Significance
Gene delivery by virus-like particles holds enormous therapeutic potential to correct inherited genetic disorders and to prevent infectious disease. However, cells express antiviral factors that prevent virus infection and, consequently, limit the success of gene therapy. Here, we reveal the mechanism by which the drug rapamycin improves lentivirus-mediated gene delivery. Rapamycin treatment led to degradation of IFITM3, a broad and potent antiviral protein which inhibits virus entry into cells. IFITM3 is selectively cleared from endosomes, the sites where viral and cellular membranes fuse, and is sorted for disposal in lysosomes. While revealing an immunosuppressive function with clinical benefits, we caution that rapamycin use in humans may facilitate infection by pathogenic viruses like Influenza A virus.
Keywords: fusion, endosome, IFITM, interferon, virus
Abstract
Rapamycin and its derivatives are specific inhibitors of mammalian target of rapamycin (mTOR) kinase and, as a result, are well-established immunosuppressants and antitumorigenic agents. Additionally, this class of drug promotes gene delivery by facilitating lentiviral vector entry into cells, revealing its potential to improve gene therapy efforts. However, the precise mechanism was unknown. Here, we report that mTOR inhibitor treatment results in down-regulation of the IFN-induced transmembrane (IFITM) proteins. IFITM proteins, especially IFITM3, are potent inhibitors of virus–cell fusion and are broadly active against a range of pathogenic viruses. We found that the effect of rapamycin treatment on lentiviral transduction is diminished upon IFITM silencing or knockout in primary and transformed cells, and the extent of transduction enhancement depends on basal expression of IFITM proteins, with a major contribution from IFITM3. The effect of rapamycin treatment on IFITM3 manifests at the level of protein, but not mRNA, and is selective, as many other endosome-associated transmembrane proteins are unaffected. Rapamycin-mediated degradation of IFITM3 requires endosomal trafficking, ubiquitination, endosomal sorting complex required for transport (ESCRT) machinery, and lysosomal acidification. Since IFITM proteins exhibit broad antiviral activity, we show that mTOR inhibition also promotes infection by another IFITM-sensitive virus, Influenza A virus, but not infection by Sendai virus, which is IFITM-resistant. Our results identify the molecular basis by which mTOR inhibitors enhance virus entry into cells and reveal a previously unrecognized immunosuppressive feature of these clinically important drugs. In addition, this study uncovers a functional convergence between the mTOR pathway and IFITM proteins at endolysosomal membranes.
Endosomes are vesicular, membrane-bound organelles that direct the internalization and sorting of fluids, macromolecules, and plasma membrane proteins via the process of endocytosis (1). Invagination of the plasma membrane and carefully regulated fission events give rise to “early” endocytic vesicles that ferry cargo into a recycling circuit, in which contents are returned to the cell surface, or a pathway terminating in highly acidified lysosomes. Protein transport to lysosomes occurs following lysosome fusion with “late” endosomes or with specialized structures known as autophagosomes. In the former, late endosomes deliver enzymes needed for lysosome function, as well as endocytosed material destined for proteolytic degradation (2, 3). Fusion between late endosomes and lysosomes gives rise to hybrid organelles known as endolysosomes, a name which is also used to generally describe the late endosome/lysosome compartment since methods to distinguish them are inadequate. Late endosomes are also known as multivesicular bodies (MVBs) due to the presence of intraluminal vesicles (ILVs). The endosomal sorting complex required for transport (ESCRT) machinery is required for sorting ubiquitinated protein cargo into ILVs, which are subsequently degraded by lysosomal hydrolases as late endosomes/MVBs fuse with lysosomes (4–6).
Overall, intricate trafficking of membrane proteins regulates specific cellular activities, such as nutrient uptake and signal transduction, and collectively allows cells to rapidly respond to their surroundings. While providing a means for large molecules to access an otherwise impermeable cell, the endocytic network is also exploited as a portal of entry for pathogens. Many pathogenic enveloped viruses infecting humans, including Influenza A virus (IAV), Dengue virus, and Ebola virus, adhere to a life cycle that relies on endosomal movement and maturation. The progressive acidification that occurs during the transition from early endosomes to late endosomes triggers fusion between viral and endosomal membranes, allowing cytoplasmic access and the initiation of viral replication (7). Endosomes are thus situated at a crucial cellular threshold, and, as a result, they are equipped with elements of the cell-intrinsic antiviral immune response (8).
The IFN-induced transmembrane (IFITM) proteins IFITM2 and IFITM3 are antiviral factors that inhibit infection by a diverse assortment of viruses at the level of virus–cell fusion and which can be found in endolysosomal membranes (9, 10). The related IFITM1 is localized rather at the plasma membrane and exhibits differential antiviral specificity. However, most studies on IFITM protein function have been performed using ectopic expression assays, and less is known about the endogenous roles they play in different cell types. While IFITM1, IFITM2, and IFITM3 expression can be up-regulated by interferons and other cytokines, there are certain tissue types in which IFITM proteins are highly expressed under basal conditions (11). For example, IFITM members are readily detectable in skin cells, including dermal and foreskin fibroblasts (12), mucosal epithelium (13, 14), and in other epithelial cells like astrocytes (15) and HeLa cells (12, 16). Additionally, they are found in hematopoietic stem cells and their progenitors, including myeloid and lymphoid cell types such as macrophages, dendritic cells, and T cells, and expression levels can be affected by differentiation and activation status (17–24). In some cases, the functional utility of IFITM proteins may not be limited to antiviral protection since they are also believed to perform cellular functions beyond restriction of virus entry (25).
Much of what is understood about the antiviral activities of IFITM proteins is the result of retroviral pseudotyping experiments. Pseudotyping is the process by which retroviruses, such as HIV or murine leukemia virus, are outfitted with heterologous envelope glycoproteins such as IAV hemagglutinin (HA) and vesicular stomatitis virus (VSV) glycoprotein (VSV-G). Since viral glycoproteins bind to distinct cell surface receptors and perform the fusion reaction at distinct cellular membranes, pseudotyping allows for customized delivery of virus particles to select cell types and select intracellular compartments, such as endosomes. This practice has extended to the clinical setting where VSV-G is currently being used to guide the delivery of retrovirus into human cells for gene therapy efforts, due to the ability of VSV-G to bind the ubiquitous LDL receptor for cell attachment (26). IFITM2 and IFITM3 overexpression inhibits virus entry mediated by both IAV HA and VSV-G while the former is considerably more sensitive to the inhibition than the latter (27–30). Furthermore, ifitm3 KO mice exhibit enhanced morbidity and mortality when challenged with sublethal doses of IAV, and polymorphisms in human IFITM3 are associated with severe disease outcomes following IAV infection (31, 32). However, it was unclear whether IFITM proteins restrict VSV-G–mediated virus entry in situations of clinical consequence to humans.
The mammalian target of rapamycin (mTOR) is a serine/threonine kinase that functions as a central regulator of cell growth and metabolism. Deregulation of the mTOR pathway contributes to many human diseases, including cancer, obesity, diabetes, and age-related pathologies (33). As its name implies, the discovery and functional characterization of mTOR is fundamentally linked to a natural compound that inhibits it. Rapamycin binds to FK506-binding protein 12 (FKBP12), and the duo acts as an allosteric inhibitor of mTOR. As a component of two complexes known as mTORC1 and mTORC2, mTOR integrates signaling from nutrients, growth factors, energy, and oxygen to promote protein and lipid synthesis, cell proliferation and survival, and macroautophagy suppression (34). Rapamycin and its derivatives inhibit these anabolic cellular processes and have been evaluated and approved for medicinal use as anticancer agents and to treat graft-versus-host disease in transplantation patients. It is generally believed that mTORC1 is sensitive to rapamycin while mTORC2 is not, but the latter can also be inhibited when the drug is used for extended periods or at high concentrations (35, 36). The localization of mTORC1 and mTORC2 to endomembranes regulates their metabolic activities (37), but it was unknown whether mTOR-associated signaling influences other events in this subcellular compartment, such as virus entry.
It was recently described that rapamycin treatment can enhance gene delivery by lentiviral vectors in human and murine hematopoietic stem and progenitor cells (HSPCs) (38–40). This effect was observed using a replication-incompetent lentivirus (lentivector) pseudotyped with VSV-G, a currently favored tool for genome modification ex vivo due to its ability to maximize infectivity and broaden cellular tropism without compromising long-term engraftment (41–43). The initial report found that rapamycin promotes lentivector fusion with cellular membranes, but the detailed mechanism remained unknown. Nonetheless, the use of rapamycin or its derivatives is already being included in gene therapy protocols for the correction or prevention of multiple human diseases, including HIV/AIDS (44). This discovery revealed a previously uncharacterized connection between mTOR, endocytic trafficking, and virus–cell fusion.
In this article, we report that mTOR inhibitors, including rapamycin, down-regulate endogenous IFITM3 through a lysosomal degradation pathway in hematopoietic and nonhematopoietic cells. Furthermore, rapamycin enhances not only lentivector transduction but also IAV infection in primary and transformed cell lines in an IFITM3-dependent manner. Our study will improve gene modification efficiency and facilitate efforts to correct a variety of human diseases. Furthermore, novel strategies that directly target cellular IFITM proteins will yield gene therapies that maximize gene modification, cell engraftment, and cell survival while bypassing unintended consequences of mTOR inhibitor use. Furthermore, these results reveal that mTOR inhibitors relieve a broadly acting, intrinsic antiviral barrier to promote viral infection ex vivo and raise the possibility that their use in some clinical settings may undermine antiviral immunity in vivo. Thus, rapamycin, its derivatives, and other mTOR inhibitors may represent a double-edged sword in the arena of public health.
Results
Endogenous IFITM3 Is Down-Regulated by Rapamycin via a Lysosomal Degradation Pathway.
