Significance
Nucleoside analog drugs are widely used to treat a variety of cancers and viral infections. With an essential role in regulating the nucleotide pool in the cell by degrading cellular nucleotides, SAMHD1 has the potential to decrease the cellular concentration of frequently prescribed nucleoside analogs and thereby decrease their clinical efficacy in cancer therapy. To improve future nucleoside analog treatments, it is important to understand SAMHD1 interactions with these drugs. Our work thoroughly examines the extent to which nucleotide analogs interact with the catalytic and allosteric sites of SAMHD1. This work contributes to the assessment of SAMHD1 as a potential therapeutic target for cancer therapy and the future design of SAMHD1 modulators that might improve the efficacy of existing therapies.
Keywords: SAMHD1, nucleotide analog drugs, dNTPase, allosteric regulation, substrate selection
Abstract
SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase (dNTPase) that depletes cellular dNTPs in noncycling cells to promote genome stability and to inhibit retroviral and herpes viral replication. In addition to being substrates, cellular nucleotides also allosterically regulate SAMHD1 activity. Recently, it was shown that high expression levels of SAMHD1 are also correlated with significantly worse patient responses to nucleotide analog drugs important for treating a variety of cancers, including acute myeloid leukemia (AML). In this study, we used biochemical, structural, and cellular methods to examine the interactions of various cancer drugs with SAMHD1. We found that both the catalytic and the allosteric sites of SAMHD1 are sensitive to sugar modifications of the nucleotide analogs, with the allosteric site being significantly more restrictive. We crystallized cladribine-TP, clofarabine-TP, fludarabine-TP, vidarabine-TP, cytarabine-TP, and gemcitabine-TP in the catalytic pocket of SAMHD1. We found that all of these drugs are substrates of SAMHD1 and that the efficacy of most of these drugs is affected by SAMHD1 activity. Of the nucleotide analogs tested, only cladribine-TP with a deoxyribose sugar efficiently induced the catalytically active SAMHD1 tetramer. Together, these results establish a detailed framework for understanding the substrate specificity and allosteric activation of SAMHD1 with regard to nucleotide analogs, which can be used to improve current cancer and antiviral therapies.
The sterile alpha motif and histidine-aspartate domain–containing protein 1 (SAMHD1) is a triphosphohydrolase that severs the triphosphate group from deoxynucleoside triphosphates (dNTPs) (1, 2). A major function of SAMHD1 is to reduce the dNTP pool in noncycling cells, making it an important regulator of dNTP levels in the cell (1). In addition to its role in regulating genome stability, SAMHD1 is best known for its ability to block infection of a broad range of retroviruses, including HIV type 1 (HIV-1). During viral infection, SAMHD1 depletes the cellular dNTPs needed for reverse transcription of the viral RNA genome (2–8). In the cell, SAMHD1 activity is modulated by allosteric activation and phosphorylation (9–19). Nucleotide binding to two allosteric sites of each subunit leads to the assembly of activated SAMHD1 tetramer. Allosteric site (Allo-site) 1 only accommodates guanosine bases (GTP or dGTP), but any canonical dNTP can bind Allo-site 2 (15, 18, 20–22). When dNTPs are needed for DNA synthesis, phosphorylation at residue T592 destabilizes the active tetramer of SAMHD1, thereby down-regulating SAMHD1 activity (9, 10, 13).
SAMHD1 is an important general sensor and regulator of the dNTP pools, and thus it is crucial to genome maintenance. All canonical dNTPs are both substrates and allosteric activators, and this promiscuity allows SAMHD1 to target therapeutic molecules that resemble nucleotides in structure. Nucleoside analogs are a large class of drugs which are used to treat viral infections and many types of cancers (23–30). These compounds interfere with viral replication or cancer cell proliferation upon incorporation into newly synthesized DNA, resulting in chain terminations, accumulation of mutations, and often cell apoptosis (24, 27, 28). Several recent reports have demonstrated that SAMHD1 reduces the efficacy of nucleotide analog drugs by depleting their cellular concentrations (31, 32). Strikingly, SAMHD1 expression levels were shown to be highly predictive of patient response to cytarabine, the primary treatment for acute myeloid leukemia (AML) (31). Although the kinetic parameters of SAMHD1 activity have been thoroughly studied for canonical dNTPs (33–35), better characterization of SAMHD1 substrate specificity is needed for the optimal administration of current nucleoside analog drugs.
Interestingly, SAMHD1 activity has been shown to increase the efficacy of some nucleotide analogs that are not substrates of SAMHD1 (36, 37). In these cases, SAMHD1 activity allows the nonsubstrate nucleotide analogs to better compete for target active sites by depleting cellular dNTPs. Therefore, depending on how SAMHD1 interacts with a particular nucleotide analog of interest, it might be desirable to either inhibit or increase SAMHD1 activity. To selectively modulate SAMHD1 activity, it is important to fully understand how nucleotide analog drugs either bind to the allosteric site to assemble an activate tetramer or bind the catalytic site to be hydrolyzed.
In this study, we characterized SAMHD1 interactions with a panel of nucleotide analogs that are used to treat a variety of cancers and viral infections (23, 26, 27, 29, 38, 39) (Fig. 1A). We found that the catalytic site of SAMHD1 is very promiscuous, allowing SAMHD1 to hydrolyze most of the analogs tested here. On the other hand, Allo-site 2 is more restrictive to modifications of the 2′ sugar moiety of the drug. These results are important for the assessment of SAMHD1 as a potential therapeutic target for cancer therapy, the design of nonhydrolyzable derivatives, and the development of modulators of SAMHD1 activity to combine with existing therapies. In addition, this work contributes to a greater understanding of the structural and biochemical principles of SAMHD1 substrate selectivity and allosteric activation.
Fig. 1.
Substrate specificity of SAMHD1 is determined by 2′ sugar moiety. (A) Chemical structures of nucleoside analogs used in this study. (B) 2Fo-Fc electron density (σ = 1.0) for the nucleotide analog drugs crystallized in the catalytic pocket of SAMHD1. Black asterisks indicate sites of modifications. (C, Top) Transparent surface view of SAMHD1 tetramer with each subunit in a different color. Selected allosteric nucleotides are shown in red sticks, and a nucleotide in a catalytic pocket is shown in blue sticks. (C, Bottom) Superposition of all of the nucleotide analogs bound to the SAMHD1 catalytic pocket. SAMHD1 backbone is shown as coils with side chains shown as sticks. Cladribine-TP (cyan), clofarabine-TP (magenta), fludarabine-TP (green), vidarabine-TP (wheat), cytarabine-TP (purple), and gemcitabine-TP (orange) are shown as sticks. (D) Water networks (shown as red spheres) observed for each nucleotide analog bound to the SAMHD1 catalytic site. Black dashed lines indicate hydrogen bonds.
Results
Crystal Structures of Nucleotide Analogs Bound to the SAMHD1 Catalytic Pocket.
