Significance
The 20 proteinogenic amino acids have physicochemical properties that allow peptides and proteins to fold and bind. However, there are numerous unnatural, nonproteinogenic amino acids that may be equally good, or even better, at folding and binding. Exploration of these alternative peptide building blocks has been limited by slow, one-at-a-time synthesis and testing. We describe how, in a single experiment, multiple nonproteinogenic amino acids can be trialed at all positions in a peptide sequence, with thousands of modifications tested in parallel. This permits detailed analysis of how chemical structure relates to function and allows for systematic comparisons of proteinogenic and nonproteinogenic chemistry. Such analysis can guide the improvement of drug-candidate peptides, including the therapeutically promising class of cyclic peptides.
Keywords: BH3 domains, noncanonical amino acids, structure–activity relationships, macrocyclic peptides, intrinsically disordered proteins
Abstract
High-resolution structure–activity analysis of polypeptides requires amino acid structures that are not present in the universal genetic code. Examination of peptide and protein interactions with this resolution has been limited by the need to individually synthesize and test peptides containing nonproteinogenic amino acids. We describe a method to scan entire peptide sequences with multiple nonproteinogenic amino acids and, in parallel, determine the thermodynamics of binding to a partner protein. By coupling genetic code reprogramming to deep mutational scanning, any number of amino acids can be exhaustively substituted into peptides, and single experiments can return all free energy changes of binding. We validate this approach by scanning two model protein-binding peptides with 21 diverse nonproteinogenic amino acids. Dense structure–activity maps were produced at the resolution of single aliphatic atom insertions and deletions. This permits rapid interrogation of interaction interfaces, as well as optimization of affinity, fine-tuning of physical properties, and systematic assessment of nonproteinogenic amino acids in binding and folding.
The chemical structure of a polypeptide determines its activity—including any folding or binding. Specific changes to chemical structure (mutants) can be analyzed in large numbers (1). Deep mutational scanning methods, in particular, allow for the analysis of many thousands of mutants (2): Mutant proteins or peptides are each coupled to their encoding DNA, libraries of pooled mutants are sorted for activity, mutants are counted via deep sequencing, and each mutant is scored. The throughput of these experiments is sufficient for exhaustive saturation mutagenesis; that is, testing all proteinogenic amino acids at all positions in a sequence and returning all effects on folding (3–5), binding (2, 5–8), or function (9). Extensive structure–activity maps are produced, but these methods are currently limited to the chemistry accessible within the universal genetic code—the 20 proteinogenic amino acids.
However, extending mutagenesis to include nonproteinogenic amino acids offers many advantages. The larger range of chemical structures allows for a finer dissection, or optimization, of molecular interactions, down to single aliphatic carbon insertions and deletions or functional group substitutions (10, 11). Certain nonproteinogenic amino acids can also improve the otherwise poor in vivo stability of short peptides and/or reduce the excessive polarity that prevents peptides crossing cell membranes (12, 13). Indeed, nonproteinogenic amino acids are abundant in peptide natural products (14) and peptides modified for in vivo use (10, 11). Nonproteinogenic amino acid mutagenesis can also address longstanding questions about the fitness of the universal genetic code and its collection of amino acids relative to plausible prebiotic alternatives (15), as well as guide development of synthetic polymers with the ability to fold (i.e., foldamers) (16, 17).
Exploration of nonproteinogenic amino acid mutants has been previously limited by the need to chemically synthesize and analyze peptides individually (10, 11). Recent advances in peptide synthesis have extended the numbers of nonproteinogenic mutants that can be constructed (18). Moreover, it is possible to combine parallel peptide synthesis with measures of function (19). However, these approaches cannot construct peptide libraries with the sequence length and numbers that deep mutational scanning can, which, at its core, uses high-fidelity nucleic acid-directed synthesis of polypeptides by the ribosome.