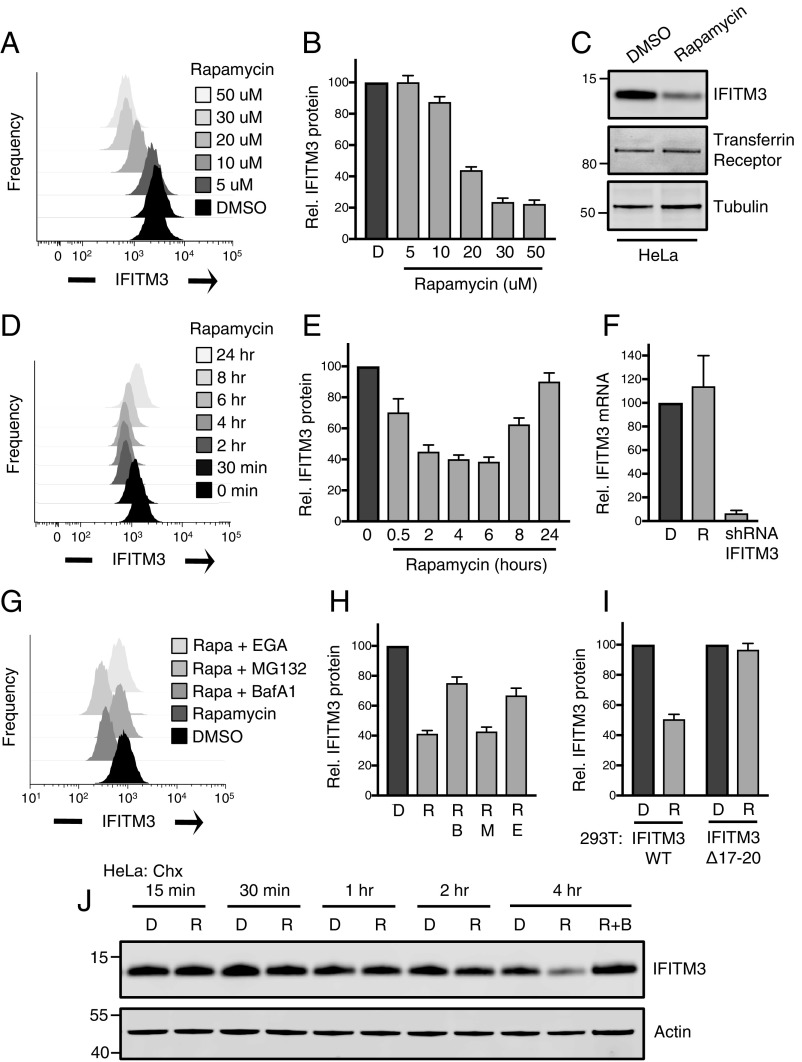
As inhibitors of virus–cell fusion in endosomes, we assessed the role played by IFITM proteins in the rapamycin-mediated enhancement of lentivector infection. We previously reported that HeLa cells and primary fibroblasts express high levels of IFITM3, which renders them refractory to infection by Zika virus (12). When HeLa were exposed to micromolar quantities of rapamycin for 4 h, we observed a dose-dependent decrease in IFITM3 protein as determined by flow cytometry and Western blot analysis (Fig. 1 A–C). The effect was selective because another transmembrane protein, transferrin receptor, was unaffected by rapamycin treatment (Fig. 1C). A time course experiment revealed that IFITM3 down-regulation was both progressive, with maximal decreases observed at 4 to 6 h posttreatment, and transient, as IFITM3 protein levels partially recovered over time (Fig. 1 D and E). We also examined how mTOR inhibition affects endogenous IFITM3 induced by type I IFN in 293T cells. Following 24 h of IFN stimulation, rapamycin addition led to decreased levels of induced IFITM3 protein in a dose-dependent manner. Here, elevated amounts of rapamycin resulted in a stable, nontransient loss of induced IFITM3 (SI Appendix, Fig. S1A).
Fig. 1.
Endogenous IFITM3 is down-regulated by rapamycin via a lysosomal degradation pathway. (A) HeLa cells were treated with indicated concentrations of DMSO or rapamycin for 4 h followed by fixation/permeabilization, immunostaining with anti-IFITM3, and analysis by flow cytometry. Univariate histograms of IFITM3 staining intensity were overlaid. (B) The mean fluorescence intensities (MFIs) from histograms in A were normalized as percentage relative to DMSO and averaged. (C) SDS/PAGE and Western blot analysis of whole cell lysates produced from HeLa treated with DMSO or 20 μM rapamycin followed by immunoblotting with anti-IFITM3. Tubulin and transferrin receptor were used as loading controls. (D) HeLa cells were treated with 20 μM rapamycin for durations indicated and immunostained with anti-IFITM3. (E) The MFIs from histograms in D were normalized and averaged. (F) HeLa cells were treated with rapamycin (20 μM) for 4 h, and total cDNA was synthesized from extracted RNA. Quantitative PCR was performed using IFITM3-specific primers. RNA from HeLa stably expressing IFITM3-specific shRNA was used as a negative control. (G) HeLa cells were treated with DMSO, rapamycin (20 μM) alone, or a combination of rapamycin and bafilomycin A1 (1 μM), MG132 (10 μM), or 4-bromobenzaldehyde N-(2,6-dimethylphenyl)semicarbazone (10 μM) for 4 h followed by immunostaining with anti-IFITM3. (H) The MFIs from histograms in G were normalized and averaged. (I) The 293T cells were transfected with pQCXIP-FLAG-IFITM3 or -IFITM3 Δ17-20, and stable expression was achieved following puromycin selection. Cells were treated with DMSO or rapamycin (20 μM) for 4 h followed by immunostaining with anti-IFITM3. MFIs were normalized and averaged. (J) SDS/PAGE and Western blot analysis of whole cell lysates produced from HeLa cells treated with 100 μg/mL cycloheximide and DMSO or rapamycin (20 μM) for the durations indicated; combined rapamycin and bafilomycin A1 (1 μM) were included for the 4-h time point. Immunoblotting was performed with anti-IFITM3. Actin was used as a loading control. Numbers and tick marks indicate size (kilodaltons) and position of protein standards in ladder. All error bars indicate SE from three to five experiments. B, bafilomycin A1; Chx, cycloheximide; D, DMSO; E, EGA; M, MG132; R, rapamycin.
We used quantitative RT-PCR to show that IFITM3 mRNA levels were unaffected by rapamycin (Fig. 1F), indicating that the down-regulation of IFITM3 manifests at the protein level. A second mTOR inhibitor, Torin 1, which was also shown to enhance lentivector transduction in CD34+ HSPCs (38), down-regulated endogenous IFITM3 in the micromolar range as well (SI Appendix, Fig. S1B). In agreement with a model of posttranslational regulation, we found that the vacuolar ATPase inhibitor bafilomycin A1 partially prevented rapamycin-mediated down-regulation (Fig. 1 G and H), indicating that endolysosomal acidification contributes to IFITM3 protein degradation during mTOR inhibition. Meanwhile, an endosomal trafficking inhibitor that functions independently of acidification also partially rescued IFITM3 protein levels while the proteasome inhibitor MG132 had no effect (Fig. 1 G and H). To explore the mechanism of IFITM3 down-regulation in molecular detail, we studied the fate of exogenous IFITM3 stably transfected into 293T cells. While rapamycin treatment resulted in decreases to WT IFITM3 protein, a mutant variant in which the proline-based PPxY site recognized by NEDD4 E3 ubiquitin ligase is disrupted (Δ17-18) (45) was partially resistant to down-regulation (SI Appendix, Fig. S1C). Similarly, a previously described ubiquitin-deficient variant of IFITM3 in which all lysines were changed to arginines (ΔLys) (46, 47) displayed a loss in sensitivity to rapamycin-mediated degradation. Furthermore, a mutant lacking both the NEDD4 recognition site and a critical tyrosine needed for AP2-mediated endocytosis (Δ17-20) (47–49) was completely resistant to the effects of rapamycin (Fig. 1I and SI Appendix, Fig. S1C). In the same experimental system, exogenous IFITM2 protein was as sensitive to rapamycin as IFITM3 while IFITM1 was resistant (SI Appendix, Fig. S1C). The differential effect of rapamycin on closely related IFITM proteins can be explained by the fact that the NEDD4 recognition and endocytosis motifs, present in the amino termini of IFITM3 and IFITM2, are absent in IFITM1. Together, these data suggest that mTOR inhibitors redirect IFITM3 protein toward a proteolytic pathway that requires endocytosis, ubiquitination, and endolysosomal acidification. To confirm that mTOR inhibitor treatment decreases IFITM3 protein half-life, we treated HeLa cells with rapamycin in the presence of cycloheximide (Chx) for up to 4 h. Rapamycin accelerated IFITM3 protein turnover under these conditions while bafilomycin A1 inhibited the loss (Fig. 1J).
IFITM3 Degradation Following Rapamycin Requires ESCRT-Dependent Endocytic Trafficking.