To better understand the structural basis for nucleotide analog binding in the catalytic pocket of SAMHD1, we cocrystallized the inactive catalytic domain of SAMHD1 (residues 113 to 626 with H206R/D207N mutations to inhibit catalysis) with six selected cancer and antiviral drugs. This SAMHD1 mutant has been previously shown to be identical in conformation and nucleotide-binding properties to the WT enzyme, but it is more amenable to crystallization (18, 19, 21, 22). The crystal structures of these SAMHD1–nucleotide analog complexes were determined at resolutions ranging from 1.7 Å to 2.5 Å (Table 1), with electron density that allows for unambiguous identification of each substrate in the catalytic pocket (Fig. 1B). The structures of SAMHD1 bound to these nucleotide analogs are similar to those obtained with canonical nucleotides (19, 21). Nucleotide analog binding does not alter the architecture of the SAMHD1 catalytic pocket (Fig. 1C), and these nucleotide analogs adopt similar conformations to the canonical nucleotides in the pocket. This suggests that the nucleotide modifications tested here do not disturb the overall integrity of the catalytic pocket or induce large structural rearrangements. The promiscuous catalytic pocket of SAMHD1 likely accommodates other nucleotide analogs with similar modifications.
Table 1.
Data collection and refinement statistics for the seven crystal structures of SAMHD1 HD bound to nucleotide analogs
| Data | Cladribine-TP | Clofarabine-TP | Fludarabine-TP | Vidarabine-TP | Cytarabine-TP | Gemcitabine-TP | [(2′R)-2′-F]-dCTP |
| Accession code | 6DW4 | 6DWD | 6DWK | 6DWJ | 6DW3 | 6DW5 | 6DW7 |
| Data collection | |||||||
| Wavelength, Å | 0.97920 | 0.97920 | 0.97920 | 0.92014 | 0.97920 | 0.97910 | 0.97912 |
| Space group | P21 | P21 | P21 | P21 | P21 | P21 | P21 |
| Cell dimensions | |||||||
| a, b, c, (Å) | 80.3, 142.3, 98.4 | 80.5, 142.2, 98.8 | 87.7, 146.7, 99.1 | 86.9, 146.7, 99.6 | 87.3, 147.3, 98.5 | 80.7, 142.9, 98.9 | 84.5, 146.4, 98.9 |
| α, β, γ, (°) | 90, 114.1, 90 | 90, 114.1, 90 | 90, 114.5, 90 | 90, 114.5, 90 | 90, 114.6, 90 | 90, 113.9, 90 | 90, 113.7, 90 |
| Molecules/asymmetric unit | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Resolution, Å | 50–2.00 (2.03–2.00) | 50–1.70 (1.73–1.70) | 50–2.30 (2.34–2.30) | 50–2.50 (2.54–2.50) | 50–2.20 (2.24–2.20) | 50–1.93 (1.96–1.93) | 50–2.5 (2.54–2.50) |
| Unique reflections | 129,994 (5,987) | 209,481 (10,532) | 100,587 (5,041) | 75,598 (3,796) | 110,334 (5,500) | 148,608 (7,362) | 76,374 (3,868) |
| Rmerge | 0.080 (0.682) | 0.086 (>1) | 0.158 (>1) | 0.107 (0.737) | 0.123 (>1) | 0.093 (0.775) | 0.126 (0.892) |
| Mean I/σ I | 12.6 (1.6) | 15.0 (1.2) | 9.3 (1.4) | 10.0 (2.2) | 7.0 (0.8) | 13.2 (1.8) | 6.3 (1.4) |
| Completeness, % | 95.1 (87.7) | 94.1 (94.7) | 99.5 (99.8) | 97.4 (97.1) | 98.1 (98.2) | 97.0 (96.4) | 97.5 (98.7) |
| Redundancy | 3.0 (2.7) | 4.2 (4.0) | 5.7 (5.6) | 3.5 (3.3) | 2.9 (2.8) | 3.5 (3.3) | 2.9 (3.0) |
| CC1/2 | 0.999 (0.518) | 0.999 (0.283) | 0.983 (0.185) | 0.989 (0.688) | 0.997 (0.252) | 0.998 (0.521) | 0.985 (0.427) |
| Refinement | |||||||
| Nonhydrogen atoms | 17,077 | 17,162 | 16,439 | 16,328 | 16,399 | 16,603 | 16,049 |
| Rwork/Rfree, % | 16.9/20.2 (26.6/26.5) | 17.6/20.2 (30.8/31.4) | 17.6/21.2 (26.2/26.0) | 17.5/20.8 (26.6/32.7) | 19.0/22.8 (37.7/38.1) | 17.3/20.5 (26.6/28.2) | 21.2/25.0 (39.0/38.6) |
| Average B factor | 32 | 29 | 46 | 74 | 43 | 37 | 67 |
| rmsd | |||||||
| Bond lengths | 0.020 | 0.019 | 0.020 | 0.019 | 0.019 | 0.023 | 0.012 |
| Bond angles | 2.0 | 2.0 | 2.1 | 2.0 | 2.0 | 2.1 | 1.6 |
| Ramachandran analysis | |||||||
| Preferred regions, % | 98.3 | 98.3 | 97.8 | 98.6 | 97.5 | 98.3 | 98.6 |
| Allowed regions, % | 1.7 | 1.7 | 2.1 | 1.4 | 2.3 | 1.7 | 1.4 |
| Outliers, % | 0 | 0 | 0.1 | 0 | 0.2 | 0 | 0 |
Statistics in parentheses indicate those for the highest resolution shell.
Base Modification Has Modest Effects on Substrate Binding.
Modifications on the Watson–Crick base edge are well-tolerated at the catalytic site of SAMHD1. Consistent with previous observations with dNTPs (19, 21), there are no base-specific interactions between SAMHD1 and the bases of the nucleotide analogs. Water networks in the active site of SAMHD1 stabilize the nucleotide analogs in the pocket, as for canonical dNTPs. In each case, three to four water molecules bridge the interaction between SAMHD1 and the Watson–Crick and sugar edges of the bound analog (Fig. 1D). Cytarabine-TP and gemcitabine-TP, each containing an unmodified cytosine base, maintained water network interactions that closely resembled those reported for dCTP (Fig. 1D) (19). In our previous structural studies, the adenosine base was not resolved in the dATP-SAMHD1 cocrystal structure due to its relatively weak binding affinity to the catalytic site, compared with other dNTPs (19). However, clear electron density for the bases of clofarabine-TP, cladribine-TP, fludarabine-TP, and vidarabine-TP was observed in the current structures (Fig. 1B). The additional chlorine atom (cladribine-TP and clofarabine-TP) or fluorine atom (fludarabine-TP) at the C2 position of the bases contributed to a more extensive water network and stabilized these molecules in the active site (Fig. 1D). These results suggest that small modifications in the base are not likely to disrupt binding to the SAMHD1 catalytic pocket because water networks that contact the base are flexible. Some nucleotide modifications may even enhance the water network surrounding the nucleotide analog and possibly increase its binding affinity.
SAMHD1 Substrate Specificity Regulated By 2′ Sugar Moiety.
It is well-established that NTPs with ribose sugars [(2′R)-2′-OH] are not substrates of SAMHD1 (2, 19). We examined how sugar modifications found in nucleotide analog drugs influence their binding to, and hydrolysis by, SAMHD1 as it has been shown that arabinose-based nucleotides with 2′S sugar modifications are substrates of SAMHD1 (31, 32). Our crystal structures corroborate the finding that arabinose-based sugars, with either [(2′S)-2′-OH] or [(2′S)-2′-F] modifications, are allowed in the catalytic site of SAMHD1. In addition to the canonical interactions observed with dNTP substrates (19), we observed van der Waals and stacking interactions between active site residues Y315 and Y374 and the [(2′S)-2′-OH/F] atoms of cytarabine-TP, clofarabine-TP, fludarabine-TP, vidarabine-TP, and gemcitabine-TP in the catalytic pocket (Figs. 1C and 2 A and B). Since the arabinose-like sugar modifications provide an additional interaction with SAMHD1, these modifications likely stabilize these analogs in the catalytic pocket. In contrast, we crystallized SAMHD1 in the presence of 10 mM [(2′R)-2′-F]-dCTP, which has a ribose-like modification, but electron density for this nucleotide analog was not observed in the catalytic pocket (SI Appendix, Fig. S1A).