Ribosomal synthesis (i.e., translation) can be manipulated to include nonproteinogenic amino acids (20). In vitro genetic code reprogramming is particularly versatile, allowing for the incorporation of amino acids with diverse chemical structures (21). Flexizymes, flexible tRNA-acylation ribozymes, can load almost any (ester-activated) amino acid onto any tRNA, and these loaded tRNAs can be added to reconstituted in vitro translation systems to replace proteinogenic amino acids in the genetic code (Fig. 1A and SI Appendix, Fig. S1). These “reprogrammed” genetic codes allow for the one-pot synthesis of trillions of unique, nonproteinogenic amino acid-containing peptides derived from a pool of mRNA sequences. Members of these peptide libraries can be coupled to their encoding mRNA/cDNA, allowing for the isolation of functional, highly nonproteinogenic peptides; most notably, de novo macrocyclic peptides (22) from the random nonstandard peptide integrated discovery (RaPID) system (23–26). Here, we use flexizyme-based genetic code reprogramming to extend the reach of deep mutational scanning to examine any number of nonproteinogenic amino acids in peptide binding and folding.
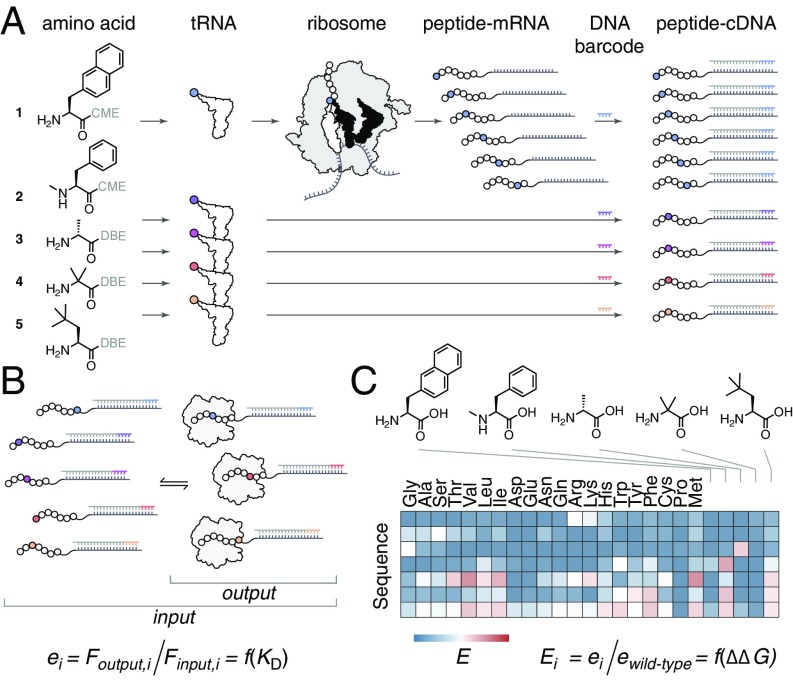
Fig. 1.
Nonproteinogenic deep mutational scanning. (A) Essentially any nonproteinogenic amino acid [activated by cyanomethyl ester (CME) or dinitrobenzyl ester (DBE)] can be loaded onto tRNA by flexizymes and delivered to the ribosome for use during in vitro translation. Translation of a site-saturation mutagenesis mRNA library using genetic code reprogramming allows for a nonproteinogenic amino acid to be incorporated at all positions in a given peptide sequence, and mRNA display can link each mutant peptide to its encoding mRNA. This can be repeated for n nonproteinogenic amino acids (n = 5 shown). Initiating reverse transcription with a barcoded DNA primer allows for the resulting peptide–cDNA products to be pooled and nonproteinogenic mutants deconvoluted after DNA sequencing. (B) The large diverse library from A, containing proteinogenic and nonproteinogenic mutants, can be sorted for binding to a partner protein. Next-generation sequencing of cDNA before (input) and after (output) binding is then performed. For each mutant, i, the fraction of DNA reads, F, can be used to calculate an enrichment score, e. (C) Enrichment scores relative to wild-type, E, provide a map of beneficial and deleterious changes to the peptide chemical structure.