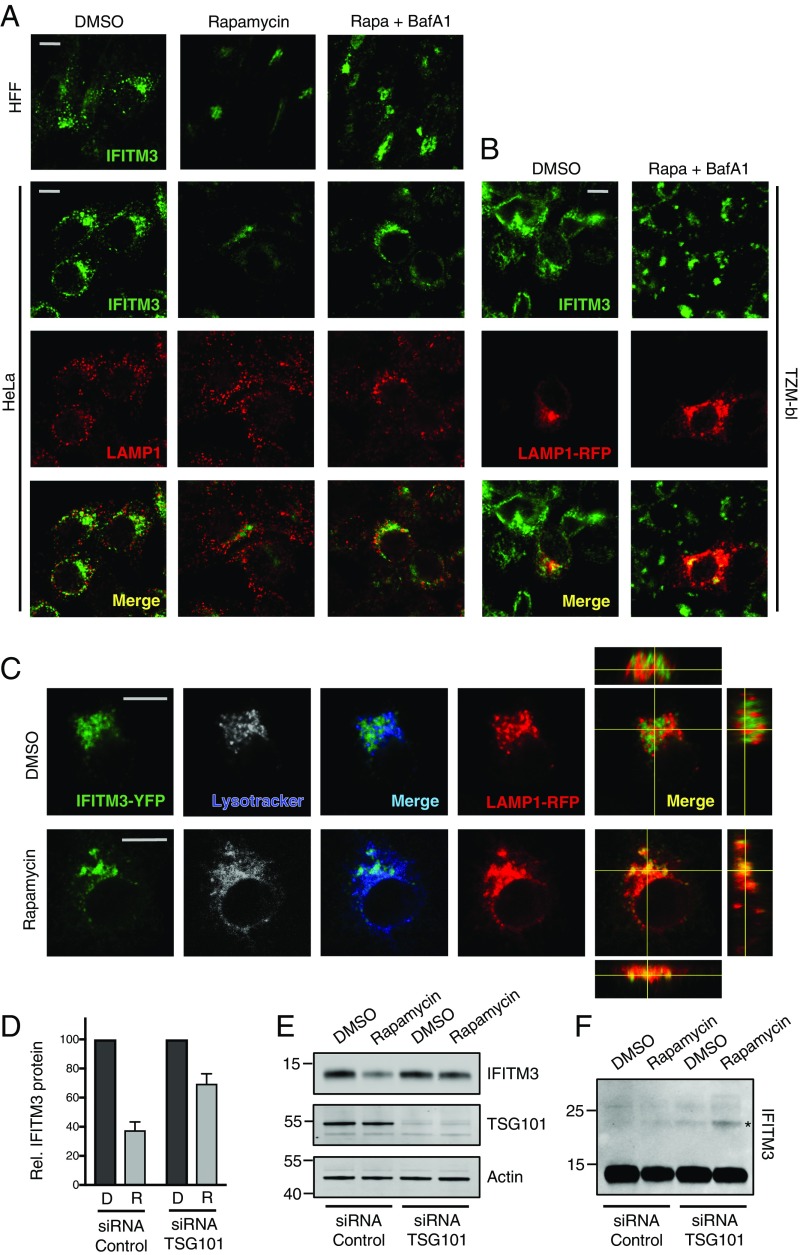
To elucidate the mechanism by which IFITM3 protein is targeted to endolysosomes for degradation, we studied the effects of rapamycin treatment in fixed cells using confocal immunofluorescence microscopy. In primary human foreskin fibroblasts (HFFs) and HeLa cells, rapamycin treatment reduced IFITM3 levels in a bafilomycin A1-sensitive manner (Fig. 2A). In mock-treated cells, endogenous IFITM3 localized to early endosomes and, to a lesser extent, endolysosomes, as determined by costaining with EEA1 and LAMP1 (SI Appendix, Fig. S2A). However, rapamycin treatment resulted in an abrupt clearance of IFITM3 from these sites without affecting levels of EEA1 or LAMP1 themselves (Fig. 2A and SI Appendix, Fig. S2 B and C). Addition of both rapamycin and bafilomycin A1 led to retention of IFITM3 in reticulated, sac-like structures surrounding the nucleus (Fig. 2A). Endolysosomal enrichment of IFITM3 in this condition was confirmed using RFP-tagged LAMP1 in the TZM-bl cell line, which also expresses high levels of endogenous IFITM3 (Fig. 2B and SI Appendix, Fig. S2D). To enable the tracking of IFITM3 in living cells, we included YFP-tagged IFITM3 in the analysis. Rapamycin treatment led to a redistribution of IFITM3-YFP to compartments positive for both LAMP1-RFP and Lysotracker reagent, further suggesting that the posttranslational degradation program terminates in acidic endolysosomes (Fig. 2C).
Fig. 2.
IFITM3 degradation following rapamycin treatment requires ESCRT-dependent trafficking through the endocytic pathway. (A) HFFs and HeLa cells were treated with DMSO or rapamycin (Rapa) (20 μM) or rapamycin plus bafilomycin A1 (BafA1) (1 μM) for 4 h followed by fixation/permeabilization, immunostaining with anti-IFITM3 and anti-LAMP1, and analysis by immunofluorescence confocal microscopy. (B) TZM-bl cells were transfected with LAMP1-RFP for 24 h and treated with DMSO or rapamycin (20 μM) or rapamycin plus bafilomycin A1 (1 μM) for 4 h followed by fixation/permeabilization, immunostaining with anti-IFITM3, and analysis by immunofluorescence confocal microscopy. (C) HeLa cells stably expressing IFITM3-YFP were transfected with LAMP1-RFP for 24 h. Cells were stained with Lysotracker Deep Red (50 nM) for 15 min, and living cells were imaged immediately by immunofluorescence confocal microscopy. Image analysis was performed using ImageJ (Fiji). Merged images are provided for IFITM3-YFP/Lysotracker (green/blue) and for IFITM3-YFP/LAMP1-RFP (green/red), and orthogonal XZ and YZ views are provided for the latter (C). All images are average Z-stacks from three to four consecutive medial sections. (D) HeLa cells were transfected with indicated siRNA for 72 h, treated with DMSO or rapamycin (20 μM) for 4 h, and then fixed/permeabilized, immunostained with anti-IFITM3, and analyzed by flow cytometry. MFIs from histograms were normalized and averaged. Error bars indicate SE from four experiments. (E) SDS/PAGE and Western blot analysis of whole cell lysates from HeLa transfected with indicated siRNA for 72 h and treated with DMSO or rapamycin (20 μM). Immunoblotting was performed with anti-IFITM3 and anti-TSG101. Actin was used as a loading control. The image provided is representative of two experiments. (F) Immunoblot with anti-IFITM3 in E was enhanced to reveal bands of higher molecular mass (marked with asterisk). (Scale bars: 10 μm.)
To resolve the mechanism by which IFITM3 is delivered to the proteolytic lumen of endolysosomes during mTOR inhibition, we considered the involvement of two cellular pathways: macroautophagy and MVB formation. Since rapamycin is commonly used to induce macroautophagy in cultured cells, we tested whether rapamycin-mediated IFITM3 degradation requires autophagosome formation. While concomitant treatment of cells with rapamycin and bafilomycin A1 resulted in IFITM3 accumulation in LAMP1-containing endolysosomes, these structures were generally not decorated with the autophagosomal marker LC3 (SI Appendix, Fig. S2D). To more directly assess the contribution of autophagy, we evaluated the effects of serum and amino acid deprivation. In TZM-bl and HFF cells, rapamycin treatment resulted in conversion of LC3-I to phosphatidylethanolamine-conjugated LC3 (LC3-II), which is recruited to autophagosomal membranes (50) (SI Appendix, Fig. S3 A and B). The conversion to and subsequent loss of LC3-II is reflective of autophagic flux, and degradation can be inhibited by bafilomycin A1. While rapamycin promotes LC3 turnover, starvation of cells for the same duration as rapamycin treatment led to a greater loss of LC3 while affecting levels of IFITM3 to a lesser extent (SI Appendix, Fig. S3D), suggesting that the two modes of mTOR inhibition differ in regard to the extent and rapidity by which IFITM3 is degraded. Furthermore, they indicate that degradation of LC3 and IFITM3 is uncoupled during mTOR inhibition, ruling out the possibility that they are both regulated by autophagosomes. To confirm the independence of IFITM3 degradation and autophagosomal processes, we tested whether rapamycin-mediated degradation of IFITM3 could occur in autophagy-deficient cells (51). In ATG9a KO cells, in which the accumulation of LC3-II and its turnover are prevented, IFITM3 degradation was apparent (SI Appendix, Fig. S3B). Together, these results suggest that mTOR inhibitors result in delivery of IFITM3 to endolysosomes in an autophagosome-independent manner. In contrast, knockdown of ESCRT-I component TSG101 partially prevented IFITM3 degradation by rapamycin, as determined by flow cytometry (Fig. 2D) and Western blot (Fig. 2E). Enhancement of the IFITM3 immunoblot revealed a covalent modification of approximately nine kilodaltons in rapamycin-treated, TSG101-depleted cells, which may represent a single ubiquitin moiety (Fig. 2F). The functional requirement for ESCRT machinery in the rapamycin-induced degradation of IFITM3 suggests that mTOR inhibition promotes the ubiquitination and sorting of IFITM3 into MVBs before disposal in lysosomes. This was supported by a strong association between IFITM3 and MVB marker CD63 in cells treated with rapamycin and bafilomycin A1 (SI Appendix, Fig. S3C).
To probe the direct involvement of mTOR and its associated complexes in events leading to IFITM3 degradation, we performed RNA interference to silence mTORC1 component Raptor and mTORC2 component Rictor. Relative to treatment with a nontargeting control siRNA, thorough silencing of Raptor had no effect on IFITM3 protein levels. In contrast, a slight reduction of Rictor resulted in decreases in IFITM3 (SI Appendix, Fig. S3 D and E). The specific, albeit modest, effect of Rictor knockdown suggests that mTORC2 inhibition may be the means by which rapamycin leads to IFITM3 degradation. A role for mTORC2 is consistent with previous observations indicating that rapamycin inhibits mTORC2-dependent activities, such as the phosphorylation of Akt and the suppression of cell proliferation, when used at micromolar concentrations (36).
Rapamycin Treatment of CD34+ HSPC Results in IFITM2/3 Down-Regulation.
