Significance
In DNA homologous recombination, recombinase-coated single-stranded DNA filament formation is the first committed step and is subject to tight regulation. Stabilization of nucleoprotein filament by accessory proteins can be achieved by enhancing filament formation, reducing filament disassembly, or both. However, the mechanism of regulation is not understood by conventional biochemical methods. This is a study of the mechanism of how accessory proteins stimulate filament assembly by applying single-molecule methods that allow us to monitor the binding of Rad51 on DNA in mouse and fission yeast. Our results show that the Swi5–Sfr1 complex demonstrates the evolutionarily conserved stimulation of Rad51 filament assembly by stabilizing Rad51 on DNA, allowing both the formation of the stable nucleus and the reduction of Rad51 dissociation.
Keywords: homologous recombination, single-molecule microscopy, Rad51, Swi5–Sfr1
Abstract
Eukaryotic Rad51 protein is essential for homologous-recombination repair of DNA double-strand breaks. Rad51 recombinases first assemble onto single-stranded DNA to form a nucleoprotein filament, required for function in homology pairing and strand exchange. This filament assembly is the first regulation step in homologous recombination. Rad51 nucleation is kinetically slow, and several accessory factors have been identified to regulate this step. Swi5–Sfr1 (S5S1) stimulates Rad51-mediated homologous recombination by stabilizing Rad51 nucleoprotein filaments, but the mechanism of stabilization is unclear. We used single-molecule tethered particle motion experiments to show that mouse S5S1 (mS5S1) efficiently stimulates mouse RAD51 (mRAD51) nucleus formation and inhibits mRAD51 dissociation from filaments. We also used single-molecule fluorescence resonance energy transfer experiments to show that mS5S1 promotes stable nucleus formation by specifically preventing mRAD51 dissociation. This leads to a reduction of nucleation size from three mRAD51 to two mRAD51 molecules in the presence of mS5S1. Compared with mRAD51, fission yeast Rad51 (SpRad51) exhibits fast nucleation but quickly dissociates from the filament. SpS5S1 specifically reduces SpRad51 disassembly to maintain a stable filament. These results clearly demonstrate the conserved function of S5S1 by primarily stabilizing Rad51 on DNA, allowing both the formation of the stable nucleus and the maintenance of filament length.
Rad51 recombinases are essential for eukaryotic homologous-recombination DNA repair (1, 2). As a replication fork encounters a lesion, collapsed forks lead to DNA double-strand breaks (DSBs). Homologous recombination is the major pathway to restart replication (3). To carry out homologous-recombination repair, the broken end of DNA is resected to reveal a 3′ protruded single-stranded DNA (ssDNA), which Rad51 binds to form a nucleoprotein filament (1, 2, 4–7). The resultant helical nucleoprotein filament is the active component responsible for homology searching and strand exchange and is essential for DSB repair and genomic maintenance.
Among the cascade of steps required for DSB repair, Rad51 nucleoprotein filament assembly is the committed step and is subject to tight regulation (1, 2, 8–10). Maintaining a stable but dynamic nucleoprotein filament is critical for recombinase function, as this filament is essential for both the homology search and strand exchange required for heteroduplex DNA formation. Filament assembly includes a rate-determining nucleation step, where several Rad51 molecules bind ssDNA to form a stable nucleus, followed by a faster extension step (11–14). The filament assembly involves monomer addition occurring both at the 3′ and 5′ ends of the recombinase filament but with different rates, leading to the overall end preference. During strand exchange, this filament likely maintains a certain length through dissociating and rebinding of Rad51. Thus, the filament overall moves dynamically with polarity (15–18). Several proteins stimulate and regulate Rad51 filament assembly (19–26). Swi5 and Sfr1 were identified by genetic studies in the fission yeast Schizosaccharomyces pombe (27, 28). Mutations in fission yeast swi5 and sfr1 decrease recombination rates and increase sensitivity to ionizing radiation and DNA-damaging chemicals (27, 28). In vitro biochemical and biophysical studies showed that both fission yeast and mouse Swi5 and Sfr1 proteins form a heterodimeric complex that physically interacts with Rad51 to facilitate Rad51-mediated recombination (25, 26, 29–31). Both fission yeast and mouse Swi5–Sfr1 (S5S1) complexes stabilize the Rad51 presynaptic filament and increase the ssDNA-dependent ATPase activity of Rad51. Moreover, mouse S5S1 (mS5S1) has been reported to enhance the release of ADP from mRAD51 presynaptic filaments (32).
How Rad51 nucleoprotein filaments are stabilized by these accessory factors is not clear. Stabilization could be achieved by increasing the Rad51 on rate or decreasing the off rate, or both. Conventional biochemical studies are based on averaged and equilibrium measurements, making it challenging to elucidate the molecular events responsible for these kinetic events. Here, we used single-molecule tethered particle motion (TPM) and fluorescence resonance energy transfer (smFRET) experiments to characterize individual Rad51 filament assembly and disassembly kinetics as well as the nucleation process in real time. Our single-molecule results support a model in which Swi5–Sfr1 stabilizes Rad51 on DNA by preventing its dissociation. This effect leads to both stable nucleus formation and a longer-lasting filament. Specifically, this stabilization decreases the nucleation size from three to two Rad51 molecules in mouse and reduces filament disassembly nearly threefold in fission yeast. Despite different kinetic properties of mouse and fission yeast Rad51, the Swi5–Sfr1 complex stimulates the Rad51 process through a similar and conserved mechanism.
Results
Mouse SWI5–SFR1 Stimulates mRAD51 Nucleoprotein Filament Assembly.
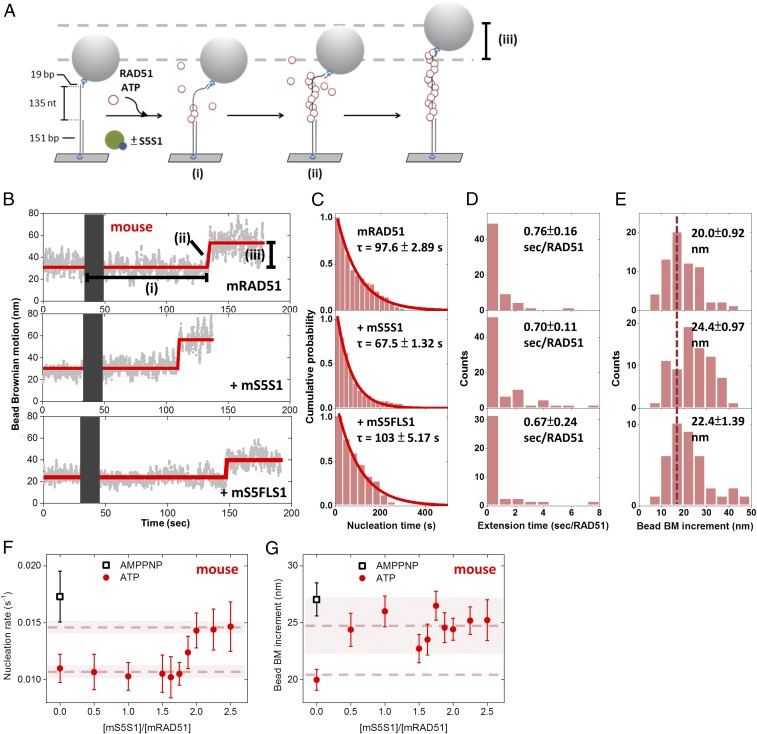
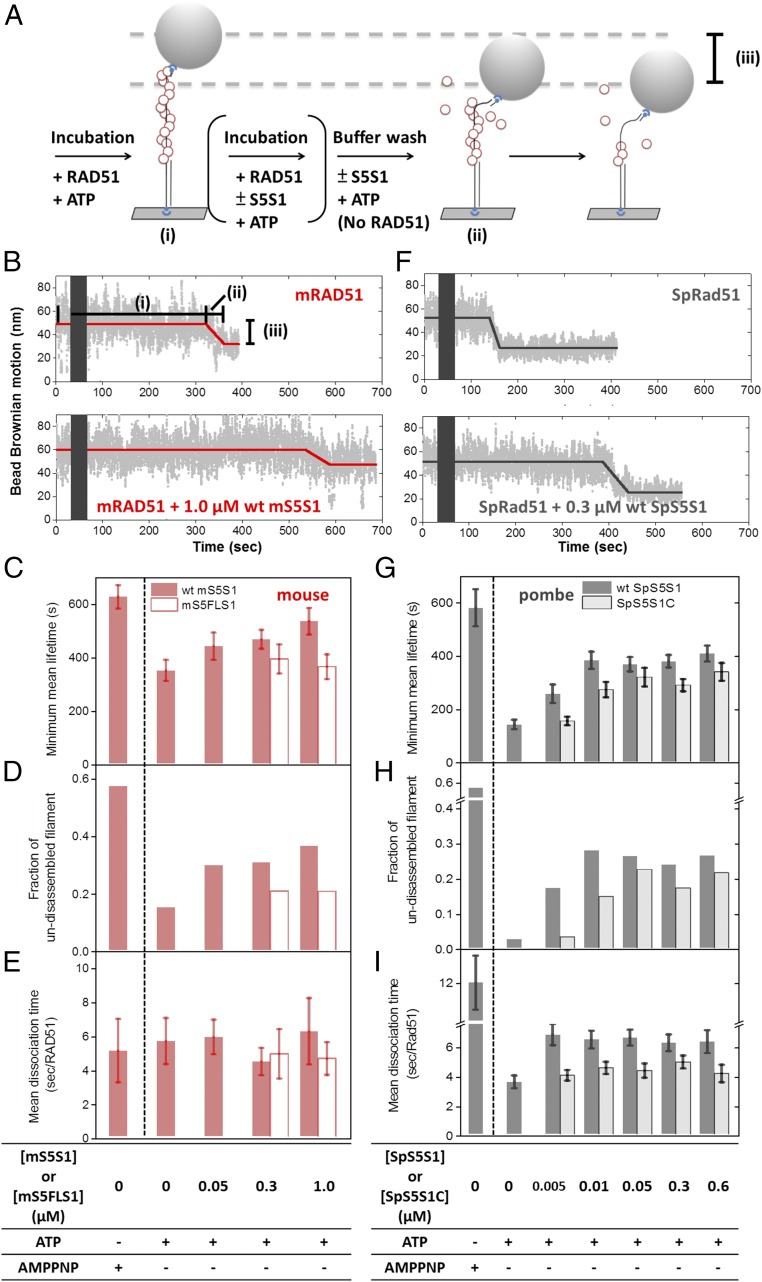