Fig. 2.
Gemcitabine-TP but not [(2′R)-2′-F]-dCTP is hydrolyzed by SAMHD1 in vitro. (A) [(2′S)-2′-OH)] of cytarabine-TP is stabilized by residues Y374 and Y315 through van der Waals interactions. Transparent surface of SAMHD1 is shown with key residues in sticks. (B) The 2′,2′-difluorine sugar modification of gemcitabine-TP is stabilized by van der Waals interactions with residues Y374 and Y315 to compensate potential close contact (yellow caution triangle) between the [(2′R)-2′-F] atom and residue L150 in the catalytic site. (C) HPLC-based activity assay measuring product produced by preassembled SAMHD1 tetramers in the presence of 25 to 1,600 μM nucleotide analog substrates. Error bars represent SEM of three independent experiments. (D, Left) dNTPase activity of SAMHD1 over the course of 15 min was measured using a malachite green assay. Product is normalized to SAMHD1 concentration (nmol PO4/nmol SAMHD1). SAMHD1 tetramers were preassembled with 250 μM GTP and dATP and then diluted 100-fold into 125 μM gemcitabine-TP, cytarabine-TP, dCTP, CTP, [(2′S)-2′-F]-dCTP, [(2′R)-2′-F]-dCTP, or buffer. Error bars represent SEM of three independent experiments. (D, Right) Chemical structures of [(2′S)-2′-F]-dCTP and [(2′R)-2′-F]-dCTP analogs. (E) A rigid body model of [(2′R)-2′-F]-dCTP (gray sticks) in the catalytic pocket potentially creates a clash (red cross) with residue L150.
Our activity assays also confirmed that the substrate specificity of SAMHD1 has a general dependency on the stereochemistry of the sugar moiety at the 2′ position. We used an HPLC-based assay (19) to measure the SAMHD1 enzyme kinetics for these substrates. Larger modifications at the 2′ sugar moiety were generally associated with higher KM and lower kcat values (Fig. 2C and Table 2). Although most 2′ sugar modifications are tolerated and the analogs can bind SAMHD1 in a similar fashion as evidenced by their structures, modest geometry perturbations due to the 2′ sugar modifications may lead to less catalytically productive binding. For example, although gemcitabine-TP is a substrate of SAMHD1, its turnover rate and specific constant are noticeably lower than cytarabine-TP (Fig. 2C and Table 2). This may explain why a previous report did not observe hydrolysis of gemcitabine-TP where the concentration of gemcitabine-TP in monocyte-derived macrophages was not dependent on SAMHD1 protein levels (32). This is perhaps due to the difficulty in detecting the low hydrolysis rate in the complex cellular environment.
Table 2.
Kinetic constants for nucleotide analog hydrolysis by SAMHD1
| Nucleotide | kcat, s−1* | KM, μM* | kcat/KM, s−1⋅μM−1 |
| dATP | 11.4 ± 0.8 | 781 ± 118 | 0.0146 |
| Cladribine-TP | 4.9 ± 0.4 | 537 ± 118 | 0.0091 |
| Clofarabine-TP | 14.2 ± 2.7 | 2,912 ± 770 | 0.0049 |
| Fludarabine-TP | 5.0 ± 0.9 | 867 ± 322 | 0.0058 |
| Vidarabine-TP | 5.6 ± 0.6 | 873 ± 180 | 0.0064 |
| dCTP | 4.7 ± 0.7 | 648 ± 230 | 0.0073 |
| Cytarabine-TP | 3.4 ± 0.4 | 446 ± 123 | 0.0076 |
| Gemcitabine-TP | 2.4 ± 0.2 | 379 ± 79 | 0.0063 |
Mean values from three independent experiments ±SEM.
To reconcile the discrepancy regarding gemcitabine-TP hydrolysis, we directly compared the hydrolysis of the doubly modified gemcitabine-TP to the singly modified [(2′S)-2′-F]-dCTP and [(2′R)-2′-F]-dCTP in vitro in a time course assay (Fig. 2D). We observed that SAMHD1 hydrolyzes [(2′S)-2′-F]-dCTP similar to cytarabine-TP, but gemcitabine-TP is hydrolyzed at a lower rate and [(2′R)-2′-F]-dCTP is unreactive (Fig. 2D). In general, these results confirmed other reports that arabinose-based nucleotide analogs are substrates of SAMHD1 whereas the ribose-based [(2′R)-2′-F]-dCTP is not (31, 32). As evidenced by the electron density of gemcitabine-TP in the catalytic pocket, both the 2′S and 2′R fluorine modifications are accommodated in the catalytic pocket of SAMHD1 (SI Appendix, Fig. S1B). However, the substructure [(2′R)-2′-F]-dCTP, with a single 2′R modification, is not accommodated. Rigid modeling of this compound into the catalytic pocket predicts that the (2′R)-2′-F atom would clash with L150 (Fig. 2D). This suggests that the 2′R modification is permitted only in the context of a 2′,2′-difluorine sugar modification, but not alone (Fig. 2 B and E). SAMHD1’s discrimination between these two substrates may arise from interactions between Y315/Y374 and the 2′S fluorine atom, which might partially compensate for the sterically unfavorable 2′R fluorine modification (Fig. 2 B and E). The presence of the 2R′ modification may lead to a less suitable positioning of the nucleotide in the catalytic pocket for catalysis, compared with 2′S modifications alone (Fig. 2 A and B). Although we captured gemcitabine-TP in the catalytic pocket at a high concentration (5 mM), the catalytically productive conformations may not be readily formed at lower concentrations used in cellular studies. This is consistent with the low kcat value of gemcitabine-TP in our measurements (Fig. 2C). These results support the notion that the catalytic pocket of SAMHD1 is highly sensitive to the stereochemistry of 2′ sugar modification.
SAMHD1 Hydrolyzes Various Nucleotide Analogs in Vivo.
Since SAMHD1 is capable of hydrolyzing cancer drugs in vitro, it has the potential for decreasing their therapeutic efficacies. We explored the extent to which the triphosphates of each of these nucleotide analogs are degraded by SAMHD1 in the cell following uptake and metabolic activation. A recent report demonstrated a strong inverse correlation between SAMHD1 expression in leukemic blasts and AML patients’ clinical response to cytarabine therapy (31). Similarly, we found that the IC50 values of fludarabine-TP and clofarabine-TP in AML cell lines were also correlated with SAMHD1 protein expression levels (Fig. 3A). However, this effect was not observed for cladribine-TP or gemcitabine-TP. We also tested whether the depletion of SAMHD1 in THP-1 cells via knock-out or targeted proteasomal degradation by Vpx-virus–like particles (VLPs) affected the IC50 values of these drugs. Although a strong effect was observed for cytarabine-TP and a moderate effect was observed for fludarabine-TP, clofarabine-TP, vidarabine-TP, and cladribine-TP (Fig. 3B and SI Appendix, Fig. S2A), there was no effect on the IC50 value for gemcitabine-TP (Fig. 3B). To directly measure SAMHD1’s effect on each nucleotide analog’s concentration in the cell, we used liquid chromatography tandem mass spectrometry (LC-MS/MS) to quantify drugs found in cells with or without SAMHD1 expression. Our results show that clofarabine-TP, fludarabine-TP, and cytarabine-TP were strongly depleted by SAMHD1 whereas cladribine-TP was depleted to a low extent and gemcitabine-TP levels remained unaffected (Fig. 3C). Although SAMHD1 hydrolyzes gemcitabine in vitro with a low activity, this effect may not be detectable under normal cellular conditions. Our cell-based assays indeed corroborate previous reports that gemcitabine-TP is not significantly degraded by SAMHD1 in vivo (31, 32).