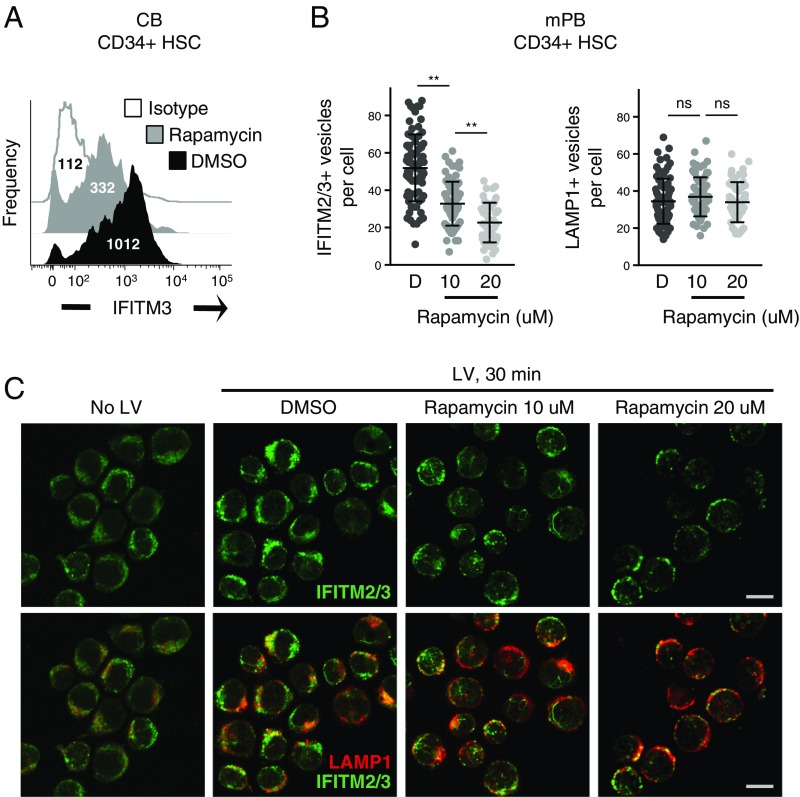
Given that rapamycin was initially described as an enhancer of lentiviral transduction in CD34+ HSPCs, we explored the effects of mTOR inhibition on endogenous IFITM2 and IFITM3 in this primary cell type. IFITM3 was readily detected in CD34+ cells derived from pooled cord blood from nine donors, and rapamycin treatment led to a marked decrease in IFITM3 levels as determined by flow cytometry (Fig. 3A). We also assessed the consequence of drug treatment on CD34+ cells from adult peripheral blood using confocal microscopy analysis (Fig. 3B). Intriguingly, both IFITM2/3 and LAMP1 signals were elevated following addition of lentivector, suggesting that virus addition mobilizes endosomes in this primary cell type. As a result, assessment of endosomal proteins necessitated the addition of lentivector. As shown in cord blood CD34+ cells, rapamycin treatment of adult CD34+ cells led to a dose-dependent down-regulation of IFITM2/3 and redistribution to perinuclear sites (Fig. 3C).
Fig. 3.
Rapamycin treatment of CD34+ HSPCs results in IFITM2/3 down-regulation. (A) CD34+ HSPCs derived from cord blood from nine donors were pooled, fixed/permeabilized, immunostained with anti-IFITM3, and analyzed by flow cytometry. Univariate histograms of IFITM3 expression are shown, with the median fluorescence intensity indicated. (B) CD34+ HSPCs derived from G-CSF–mobilized peripheral blood from adults were thawed, prestimulated in stem cell growth medium for 48 h, treated for 4 h with the indicated doses of rapamycin, and exposed to lentivector for 30 min. Actual concentrations of rapamycin used were 9.8 μM and 18.1 μM. Cells were then fixed/permeabilized, stained with anti-IFITM2/3 and anti-LAMP1, and analyzed by immunofluorescence confocal microscopy. The number of IFITM2/3+ and LAMP1+ vesicles was determined using Imaris software. At least 50 cells were imaged for each condition. (C) Representative images from each treatment condition in B are shown. (Scale bars: 10 μm.) Statistical analysis of vesicle number was conducted with the Kruskal–Wallis test using Dunn’s multiple comparison correction. ns, not significant, P > 0.05; **P < 0.0001. D, DMSO; HSC, hematopoietic stem cells; LV, lentivector.
IFITM Silencing Abrogates the Rapamycin-Mediated Enhancement of Lentivector Transduction.
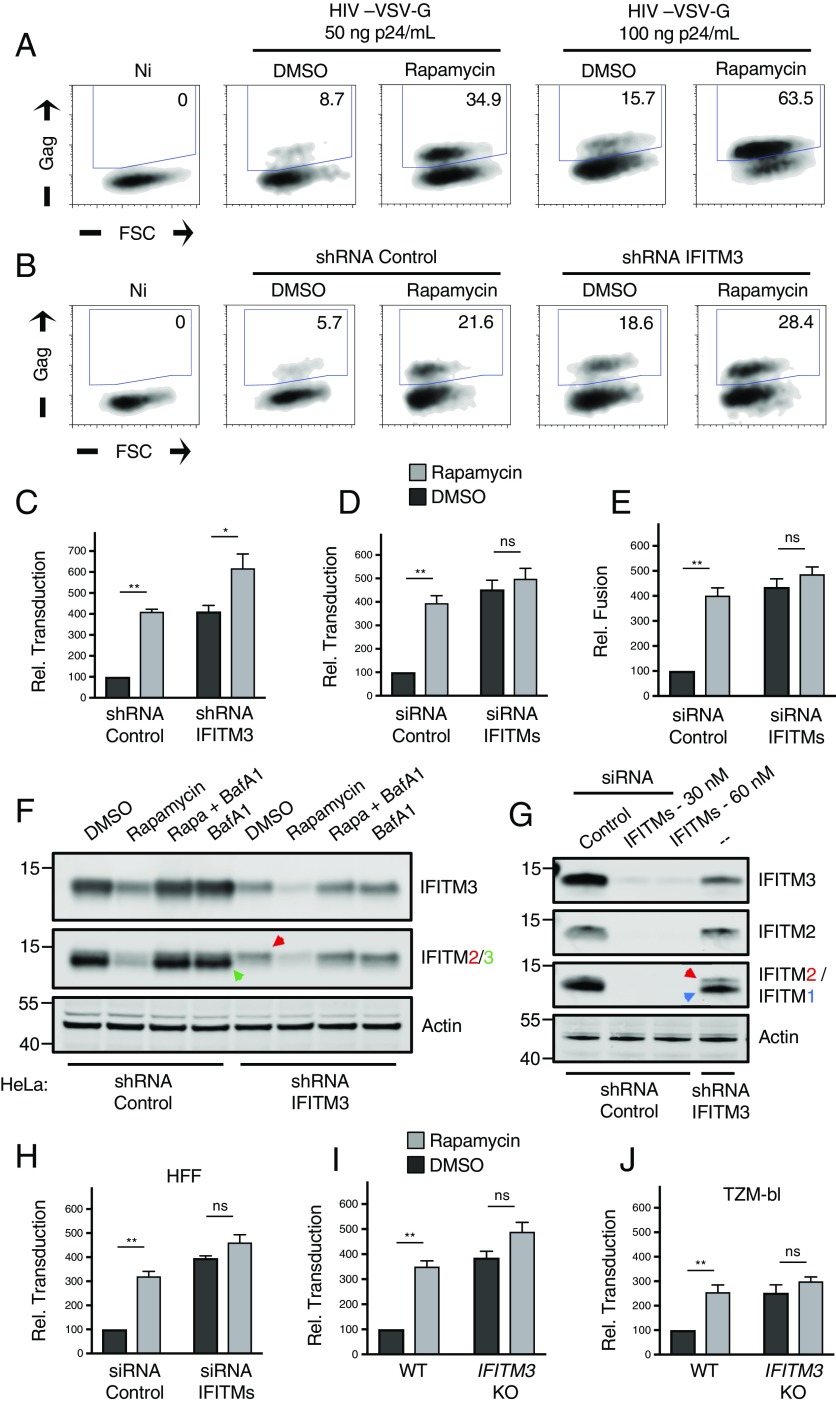
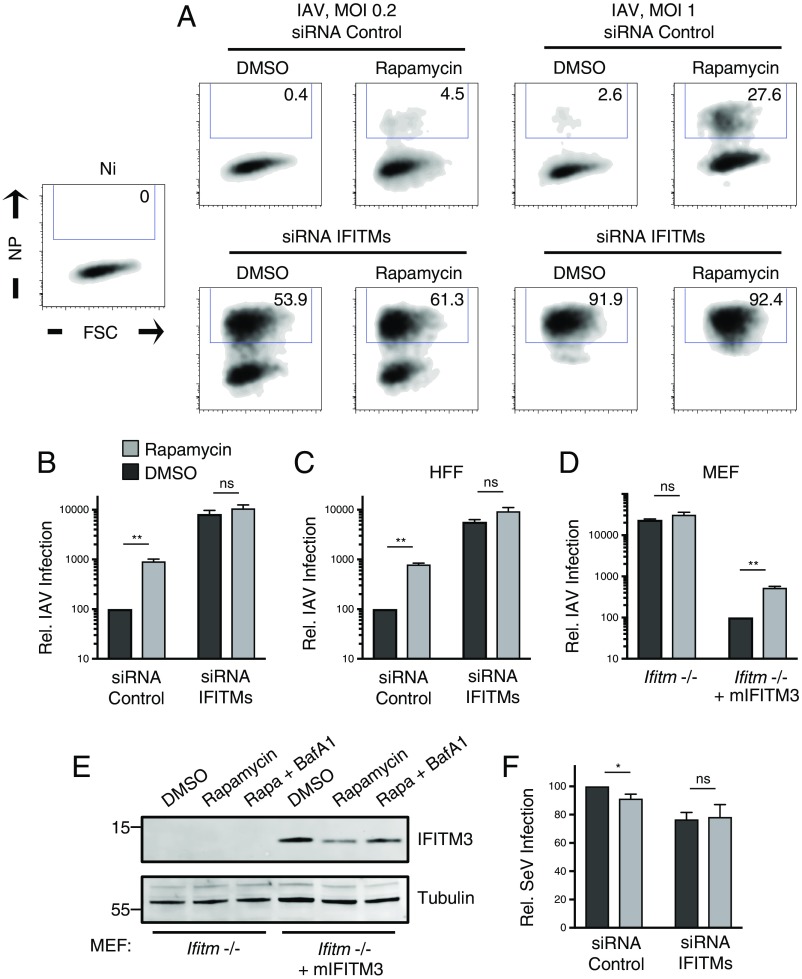
While the preceding experiments collectively demonstrate that mTOR inhibition negatively regulates IFITM2 and IFITM3 in various cell types, it remained to be tested whether this phenomenon accounts for the rapamycin-mediated enhancement of lentiviral transduction reported previously in CD34+ HSPCs (52). To reproduce the conditions used in CD34+ HSPCs, HeLa cells were incubated with 20 micromolar rapamycin 4 h before and during lentivector exposure, and productive transduction was scored by de novo HIV-1 Gag expression. We found that rapamycin treatment of HeLa cells increased permissiveness to an HIV-1–based lentivector bearing VSV-G by three- to fourfold (Fig. 4A), and partial knockdown of IFITM3 using shRNA resulted in transduction rates similar to those following drug treatment (Fig. 4 B and C). Importantly, rapamycin treatment in IFITM3 knockdown cells led to an even higher transduction rate, but the enhancing effect of the drug was relatively diminished (less than twofold) (Fig. 4C). This result suggested that the function of rapamycin as a transduction enhancer depends on levels of endogenous IFITM3. To fully suppress protein production of IFITM3, as well as IFITM2 and IFITM1, we targeted the three gene products with a mixture of siRNA as previously described (12, 18). In HeLa cells in which IFITM1-3 are silenced, the influence of rapamycin on transduction was absent (Fig. 4D). Since IFITM proteins are known to inhibit infection during virus–cell fusion, we used a FRET-based assay to measure early steps of virus entry into cells (53). The results indicated that rapamycin enhances lentivector transduction in HeLa by promoting virus–cell fusion, consistent with findings in CD34+ HSPCs (38) (Fig. 4E). Moreover, while depletion of IFITM1-3 led to increases in virus–cell fusion, no further enhancement was observed with rapamycin (Fig. 4E). Therefore, rapamycin enhances virus–cell fusion and does so, to a large extent, by down-regulating IFITM protein expression. Western blot analysis was used to establish the efficiency and specificity by which RNAi reagents down-regulated IFITM proteins in these experiments (Fig. 4 F and G and SI Appendix, Fig. S4A) and also provided evidence that endogenous IFITM2, in addition to IFITM3, is negatively regulated by rapamycin (Fig. 4F). We again detected the likely ubiquitinated form of IFITM3 in cells treated with both rapamycin and bafilomycin A1, and this form was absent in IFITM3-silenced cells (SI Appendix, Fig. S4B). Combining rapamycin with RNAi in primary HFFs confirmed that the rapamycin-mediated lentiviral transduction enhancement requires IFITM proteins (Fig. 4H). Furthermore, by achieving a robust and specific gene knockout of IFITM3 using CRISPR-Cas9, we found that IFITM3 is the major determinant of rapamycin sensitivity in HeLa (Fig. 4I) and TZM-bl cells (Fig. 4J and SI Appendix, Fig. S4 C–E).