Results
Selection of Nonproteinogenic Amino Acids for Mutagenesis.
Hundreds of nonproteinogenic amino acids are accepted by flexizymes and the ribosome (23); a sample set was chosen for nonproteinogenic mutagenesis, focusing on simple, largely nonpolar structures absent from the universal genetic code. This set of 21 amino acids included both alternative side chains and backbone modifications such as N-methyl substitution, disubstitution, and d- stereochemistry (Fig. 2A). We chose initially to test these in a natural protein–protein interaction that has already been studied using traditional mutagenesis: the interaction between the apoptosis regulatory BH3 domain of PUMA and the folded protein MCL1 (27, 28). BH3 domains are relatively short and unstructured in isolation but will fold to a single α-helix upon binding (Fig. 2B). This simple structure has made PUMA and its homologs model systems for protein folding upon binding (29, 30) and for exploring the potential of nonproteinogenic amino acids in druglike peptides (16, 31).
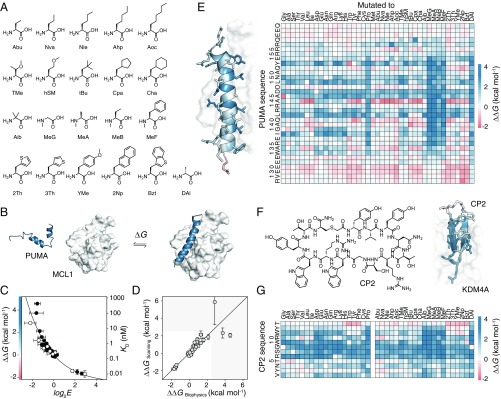
Fig. 2.
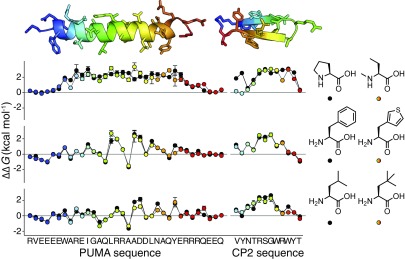
Nonproteinogenic deep mutational scanning of linear PUMA and cyclic CP2. (A) Test set of 21 nonproteinogenic amino acids used in this study. (B) PUMA (blue), an intrinsically disordered protein, folds to an α-helix upon binding with MCL1 (white) [Protein Data Bank (PDB) ID code 2ROC]. (C) Deep mutational scanning scores (log2E) report on free energy changes of binding upon mutation, ∆∆G. Proteinogenic (black) and nonproteinogenic mutants (white) overlay. Errors in log2E represent SDs of repeats of library binding and DNA recovery. (D) Leave-one-out cross-validation of empirical fit of ∆∆G and log2E showing agreement between calculated ∆∆G of the left-out mutant (∆∆GScanning) and expected experimental value (∆∆GBiophysics). Greater than 90% of PUMA mutant ∆∆G values fall within the white region. (E) ∆∆G for all mutants of PUMA; stabilizing (red) and destabilizing (blue). (Left) Average ∆∆G projected onto PUMA/MCL1 structure. (F) De novo macrocyclic peptide CP2 (Left). Average ∆∆G projected onto CP2/KDM4A structure (Right) (PDB ID code 5LY1). (G) ∆∆G for all mutants of CP2 binding to KDM4A.
Nonproteinogenic Deep Mutational Scanning.