Fig. 3.
SAMHD1 depletes several TPs of nucleoside analogs in vivo. (A) Correlations of cytarabine, clofarabine, fludarabine, cladribine, and gemcitabine concentrations inhibiting 50% of cell viability (IC50) and relative protein expression levels of SAMHD1 in 13 AML cell lines. Relative expression levels (ratios of a SAMHD1/β-actin) are shown as arbitrary units (a.u.). Ratio of SAMHD1/β-actin for parental THP-1 cells is set to 1, and ratios of other cell lines are set relative to it. Filled circles represent mean values, and error bars indicate SD of three independent experiments. Correlations were analyzed using linear or log-linear regression models. (B) Cytarabine, fludarabine, clofarabine, cladribine, or gemcitabine IC50 values of THP-1 KO, THP-1 Control cells, or parental THP-1 cells exposed to VSV-G pseudotyped VLPs carrying either lentiviral Vpr (Vpr-VLP, control) or SAMHD1-degrading Vpx proteins (Vpx-VLP). The bars represent mean values, and the error bars are SD of three independent experiments. The numbers above indicate factor of decrease of the IC50 values in the absence of SAMHD1. Statistical analyses were performed using unpaired two-tailed Student’s t test comparing treated samples with untreated control. n.s., no statistical significance. ***P < 0.001. (C) Representative liquid chromatography tandem mass spectrometry (LC-MS/MS) measurements of cytarabine-TP, fludarabine-TP, clofarabine-TP, cladribine-TP, or gemcitabine-TP in THP-1 KO cells (black) and THP-1 control cells (red).
The structural and biochemical framework established in this study also allows for the rational modeling of other known cancer drugs into the catalytic pocket of SAMHD1. For example, the IC50 of nelarabine-TP in AML cell lines relies on the expression level of SAMHD1, indicating that it is a substrate of SAMHD1, too (SI Appendix, Fig. S2B). Modeling the metabolite araGTP into the catalytic pocket of SAMHD1 predicts that it would fit into the catalytic pocket like any other arabinose-based nucleotide analog, such as vidarabine-TP (SI Appendix, Fig. S2C).
Allosteric Site 2 of SAMHD1 Is More Restrictive than Its Catalytic Site.
In addition to establishing SAMHD1’s substrate specificity for nucleotide analogs, we also tested whether these drugs were capable of binding to the allosteric sites to induce the catalytically active SAMHD1 tetramer. None of the nucleotide analogs tested here contained the guanosine base required for Allo-site 1 binding; thus, they alone were not sufficient for SAMHD1 activation (SI Appendix, Fig. S1C). To test which analogs bind Allo-site 2, we monitored the oligomerization state of SAMHD1 in the presence of GTP and each of the analogs. Previous studies indicated that clofarabine-TP is an activator of SAMHD1 (40), but cytarabine-TP is not (31, 32). Consistent with these reports, our size-exclusion chromatography (SEC) assays showed that only clofarabine-TP and cladribine-TP caused a shift in the elution profile of SAMHD1 toward a higher molecular weight species (Fig. 4A), with cladribine-TP being more effective at inducing SAMHD1 oligomerization. Sedimentation velocity analytical ultracentrifugation (SV-AUC) also showed that cladribine-TP induced SAMHD1 oligomerization (Fig. 4B). However, clofarabine-TP–induced SAMHD1 oligomerization was not detected by SV-AUC, likely due to experimental constraints requiring about 40-fold fewer nucleotide analogs compared with SEC. When mixed with GTP, cladribine-TP and, to some extent, clofarabine-TP both induce SAMHD1 activity (Fig. 4C). To compare relative affinities of each drug for Allo-site 2, we measured the activity of SAMHD1 preassembled with GTP and increasing concentrations of dATP, cladribine-TP, clofarabine-TP, and ATP activators (17). Hydrolysis of the dATP substrate was measured as readout for the effective affinity of the activators in the Allo-site 2. While cladribine-TP assembled active SAMHD1 tetramers with a modest efficiency, the apparent affinity of clofarabine-TP for Allo-site 2 was drastically reduced (Fig. 4D). Together, these results suggest that only minor sugar modifications are permitted for SAMHD1 allosteric activators.
Fig. 4.
The allosteric sites of SAMHD1 are highly restrictive. (A) Size-exclusion chromatography elution profile of SAMHD1 in the presence of 0.5 mM GTP and 4 mM of color-coded nucleotide analog. (B) SV-AUC analysis of SAMHD1 in the absence of nucleotides or the presence of dGTP, GTP with clofarabine-TP, or GTP with cladribine-TP at a final concentration of 150 μM. (C) Malachite green activity assay performed in the presence of 125 μM GTP and 125 μM dCTP, dATP, nucleotide analog, or buffer. Error bars represent SEM of three independent experiments. (D) Malachite green activity assay measuring the hydrolysis of dATP by SAMHD1 tetramers preassembled in the presence of 125 μM GTP and 6.3 to 3,200 μM dATP, cladribine-TP, clofarabine-TP, or ATP. Error bars represent SEM of three independent experiments.
2′ Sugar Modifications Are Highly Restrictive at the Allo-Site 2.
To examine how SAMHD1 accommodates nucleotide analogs in the allosteric pocket, we attempted to crystalize SAMHD1 with GTP and each of the nucleotide analogs assayed above. Consistent with our activity assay and oligomerization measurements, only the two allosteric activators, cladribine-TP and clofarabine-TP, resulted in SAMHD1 tetramer crystals. The resulting structures revealed unambiguous electron density for each nucleotide in the Allo-site 2 (Fig. 5A). As predicted, the nucleotide analogs bind to Allo-site 2, and GTP binds to the adjacent Allo-site 1. The allosteric sites are not disturbed by cladribine-TP or clofarabine-TP as the chlorine atom modification at the C2 position of the base does not interfere with the allosteric pocket interactions (Fig. 5B). Hydrogen bonds between the adenosine base and residues N119 and N358 (19) are preserved in the cladribine-TP and clofarabine-TP structures, allowing for the correct positioning of each nucleotide in the Allo-site 2 pocket. These results suggest that nucleotide analogs with some similar modifications at the Watson–Crick edge of the base may also be permitted in Allo-site 2.
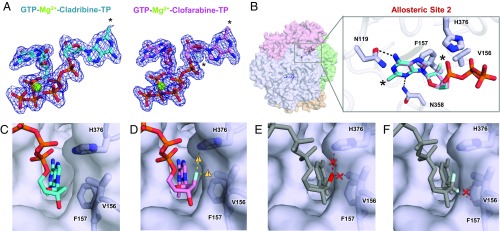
Fig. 5.