Among many accessory proteins regulating homologous recombination, the Swi5–Sfr1 complex stimulates various stages of the process, including filament assembly and strand exchange (25, 26, 28–35). Here, we took advantage of previously developed single-molecule TPM experiments (36–38) to monitor Rad51 nucleoprotein filament assembly in real time (Fig. 1A). We designed a gapped DNA substrate containing a 135-nt secondary structure-free, poly(dT) ssDNA region [(dT)135 gapped DNA; Fig. 1A] immobilized on the surface of a glass slide via a 5′ digoxigenin–anti-digoxigenin linkage. The substrate was annealed to a short oligo labeled with biotin at its 5′ end, which was, in turn, attached to a streptavidin-coated polystyrene bead for visualization purposes. The 151-bp double-stranded handle in the gapped DNA was used to prevent potential Rad51–surface interaction during TPM experiments. Bead Brownian motion (BM) of the DNA tether is constrained to a small region near the glass surface. The bead BM of bare (dT)135 gapped DNA for assembly experiments and 301-bp duplex DNA used for control experiments was measured to be 21.3 ± 5.78 and 35.2 ± 3.77 nm (SI Appendix, Fig. S1 A and B). In the presence of either ATP or AMPPNP, mRAD51 preferentially assembles onto the ssDNA region of the gapped substrate at 150 mM KCl (39) (SI Appendix, Fig. S1 A and C–G). Assembly experiments were initiated by introducing an mRAD51/ATP mixture into the reaction chamber containing the surface-anchored gapped substrates. Increases in bead Brownian motion reveal nucleoprotein filament formation as mRAD51 assembling onto DNA increases ssDNA length and stiffness of the DNA tether. This results in a change in the spatial extent of bead BM (32, 40, 41). Analysis of individual single-molecule TPM time courses revealed several kinetic parameters, including the (i) dwell time before bead BM increase (nucleation time, the time needed for RAD51 to form stable nuclei on ssDNA; Fig. 1 B and C), (ii) extension time (Fig. 1D; the time required to add RAD51 monomers to ssDNA), and (iii) amount of BM increase (Fig. 1E; reflecting the length of the nucleoprotein filament). Our results show that mRAD51 alone (0.8 μM) exhibits a nucleation time of 97.6 ± 2.89 s (Fig. 1C). Interestingly, a shorter nucleation time (67.5 ± 1.32 s) was observed when mRAD51 was preincubated with more than twofold excess mS5S1 (Fig. 1C). This observation demonstrates that mS5S1 stimulates the nucleation step of mRAD51 filament formation, which is rate-limiting. mS5S1 has neither dsDNA nor ssDNA affinity (25), so adding mS5S1 in the absence of mRAD51 did not change BM values (SI Appendix, Fig. S1H). Control experiments using an mS5S1 mutant that is defective for mRAD51 interaction, mSWI5FL/AA–SFR1 (mS5FLS1) (31), showed no stimulation of nucleation times (Fig. 1C). Collectively, these findings suggest that the interaction of mRAD51–mS5S1 in solution stimulates mRAD51 nucleation. Within our experimental resolution, we did not observe any change in mRAD51 extension times in the presence of mS5S1 (Fig. 1D and SI Appendix, Fig. S2A). On the other hand, the mean BM increment increases in the presence of mS5S1 (Fig. 1E), indicating that the mRAD51–mS5S1 complex forms longer filaments than mRAD51 alone.
Fig. 1.
Mouse SWI5–SFR1 stimulates mRAD51 nucleoprotein filament assembly. (A) Schematic illustration of the RAD51 nucleoprotein assembly experiments. (B) Representative bead BM time courses of mRAD51 (0.8 μM) assembly on (dT)135 DNA substrates without mS5S1 (Upper) or with 1.6 μM mS5S1 (Middle) or 1.6 μM mS5FLS1 mutant (Lower). Gray bars correspond to the dead time when recombinase mixtures with 2 mM ATP were introduced. (C–E) Histograms of nucleation time (C), mean extension time (seconds per RAD51) (D), and bead BM increment (E) of mRAD51 assembling. All experiments were carried out at 2 mM ATP. Error bars of nucleation rate are the SD of the mean by bootstrapping 5,000 times, and error bars of extension time are 1 SEM. (F) mRAD51 (0.8 μM) was preincubated with various stoichiometric ratios of mS5S1, and the mixture was introduced into a reaction chamber containing surface-bound (dT)135 gapped DNA. Nucleation rates are about constant (∼0.010 s−1) when the [mS5S1]/[mRAD51] ratio is less than 1.625. The nucleation rates of mRAD51 increase and achieve maximum value (∼0.015 s−1) when the ratio of [mS5S1]/[mRAD51] is larger than 2, suggesting that maximum nucleation stimulation occurs in the mixture of one mRAD51 and two mS5S1s. Individual nucleation rates were obtained based on maximum-likelihood estimation. (G) Bead BM increment of mRAD51 assembly on (dT)135 DNA substrates in the presence of the indicated ratios of mS5S1 to mRAD51. Bead BM increment reflects the coverage of the RAD51 nucleoprotein filaments. mRAD51 forms longer and more stable filaments in the presence of mS5S1. All experiments were carried out at 2 mM ATP. Black open squares represent the nucleation rate of mRAD51 in the presence of the nonhydrolyzable ATP analog AMPPNP in the absence of mS5S1. Dashed lines are the mean, and the shaded regions span 2 SDs. Error bars of bead BM increment are 1 SEM.
Stimulation of mRAD51 Nucleation Depends on mS5S1 Concentration.
We next asked how much mS5S1 is required for maximum stimulation. We incubated various concentrations of mS5S1 (0 to 2.0 μM) with 0.8 μM mRAD51 in solution and observed filament assembly kinetics as in Fig. 1A. Interestingly, a sigmoidal dependence of mS5S1/mRAD51 ratios is shown for nucleation rates (Fig. 1F), with nucleation rates starting to increase at a ratio larger than 1.5 and reaching a constant maximum nucleation rate of ∼0.015 s−1 when the ratio is larger than 2. This finding implies that two mS5S1 per one mRAD51 are required for maximum nucleation stimulation. For comparison purposes, we measured nucleation rates in the presence of the nonhydrolyzable ATP analog AMPPNP (black open square in Fig. 1F). Under these conditions, the nucleation rate for mRAD51 alone is fast (∼0.017 s−1), suggesting that mRAD51 nucleating clusters are more stable in the absence of ATP hydrolysis, consistent with previous reports on bacterial RecA proteins (12, 42). It is possible that mS5S1 stimulates mRAD51 nucleation by stabilizing mRAD51 nucleating clusters.