Fig. 4.
IFITM silencing abrogates the rapamycin-mediated enhancement of lentivector transduction. (A) HeLa cells were seeded in 24-well plates (50,000 per well) overnight. Cells were treated with DMSO or rapamycin (20 μM) for 4 h and then with fresh media containing DMSO or rapamycin (20 μM) and the indicated quantity of lentivector (HIV-VSV-G). Approximately 24 h later, cells were fixed/permeabilized, stained with anti-Gag, and analyzed by flow cytometry. Numbers inside gates indicate percent of Gag+ cells. (B) HeLa cells stably expressing shRNA (scrambled control or IFITM3) were treated with DMSO or rapamycin (20 μM) and exposed to HIV-VSV-G (50 ng p24/mL). Transduction was scored 24 h later. (C) Relative transduction scores from B were normalized to DMSO-treated shRNA control cells and averaged. (D) HeLa cells were transfected with siRNA (nontargeting control or a mixture targeting IFITM1, IFITM2, and IFITM3) for 48 h, treated with DMSO or rapamycin (20 μM) for 4 h, and exposed to HIV-VSV-G (50 ng p24/mL). (E) HeLa cells were transfected with siRNA as in D, treated with DMSO or rapamycin (20 μM) for 4 h, and exposed to HIV-VSV-G for 2 h. Cells were resuspended in media containing CCF2-AM for 1 h, washed, and fixed/permeabilized for flow cytometry. (F) Western blot analysis of whole cell lysates derived from HeLa cells stably expressing shRNA treated with DMSO, rapamycin (20 μM) alone, rapamycin and bafilomycin A1 (1 μM), or bafilomycin A1 alone. Immunoblotting was performed with anti-IFITM3 and anti-IFITM2/3 antibodies (IFITM2 and IFITM3 are indicated by red and green arrows, respectively). (G) Western blot analysis of whole cell lysates derived from HeLa cells transfected with siRNA for 48 h. Lysates from HeLa IFITM3 shRNA are provided for comparison. Immunoblotting was performed with anti-IFITM3, anti-IFITM2, and anti-IFITM1 (IFITM2 and IFITM1 are indicated by red and blue arrows, respectively). Actin was used as loading control. (H) Primary HFFs were seeded in 24-well plates (50,000 per well) overnight and treated and infected as in D. (I) HeLa (WT or IFITM3 KO) were seeded in 24-well plates (50,000 per well) and treated and infected as in C. Relative transduction scores were normalized to DMSO-treated WT cells and averaged. (J) TZM-bl (WT or IFITM3 KO) were seeded in 24-well plates (50,000 per well) overnight and treated and infected as in C. Relative transduction scores were normalized and averaged. All error bars indicate SE from three experiments. Statistical analysis was conducted with the Student’s t test. ns, not significant, P > 0.05; *P < 0.05; **P < 0.005. FSC, forward scatter; Ni, noninfected.
Rapamycin Promotes IAV Infection in an IFITM3-Dependent Manner.
While IFITM3 functions broadly to inhibit many pathogenic RNA viruses in cell culture experiments, the physiological impact of IFITM3 is best characterized during IAV infection in mice and humans (32, 54). Consistent with its ability to coordinate the selective clearance of IFITM3 and IFITM2 from cells, we found that rapamycin also promotes infection by IAV in HeLa and HFFs in an IFITM-dependent manner (Fig. 5 A–C). Relative to VSV-G–mediated virus entry, IAV infection is more sensitive to inhibition by IFITM3, and rapamycin treatment led to a greater degree of infection enhancement (9- to 10-fold). Our silencing and knockout experiments indicated that IFITM3 expression is required for the observed enhancement of viral infections by rapamycin. To establish whether it is sufficient to confer rapamycin sensitivity to cells, we performed experiments in mouse embryonic fibroblasts (MEFs) from ifitm-deficient mice (55, 56). IAV infection in cells lacking murine IFITM proteins was not significantly boosted by rapamycin treatment (Fig. 5D). In contrast, complementation of cells with murine IFITM3 resulted in a more than 100-fold inhibition of infection, and this restriction was partially relieved following rapamycin treatment (Fig. 5D). Analysis of protein levels in complemented cells revealed that murine IFITM3 is partially down-regulated by rapamycin and that levels are partially restored by the presence of bafilomycin A1 (Fig. 5E). These results suggest that IFITM3 expression alone confers sensitivity to the rapamycin-mediated infection enhancement effect. Further solidifying the functional relationship between endogenous IFITM proteins and the effects of mTOR inhibition on virus infections is the finding that Sendai virus, a paramyxovirus, is resistant to the effects of both rapamycin treatment and IFITM silencing (Fig. 5F). That is, neither rapamycin treatment nor IFITM knockdown led to an enhancement of Sendai virus infection. Therefore, rapamycin enhances infection by IFITM-sensitive viruses, but not IFITM-resistant viruses, demonstrating that the primary mechanism by which mTOR inhibition promotes infection is down-modulation of IFITM3 protein.
Fig. 5.