The flexizyme protocol allows for facile reprogramming of genetic codes (Fig. 1A). We assembled in vitro translation systems in which methionine was replaced with one of the 21 nonproteinogenic amino acids (Fig. 2A), with high-fidelity incorporation at AUG codons (SI Appendix, Fig. S2). A site-saturation mutagenesis mRNA library for PUMA was added to each of these systems and translated into PUMA peptides. Similar to previous studies (29, 30), a pseudo wild-type sequence was used containing the mutation M144A (henceforth referred to as the wild-type); this mutation prevents oligomerization of PUMA peptides at high (micromolar) concentrations. Each peptide was covalently attached to its encoding mRNA via a puromycin linker, and the pool of peptide–mRNA fusions was reverse transcribed into noncovalent cDNA complexes using barcoded DNA primers (Fig. 1A) (32). Barcoding, which encoded the reprogrammed genetic codes themselves, permitted an outsized 41 amino acid alphabet.
The resulting diverse PUMA peptide library was incubated with immobilized MCL1 and washed, and the bound fraction was recovered (Fig. 1B). Populations before and after binding were enumerated by deep sequencing, and enrichment scores for binding (E) were calculated for every mutant (2) (Fig. 1C). To validate this approach, previously reported KD/∆∆G values (30) were compared with E scores, which revealed that E is a smooth function of MCL1 binding affinity (Fig. 2C). To validate that E is a function of binding for nonproteinogenic mutants, we chemically synthesized additional PUMA mutants and measured binding to MCL1 (SI Appendix, Figs. S3 and S4 and Table S1). For synthetic convenience, the PUMA peptides in this collection were 27 amino acids in length, shorter than the 35-aa peptides used in previous studies (29, 30), and ∆∆G values were calculated relative to the binding of an equivalent 27-aa wild-type. The link between ∆∆G and E is maintained for this collection and, importantly, the proteinogenic and nonproteinogenic mutant data overlay (Fig. 2C).
E scores alone are sufficient for analysis of deep mutational scanning data (2–9). However, to aid interpretation of the mutant data and any structure–activity relationships that follow, we chose to calibrate log2E against a handful of experimentally validated mutants and calculate true thermodynamic parameters, ∆∆G/KD, for each mutant. Rather than make assumptions about the function linking log2E and KD (1, 5), we chose to use an empirical fit of E and ∆∆G to calibrate the deep mutational scanning data. The predictive power of our approach was assessed using leave-one-out cross-validation (Fig. 2D).
An exhaustive map of PUMA mutant ∆∆G was constructed, including all mutations to all 41 proteinogenic and nonproteinogenic amino acids (Fig. 2E). As expected for α-helical folding, mutations to proline or backbone N-methyl amino acids were highly destabilizing (29) (Fig. 2E and SI Appendix, Fig. S5). As expected for a BH3 domain (27, 28), mutations to the highly conserved D146 were not tolerated (Fig. 2D and SI Appendix, Fig. S6). A145G, which brings PUMA closer to the BH3 consensus of LXXXGD (28), was highly stabilizing. Interestingly, a nonproteinogenic amino acid substitution was the most favorable at many positions in the PUMA sequence (SI Appendix, Fig. S6).
Nonproteinogenic Deep Mutational Scanning of the de Novo Macrocyclic Peptide CP2.
In principal, this scanning approach can be applied to sequences that already contain nonproteinogenic amino acids, such as the small de novo macrocyclic peptides discovered using the RaPID system (23–26). We reasoned that saturation mutagenesis could help understand the molecular details behind the potent activities of these macrocycles and might suggest modifications to amino acids known to improve protease resistance or membrane permeability (13). We chose to investigate the peptide CP2 (Fig. 2F), a potent and isoform-selective inhibitor of the KDM4A histone demethylase, which contains a nonproteinogenic d-tyrosine and is cyclized via a nonreducible thioether bond. CP2 binds KDM4A with a 30 nM KD, forms a small β-sheet–like secondary structure when bound (Fig. 2F and SI Appendix, Fig. S7), and exhibits inhibitory activity with ∼40 nM IC50 (26).