Bead BM increment, indicative of the length of the mRAD51 filament, on (dT)135 gapped DNA without mS5S1 is 20.0 ± 0.92 nm (Fig. 1 E and G), likely reflecting the equilibrium filament length of disassembly and assembly dynamics of the filament (14). The longer filament of 27.3 ± 1.51 nm is seen in the presence of AMPPNP (black open square in Fig. 1G), consistent with mRAD51 disassembly requiring ATP hydrolysis. Notably, in the presence of just 0.5-fold mS5S1, mRAD51 forms longer, more stable filaments, which is consistent with previous biochemical studies (25, 31, 32). Control experiments confirm that longer filaments in the presence of either S5S1/ATP or AMPPNP do not result from mRAD51 binding to the duplex handle of gapped DNA substrates (SI Appendix, Fig. S1 A and C–G).
Molecular Determinants of mSWI5–SFR1 Stimulation on mRAD51 Nucleation.
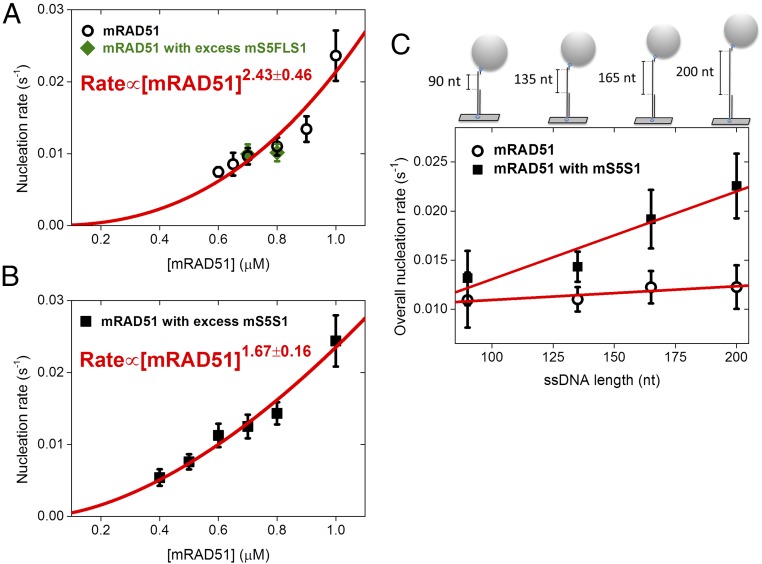
In the rate-limiting nucleation step of filament assembly, recombinases first form a stable nucleating cluster before extending into a longer functional filament. We used TPM experiments to measure how filament nucleation rates change with mRAD51 concentrations. The results are fitted to a power-law dependence, providing information about the mRAD51 nucleation unit of this rate-limited step. In the case of mRAD51 only, the fit returns n = 2.43 ± 0.46 (Fig. 2A). As the fit renders the lower limit, our result suggests that three mRAD51 monomers are required for stable nucleating cluster formation. This is consistent with earlier work showing that human RAD51 (hRAD51) has a nucleation unit of three monomers (14). Adding a twofold molar excess of mS5FLS1 to mRAD51 (green solid diamonds, Fig. 2A) does not change the mRAD51 nucleation unit. However, adding more than a twofold molar excess of mS5S1 to mRAD51 returned a power-law dependence of n = 1.67 ± 0.16 (Fig. 2B). Thus, the mRAD51–mS5S1 complex only needs two mRAD51 molecules to form a stable nucleation cluster. This reduction in the number of mRAD51 molecules required to form a stable nucleation cluster in the presence of mS5S1 provides a clear mechanism for increasing the rate of the mRAD51 nucleoprotein assembly process.
Fig. 2.
Mouse mS5S1 reduces the nucleation unit of mRAD51 and increases ssDNA affinity. (A and B) RAD51 concentration dependence of filament nucleation obtained by TPM experiments. Power-law fitting to the observed nucleation rates suggests the nucleation unit of RAD51: 2.43 ± 0.46 for mRAD51 (A) and 1.67 ± 0.16 for the mRAD51–mS5S1 complex (B). Green diamonds in A are nucleation rates of mRAD51 in the presence of excess mS5FLS1 mutant defective in stimulating mRAD51. mS5S1 and mS5FLS1 are in twofold excess in A and B. (C) ssDNA length dependence of mRAD51 filament nucleation rate obtained by TPM experiments. Gapped DNA substrates contain only one 5′ ds/ss junction but various lengths of ssDNA gaps (90 to 200 nt). As the gapped DNA substrate structure, overall nucleation rates are fitted to kssDNAapp(LssDNA) + kjunctionapp, where kssDNAapp and kjunctionapp are apparent ssDNA-dependent nucleation rate constant and apparent ds/ss junction-dependent nucleation rate constant. Circle, 0.8 μM mRAD51 only; square, mixture of 0.8 μM mRAD51 and 1.6 μM mS5S1. All experiments were carried out at 2 mM ATP.
During DNA DSB repair, the ends of the DNA breaks are resected to produce 3′ ssDNA overhangs. Thus, the repair substrate possesses a double-strand–single-strand junction (ds/ss junction) and a 3′ protruding ssDNA tail. Rad51 nucleation clusters could initiate filament assembly on ssDNA either near the ds/ssDNA junction or onto the free ssDNA sites along the tail. We compared the nucleation rates of mRAD51 in four gapped substrates containing one ds/ssDNA junction but different lengths of ssDNA tail (90, 135, 165, and 200 dT). Nucleation rates can be fitted linearly to ssDNA tail lengths, with the slope corresponding to the apparent nucleation rate on free ssDNA sites (Fig. 2C). For mRAD51 only, the dependence on ssDNA length was small, suggesting that mRAD51 alone prefers to nucleate near the junction. On the other hand, in the presence of a twofold excess of mS5S1, nucleation rates of mRAD51 showed a much stronger dependence on ssDNA lengths (filled squares, Fig. 2C), with an ∼6.5-fold increase in the slope [(8.95 ± 1.93) × 10−5 s−1⋅nt−1 for mS5S1–mRAD51 and (1.40 ± 0.52) × 10−5 s−1⋅nt−1 for mRAD51 alone; Fig. 2C and SI Appendix, Table S1]. These data indicate that mS5S1 stimulates mRAD51 binding by increasing its ssDNA affinity. Previous studies showed that DSB ends are resected to lengths of up to several kilobases in cells (4, 5); therefore, by dramatically increasing the ssDNA affinity of mRAD51, mS5S1 can effectively stimulate mRAD51 filament assembly.
smFRET Experiments Reveal Rad51 Binding and Dissociation Dynamics During Nucleating Events.