Structures of cladribine-TP and clofarabine-TP bound to Allo-site 2 of SAMHD1. (A) 2Fo-Fc electron density (σ = 1.0) for GTP, cladribine-TP, and clofarabine-TP in the allosteric pocket of SAMHD1. Black asterisks indicate sites of modifications, and black dotted lines indicate hydrogen bonds. (B, Left) Transparent surface view of the SAMHD1 tetramer. (B, Right) Overlay of cladribine-TP (cyan) and clofarabine-TP (pink) in Allo-site 2. The main chain of SAMHD1 is shown as tubes, with selected residues and nucleotides represented as sticks. Residues important for gating the 2′-atom are highlighted in thicker sticks. Black asterisks indicate sites of modification. (C) The structure of cladribine-TP (cyan, sticks) in Allo-site 2 with V156, F157, and H376 shown as sticks under semitransparent surface of SAMHD1. (D) The structure of clofarabine-TP (pink, sticks) in Allo-site 2 with close contacts between the [(2′S)-2′-F] atom and V156, F157, and H376 highlighted with caution triangles and yellow dashed lines. (E) A rigid body model of cytarabine-TP in Allo-site 2 with potential steric clashes between the [(2′S)-2′-OH] group and V156 and H376 highlighted with a red cross and red dashed lines. (F) A rigid body model of gemcitabine-TP in Allo-site 2 with a potential steric clash between the [(2′R)-2′-F] atom and F157 highlighted with a red cross and red dashed lines.
While modest base modifications do not affect Allo-site 2 binding, modifications to the sugar at the 2′ position are restricted. We found that Allo-site 2 excludes all arabinose-based analogs with the [(2′S)-2′-OH] group, which is consistent with previous observations (31). The ribose-based [(2′R)-2′-F]-dCTP is also excluded from the site, the same as the NTP molecules reported before (19). As expected, Allo-site 2 does not tolerate a nucleotide with 2′,2′-difluorine modifications. The exclusion of nucleotides with these sugar modifications is likely due to SAMHD1 residues H376, V156, and F157, which form a tight pocket around the 2′ carbon of the dNTPs (Fig. 5 B and C). While clofarabine-TP, which contains a [(2′S)-2′-F] modification in this position, is accommodated at this site, the structure shows that the fluorine atom may still cause some steric constraints (Fig. 5D). This is consistent with our oligomerization and activity assays, which showed reduced SAMHD1 activation by clofarabine-TP (Fig. 4C). Modeling cytarabine-TP in the Allo-site 2 pocket suggests that steric clashes arise between the [(2′S)-2′-OH] group and residues H376 and V156 of SAMHD1 (Fig. 5E). On the other side of the sugar ring, residue F157 limits the accessibility of nucleotides with [(2′R)-2′-OH] or [(2′R)-2′-F] modifications, such as gemcitabine-TP (Fig. 5F). Cladribine-TP is the only deoxyribonucleotide analog tested here, and most likely the lack of a 2′ sugar modification allows it to be the strongest allosteric activator of SAMHD1. Overall, Allo-site 2 in SAMHD1 was highly sensitive to modifications to the 2′ position of the sugar, and this moiety is a major binding determinant of nucleotide analog binding to the allosteric site.
Discussion
The study presented herein provides a comprehensive structural and biochemical framework for understanding how a wide range of nucleotide analog drugs interact with SAMHD1. Our biochemical and structural analyses of a panel of nucleotide analogs, with a variety of 2′ sugar modifications, reveal the detailed binding determinants for the catalytic site of SAMHD1 (Figs. 1 and 6). SAMHD1 selects substrates through indirect interactions between water molecules in the catalytic pocket and the base of the analog drugs. The network of water molecules changes to adapt to variations found in the nucleotide analogs. Water-mediated interactions between the substrate and the enzyme allow for binding pocket plasticity and accommodation of different modifications. In addition, the catalytic pocket is accessible to the arabinose-like 2′ sugar modifications with the 2′S geometry. This provides a mechanistic understanding of the findings that arabinose-based nucleotides are substrates of SAMHD1 (31, 32). Interestingly, we were able to observe electron density for analogs with modified adenosine bases, which was previously unresolved in our dATP-SAMHD1 structure (19). This indicates that the nucleotide analog modifications may provide additional stabilizing interactions with the catalytic pocket of SAMHD1.
Fig. 6.
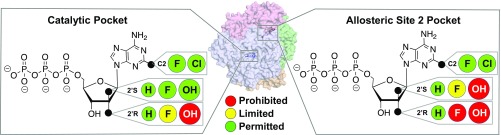
Summary of the effects of the 2′ sugar moiety on nucleotide analog binding to the catalytic and allosteric sites of SAMHD1. (Middle) Transparent surface view of SAMHD1 tetramer. (Left) In the catalytic pocket of SAMHD1, while small substitutions such as fluorine atoms at the 2′R and 2′S positions of the sugar are permitted (green circle), their access to the 2′R position is limited (yellow circle). Larger modifications, such as hydroxyl groups, are permitted in the 2′S position, but not the 2′R position (red circle). (Right) In Allo-site 2 of SAMHD1, hydroxyl groups are prohibited in both 2′R and 2′S positions of the sugar moiety. Fluorine atoms have limited access to the 2′S position, but they are prohibited from the 2′R position. Small base modifications, such as fluorine or chlorine atoms, are tolerated in both Allo-site 2 and the catalytic site.
Our data also demonstrate that a nucleotide analog can bind to the allosteric sites of SAMHD1 to activate the enzyme, depending on modifications to its sugar moiety (Figs. 5 and 6). We show that modest base modifications, such as a chlorine atom at C2 of adenine, are tolerated at the Allo-site 2. In contrast, Allo-site 2 does not allow arabinose-based or ribose-based nucleotides. Only deoxyribose-based nucleotides, such as cladribine-TP, can efficiently enter Allo-site 2 (Figs. 5 and 6). The results suggest that any modifications to the 2′ carbon of the sugar ring may affect the drug’s binding affinity to the allosteric pocket. These findings may help advance the understanding of the effect of current nucleotide analog drugs and guide the design of nonhydrolyzable analogs that can activate SAMHD1 for targeted depletion of cellular dNTP pools.
Interestingly, we found that cytarabine-TP is particularly dependent on SAMHD1 cellular expression, compared with other drugs tested in this study. Although many of the drugs tested were hydrolyzed by purified SAMHD1 in our biochemical assays, the effect of SAMHD1 expression on drug efficacy varied in cells. This may be due to different interactions between these drugs with other components of the nucleotide metabolism pathway in the cell. For example, even though clofarabine, fludarabine, and cladrabine are similar in structure, these drugs have been reported to have varying degrees of interactions with cellular factors that affect their ability to be phosphorylated by dCK and to inhibit ribonucleotide reductase activity and DNA/RNA chain elongation (26). Moreover, nucleoside analogs may differentially influence activity and expression of cell cycle regulatory proteins, such as cyclins and cyclin-dependent kinases (41, 42), which in turn may interact with SAMHD1 and influence its dNTPase activity in leukemic cells (43). It remains possible that, although these drugs are hydrolyzed to a similar extent by SAMHD1 in vitro, their metabolism in the cell might be influenced by other cellular factors. In the case of cytarabine, our in vivo data strongly support that SAMHD1 is one of the main determinants of intracellular cytarabine-TP concentrations. On the other hand, we demonstrated that SAMHD1 strongly hydrolyzed cladribine-TP in vitro but had little influence on cladribine-TP concentrations and activity in AML cells. Combining biochemical, biophysical, and cellular studies offers a comprehensive approach to evaluate the response of nucleoside analog drugs to SAMHD1 both at the mechanistic level and in application.