Nonproteinogenic deep mutational scanning was performed for CP2. The raw E scores for binding KDM4A correlated well with previously reported inhibitory IC50 values (SI Appendix, Fig. S7). A small collection of CP2 mutants was synthesized, and measured ∆∆G values for binding (SI Appendix, Fig. S8 and Table S2) were used to calibrate E and calculate ∆∆G for all proteinogenic and nonproteinogenic CP2 mutants (Fig. 2G and SI Appendix, Fig. S7). The map of ∆∆G shows that a critical RSG motif was intolerant to mutation (Fig. 2G and SI Appendix, Fig. S9) and confirms many of the observations from previous structure–activity studies (33). The RSG motif forms the turn of the β-sheet and is deeply buried in KDM4A (Fig. 2F and SI Appendix, Fig. S7).
Structure–Activity Relationships at Single-Atom Resolution.
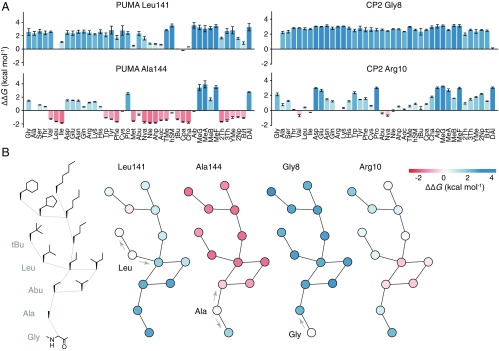
The expanded range of amino acid structures and the systematic nature of this mutagenesis allowed for the extraction of detailed structure–activity relationships at every position in each peptide sequence. As an example, progressive deletion of aliphatic carbon atoms from the side chain L141 of PUMA increasingly destabilizes the interaction with MCL1 (Fig. 3), whereas some atom insertion was stabilizing (e.g., the mutant L141Cpa; Fig. 3) (31). At position A144, where there is a nonnatural feature in the sequence of PUMA (M144A, see above), mutants with longer aliphatic side chains were increasingly stabilizing (Fig. 3). Thus, deep mutational scanning was able to identify the vacant hydrophobic pocket left by the M144A mutation. In the cyclic peptide CP2, R10 could be truncated to aliphatic side chains such as Nva without affecting affinity for KDM4A (Fig. 3), suggesting that it is the hydrophobic chain of the arginine that interacts favorably with the partner protein, not the charged head group. Strikingly, at G8, addition of an (l-) side chain (Fig. 3) is not tolerated, but d-alanine is accepted (Fig. 2F and SI Appendix, Figs. S9 and S10). This led us to inspect the phi and psi angles of bound CP2, which identified G8 as being in a region disallowed for l- but permissible for d- amino acids (SI Appendix, Fig. S11).
Fig. 3.
Structure–activity relationships at single-atom resolution. (A) ∆∆G for each proteinogenic and nonproteinogenic mutation at position L141 and A144 in PUMA (Left), and G8 and R10 in CP2 (Right). (B, Left) Graph of amino acid side chains in which edges represent the insertion or deletion of a single aliphatic carbon atom. Other graphs, Left to Right, show nodes colored according to ∆∆G [stabilizing (red) and destabilizing (blue)] for mutations at L141 and A144 of PUMA, and G8 and R10 of CP2. Arrows highlight single carbon atom changes to the wild-type side chain.
Deep Mutational Scanning-Guided Redesign for Improved Affinity.