Rapamycin promotes IAV infection in an IFITM3-dependent manner. (A) HeLa were seeded in 24-well plates (50,000 per well) overnight and transfected with siRNA (nontargeting control or a mixture targeting IFITM1, IFITM2, and IFITM3) for 48 h. Cells were treated with DMSO or rapamycin (20 μM) for 4 h and then with fresh media containing DMSO or rapamycin (20 μM) and the indicated quantity of IAV PR8. Approximately 18 h after virus exposure, cells were fixed/permeabilized, stained with anti-NP, and analyzed by flow cytometry. Numbers inside gates indicate percent of NP+ cells. (B) Relative infection scores from A using a multiplicity of infection (MOI) of 0.2 were normalized to DMSO-treated siRNA control cells and averaged. (C) Primary HFFs were seeded in 24-well plates overnight and subsequently transfected, treated, and infected as in A with an MOI of 0.2. (D) MEFs derived from ifitm−/− mice and the same cells transduced with murine IFITM3 (mIFITM3) were seeded in 24-well plates (50,000 per well) overnight. Cells were treated with DMSO or rapamycin (20 μM) for 4 h and then with fresh media containing DMSO or rapamycin (20 μM) and IAV PR8 at an MOI of 0.2. Relative infection scores were normalized and averaged, with infection of DMSO-treated ifitm−/− plus mIFITM3 cells set to 100. (E) Western blot analysis of whole cell lysates derived from ifitm−/− and ifitm−/− plus mIFITM3 MEF treated with DMSO, rapamycin (20 μM), or rapamycin and bafilomycin A1 (1 μM) for 4 h. Immunoblotting was performed with anti-IFITM3. Tubulin served as loading control. (F) HeLa were seeded and transfected as in A and exposed to Sendai virus (Cantell) at an MOI of 0.1. Approximately 18 h after virus exposure, cells were fixed/permeabilized, stained with polyclonal anti-SeV, and analyzed by flow cytometry. Relative infection scores were normalized to DMSO-treated siRNA control and averaged. All error bars indicate SE from three experiments. Statistical analysis was carried out with a Student’s t test. ns, not significant, P > 0.05; *P < 0.05; **P < 0.005. NP, nucleoprotein.
Discussion
The results outlined here describe the causes and consequences of a selective protein degradation program following mTOR inhibition. Within a manner of minutes, rapamycin treatment results in the negative regulation of IFITM3 (and IFITM2) at the posttranslational level in diverse cell types. This finding is reminiscent of a recent report describing that mTOR inhibitors activate extensive degradation of cellular proteins via both proteasomal and autophagy pathways (57). While it is possible that IFITM3 belongs to a suite of proteins subject to regulation by mTOR inhibition, we show that certain other endosomal proteins (transferrin receptor, LAMP1, and EEA1) are spared during this process. Most importantly, our experiments demonstrate that IFITM3 protein down-regulation accounts for the majority of the infection enhancement effect.
Our observations indicate that IFITM2 and IFITM3 are degraded following rapamycin treatment in cells under basal conditions or following ectopic or induced expression while IFITM1 is unaffected. IFITM3 was the dominant source of restriction in cells assessed here, but the relative abundance of IFITM proteins may vary by tissue and by the microenvironmental cues present therein. In addition to enhancing infections mediated by VSV-G and IAV HA, it is likely that mTOR inhibition promotes infection by a number of other viruses pathogenic to humans. Of particular interest is the impact on HIV-1, which is sensitive to IFITM-mediated restriction at early stages (entry) and late stages (virion infectivity) of its life cycle in a strain-dependent manner (17, 18, 20, 58, 59). Furthermore, drugs and conditions that lead to mTOR inhibition are expected to impact other cellular processes in which IFITM2 and IFITM3 are involved, such as cell–cell adhesion and cytokine regulation (25). Indeed, further studies into these effects may reveal the physiological basis for the interconnectedness of mTOR and IFITM proteins.
The localization of IFITM proteins to endomembranes has led others to conclude that they feature in processes of macroautophagy. IFITM3 overexpression can result in elevation of the autophagosomal marker LC3-II (60), and, more recently, IFITM3 has been shown to shuttle the cellular proteins IRF3 and Ambra1 to autophagosomes for degradation (61, 62). These latter findings reveal how IFITM3 inhibits IFN signaling and cell migration, respectively, and provide examples of how IFITM proteins influence immunity in many ways. Nonetheless, our results suggest that IFITM3 is not a substrate regulated by macroautophagy itself, at least not in the contexts explored in this study. Instead, we show that the negative regulation of IFITM3 by rapamycin involves endosomal trafficking events likely guided by ubiquitination and ESCRT-mediated sorting into MVBs.
Earlier reports discovered that IFITM3 abundance is fine-tuned by the E3 ubiquitin ligase NEDD4. By recognizing a tyrosine-based motif (PPxY) in the amino terminus, NEDD4 was shown to ubiquitinate and accelerate the turnover of IFITM3 in human and murine cells (45). NEDD4 is known to ubiquitinate cargo proteins sorted by ESCRT (63–65). We did not explicitly rule in a role for NEDD4 in the rapamycin-mediated degradation of IFITM3, but partial escape from the effects of mTOR inhibition were displayed by mutant IFITM3 lacking the PPxY motif or lacking lysine residues previously shown to be targeted for ubiquitination. However, since NEDD4-like ubiquitin ligases including ITCH also recognize the PPxY motif in the protein substrates they regulate (66, 67), IFITM3 turnover may be controlled by multiple E3 ligases. Our results using TSG101 depletion link IFITM proteins with the ESCRT machinery. It remains to be seen whether ESCRT governs the relative amounts and localization of IFITM3 during basal homeostasis or during cellular stresses other than mTOR inhibition.
Irrespective of the E3 ligase responsible, the redirection of IFITM3 to acidic endolysosomes and the functional requirement for ESCRT imply that the MVB represents an intermediate through which IFITM3 passes before lysosomal delivery and degradation. Consistent with a role for MVBs, when degradation is interrupted with bafilomycin A1, IFITM3 accumulates in multivesicular sac-like structures. To arrive at this transient compartment, ESCRT may coordinate the sorting of IFITM3 from late endosomal and/or lysosomal membranes into membrane invaginations known as ILVs (5, 68). Fusion between MVB and terminal lysosomes at perinuclear sites, which is bafilomycin A1-sensitive (69), may facilitate the transfer of IFITM3-containing ILVs into the lysosomal lumen for degradation by hydrolases. Of note, ILVs are also released from cells when MVBs fuse with the plasma membrane in a process known as exocytosis (4, 70). Since IFITM3 protein is regularly detected in virus particles and extracellular vesicles when expressed at high levels in cells (17, 71), it is possible that mTOR inhibition augments the pool of IFITM3 ejected into the extracellular space.
In addition to the lysosomal lumen or cell exterior, cargo within ILV can also gain access to the cytoplasm by undergoing “back-fusion” with the limiting membrane of the MVB (7). In fact, while initially believed to undergo virus–cell fusion in early endosomes, evidence exists that VSV-G uses a back-fusion step in the MVB to complete cytoplasmic entry (72). Thus, together with the well-characterized low pH trigger required for IAV HA-mediated fusion, both viral glycoproteins utilized in this study display a dependence on late endosomal/MVB membranes. Most previous studies employing overexpression of IFITM3 have documented an endolysosomal expression pattern in cells, suggesting that subcellular localization underlies its ability to restrict viruses entering at these sites (73, 74). However, we observed that endogenous IFITM3 localizes mainly to early endosomes in HeLa cells. It is possible that virus restriction occurs as IFITM3 and internalized virus particles overlap in trafficking endosomes maturing toward a late phenotype. Furthermore, virus infection driven by VSV-G is less sensitive to restriction by IFITM3 relative to IAV HA (27, 75), which may reflect differential dependence on late endosomal/MVB membranes for entry or qualitatively different interactions with said membranes. The insensitivity of Sendai virus to both IFITM proteins and rapamycin treatment is likely attributed to its ability to fuse at the plasma membrane (63). It will be worthwhile to assess how virus infection or IFN signaling impacts IFITM3 abundance in different endosomal compartments.
The data presented herein provide a mechanistic understanding of how rapamycin facilitates lentiviral transduction in cells ex vivo, but they also raise important questions regarding their uses in vivo. Pharmacologic mTOR inhibition is currently being used to inhibit metabolic disorders, neurodegeneration, aging, graft rejection, and tumorigenesis, and certain applications have already received Food and Drug Administration (FDA) approval (76). However, by limiting IFITM2 and IFITM3 levels in endosomes, exposure to rapamycin may undermine an important component of cell-intrinsic antiviral immunity. Indeed, humans immunocompromised by mTOR inhibitors during cancer treatment or following organ transplant exhibit increased incidence of respiratory tract infections, including those caused by viruses such as IAV (77–80). Furthermore, administration of rapamycin in vivo shortly after infection with IAV led to exacerbated disease and mortality in mice (81). In humans, a naturally occurring single nucleotide polymorphism resulting in decreased IFITM3 mRNA expression is associated with severe outcomes following IAV infection (54). That reduced IFITM3 expression can have physiological impact highlights the need for further investigation into the molecular players that link IFITM3 to the mTOR pathway. Our work suggests that the complex responsible for regulating IFITM3 is mTORC2 since the silencing of Rictor alone led to detectable decreases in IFITM3 protein. However, the transient nature of IFITM3 down-regulation following mTOR inhibition and the existence of feedback loops imply that genetic approaches to identify the genes responsible will be challenging. It would be beneficial to determine whether and how caloric restriction, like exposure to rapamycin, influences the expression of IFITM3 and modulates vulnerability to virus infection.
Enhanced gene transduction rates in CD34+ HSPCs are highly valued in the field of gene therapy as even incremental improvements raise the probability of achieving therapeutic benefit in patients (52). However, the use of mTOR inhibitors carries the risk that other mTOR-dependent processes may be interrupted, such as cell proliferation and survival. Methods that enable specific targeting of IFITM3 abundance or function ex vivo are likely to provide a greater degree of transduction enhancement without impairing reconstitution potential in vivo.
Materials and Methods
Cells from Human Donors.
Cord blood (CB) was generously donated from the Cleveland Cord Blood Center, Cleveland, for isolation of CD34+ cells and adult mobilized peripheral blood CD34+ cells were obtained from Fred Hutchinson Cancer Research Center Processing and Procurement Core under the approved institutional protocol of the Scripps Research Institute (no. IRB-13-6173).
Viruses and Infections.
