gleizes PE, et al. Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J Cell Biol. 2001;155:923–936. doi: 10.1083/jcb.200108142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen BL, Taatjes DJ. The mediator complex: A central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oesterreich FC, et al. Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell. 2016;165:372–381. doi: 10.1016/j.cell.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engel BD, et al. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife. 2015;4:e04889, and erratum (2015) 4:e11383. doi: 10.7554/eLife.04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai S, Song Y, Chen C, Shi J, Gan L. Natural chromatin is heterogeneous and self-associates in vitro. Mol Biol Cell. 2018;29:1652–1663. doi: 10.1091/mbc.E17-07-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai S, Bock D, Pilhofer M, Gan L. The in situ structures of mono-, di-, and tri-nucleosomes in human heterochromatin. Mol Biol Cell. August 9, 2018 doi: 10.1091/mbc.E18-05-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eltsov M, et al. Nucleosome conformational variability in solution and in interphase nuclei evidenced by cryo-electron microscopy of vitreous sections. Nucleic Acids Res. July 25, 2018 doi: 10.1093/nar/gky670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harlen KM, Churchman LS. The code and beyond: Transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat Rev Mol Cell Biol. 2017;18:263–273. doi: 10.1038/nrm.2017.10. [DOI] [PubMed] [Google Scholar]
- 41.Bilokapic S, Strauss M, Halic M. Histone octamer rearranges to adapt to DNA unwrapping. Nat Struct Mol Biol. 2018;25:101–108. doi: 10.1038/s41594-017-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bilokapic S, Strauss M, Halic M. Structural rearrangements of the histone octamer translocate DNA. Nat Commun. 2018;9:1330. doi: 10.1038/s41467-018-03677-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collepardo-Guevara R, Schlick T. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc Natl Acad Sci USA. 2014;111:8061–8066. doi: 10.1073/pnas.1315872111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouchet-Marquis C, Dubochet J, Fakan S. Cryoelectron microscopy of vitrified sections: A new challenge for the analysis of functional nuclear architecture. Histochem Cell Biol. 2006;125:43–51. doi: 10.1007/s00418-005-0093-x. [DOI] [PubMed] [Google Scholar]
- 45.Fussner E, et al. Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J. 2011;30:1778–1789. doi: 10.1038/emboj.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fussner E, et al. Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep. 2012;13:992–996. doi: 10.1038/embor.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishino Y, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eltsov M, Sosnovski S, Olins AL, Olins DE. ELCS in ice: Cryo-electron microscopy of nuclear envelope-limited chromatin sheets. Chromosoma. 2014;123:303–312. doi: 10.1007/s00412-014-0454-0. [DOI] [PubMed] [Google Scholar]
- 49.Kieffer-Kwon KR, et al. Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol Cell. 2017;67:566–578.e10. doi: 10.1016/j.molcel.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hihara S, et al. Local nucleosome dynamics facilitate chromatin accessibility in living mammalian cells. Cell Rep. 2012;2:1645–1656. doi: 10.1016/j.celrep.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 51.Nozaki T, et al. Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol Cell. 2017;67:282–293.e7. doi: 10.1016/j.molcel.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 52.Fang K, et al. Super-resolution imaging of individual human sub-chromosomal regions in situ reveals nanoscopic building blocks of higher-order structure. ACS Nano. 2018;12:4909–4918. doi: 10.1021/acsnano.8b01963. [DOI] [PubMed] [Google Scholar]
- 53.Taylor JH. Nucleic acid synthesis in relation to the cell division cycle. Ann N Y Acad Sci. 1960;90:409–421. doi: 10.1111/j.1749-6632.1960.tb23259.x. [DOI] [PubMed] [Google Scholar]
- 54.Prescott DM, Bender MA. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- 55.Martínez-Balbás MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 56.McGinty RK, Tan S. Nucleosome structure and function. Chem Rev. 2015;115:2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gruska M, Medalia O, Baumeister W, Leis A. Electron tomography of vitreous sections from cultured mammalian cells. J Struct Biol. 2008;161:384–392. doi: 10.1016/j.jsb.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Bharat TAM, Hoffmann PC, Kukulski W. Correlative microscopy of vitreous sections provides insights into BAR-domain organization in situ. Structure. 2018;26:879–886.e3. doi: 10.1016/j.str.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsiung CC, et al. Genome accessibility is widely preserved and locally modulated during mitosis. Genome Res. 2015;25:213–225. doi: 10.1101/gr.180646.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teves SS, et al. A dynamic mode of mitotic bookmarking by transcription factors. eLife. 2016;5:e22280. doi: 10.7554/eLife.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palozola KC, et al. Mitotic transcription and waves of gene reactivation during mitotic exit. Science. 2017;358:119–122. doi: 10.1126/science.aal4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 63.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 64.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.