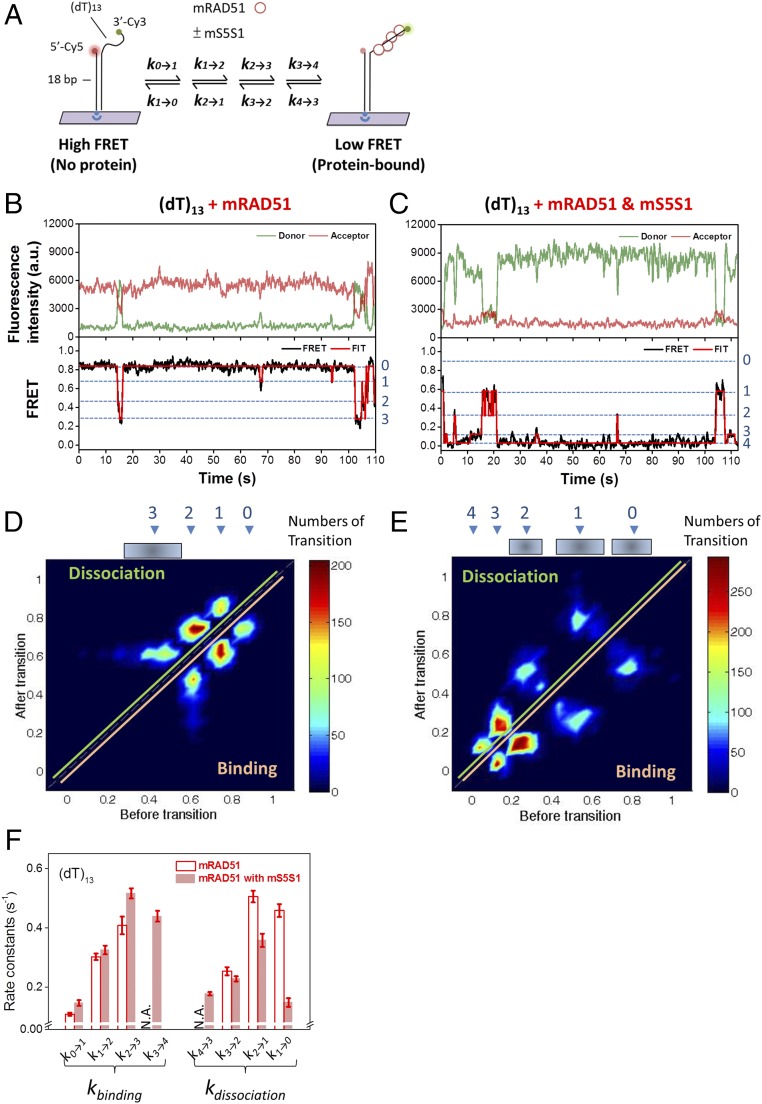
In TPM assembly experiments, we monitored the filament assembly kinetics that led to the successful assembly of recombinase nucleoprotein filaments. Due to their limited spatiotemporal resolution, TPM experiments cannot detect dynamics during nonproductive assembly events. For example, transient recombinase binding events are likely taking place but would be difficult to detect by TPM. To monitor transient binding events, we used smFRET to characterize nucleation dynamics of mRAD51 at high spatiotemporal resolution. The DNA substrates used in the smFRET experiments were short and composed of an 18-bp dsDNA handle and a short 3′ terminating dT overhang (13 or 18 nt; Fig. 3A and SI Appendix, Fig. S3 A–D). The fluorophore donor (Cy3) and acceptor (Cy5) dyes were positioned so that mRAD51 monomer binding and dissociation on the ssDNA region could be monitored. In the absence of mRAD51, ssDNA is flexible, and separation between the dye pairs is short, resulting in a high FRET efficiency [∼0.85 for (dT)13 and ∼0.8 for (dT)18; SI Appendix, Fig. S3 A–D]. When mRAD51 assembles onto ssDNA, the distance between the dye pair increased, resulting in reduced FRET efficiency (Fig. 3 B and C and SI Appendix, Fig. S3 C and D). Therefore, each high-to-low FRET transition represents one or more mRAD51 binding events. In the case of mRAD51 alone, using a shorter (dT)13 substrate, the percentage of DNA molecules with FRET alternation observed within 3 min (binding fraction) was 18.5 ± 3.62% (SI Appendix, Fig. S3E), with the time traces dominated by the protein-free, high-FRET state (Fig. 3B and SI Appendix, Figs. S3C and S4A). The low-binding fraction and the transience of the low-FRET states indicate that the (dT)13 substrate is too short to form stable nucleating clusters of mRAD51. On the other hand, in the presence of mS5S1, the binding fraction of mRAD51 dramatically increases to 66.6 ± 14.0% (SI Appendix, Fig. S3E) and the time traces are dominated by low-FRET states (Fig. 3C and SI Appendix, Figs. S3C and S4B), reflecting more RAD51 binding. This observation is consistent with our previous finding that mS5S1 reduces the nucleating unit from three mRAD51 molecules to two (Fig. 2 B and C) and mS5S1 increases the ssDNA affinity of mRAD51 and stabilizes mRAD51 nucleating clusters on ssDNA (Fig. 2C). Adding mS5FLS1 leads to similar consequences as the case of mRAD51 alone (SI Appendix, Figs. S3 C and E, S4C, and S5A), consistent with the inability of mS5FLS1 to stimulate mRAD51 assembly. For the longer (dT)18 ssDNA, mRAD51 alone results in stable binding with many more bound ssDNA molecules (58.0 ± 7.51%), and the middle-to-low FRET signal is dominant (SI Appendix, Figs. S3D and S4C). We identified seven FRET states (SI Appendix, Fig. S5) and corresponding binding/dissociation rate constants (SI Appendix, Fig. S6) using the longer (dT)18 substrates, reflecting the binding of up to six mRAD51 monomers.
Fig. 3.
Single-molecule FRET experiments demonstrate that mS5S1 stabilizes mRAD51 nucleating clusters. (A) Schematic illustration of the single-molecule fluorescence resonance energy transfer experimental setup. mRAD51 assembling onto (dT)13 ssDNA results in FRET decrease due to the increase of dye pair separation. (B–F) Single-molecule FRET observation of mRAD51 nucleating cluster dynamics. Exemplary FRET time traces of mRAD51 (B) and the mRAD51–mS5S1 complex (C) assembling on (dT)13 ssDNA substrate. High-FRET state (∼0.8) corresponds to a DNA-only state, and low-FRET state (0.0∼0.6) corresponds to the mRAD51-bound state. (D and E) Transition density plots clearly reflect four states (without mS5S1) and five states (with mS5S1) in mRAD51 nucleating cluster dynamics. (F) Rate constants of mRAD51 nucleating cluster dynamics in the absence (empty bars) and presence (solid bars) of mS5S1. Error bars of binding and dissociation rates are the SD of the mean by bootstrapping 5,000 times. N.A., data not available.
The (dT)13 substrates allowed at most four mRAD51 molecules to bind the ssDNA, but these binding events were not long enough to form stable filaments. We observed multiple FRET states, as well as alternations among these states, reflecting dynamics among multiple mRAD51-bound states. These intermediate-FRET states are identified by FRET histograms (SI Appendix, Fig. S3 C and D). Previous work on budding yeast Saccharomyces cerevisiae Rad51 (ScRad51) on the same (dT)13 substrates resulted in five different FRET states, corresponding to between zero and four ScRad51 molecules bound (17). We used Bayesian analysis (43) to globally fit all of the FRET time courses, and a total of four FRET states was the best found in mRAD51-only experiments (Fig. 3B). The four identified FRET states in mRAD51-only experiments match the zero, one, two, and three Rad51-bound states seen in ScRad51 experiments (17), confirming the validity of our analysis. We did not observe the lowest FRET state seen in the ScRad51 studies (∼0.1, the four Rad51-bound state; Fig. 3B), reflecting that four mRAD51 oligomers in (dT)13 substrates are not stable enough to be seen. On the other hand, in the mixture of the mRAD51–mS5S1 complex, we identified five FRET states, even though the highest FRET state (protein-free state, state 0; Fig. 3C) is less populated. To confirm these FRET-state assignments, we analyzed the FRET time courses to generate transition density plots (TDPs) (18, 44) (Fig. 3 D and E) in both cases. TDP analysis identifies the FRET states before and after each transition, and the heatmaps allow the identification of the distribution of multiple FRET states. For example, for the mRAD51-only case, a transition from a FRET value of ∼0.85 (state 0) to ∼0.75 (state 1) will score in the “binding” section in the TDP, as it reflects the binding of the first mRAD51 onto the ssDNA. The mirror symmetry along the diagonal of the TDP indicates the reversible changes between these FRET states.
The transitions seen in time courses and TDPs suggest that the up to five FRET states observed represent the zero to four mRAD51-bound states in the (dT)13 substrates. The intermediate FRET values identified for the mRAD51–mS5S1 complex are different from the ones seen in ScRad51- and mRAD51-only cases. This difference likely reflects the steric effect of the large mRAD51–mS5S1 complex (∼80 kDa), driving separation between the donor and acceptor dye. The large mRAD51–mS5S1 complex could also account for wider FRET values corresponding to zero, one, and two mRAD51-bound states (Fig. 3E). We also noted that total fluorescence signal increases upon mRAD51 binding (Fig. 3 B and C), suggesting a protein-induced fluorescence enhancement (PIFE) effect, consistent with the previous observation in ScRad51 (16, 17, 45). This PIFE effect is more apparent in the presence of mS5S1 (Fig. 3C and SI Appendix, Fig. S7) or on (dT)18 substrates, potentially because mRAD51 is able to fully extend toward the Cy3-tagged 3′ terminating end with mS5S1 or on the longer ssDNA.
Identification of these FRET states in time courses allows us to determine the evolution of these FRET states and the dwell time associated with individual states. The FRET time courses are best described by a consecutive and reversible kinetic model, where individual mRAD51 monomers can bind and dissociate during nucleating cluster formation and the extension also occurs in the monomer, with or without mS5S1 (Fig. 3 B and C). The kinetics of each binding (ki→i+1; i is the number of mRAD51 molecules bound to ssDNA) and dissociation (ki→i−1) can be determined in both mRAD51-only and mRAD51/S5S1 mixtures (Fig. 3F). Surprisingly, when comparing mRAD51-only (open bars; Fig. 3F) and the mRAD51–mS5S1 mixture (filled bars; Fig. 3F), mS5S1 significantly reduces the mRAD51 dissociation rates during the nucleating cluster formation. Although mS5S1 could possibly increase mRAD51 binding rates, the change is not significant in our experimental resolution. Therefore, the major stabilization effort of mS5S1 in mRAD51 nucleating cluster formation comes from the reduction in dissociation rates.