Replication-incompetent lentivector (HIV-VSV-G) was produced by transfecting 293T with 15 μg of pNL4-3ΔEnv (82) (a gift from O. Schwartz, Institut Pasteur, Paris) and 5 μg of pMD2.G (VSV-G, 12259; Addgene) using the calcium phosphate method. Briefly, six million 293T cells were seeded in a T75 flask. Plasmid DNA was mixed with sterile H2O, CaCl2, and Tris-EDTA (TE) buffer, and the totality was combined with Hepes-buffered saline (HBS). The transfection volume was added dropwise, and cells were incubated at 37 °C for 48 h. Supernatants were clarified by centrifugation, passed through a 0.45-μm filter, and stored at −80 °C. Lentivector titers were measured using an HIV-1 p24 ELISA kit (XpressBio). To produce lentivector for FRET-based virus–cell fusion measurements, 293Ts were transfected with 15 μg of pNL4-3ΔEnv, 5 μg of pMD2.G (VSV-G), and 5 μg of pCMV4-BlaM-Vpr (21950; Addgene). HeLa cells were seeded in 24-well plates (50,000 per well) overnight and overlaid with 50 ng of p24 equivalent of BlaM-Vpr–containing HIV-VSV-G for 2 h. Cells were washed and labeled with the CCF2-AM β-lactamase Loading Kit (Invitrogen) for an additional 2 h and analyzed for virus–cell fusion as described (18, 83). Influenza A virus [A/PR/8/34 (PR8), H1N1] and Sendai virus (Cantell strain) supplied as clarified allantoic fluid were purchased from Charles River Laboratories. Infectious virus titers were calculated using a flow cytometry-based method in HeLa cells (84), and infections were performed as follows: Cells were seeded in 24-well plates (50,000 per well) overnight and overlaid with indicated amounts of virus in a final volume of 225 μL for ∼18 h. Cells were washed with 1× PBS, detached with Trypsin-EDTA, fixed/permeabilized with Cytofix/Cytoperm, immunostained with antibodies to IAV NP or SeV proteins, respectively, and analyzed by flow cytometry.
Flow Cytometry.
Cells were fixed/permeabilized with Cytofix/CytoPerm reagent (BD) for 20 min and washed in Perm/Wash buffer (BD). Cells were pelleted and resuspended in primary antibodies diluted in Perm/Wash buffer, incubated at room temperature for 30 min, and washed in Perm/Wash buffer. Cells were pelleted and resuspended in Alexa Fluor-conjugated secondary antibodies diluted in Perm/Wash buffer, incubated at room temperature for 30 min, and washed in Perm/Wash buffer, and cells were acquired and analyzed on a LSRFortessa (BD). The following primary antibodies were used to score virus infections: anti-HIV Gag KC57-FITC (BD), anti-p24 Gag (3537; NIH AIDS Reagent Resource), anti-IAV-NP (AA5H; Abcam), and anti-SeV (PD029; MBL). Endogenous IFITM3 was measured by anti-IFITM3 (EPR5242; Abcam) while exogenous IFITM3 WT and mutants, IFITM2, and IFITM1 expressed from pQCXIP were measured by anti-FLAG M2 (Sigma). Endogenous LAMP1 and EEA1 were measured by anti-LAMP1 (H5G11, sc-18821; Santa Cruz Biotechnology) and anti-EEA1 (clone 14, 610456; BD). The following reagent was obtained through BEI Resources, NIAID, NIH: Human Recombinant IFN Beta, rHuIFN-βser17 (NR-3085).
Plasmid Transfection, RNA Interference, and CRISPR-Cas9–Generated Knockouts.
LAMP1-RFP (carboxyl-terminal tag, a gift from V. Pathak, National Cancer Institute, Frederick, MD) was transfected into TZM-bl and HeLa cells using Mirus TransIT-LT1. IFITM3-YFP (amino-terminal tag; Genecopoeia) was transfected into HeLa cells using Mirus TransIT-LT1. Stably expressing cells were created following selection with Geneticin/G418 for 3 wk. pQCXIP-IFITM1, -IFITM2, -IFITM3 (a gift from C. Liang, McGill University, Montreal), and -IFITM3 mutants containing amino-terminal FLAG (previously described) (47, 83) were transfected into 293T cells with Mirus TransIT-LT1. Stably expressing cells were created following selection with puromycin for more than 2 wk. HeLa cells stably expressing shRNA were created by transduction with a pGIPZ-GFP–based lentivector expressing shRNA (scrambled control and IFITM3) (V3LHS_325106; Thermo Fisher). HeLa and TZM-bl IFITM3 KO cells were created by transfection with a set of plasmids encoding Cas9 and three IFITM3-specific guide RNAs (sc-403281; Santa Cruz Biotechnology) and a set of three plasmids providing templates for homology-directed repair (sc-403281-HDR; Santa Cruz Biotechnology). A population of modified cells was selected following puromycin treatment for 3 wk. HeLa ATG9a KO were a gift from J. Bonifacino, National Institute of Child Health and Human Development, Bethesda, MD. Transient siRNA transfection of cells was performed with Lipofectamine RNAiMAX. Knockdown efficiency was assessed by flow cytometry or Western blot analysis at times indicated posttransfection. The following siRNAs were Silencer Select (Ambion): IFITM1 (s16192), IFITM2 (s20771), IFITM3 (s195035), and negative control no. 1 (4390844). Silencing of human IFITM1, IFITM2, and IFITM3 was performed by transfecting a mixture of siRNA (20 nM each), compared with negative control siRNA no. 1 (60 nM). The following siRNAs were ON-TARGETplus SMARTpool (GE Dharmacon): TSG101 (7251), Raptor (57521), Rictor (253260), and nontargeting siRNA pool (D-001810-10-05). Silencing of human TSG101, Raptor, and Rictor (20 nM) was compared with nontargeting siRNA pool (20 nM). Transfection reagents and plasmid DNA, shRNA, and siRNA were diluted in Opti-MEM for delivery.
SDS/PAGE and Western Blot Analysis.
Whole cell lysis was performed with radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher) supplemented with Halt Protease Inhibitor mixture EDTA-free (Thermo Fisher). Lysates were clarified by centrifugation, and supernatants were collected and stored at −80 °C. Protein concentration was determined with the Protein Assay Kit II (Bio-Rad), and 5 to 15 μg of protein were loaded into 12% acrylamide Criterion XT Bis-Tris Precast Gels (Bio-Rad). Electrophoresis was run with NuPage Mes SDS Running Buffer (Invitrogen), and proteins were transferred to Amersham Protrain Premium Nitrocellulose Membrane, pore size 0.20 μm (GE Healthcare). Membranes were blocked with Odyssey blocking buffer in PBS (Li-Cor) and incubated with dilutions of the following primary antibodies: anti-IFITM1 (60074-1-Ig; Proteintech), anti-IFITM2 (66137-1-Ig; Proteintech), anti-IFITM3 (ab109429; Abcam), anti-IFITM2/3 (66081-1-Ig; Proteintech), anti-transferrin receptor (ab1086; Abcam), anti-LC3 (PM036; MBL), anti-TSG101 (14497-1-AP; Proteintech), anti-ATG9a (ab108338; Abcam), anti-Raptor (42-4000; Thermo Fisher), anti-Rictor (D16H9, 9486; Cell Signaling Technology), anti-actin (C4, sc-47778; Santa Cruz Biotechnology), and anti-tubulin (B-7, sc-5286; Santa Cruz Biotechnology). Secondary antibodies conjugated to DyLight 800 or 680 (Li-Cor) and the Li-Cor Odyssey imaging system were used to reveal specific protein detection. Images were analyzed and assembled using ImageStudioLite (Li-Cor).
Supplementary Material
Acknowledgments
We thank Eric O. Freed for advice and Kip Hermann for critical reading of the manuscript. We thank the following entities and individuals for reagents: NIH AIDS Reagent Resource, Eric O. Freed, Vinay Pathak, Chen Liang, Olivier Schwartz, Vineet KewalRamani, Michael S. Diamond, Juan Bonifacino, and Jing Pu. We thank the following individuals for assistance with flow cytometry and confocal microscopy: Mac Trubey, Stephen Lockett, and David Scheiblin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811892115/-/DCSupplemental.