SAMHD1’s substrate promiscuity helps it function as a general sensor and regulator of nucleotide pools, but it also allows SAMHD1 to facilitate cancer cells to escape from nucleotide analog treatments. Thus, it is important to consider how different modifications affect a nucleotide analog drug’s access to the catalytic and allosteric sites of SAMHD1 (Fig. 6) when developing new therapies. Defining SAMHD1 interactions with nucleotide analog drugs may be critical for better predicting patient response to the current and future therapies. More detailed future studies may focus on how each of these analogs competes for relevant active sites. It will shed light onto the extent to which nucleotide analogs can be combined to improve current treatments.
Materials and Methods
Nucleotide Analog Compounds.
GTP, dATP, and dCTP were purchased from Thermo Scientific. Cytarabine-TP, cladribine-TP, clofarabine-TP, fludarabine-TP, gemcitabine-TP, [(2′R)-2′-F]-dCTP, and [(2′S)-2′-F]-dCTP for in vitro assays were purchased from Jena Biosciences. The precursor nucleoside analogs (unphosphorylated) used for all cell culture assays were purchased from the following sources: Tocris (cytarabine, fludarabine, clofarabine, and cladribine), Accord Healthcare GmbH (gemcitabine), and Jena Bioscience (vidarabine and nelarabine). All nucleotide standards, internal standards, and dNs for the LC-MS/MS analysis were obtained from Sigma-Aldrich, Silantes, or Alsachim (44). Cytarabine-13C3 was purchased from Santa Cruz and used for LC-MS/MS analysis.
Protein Expression and Purification.
N-terminal 6×His-tagged SAMHD1 (residues 113 to 626) was expressed in Escherichia coli and purified using nickel-nitrilotriacetic acid (Ni-NTA) affinity and size-exclusion chromatography as previously described (19).
Analytical Size-Exclusion Chromatography.
Purified samples of SAMHD1 (2 mg/mL, 50 μL) mixed with a final concentration of 500 μM GTP and 4 mM nucleotide analog were applied to a Superdex 200 5/150 GL column (GE Healthcare) preequilibrated in 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, and 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP). The UV absorbance at 280 nm was measured as the protein sample eluted from the column.
Analytical Ultracentrifugation.
Sedimentation velocity experiments were performed with a Beckman XL-I analytical ultracentrifuge. Samples were prepared with protein concentration of 0.8 to 1.3 mg/mL in the buffer containing 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, and 0.5 mM TCEP and equilibrated with a final nucleotide concentration of 150 μM. AUC was performed at 26,892 × g and 20 °C with an An60-Ti rotor. The experimental parameters, including sample partial specific volume, buffer density, and viscosity, were calculated with SEDNTERP (www.jphilo.mailway.com). Velocity data were analyzed using the program SEDFIT (45).
Crystallization and Data Collection.
Purified SAMHD1 protein in buffer (50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, and 0.5 mM TCEP) was mixed with 1 mM GTP and 10 mM analog nucleotides (5 mM or 0.5 mM for gemcitabine-TP and vidarabine-TP, respectively) with or without 100 μM dATP (or 2.5 mM dATP for vidarabine-TP) and incubated at 4 °C for 15 min before crystallization. The small amount of dATP was included to ensure the formation of the SAMHD1 tetramer as most nucleotide analogs do not bind Allo-site 2. All crystals were grown at 25 °C using the microbatch under-oil method by mixing 1 μL of protein (5 mg/mL) with 1 μL of crystallization buffer (100 mM succinate–phosphate–glycine (SPG) buffer, pH 7.4, 25% PEG 1500; Qiagen). Crystals were cryoprotected by crystallization buffer supplemented with 25% (vol/vol) glycerol before being frozen in liquid nitrogen. Diffraction data were collected at Brookhaven National Laboratory (BNL) beamline AMX and the Advanced Photon Source beamline 24-ID. The data statistics are summarized in Table 1.
Structure Determination and Refinement.
The structures were solved by molecular replacement using PHASER (46). We used the previously published SAMHD1 tetramer structure (PDB ID code 4BZB), with the bound nucleotides removed, as the search model. The model was refined with iterative rounds of translation/libration/screw (TLS) and restrained refinement using Refmac5 (47), followed by rebuilding the model to the 2Fo-Fc and the Fo-Fc maps using Coot (48). Refinement statistics are summarized in Table 1. Coordinates and structure factors have been deposited in the Protein Data Bank, with accession codes listed in Table 1.
Malachite Green Activity Assay.
The enzymatic activity assay was adapted from ref. 49. All assays were performed with purified catalytic domain of SAMHD1 (residues 113 to 626) at 25 °C in a reaction buffer containing 50 mM Tris⋅HCl, pH 8, 150 mM NaCl, 5 mM MgCl2, and 0.5 mM TCEP. Each reaction, containing 10 μM E. coli inorganic pyrophosphatase, 0.5 μM SAMHD1, and 125 μM substrate or allosteric activator, was quenched with 20 mM EDTA after 5 or 15 min. Then, Malachite Green reagent was added to the solution and developed for 15 min before the absorbance at 650 nm was measured.
HPLC-Based Kinetics Assay.
Reactions were initiated by the addition of preassembled SAMHD1 (final concentration of 500 nM) to 12 to 1,600 μM nucleotide analog substrates and incubated at room temperature. Reactions were terminated by a 10× dilution into 20 mM EDTA after 5 min. Samples were deproteinized by spinning through an Amicon Ultra 0.5-mL 10-kDa filter (Millipore) for 10 min at 16,000 × g. Samples were analyzed by HPLC with 100 μL of sample loaded on the Synergi C18 column 150 × 4.6 mm (Phenomenex). The column was preequilibrated in 20 mM ammonium acetate, pH 4.5 (buffer A), and samples were eluted at a flow rate of 1 mL/min with a gradient of methanol (buffer B) over 19 min. UV absorption was recorded at 260 nm.
Kinetic constants describing nucleotide analog hydrolysis were calculated with the Michaelis–Menten equation (Eq. 1) using Prism software (version 7.0a; GraphPad Software),
| [1] |
where V is the enzyme velocity, Et is the total concentration of enzyme, and X is the concentration of nucleotide analog. Et was constrained to a constant value of 0.005 nanomoles. Data shown in Table 2 indicate the mean of three independent experiments with SE of three independent experiments.
Cells and Cell Culture.
Human AML cell lines, including THP-1 [DSMZ no. ACC16; French–American–British classification (FAB) M6], OCI-AML2 (DSMZ no. ACC 99; FAB M4), OC-AML3 (DSMZ no. ACC 582; FAB M4), Molm13 (DSMZ no. ACC 554; FAB M5a), PL-21 (DSMZ no. ACC 536; FAB M3), HL-60 (DSMZ no. ACC 3; FAB M2), MV4-11 (DSMZ no. ACC 102; FAB M5), SIG-M5 (DSMZ no. ACC 468; FAB M5a), ML2 (DSMZ no. ACC 15; FAB M4), NB4 (DSMZ no. ACC 207; FAB M3), KG1 (DSMZ no. ACC 14; FAB not indicated), MonoMac6 (DSMZ no. ACC 124; FAB M5), and HEL (DSMZ No. ACC 11; FAB M6), were obtained from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH). A THP-1 cell clone engineered for SAMHD1 deficiency (THP-1 KO) and a corresponding SAMHD1-positive control cell clone (THP-1 Control) have been reported (50). All cell lines were cultured in Iscove’s modified Dulbecco’s medium (IMDM) (Biochrom) supplemented with 10% FBS (SIG-M5 20% FBS), 4 mM l-glutamine, 100 IU/mL penicillin, and 100 mg/mL streptomycin at 37 °C in a humidified 5% CO2 incubator. Cells were routinely tested for mycoplasma contamination (LT07-710; Lonza) and authenticated by short tandem repeat profiling, as reported (51).