Fission Yeast Rad51 Filament Assembly Is Fast, with No Apparent Stimulation from S5S1.
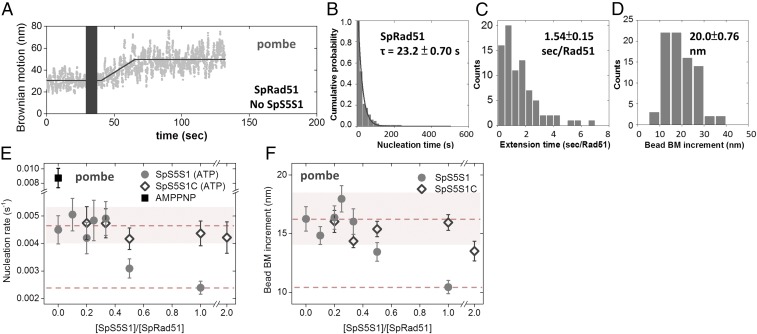
Mouse S5S1 stimulates mRAD51 activity by accelerating the nucleation step during nucleoprotein filament assembly. Fission yeast Swi5–Sfr1 (SpS5S1) is also known to stimulate SpRad51 activity (26, 29, 30, 33, 35). To examine whether the activation mechanism by S5S1 is evolutionarily conserved, we used the same TPM approach toward SpS5S1. Under the same reaction condition (150 mM KCl, pH 7.5), SpRad51 does not assemble on dsDNA in either ATP or AMPPNP (SI Appendix, Fig. S1 I, J, L, and M). At the same recombinase concentration (0.8 μM), SpRad51 displayed much faster assembly kinetics than mRAD51 (Figs. 1 B and C and 4 A and B). In the absence of S5S1, SpRad51 nucleation time is 23.2 ± 0.70 s (Fig. 4B), fourfold faster than that observed for mRAD51 (97.6 ± 2.89 s) (Fig. 1C). To see if the SpS5S1 complex further stimulates SpRad51 assembly, we used a reduced recombinase concentration (0.3 μM) to allow nucleation rates to be determined accurately (Fig. 4E and SI Appendix, Fig. S8). Interestingly, SpS5S1 did not stimulate SpRad51 nucleation at low concentrations and even inhibited SpRad51 nucleation at higher concentrations (solid circles, Fig. 4E and SI Appendix, Fig. S8B). It is possible that SpRad51 nucleation is sufficiently fast that no additional stimulation is necessary, unlike the slow nucleation observed for mRAD51. As to the inhibition effect seen at higher SpS5S1 concentrations, it likely results from the ssDNA binding property of SpS5S1 (26, 30). Even though SpS5S1 has DNA affinity, its binding to DNA substrates does not alter bead BM (SI Appendix, Fig. S1 K–N). The control experiments verify that a decrease in bead BM increment with increasing SpS5S1 concentration derives from reduced amounts of SpRad51 binding to the gapped ssDNA substrate (gray solid circles in Fig. 4F). To test whether or not the ssDNA binding property of SpS5S1 inhibits SpRad51 filament assembly, we used an N-terminal truncation mutant of SpSfr1 in a complex with Swi5 (SpS5S1C). The N-terminal region of SpSfr1 possesses an initial interaction site with Rad51 to serve as an anchor and DNA binding site, which are overlapped with each other. Thus, SpS5S1C is deficient in DNA binding but retains SpRad51 filament stabilization (30). This DNA binding-deficient mutant showed neither inhibition nor stimulation of nucleoprotein filament formation even when twofold excess amounts were included (open diamonds, Fig. 4E). Therefore, SpS5S1 and SpRad51 compete for ssDNA binding, and we conclude that SpS5S1 has no stimulatory effects on SpRad51 nucleation. In addition, the magnitude of BM increment (Fig. 4F and SI Appendix, Fig. S8D) drops with increasing amounts of wild-type SpS5S1, while the SpS5S1C mutant protein induced no apparent change. Thus, we confirmed that higher SpS5S1 concentrations can compete with SpRad51 for substrates.
Fig. 4.
Fission yeast SpS5S1 does not stimulate SpRad51 filament assembly. (A) Representative bead BM time courses of fission yeast Rad51 (0.8 μM) assembly on (dT)135 DNA substrates without S5S1. (B–D) Nucleation time (B), mean extension time (C) (seconds per Rad51), and bead BM increment (D) analyzed from individual assembly time courses. Nucleation time histograms are fitted by single exponential decay. (E) Nucleation rates of various concentrations of SpS5S1 at constant 0.3 μM SpRad51. (F) Bead BM increments of SpRad51 assembly at various ratios of SpS5S1 and SpRad51 mixtures also decreased at higher SpS5S1 concentrations. All experiments were carried out at 2 mM ATP and 0.3 μM SpRad51. Gray solid circles are from wild-type SpS5S1 experiments. Black diamonds are from the N-terminal truncation mutant of SpS5S1 (SpS5S1C) deficient in ssDNA binding. Dashed lines are the mean, and the shaded regions span 2 SDs.
With the fast nucleation rates observed, SpRad51 displays more apparent dependence on ssDNA length than mRAD51 (SI Appendix, Fig. S9A). An ∼40-fold difference in slope observed and similar y intercepts here suggest that SpRad51 has higher ssDNA binding affinity than mRAD51, and likely contributes to the faster nucleation rates. SpRad51 concentration dependence of nucleation rates returns a power-law fitting of 2.70 ± 0.29, suggesting that three SpRad51 monomers are required for stable nucleation events (SI Appendix, Fig. S9B), similar to that of mRAD51 (Fig. 2A).
S5S1 Prevents Rad51 Filament Disassembly.
In addition to accelerating the binding event, nucleoprotein filament stability can also be achieved by the prevention of filament dissociation. In the TPM-based disassembly experiments (Fig. 5), surface-bound (dT)135 gapped DNA substrates were first incubated with Rad51/ATP mixture, and then another mixture of Rad51, ATP, and S5S1 was added to the microscope slide. This set of disassembly experiments was done for mouse (Fig. 5B) and fission yeast (Fig. 5F) proteins. The two-stage incubation avoids potential ssDNA substrate competition between SpS5S1 and SpRad51. An extensive buffer wash containing no Rad51 but all other components removed free Rad51 from the reaction chamber. This was defined as time 0 of the disassembly reaction. The bead Brownian motions are measured in real time to monitor the filament length. Several kinetic parameters were determined, including (i) lifetime of the stable filament (dwell time before BM decrease), (ii) dissociation time, time for individual RAD51 monomer dissociation continuously, and (iii) net BM decrease, a decrease in filament coverage. Our results showed that the mRAD51 filament is quite stable in ATP even without mS5S1, with a minimum mean lifetime around 350 ± 39.3 s (Fig. 5C). In the presence of 1.0 μM mS5S1, the lifetime of the mRAD51 filament was further stabilized to 535 ± 49.9 s (∼1.53-fold). Using low concentrations of mS5S1 (0.05 or 0.3 μM) also resulted in longer lifetimes (441 ± 51.1 and 436 ± 45.1 s, respectively, ∼1.27-fold). This suggests that mS5S1 prevents mRAD51 filament disassembly. Control experiments using the mS5FLS1 mutant return no additional stabilization, confirming that mRAD51–mS5S1 interaction is essential for mRAD51 nucleoprotein filament stabilization. We also noticed that the fraction of full-length mRAD51 filaments retained at the end of the 15-min reaction, undisassembled filament (Fig. 5D), increases in the presence of mS5S1, correlating with the increased filament lifetime with mS5S1 (Fig. 5C). However, once dissociation was initiated (as BM starts to decrease), the dissociation rate is similar either with or without mS5S1, within the resolution of our TPM measurements (Fig. 5E). Compared with mRAD51, the SpRad51-alone filament is less stable (lifetime of 140 ± 17.9 s; Fig. 5 F and G). Essentially all SpRad51 filaments were disassembled within 15 min (Fig. 5H). In the presence of SpS5S1, the lifetime of the SpRad51 filament is significantly increased, as was the fraction of filaments retained at the end of 15-min observations. For example, 0.01 μM SpS5S1 increases the lifetime to 382 ± 32.6 s (∼2.73-fold). SpS5S1 increases the dissociation time as well, making the disassembly events slower (Fig. 5I). The SpS5S1C mutant also protects SpRad51 filaments (empty bars in Fig. 5G), but is less effective than wild-type SpS5S1 (gray solid bars), especially at lower concentrations. This may be due to the lower affinity to SpRad51 (35), because higher amounts of SpS5S1C show a similar ability to protect filament disassembly. Therefore, we conclude that both S5S1 heterodimers act as a stabilizer of Rad51 filament via direct interaction with the recombinase.