References
- 1.Alberts B, et al. Molecular Biology of the Cell. 6th Ed Garland Science; New York: 2014. [Google Scholar]
- 2.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luzio JP, Pryor PR, Bright NA. Lysosomes: Fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 4.McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu L, Jorgensen JR, Li M, Chuang Y-S, Emr SD. ESCRTs function directly on the lysosome membrane to downregulate ubiquitinated lysosomal membrane proteins. eLife. 2017;6:3365. doi: 10.7554/eLife.26403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Le Blanc I, et al. Endosome-to-cytosol transport of viral nucleocapsids. Nat Cell Biol. 2005;7:653–664. doi: 10.1038/ncb1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. 2013;425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith S, Weston S, Kellam P, Marsh M. IFITM proteins-cellular inhibitors of viral entry. Curr Opin Virol. 2014;4:71–77. doi: 10.1016/j.coviro.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegrist F, Ebeling M, Certa U. The small interferon-induced transmembrane genes and proteins. J Interferon Cytokine Res. 2011;31:183–197. doi: 10.1089/jir.2010.0112. [DOI] [PubMed] [Google Scholar]
- 12.Monel B, et al. Zika virus induces massive cytoplasmic vacuolization and paraptosis-like death in infected cells. EMBO J. 2017;36:1653–1668. doi: 10.15252/embj.201695597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibi D, et al. Astroglial IFITM3 mediates neuronal impairments following neonatal immune challenge in mice. Glia. 2013;61:679–693. doi: 10.1002/glia.22461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alteber Z, et al. The anti-inflammatory IFITM genes ameliorate colitis and partially protect from tumorigenesis by changing immunity and microbiota. Immunol Cell Biol. 2018;96:284–297. doi: 10.1111/imcb.12000. [DOI] [PubMed] [Google Scholar]
- 15.Nakajima A, et al. Induction of interferon-induced transmembrane protein 3 gene expression by lipopolysaccharide in astrocytes. Eur J Pharmacol. 2014;745:166–175. doi: 10.1016/j.ejphar.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 16.Handfield C, Kwock J, MacLeod AS. Innate antiviral immunity in the skin. Trends Immunol. 2018;39:328–340. doi: 10.1016/j.it.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tartour K, et al. IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology. 2014;11:103. doi: 10.1186/s12977-014-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compton AA, et al. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe. 2014;16:736–747. doi: 10.1016/j.chom.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, et al. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172:423–438.e25. doi: 10.1016/j.cell.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, et al. The IFITM proteins inhibit HIV-1 infection. J Virol. 2011;85:2126–2137, and erratum (2011) 85:4043. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjbar S, Haridas V, Jasenosky LD, Falvo JV, Goldfeld AE. A role for IFITM proteins in restriction of Mycobacterium tuberculosis infection. Cell Rep. 2015;13:874–883. doi: 10.1016/j.celrep.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakim LM, Gupta N, Mintern JD, Villadangos JA. Enhanced survival of lung tissue-resident memory CD8+ T cells during infection with influenza virus due to selective expression of IFITM3. Nat Immunol. 2013;14:238–245. doi: 10.1038/ni.2525. [DOI] [PubMed] [Google Scholar]
- 24.Wu W-L, et al. Δ20 IFITM2 differentially restricts X4 and R5 HIV-1. Proc Natl Acad Sci USA. 2017;114:7112–7117. doi: 10.1073/pnas.1619640114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi G, Schwartz O, Compton AA. More than meets the I: The diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology. 2017;14:53. doi: 10.1186/s12977-017-0377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finkelshtein D, Werman A, Novick D, Barak S, Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci USA. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feeley EM, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alber D, Staeheli P. Partial inhibition of vesicular stomatitis virus by the interferon-induced human 9-27 protein. J Interferon Cytokine Res. 1996;16:375–380. doi: 10.1089/jir.1996.16.375. [DOI] [PubMed] [Google Scholar]
- 29.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weidner JM, et al. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol. 2010;84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everitt AR, et al. GenISIS Investigators; MOSAIC Investigators IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey CC, Huang IC, Kam C, Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8:e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blenis J. TOR, the gateway to cellular metabolism, cell growth, and disease. Cell. 2017;171:10–13. doi: 10.1016/j.cell.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 35.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 36.Foster DA, Toschi A. Targeting mTOR with rapamycin: One dose does not fit all. Cell Cycle. 2009;8:1026–1029. doi: 10.4161/cc.8.7.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pu J, Guardia CM, Keren-Kaplan T, Bonifacino JS. Mechanisms and functions of lysosome positioning. J Cell Sci. 2016;129:4329–4339. doi: 10.1242/jcs.196287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang CX, et al. Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. 2014;124:913–923. doi: 10.1182/blood-2013-12-546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrillo C, et al. Cyclosporin a and rapamycin relieve distinct lentiviral restriction blocks in hematopoietic stem and progenitor cells. Mol Ther. 2015;23:352–362. doi: 10.1038/mt.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohrabaugh SL, Campbell TB, Hangoc G, Broxmeyer HE. Ex vivo rapamycin treatment of human cord blood CD34+ cells enhances their engraftment of NSG mice. Blood Cells Mol Dis. 2011;46:318–320. doi: 10.1016/j.bcmd.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandez M, Porosnicu M, Markovic D, Barber GN. Genetically engineered vesicular stomatitis virus in gene therapy: Application for treatment of malignant disease. J Virol. 2002;76:895–904. doi: 10.1128/JVI.76.2.895-904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akkina RK, et al. High-efficiency gene transfer into CD34+ cells with a human immunodeficiency virus type 1-based retroviral vector pseudotyped with vesicular stomatitis virus envelope glycoprotein G. J Virol. 1996;70:2581–2585. doi: 10.1128/jvi.70.4.2581-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi JG, et al. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 2016;23:627–633. doi: 10.1038/gt.2016.27. [DOI] [PubMed] [Google Scholar]
- 44.Li M-J, et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 45.Chesarino NM, McMichael TM, Yount JS. E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 2015;11:e1005095. doi: 10.1371/journal.ppat.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yount JS, Karssemeijer RA, Hang HC. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J Biol Chem. 2012;287:19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chesarino NM, McMichael TM, Hach JC, Yount JS. Phosphorylation of the antiviral protein interferon-inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J Biol Chem. 2014;289:11986–11992. doi: 10.1074/jbc.M114.557694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia R, et al. Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell Microbiol. 2014;16:1080–1093. doi: 10.1111/cmi.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia R, et al. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol. 2012;86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galluzzi L, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hurley JH, Young LN. Mechanisms of autophagy initiation. Annu Rev Biochem. 2017;86:225–244. doi: 10.1146/annurev-biochem-061516-044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang CX, Torbett BE. Role of the mammalian target of rapamycin pathway in lentiviral vector transduction of hematopoietic stem cells. Curr Opin Hematol. 2015;22:302–308. doi: 10.1097/MOH.0000000000000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones DM, Padilla-Parra S. The β-lactamase assay: Harnessing a FRET biosensor to analyse viral fusion mechanisms. Sensors (Basel) 2016;16:950–914. doi: 10.3390/s16070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allen EK, et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat Med. 2017;23:975–983. doi: 10.1038/nm.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lange UC, et al. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol Cell Biol. 2008;28:4688–4696. doi: 10.1128/MCB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gorman MJ, Poddar S, Farzan M, Diamond MS. The interferon-stimulated gene Ifitm3 restricts West Nile virus infection and pathogenesis. J Virol. 2016;90:8212–8225. doi: 10.1128/JVI.00581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci USA. 2015;112:15790–15797. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu J, et al. IFITM proteins restrict HIV-1 infection by antagonizing the envelope glycoprotein. Cell Rep. 2015;13:145–156. doi: 10.1016/j.celrep.2015.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foster TL, et al. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe. 2016;20:429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McMichael TM, Chemudupati M, Yount JS. A balancing act between IFITM3 and IRF3. Cell Mol Immunol. 2017;14:1–2. doi: 10.1038/cmi.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang L-Q, et al. IFITM3 inhibits virus-triggered induction of type I interferon by mediating autophagosome-dependent degradation of IRF3. Cell Mol Immunol. 2017;14:1–10. doi: 10.1038/cmi.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoenherr C, et al. Ambra1 spatially regulates Src activity and Src/FAK-mediated cancer cell invasion via trafficking networks. eLife. 2017;6:452. doi: 10.7554/eLife.23172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mudhakir D, Harashima H. Learning from the viral journey: How to enter cells and how to overcome intracellular barriers to reach the nucleus. AAPS J. 2009;11:65–77. doi: 10.1208/s12248-009-9080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol. 2010;84:8181–8192. doi: 10.1128/JVI.00634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blot V, et al. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J Cell Sci. 2004;117:2357–2367. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- 66.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 67.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li M, Rong Y, Chuang Y-S, Peng D, Emr SD. Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol Cell. 2015;57:467–478. doi: 10.1016/j.molcel.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 69.Mauvezin C, Nagy P, Juhász G, Neufeld TP. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. 2015;6:7007. doi: 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Théry C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu X, et al. IFITM3-containing exosome as a novel mediator for anti-viral response in dengue virus infection. Cell Microbiol. 2015;17:105–118. doi: 10.1111/cmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uchil P, Mothes W. Viral entry: A detour through multivesicular bodies. Nat Cell Biol. 2005;7:641–642. doi: 10.1038/ncb0705-641. [DOI] [PubMed] [Google Scholar]
- 73.Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: The cell’s first line of antiviral defense. Annu Rev Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang IC, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Desai TM, et al. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014;10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Kim SG, Blenis J. Rapamycin: One drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaymakcalan MD, et al. Risk of infections in renal cell carcinoma (RCC) and non-RCC patients treated with mammalian target of rapamycin inhibitors. Br J Cancer. 2013;108:2478–2484. doi: 10.1038/bjc.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Englund J, Feuchtinger T, Ljungman P. Viral infections in immunocompromised patients. Biol Blood Marrow Transplant. 2011;17(Suppl 1):S2–S5. doi: 10.1016/j.bbmt.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choueiri TK, et al. Incidence and risk of treatment-related mortality in cancer patients treated with the mammalian target of rapamycin inhibitors. Ann Oncol. 2013;24:2092–2097. doi: 10.1093/annonc/mdt155. [DOI] [PubMed] [Google Scholar]
- 80.Kunisaki KM, Janoff EN. Influenza in immunosuppressed populations: A review of infection frequency, morbidity, mortality, and vaccine responses. Lancet Infect Dis. 2009;9:493–504. doi: 10.1016/S1473-3099(09)70175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang C-T, et al. Rapamycin adjuvant and exacerbation of severe influenza in an experimental mouse model. Sci Rep. 2017;7:4136. doi: 10.1038/s41598-017-04365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Borman AM, Quillent C, Charneau P, Dauguet C, Clavel F. Human immunodeficiency virus type 1 Vif- mutant particles from restrictive cells: Role of Vif in correct particle assembly and infectivity. J Virol. 1995;69:2058–2067. doi: 10.1128/jvi.69.4.2058-2067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Compton AA, et al. Natural mutations in IFITM3 modulate post-translational regulation and toggle antiviral specificity. EMBO Rep. 2016;17:1657–1671. doi: 10.15252/embr.201642771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grigorov B, Rabilloud J, Lawrence P, Gerlier D. Rapid titration of measles and other viruses: Optimization with determination of replication cycle length. PLoS One. 2011;6:e24135. doi: 10.1371/journal.pone.0024135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.