Cell Viability Assay.
Viability of AML cell lines treated with various drug concentrations was determined by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye reduction assay after 96 h of incubation as described previously (52). IC50 values were determined using CalcuSyn (Biosoft).
Immunoblotting.
Cells were lysed in Triton X-100 sample buffer, and proteins were separated by SDS-polyacrylamide gel electrophoresis. Proteins were blotted onto a nitrocellulose membrane (Thermo Scientific). The membrane was incubated overnight at 4 °C with primary antibodies used at the indicated dilutions: SAMHD1 (12586-1-AP, 1:1,000; Proteintech), β-actin (3598R-100, 1:2,000; BioVision via BioCat). Visualization and quantification was performed using fluorescently labeled secondary antibodies (926-32210 IRDye 800CW goat anti-mouse and 926-32211 IRDye 800CW goat anti-rabbit, 1:20000; LI-COR) and Odyssey LICOR.
LC-MS/MS Analysis.
Cells (1 × 106) were treated with 10 µM of the specific drug and incubated at 37 °C in a humidified 5% CO2 incubator for 6 h. Subsequently, cells were washed twice in 1 mL of PBS, pelleted, and stored at −20 °C until measurement. The concentrations of dNTPs 13C3- cytarabine-TP, fludarabine-TP, clofarabine-TP, cladribine-TP, and gemcitabine-TP in the samples were measured by liquid chromatography-electrospray ionization-tandem mass spectrometry as previously described (44). Briefly, the analytes were extracted by protein precipitation with methanol. An anion exchange HPLC column (BioBasic AX, 150 × 2.1 mm; Thermo) was used for the chromatographic separation, and a 5500 QTrap (Sciex) was used as analyzer, operating as triple quadrupole in positive multiple reaction monitoring (MRM) mode. The analysis of the dNTP was performed as previously described (44). Additionally, 13C3- cytarabine-TP, fludarabine-TP, clofarabine-TP, cladribine-TP, and gemcitabine-TP were quantified using cytidine-13C9-15N3-5′-triphosphate as internal standard (IS). The precursor-to-product ion transitions used as quantifiers were as follows: m/z 487.0 → 115.1 for 13C3-cytarabine-TP, m/z 525.7 → 154.1 for fludarabine-TP, m/z 543.7 → 134.0 for clofarabine-TP, m/z 526.0 → 170.0 for cladribine-TP, and m/z 504.0 → 326.0 for gemcitabine-TP. Due to the lack of commercially available standards, relative quantification was performed by comparing the peak area ratios (analyte/IS) of the differently treated samples.
Production of Virus-Like Particles.
VLPs, carrying either Vpx or Vpr from SIVmac251, were produced by cotransfection of 293T cells with pSIV3+ gag pol expression plasmids and a plasmid encoding vesicular stomatitis virus glycoprotein (VSV-G). The SIVmac251-based gag-pol expression constructs pSIV3+R− (Vpr-deficient) and pSIV3+X− (Vpx-deficient) were previously reported (53). The SAMHD1 degradation capacity of Vpx-VLPs was determined in THP-1 cells 24 h posttransduction by intracellular SAMHD1 staining. AML cell lines were spinoculated with VSV-G pseudotyped VLPs carrying either Vpx or Vpr. Expression of SAMHD1 was monitored by Western blotting.
Statistical Information.
The average SEs and SDs were calculated from multiple separate experiments as indicated in each figure legend, and the results are shown in each graph.
Supplementary Material
Acknowledgments
We thank J. Wang, T. Sasaki, V. Duong, and K. Anderson for technical assistance and discussions. This work was supported in part by NIH Grants AI136737 (to Y.X.) and AI120845 (to X.J.), by a Yale Cancer Center Pilot Grant (to Y.X.), by Hilfe für Krebskranke Kinder Frankfurt e.V. and the Frankfurter Stiftung für Krebskranke Kinder (J.C. and C.S.), and by Deutsche Forschungsgemeinschaft Grant KE 742/4-1 (to O.T.K.). O.B. was supported by NIH Predoctoral Program in Cellular and Molecular Biology Grant T32 GM007223 and by National Science Foundation Graduate Research Fellowship DGE1122492. K.M.K. was supported by NIH Predoctoral Program in Biophysics Grant T32 GM008283. G.G. was supported by the Hessischen Landesoffensive zur Entwicklung Wissenschaftlicher und Ökonomischer Exzellenz (LOEWE-Center) Translational Medicine and Pharmacology. This work is in part based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from NIH Grant P41 GM103403. The Pilatus 6M detector on the 24-ID-C beamline is funded by NIH-Office of Research Infrastructure Programs High-End Instrumentation Grant S10 RR029205. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.A.S. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6DW4, 6DWD, 6DWK, 6DW3, 6DW5, and 6DW7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805593115/-/DCSupplemental.