Fig. 5.
Nucleoprotein filament disassembly experiments showed that S5S1 prevents Rad51 filament disassembly. (A) Schematic illustration of nucleoprotein filament disassembly experiments using the TPM setup. (B) Representative bead BM time courses of mouse mRAD51 disassembly on (dT)135 DNA substrates without mS5S1 (Upper) and in the presence of 1.0 μM mS5S1 (Lower). mRAD51 filaments were preassembled in the presence of ATP. Dark gray bars stand for void time for extensive buffer wash to remove free mRAD51. A lifetime of the preassembled filament before the BM decrease dictates the mRAD51 disassembly kinetics. (C–E) Mean lifetime of mouse mRAD51 nucleoprotein filament (C), fraction of the undisassembled filament within 15 min (D), and mean dissociation time per mRAD51 (E) in the presence of various mS5S1 concentrations and nucleotide conditions. (F) Representative bead BM time courses of SpRad51 disassembly without SpS5S1 (Upper) and in the presence of 0.3 μM SpS5S1 (Lower). (G–I) Kinetic parameters for fission yeast. The fraction of undisassembled tethers is correlated with the mean lifetime of the SpRad51 filament in both species. Here, the N-terminal truncation S5S1C mutants (open bars) are deficient in DNA binding. All experiments were carried out at 2 mM ATP. Error bars are 1 SEM.
Comparing S5S1 from these two species, we found that 1.0 μM mS5S1 only achieves an ∼1.53-fold increase in filament lifetime while SpS5S1 increases to ∼2.73-fold. Considering the fast disassembly kinetics of SpRad51, SpS5S1 predominantly acts on this filament koff step. In addition, comparing bead BM changes between the assembly and disassembly experiments (SI Appendix, Fig. S10), we found that not all mRAD51 dissociated from the gapped DNA substrates, reflecting the incomplete disassembly of mRAD51 filaments, as seen in human RAD51 (14). On the contrary, SpRad51 was almost entirely released from DNA, as filaments initiate disassembly even in the presence of SpS5S1, confirming that the SpRad51 filament is more susceptible to disassembly. The reason the apparent function of S5S1 seems to be different is due to the different stabilities of Rad51 filaments between yeast and mouse proteins. Thus, we conclude that the primary conserved function of S5S1 is to stabilize Rad51 on DNA, allowing both the formation of the stable nucleus and the maintenance of filament length.
Discussion
Nucleoprotein filament assembly is the first committed step in homologous recombination and is targeted for regulation (1, 2, 23, 26, 30, 32, 46, 47). A nucleoprotein filament with sufficient length is advantageous for initial homology search. The filament is also expected to be dynamic during the directional exchange of different parts of duplex homologous DNA. Accessory proteins have been found to stimulate the recombination process by maintaining a stabilized yet dynamic recombinase nucleoprotein filament. Stabilization of the nucleoprotein filament can be achieved by speeding up the filament assembly, reducing filament disassembly, or both. However, these kinetic parameters are typically obscured in ensemble biochemical experiments. In this study, we used two different single-molecule tools to determine these kinetic parameters that allow us to characterize the mechanism of S5S1 regulation in mouse and fission yeast proteins. Several kinetic steps are involved in filament assembly: initial Rad51 binding and dissociation events leading to a stable nucleus, fast extension steps, and dissociation events within growing filaments. S5S1 of both species stabilized Rad51 nucleoprotein filaments by preventing Rad51 dissociation from nucleation clusters and from the assembled filaments. Although the S5S1 complex of these species alters the kinetic steps differentially due to intrinsic characteristics of Rad51 recombinases in different species, they both prevent Rad51 dissociation to facilitate efficient recombination progression. Here, we used two complementary single-molecule tools to characterize the assembly. TPM experiments allowed us to characterize the formation and disassembly of individual stable filaments, and FRET experiments capture the dynamics of individual Rad51 binding and dissociation before a stable nucleus is formed. An apparent stimulation of the Rad51 filament nucleation by S5S1 seen in TPM experiments is attributed to the reduction of the dissociation rate in nucleus formation observed in FRET. A nearly fourfold reduction of dissociation rate (k1→0) of mRAD51 is seen in the presence of mS5S1, but no apparent change in binding rates is seen (Fig. 3F). This then leads to a reduction in nucleation size of three mRAD51 monomers to two mRAD51 monomers in the presence of mS5S1. Fission yeast Rad51 is fast in filament assembly, but is prone to disassembly. SpS5S1 stabilizes the SpRad51 filaments by specifically preventing SpRad51 dissociation. Therefore, S5S1 of two species stabilizes Rad51 filaments by using the same strategy of preventing Rad51 dissociation. Our single-molecule experiments speak specifically to the Rad51 state when it nucleates and extends the DNA substrates, and our data are consistent with those previously shown in the literature (14, 17). Previous work has suggested that recombinases from various species can exist in oligomers in solution (48–52). It is possible that a structural transition is made in solution before or during the DNA binding, so smaller units of Rad51 oligomers are responsible for nucleation and an individual Rad51 molecule is added during filament growth.
S5S1 association with Rad51 could either expose the DNA-binding domain of Rad51 or stabilize the oligomeric interface of Rad51. SpS5S1 has two Rad51 binding sites: a high-affinity one in the N terminus of Sfr1 and a low-affinity one in the C terminus (30). Mouse S5S1 has only one mRAD51 binding site, residing in the C terminus (31). The difference in Rad51 binding affinity between mouse and fission yeast S5S1 is responsible for the observation that more S5S1 is required for Rad51 stability in the cases of mS5S1 and SpS5S1C mutants (Fig. 5 C and G). Considering its large effect on both dissociation rates (FRET experiments) and disassembly rates (TPM disassembly experiments), we suggest that S5S1 acts by holding adjacent Rad51 molecules together, reducing dissociation. The interaction between S5S1 and RAD51 is thus essential for stimulation, confirmed by the abolished stimulation seen in mS5FLS1 mutants (Figs. 1C, 2A, and 5C and SI Appendix, Figs. S3–S5).
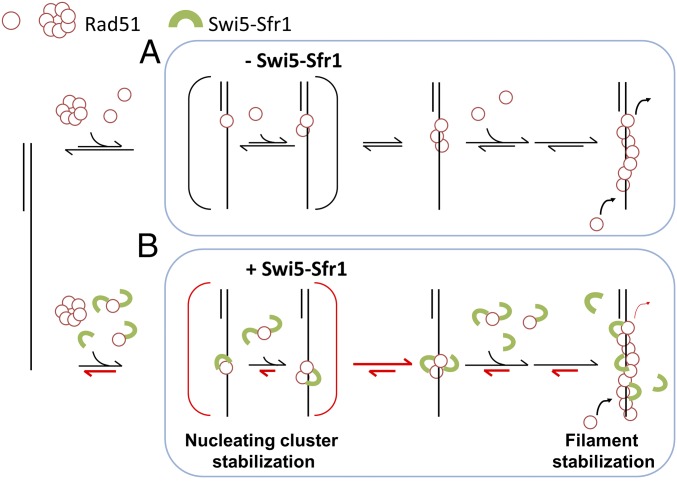
The different amounts of S5S1 required to stimulate nucleation and reduce filament disassembly provide hints as to how S5S1 interacts with Rad51 molecules to achieve filament stabilization. For the mouse proteins, a twofold excess of mS5S1 maximally stimulates mRAD51 nucleation (Fig. 1F). However, stabilization in the disassembly experiments can be seen with the low coverage of the mS5S1 complex (∼17%; Fig. 5C; 0.05 μM vs. 0.3 μM mRAD51 added). Previous structural studies suggested that S5S1 is accommodated within the groove of the Rad51 filament (30). The different concentration requirements of S5S1 point to a model that S5S1 binding stabilizes adjacent Rad51 molecules through binding within the filament groove to prevent Rad51 dissociation (Fig. 6 and SI Appendix, Fig. S11). During nucleus formation, more mS5S1 is required so most of Rad51 is stabilized by S5S1. As Rad51 dissociation is inhibited by S5S1, a stable Rad51 nucleus can be formed more easily. On the other hand, as filament disassembly could take place at the filament end more frequently, filament stabilization can be achieved by S5S1 binding to terminal Rad51. Therefore, low-coverage S5S1 is sufficient. As S5S1 has also been shown to activate Rad51 filaments (30, 32), S5S1 binding within Rad51 filaments also contributes to the overall Rad51 activity stimulation. During the dynamic progression of strand exchange, low S5S1 coverage allows efficient budgeting of S5S1, so S5S1 can be available for stimulation at later stages of recombination progression.
Fig. 6.
Proposed models for regulating Rad51 nucleoprotein filament formation by the S5S1 complex. Swi5–Sfr1 stabilizes Rad51 on ssDNA primarily by preventing its dissociation. This stabilization effect leads to a stable nucleating cluster formation and a reduction in filament disassembly. Despite different kinetic properties of mouse and fission yeast Rad51, the Swi5–Sfr1 complex stimulates the Rad51 process through a general, evolutionarily conserved mechanism. Red half-arrows indicate the kinetic steps affected by S5S1.
There exist several heterodimeric complexes regulating recombinase nucleoprotein filament stability (2–4, 53, 54). The elongated crescent-like structure of Hop2–Mnd1 shares a similar structural motif with S5S1. In both cases, the heterodimeric complex interacts with the groove of the Dmc1 and/or Rad51 filament to stabilize the nucleoprotein filaments (1, 55–58). A Rad51 paralog complex in Caenorhabditis elegans, the RFS-1–RIP-1 complex, also has been proposed to act similarly on Rad51 filament remodeling (54). The S. cerevisiae Psy3–Csm2 dimer has been suggested to bind to the end of Rad51 filaments to achieve filament stabilization (53). Together with the structural evidence, it is possible that the mechanism proposed here likely serves as a general principle for these heterodimeric complexes involved in filament stabilization, for example, whether these accessory factors, such as BRCA2, the PCSS complex, and RAD51 paralogs (24, 53, 54), work synergistically on filament stabilization. It is possible that these accessory proteins allow better modulation on filament dynamics for efficient strand exchange progression.
Differences in kinetics are seen between mouse RAD51 and SpRad51 recombinases. Mouse RAD51 is slower in nucleation, but SpRad51 is more prone to disassembly. We have found that mS5S1 acts effectively on the nucleation step while SpS5S1 acts primarily on the disassembly step to achieve nucleoprotein filament stabilization. However, DNA binding of SpS5S1 is shown to inhibit SpRad51 filament assembly (Fig. 4 E and F) and strand exchange (26, 33). It is likely that accessory proteins evolve to accommodate different kinetic characteristics of recombinases during speciation and play different roles in the progression of the homologous-recombination process. For example, small-angle X-ray scattering and X-ray crystallographic studies showed that SpS5S1 fits into the helical groove of the SpRad51 filament and also extends onto ssDNA (29, 30, 35, 59). In addition to stabilizing SpRad51, SpS5S1 interacting with ssDNA provides an additional safe latch on SpRad51. The budding yeast Mei5–Sae3 complex (ScM5S3), orthologs of Sfr1 and Swi5, has been shown to stabilize ScDmc1 filament and to stimulate ScDmc1-mediated strand exchange during meiosis (19, 60, 61). ScM5S3 possesses both ssDNA and dsDNA affinities, and it is possible that budding yeast M5S3 stabilizes ScDmc1 filament in a similar, general mechanism found in mouse and fission yeast.
ssDNA is likely bound by single-stranded DNA-binding proteins, for example, replication protein A (RPA) to prevent nucleolytic cleavage. RPA binding to ssDNA is a physical barrier for Rad51 assembly. Previous findings demonstrate that fission yeast S5S1 works synergistically with Rad52 to stimulate Rad51 assembly on RPA-coated ssDNA (28, 35). It would be interesting to see whether SpS5S1 also primarily acts on preventing SpRad51 disassembly in the presence of RPA.
Both mouse and fission yeast S5S1 have been shown to activate Rad51 filament by stimulating ssDNA-dependent ATPase activity of Rad51 (30, 32). Moreover, mouse S5S1 enhances ATPase activities of mRAD51 by stimulating the release of ADP to maintain the filament in an active form (30, 32). Escherichia coli RecA recombinases have also been shown to continuously hydrolyze ATP and bind new ATP molecules without dissociating from DNA (62). It is possible that mS5S1 stabilizes interfaces between adjacent mRAD51 molecules within the filament during ATPase turnover events, and thus maintains active nucleoprotein filaments. It is important to note that cofactors like AMPPNP and Ca2+ increase the stability of Rad51 filaments by inhibiting ATP hydrolysis and maintaining the ATP-bound form of Rad51, and then these cofactors promote strand-exchange activity (25, 33, 35, 63, 64). Therefore, in contrast to AMPPNP and Ca2+ effects, S5S1 stimulates Rad51 progression in a unique mechanism. Maintaining a dynamic nucleoprotein filament homeostasis requires Rad51 dissociation at appropriate rates. Swi5–Sfr1 and other accessory proteins serve the purpose of modulating dissociation rates of Rad51 to fine-tune recombinase progression.
Materials and Methods
Supporting Information.
A detailed description of the DNA substrate preparations, proteins, and buffer conditions and detailed experimental procedures of the single-molecule tethered particle motion assembly and disassembly experiments are provided in SI Appendix.
Single-Molecule TPM Assembly Experiments and Data Analysis.
Streptavidin-labeled beads were prepared as previously described (65). In filament assembly experiments, a glass slide was coated with 5 μg/mL anti-digoxigenin and blocked with 2 mg/mL BSA sequentially. DNA substrates (4 nM) were incubated on the anti-digoxigenin–coated slide for 30 min followed by buffer washing to remove unbound DNA. Streptavidin-decorated polystyrene beads (220 nm) were then attached to the DNA substrates for microscopic visualization. All TPM reactions were performed in either mouse or fission yeast buffers supplemented with 1 mM DTT and 2 mM ATP. TPM assembly experiments were initiated by introducing a Rad51/ATP (or with S5S1) mixture into the reaction chamber containing the surface-anchored gapped substrates.
We used an inverted optical microscope (IX71; Olympus) with a differential interference contrast imaging mode to visualize tethers and measure bead Brownian motion. Images of assembly experiments were acquired at 30 Hz using a Newvicon camera (Dage-MTI) and analyzed using software written in LabVIEW. The amplitude of tether Brownian motion is defined by the SD of the bead centroid positions of 20 images using sliding windows. In addition to the DNA contour length change, polymer stiffness and camera exposure time alter Brownian motion amplitudes in practice (42). For each independent TPM assembly experiment, images were first recorded for around 30 s (∼1,000 image frames) before the addition of Rad51 or Rad51/S5S1 mixture and then for about 15 min (∼30,000 image frames) after the addition of the mixtures. For snapshot control experiments, 1,000 image frames were recorded at five or six different fields of view on the coverslip after a 5-min reaction (SI Appendix, Fig. S1).
Single-Molecule Fluorescence Resonance Energy Transfer Experiments and Data Analysis.
In smFRET experiments, PEGylated glass slides and coverslips were prepared as previously described (66). To perform smFRET experiments, reaction chambers were incubated with 20 μg/mL streptavidin for 5 min. Excess streptavidin was washed away with buffer containing 20 mM Tris and 50 mM NaCl. The 3′-biotinylated fluorophore-labeled hybrid DNA (15 pM) was then immobilized on the surface for 5 min. After a 5-min incubation, free DNA was removed by flowing in mRAD51 imaging buffer containing 1 mM Trolox (Sigma-Aldrich), 2.6 mM protocatechuic acid (Sigma-Aldrich), 0.21 units per milliliter protocatechuate 3,4-dioxygenase (OYC Americas), 30 mM Tris, 2.5 mM magnesium chloride, and 150 mM potassium chloride at pH 7.5. The reaction included a mixture of 1 μM mRAD51 and 2 mM ATP in mRAD51 imaging buffer in reaction chambers. mS5S1 (or mS5FLS1)-containing experiments included 1 μM mRAD51, 2 μM mS5S1 (or mS5FLS1), and 2 mM ATP in mRAD51 imaging buffer. Under our imaging conditions, ∼78% of fluorophores in hybrid DNA survive for more than 200 s.
We utilized an objective-type total internal reflection fluorescence microscope (IX2; Olympus) and 532-nm laser as excitation light source in smFRET experiments. Fluorescence intensity signals of both Cy3 and Cy5 were acquired with an electron-multiplying CCD (ProEM 512B; Princeton Instruments) at 20 Hz using a dual-view system. Emission movies of Cy3 and Cy5 fluorescence were recorded using a software program written in LabVIEW 8.6. Colocalized Cy3 and Cy5 spots were analyzed using a mapping software program written in IDL. Fluorescence intensity time traces of each individual mapped DNA molecule were analyzed using MATLAB (MathWorks). Alternation in FRET values was analyzed using variational Bayesian analysis (vbFRET) to globally fit all time courses (43).