References
- 1.Franzolin E, et al. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci USA. 2013;110:14272–14277. doi: 10.1073/pnas.1312033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 3.Lahouassa H, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Descours B, et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bermejo M, et al. Dasatinib inhibits HIV-1 replication through the interference of SAMHD1 phosphorylation in CD4+ T cells. Biochem Pharmacol. 2016;106:30–45. doi: 10.1016/j.bcp.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger A, et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutières syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cribier A, Descours B, Valadão AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Tang C, Ji X, Wu L, Xiong Y. Impaired dNTPase activity of SAMHD1 by phosphomimetic mutation of Thr-592. J Biol Chem. 2015;290:26352–26359. doi: 10.1074/jbc.M115.677435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauls E, et al. Palbociclib, a selective inhibitor of cyclin-dependent kinase4/6, blocks HIV-1 reverse transcription through the control of sterile α motif and HD domain-containing protein-1 (SAMHD1) activity. AIDS. 2014;28:2213–2222. doi: 10.1097/QAD.0000000000000399. [DOI] [PubMed] [Google Scholar]
- 12.Pauls E, et al. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol. 2014;193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- 13.Welbourn S, Dutta SM, Semmes OJ, Strebel K. Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J Virol. 2013;87:11516–11524. doi: 10.1128/JVI.01642-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Gelais C, et al. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J Virol. 2014;88:5834–5844. doi: 10.1128/JVI.00155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J, et al. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J Biol Chem. 2013;288:10406–10417. doi: 10.1074/jbc.M112.443796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amie SM, Bambara RA, Kim B. GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J Biol Chem. 2013;288:25001–25006. doi: 10.1074/jbc.C113.493619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen EC, Seamon KJ, Cravens SL, Stivers JT. GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc Natl Acad Sci USA. 2014;111:E1843–E1851. doi: 10.1073/pnas.1401706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji X, et al. Mechanism of allosteric activation of SAMHD1 by dGTP. Nat Struct Mol Biol. 2013;20:1304–1309. doi: 10.1038/nsmb.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji X, Tang C, Zhao Q, Wang W, Xiong Y. Structural basis of cellular dNTP regulation by SAMHD1. Proc Natl Acad Sci USA. 2014;111:E4305–E4314. doi: 10.1073/pnas.1412289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu C, et al. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat Commun. 2013;4:2722. doi: 10.1038/ncomms3722. [DOI] [PubMed] [Google Scholar]
- 21.Koharudin LM, et al. Structural basis of allosteric activation of sterile α motif and histidine-aspartate domain-containing protein 1 (SAMHD1) by nucleoside triphosphates. J Biol Chem. 2014;289:32617–32627. doi: 10.1074/jbc.M114.591958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu CF, et al. The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP. Acta Crystallogr D Biol Crystallogr. 2015;71:516–524. doi: 10.1107/S1399004714027527. [DOI] [PubMed] [Google Scholar]
- 23.Liliemark J. The clinical pharmacokinetics of cladribine. Clin Pharmacokinet. 1997;32:120–131. doi: 10.2165/00003088-199732020-00003. [DOI] [PubMed] [Google Scholar]
- 24.Balzarini J. Effect of antimetabolite drugs of nucleotide metabolism on the anti-human immunodeficiency virus activity of nucleoside reverse transcriptase inhibitors. Pharmacol Ther. 2000;87:175–187. doi: 10.1016/s0163-7258(00)00050-4. [DOI] [PubMed] [Google Scholar]
- 25.Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues: Mechanisms of drug resistance and reversal strategies. Leukemia. 2001;15:875–890. doi: 10.1038/sj.leu.2402114. [DOI] [PubMed] [Google Scholar]
- 26.Bonate PL, et al. Discovery and development of clofarabine: A nucleoside analogue for treating cancer. Nat Rev Drug Discov. 2006;5:855–863. doi: 10.1038/nrd2055. [DOI] [PubMed] [Google Scholar]
- 27.Ewald B, Sampath D, Plunkett W. Nucleoside analogs: Molecular mechanisms signaling cell death. Oncogene. 2008;27:6522–6537. doi: 10.1038/onc.2008.316. [DOI] [PubMed] [Google Scholar]
- 28.Jordheim LP, Durantel D, Zoulim F, Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat Rev Drug Discov. 2013;12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- 29.Fridle C, et al. Cladribine, cytarabine and idarubicin (CLA-Ida) salvage chemotherapy in relapsed acute myeloid leukemia (AML) Leuk Lymphoma. 2017;58:1068–1075. doi: 10.1080/10428194.2016.1235274. [DOI] [PubMed] [Google Scholar]
- 30.Tamamyan G, et al. Frontline treatment of acute myeloid leukemia in adults. Crit Rev Oncol Hematol. 2017;110:20–34. doi: 10.1016/j.critrevonc.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider C, et al. SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat Med. 2017;23:250–255. doi: 10.1038/nm.4255. [DOI] [PubMed] [Google Scholar]
- 32.Hollenbaugh JA, et al. Substrates and inhibitors of SAMHD1. PLoS One. 2017;12:e0169052. doi: 10.1371/journal.pone.0169052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miazzi C, et al. Allosteric regulation of the human and mouse deoxyribonucleotide triphosphohydrolase sterile α-motif/histidine-aspartate domain-containing protein 1 (SAMHD1) J Biol Chem. 2014;289:18339–18346. doi: 10.1074/jbc.M114.571091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold LH, et al. Phospho-dependent regulation of SAMHD1 oligomerisation couples catalysis and restriction. PLoS Pathog. 2015;11:e1005194. doi: 10.1371/journal.ppat.1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Bhattacharya A, Villacorta J, Diaz-Griffero F, Ivanov DN. Allosteric activation of SAMHD1 protein by deoxynucleotide triphosphate (dNTP)-dependent tetramerization requires dNTP concentrations that are similar to dNTP concentrations observed in cycling T cells. J Biol Chem. 2016;291:21407–21413. doi: 10.1074/jbc.C116.751446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amie SM, et al. Anti-HIV host factor SAMHD1 regulates viral sensitivity to nucleoside reverse transcriptase inhibitors via modulation of cellular deoxyribonucleoside triphosphate (dNTP) levels. J Biol Chem. 2013;288:20683–20691. doi: 10.1074/jbc.M113.472159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ordonez P, et al. SAMHD1 enhances nucleoside-analogue efficacy against HIV-1 in myeloid cells. Sci Rep. 2017;7:42824. doi: 10.1038/srep42824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitley R, Alford C, Hess F, Buchanan R. Vidarabine: A preliminary review of its pharmacological properties and therapeutic use. Drugs. 1980;20:267–282. doi: 10.2165/00003495-198020040-00002. [DOI] [PubMed] [Google Scholar]
- 39.Whitley RJ, et al. Pharmacology, tolerance, and antiviral activity of vidarabine monophosphate in humans. Antimicrob Agents Chemother. 1980;18:709–715. doi: 10.1128/aac.18.5.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold LH, Kunzelmann S, Webb MR, Taylor IA. A continuous enzyme-coupled assay for triphosphohydrolase activity of HIV-1 restriction factor SAMHD1. Antimicrob Agents Chemother. 2015;59:186–192. doi: 10.1128/AAC.03903-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radosevic N, Delmer A, Tang R, Marie JP, Ajchenbaum-Cymbalista F. Cell cycle regulatory protein expression in fresh acute myeloid leukemia cells and after drug exposure. Leukemia. 2001;15:559–566. doi: 10.1038/sj.leu.2402092. [DOI] [PubMed] [Google Scholar]
- 42.Bastin-Coyette L, et al. Nucleoside analogs induce proteasomal down-regulation of p21 in chronic lymphocytic leukemia cell lines. Biochem Pharmacol. 2011;81:586–593. doi: 10.1016/j.bcp.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 43.St Gelais C, et al. A cyclin-binding motif in human SAMHD1 is required for its HIV-1 restriction, dNTPase activity, tetramer formation, and efficient phosphorylation. J Virol. 2018;92:e01787-17. doi: 10.1128/JVI.01787-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas D, Herold N, Keppler OT, Geisslinger G, Ferreirós N. Quantitation of endogenous nucleoside triphosphates and nucleosides in human cells by liquid chromatography tandem mass spectrometry. Anal Bioanal Chem. 2015;407:3693–3704. doi: 10.1007/s00216-015-8588-3. [DOI] [PubMed] [Google Scholar]
- 45.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vagin AA, et al. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Seamon KJ, Stivers JT. A high-throughput enzyme-coupled assay for SAMHD1 dNTPase. J Biomol Screen. 2015;20:801–809. doi: 10.1177/1087057115575150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wittmann S, et al. Phosphorylation of murine SAMHD1 regulates its antiretroviral activity. Retrovirology. 2015;12:103. doi: 10.1186/s12977-015-0229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capes-Davis A, et al. Match criteria for human cell line authentication: Where do we draw the line? Int J Cancer. 2013;132:2510–2519. doi: 10.1002/ijc.27931. [DOI] [PubMed] [Google Scholar]
- 52.Michaelis M, et al. Identification of flubendazole as potential anti-neuroblastoma compound in a large cell line screen. Sci Rep. 2015;5:8202. doi: 10.1038/srep08202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gramberg T, Sunseri N, Landau NR. Evidence for an activation domain at the amino terminus of simian immunodeficiency virus Vpx. J Virol. 2010;84:1387–1396. doi: 10.1128/JVI.01437-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.