Supplementary Material
Acknowledgments
We thank Steve Bell (MIT, Biology) for helpful comments and suggestions, and Kai-Chun Chang (NTU) for discussion of the smFRET data analysis. This work was supported by grants from the Ministry of Science and Technology of Taiwan (MOST 104-2628-M-002-008-MY3 to H.-W.L.; MOST 105-2314-B-002-073-MY4 to P.C.), Academia Sinica (P.C.), the career development program of National Taiwan University (H.-W.L. and P.C.), and partly by Grants-in-Aid for Scientific Research on Innovative Areas (15H05974) and for Scientific Research-A (18H03985) from the Japan Society for the Promotion of Science (to H.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812753115/-/DCSupplemental.
References
- 1.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 3.Cox MM, et al. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, Caron P, Legube G, Paull TT. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kowalczykowski SC. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2015;7:a016410. doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrical SW. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol. 2015;7:a016444. doi: 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature. 2012;491:274–278. doi: 10.1038/nature11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candelli A, et al. Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc Natl Acad Sci USA. 2014;111:15090–15095. doi: 10.1073/pnas.1307824111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma CJ, Gibb B, Kwon Y, Sung P, Greene EC. Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament. Nucleic Acids Res. 2017;45:749–761. doi: 10.1093/nar/gkw1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antony E, et al. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu Y, et al. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat Commun. 2013;4:2281. doi: 10.1038/ncomms3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 19.Say AF, et al. The budding yeast Mei5-Sae3 complex interacts with Rad51 and preferentially binds a DNA fork structure. DNA Repair (Amst) 2011;10:586–594. doi: 10.1016/j.dnarep.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun J, Buechelmaier ES, Powell SN. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol Cell Biol. 2013;33:387–395. doi: 10.1128/MCB.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, et al. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 24.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 25.Tsai SP, et al. Rad51 presynaptic filament stabilization function of the mouse Swi5-Sfr1 heterodimeric complex. Nucleic Acids Res. 2012;40:6558–6569. doi: 10.1093/nar/gks305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haruta N, et al. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- 27.Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc Natl Acad Sci USA. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Argunhan B, Murayama Y, Iwasaki H. The differentiated and conserved roles of Swi5-Sfr1 in homologous recombination. FEBS Lett. 2017;591:2035–2047. doi: 10.1002/1873-3468.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kokabu Y, et al. Fission yeast Swi5-Sfr1 protein complex, an activator of Rad51 recombinase, forms an extremely elongated dogleg-shaped structure. J Biol Chem. 2011;286:43569–43576. doi: 10.1074/jbc.M111.303339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuwabara N, et al. Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex. Structure. 2012;20:440–449. doi: 10.1016/j.str.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Su GC, et al. Role of the RAD51-SWI5-SFR1 ensemble in homologous recombination. Nucleic Acids Res. 2016;44:6242–6251. doi: 10.1093/nar/gkw375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su GC, et al. Enhancement of ADP release from the RAD51 presynaptic filament by the SWI5-SFR1 complex. Nucleic Acids Res. 2014;42:349–358. doi: 10.1093/nar/gkt879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito K, Murayama Y, Takahashi M, Iwasaki H. Two three-strand intermediates are processed during Rad51-driven DNA strand exchange. Nat Struct Mol Biol. 2018;25:29–36. doi: 10.1038/s41594-017-0002-8. [DOI] [PubMed] [Google Scholar]
- 34.Akamatsu Y, Jasin M. Role for the mammalian Swi5-Sfr1 complex in DNA strand break repair through homologous recombination. PLoS Genet. 2010;6:e1001160. doi: 10.1371/journal.pgen.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurokawa Y, Murayama Y, Haruta-Takahashi N, Urabe I, Iwasaki H. Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biol. 2008;6:e88. doi: 10.1371/journal.pbio.0060088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu YW, et al. Using single-molecule approaches to study archaeal DNA-binding protein Alba1. Biochemistry. 2013;52:7714–7722. doi: 10.1021/bi4010478. [DOI] [PubMed] [Google Scholar]
- 37.Piechura JR, et al. Biochemical characterization of RecA variants that contribute to extreme resistance to ionizing radiation. DNA Repair (Amst) 2015;26:30–43. doi: 10.1016/j.dnarep.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H-Y, Lu C-H, Li H-W. RecA-SSB interaction modulates RecA nucleoprotein filament formation on SSB-wrapped DNA. Sci Rep. 2017;7:11876. doi: 10.1038/s41598-017-12213-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, et al. Conformational changes modulate the activity of human RAD51 protein. J Mol Biol. 2004;337:817–827. doi: 10.1016/j.jmb.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 40.Ristic D, et al. Human Rad51 filaments on double- and single-stranded DNA: Correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towles KB, Beausang JF, Garcia HG, Phillips R, Nelson PC. First-principles calculation of DNA looping in tethered particle experiments. Phys Biol. 2009;6:025001. doi: 10.1088/1478-3975/6/2/025001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu HF, Ngo KV, Chitteni-Pattu S, Cox MM, Li HW. Investigating Deinococcus radiodurans RecA protein filament formation on double-stranded DNA by a real-time single-molecule approach. Biochemistry. 2011;50:8270–8280. doi: 10.1021/bi200423t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bronson JE, Fei J, Hofman JM, Gonzalez RL, Jr, Wiggins CH. Learning rates and states from biophysical time series: A Bayesian approach to model selection and single-molecule FRET data. Biophys J. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci USA. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lusetti SL, et al. The RecF protein antagonizes RecX function via direct interaction. Mol Cell. 2006;21:41–50. doi: 10.1016/j.molcel.2005.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Passy SI, et al. Human Dmc1 protein binds DNA as an octameric ring. Proc Natl Acad Sci USA. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 50.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 51.McIlwraith MJ, et al. RadA protein from Archaeoglobus fulgidus forms rings, nucleoprotein filaments and catalyses homologous recombination. Nucleic Acids Res. 2001;29:4509–4517. doi: 10.1093/nar/29.22.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang S, Yu X, Seitz EM, Kowalczykowski SC, Egelman EH. Archaeal RadA protein binds DNA as both helical filaments and octameric rings. J Mol Biol. 2001;314:1077–1085. doi: 10.1006/jmbi.2000.5213. [DOI] [PubMed] [Google Scholar]
- 53.Sasanuma H, et al. A new protein complex promoting the assembly of Rad51 filaments. Nat Commun. 2013;4:1676. doi: 10.1038/ncomms2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor MRG, et al. A polar and nucleotide-dependent mechanism of action for RAD51 paralogs in RAD51 filament remodeling. Mol Cell. 2016;64:926–939. doi: 10.1016/j.molcel.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bugreev DV, et al. HOP2-MND1 modulates RAD51 binding to nucleotides and DNA. Nat Commun. 2014;5:4198. doi: 10.1038/ncomms5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chi P, San Filippo J, Sehorn MG, Petukhova GV, Sung P. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–1757. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao W, Sung P. Significance of ligand interactions involving Hop2-Mnd1 and the RAD51 and DMC1 recombinases in homologous DNA repair and XX ovarian dysgenesis. Nucleic Acids Res. 2015;43:4055–4066. doi: 10.1093/nar/gkv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang HA, et al. Crystal structure of Hop2-Mnd1 and mechanistic insights into its role in meiotic recombination. Nucleic Acids Res. 2015;43:3841–3856. doi: 10.1093/nar/gkv172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saikusa K, et al. Characterisation of an intrinsically disordered protein complex of Swi5-Sfr1 by ion mobility mass spectrometry and small-angle X-ray scattering. Analyst (Lond) 2013;138:1441–1449. doi: 10.1039/c2an35878f. [DOI] [PubMed] [Google Scholar]
- 60.Ferrari SR, Grubb J, Bishop DK. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J Biol Chem. 2009;284:11766–11770. doi: 10.1074/jbc.C900023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayase A, et al. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940. doi: 10.1016/j.cell.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 62.Kim SH, et al. Cooperative conformational transitions keep RecA filament active during ATPase cycle. J Am Chem Soc. 2014;136:14796–14800. doi: 10.1021/ja506363y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 65.Chung C, Li HW. Direct observation of RecBCD helicase as single-stranded DNA translocases. J Am Chem Soc. 2013;135:8920–8925. doi: 10.1021/ja401626t. [DOI] [PubMed] [Google Scholar]
- 66.Lu C-H, Chang T-T, Cho C-C, Lin H-C, Li H-W. Stable nuclei of nucleoprotein filament and high ssDNA binding affinity contribute to enhanced RecA E38K recombinase activity. Sci Rep. 2017;7:14964. doi: 10.1038/s41598-017-15088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.