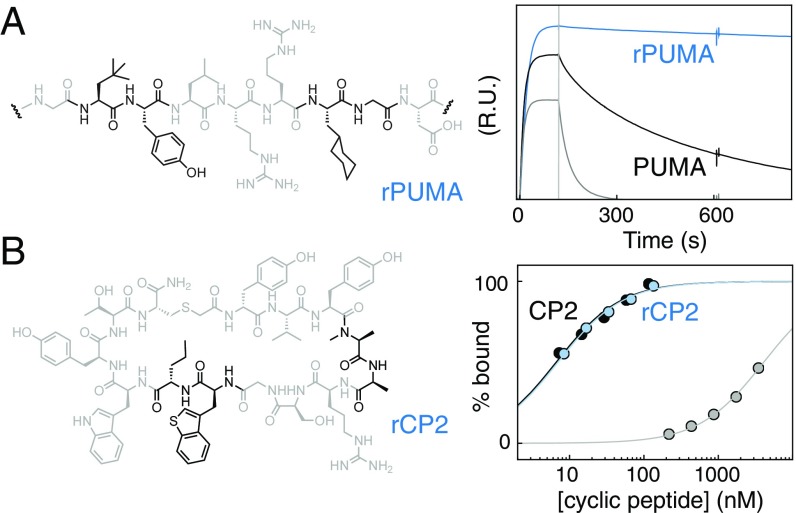
The information from these nonproteinogenic mutant scans can be used to engineer peptides for increased affinity, assuming that the effects of multiple mutants are, to some extent, additive. For example, five individual affinity-enhancing mutants of PUMA were combined into one redesigned peptide, “rPUMA” (Fig. 4A). rPUMA bound MCL1 with improved affinity (KD < 0.16 vs. 4 nM for an equivalent 27-aa wild-type peptide), stronger than any of the individual mutants, and with markedly slower dissociation compared with the unmodified peptide (Fig. 4A).
Fig. 4.
Nonproteinogenic deep mutational scanning-guided peptide optimization. (A, Left) Five PUMA mutants identified as stabilizing the interaction with MCL1 (A139tBu, Q140Y, A144Cha, A145G, and Y152Bzt) were combined to make rPUMA (segment shown, mutated amino acids in black). (A, Right) By surface plasmon resonance (SPR), rPUMA (blue) bound tighter to MCL1, with markedly slower dissociation than the equivalent PUMA wild-type (black). This result is notable, because even one poorly chosen mutation can significantly weaken the interaction; for example, I137hSM (gray). (B, Left) Four mutants of CP2 that reduce polarity or steric bulk but individually do not affect binding to KDM4A (N4MeA, T5A, W9Bzt, and R10Nva), were combined to make rCP2 (structure shown, mutants in black). (B, Right) SPR amplitude analysis shows that rCP2 (blue) binds to KDM4A with similar affinity as CP2 (black). Compare with a single poorly chosen mutant, R6G (gray).
Deep Mutational Scanning-Guided Tuning of Peptide Physical Properties.
Mutations to nonproteinogenic amino acids can engender peptides with beneficial physical properties that can increase in vivo potency (13), provided they do not disrupt the main function of the molecule. For CP2, this has been attempted before using structure-based design (26) (SI Appendix, Fig. S12), whereby a number of nonproteinogenic mutants were chosen to increase protease resistance and/or cellular activity without impacting the bound state, but modest reductions in affinity for KDM4A were observed (26). The deep mutational scanning data corroborated the structure-based design but also highlighted additional beneficial mutations (SI Appendix, Fig. S12): mutants that reduced steric bulk, added N-methylation, removed charges or hydrogen bonding groups, and did not affect KDM4A binding. These mutations were combined to produce “rCP2” (Fig. 4B) and remained energetically neutral in combination: rCP2 bound KDM4A with equivalent affinity to CP2 (Fig. 4B) (KD = 7.0 vs. 6.6 nM for the wild-type), in contrast to the modified CP2 peptides generated through structure-based design.
Analysis of Nonproteinogenic Amino Acids in Binding and Folding.
The systematic nature of deep mutational scanning has allowed for global analysis of proteinogenic amino acid substitutions (34) (i.e., comparing the amino acids themselves). Here, we could extend this analysis to include nonproteinogenic amino acids. As an example, scanning the α-helical PUMA with mutations to the nonproteinogenic MeB showed a pattern of ∆∆G almost identical to the scan with proteinogenic Pro (Fig. 5). As MeB is a noncyclic homolog of Pro, this emphasizes that the N-substitution, rather than cyclic structure, is the root cause of Pro destabilization of α-helices. Interestingly, certain nonproteinogenic amino acids closely mimic the behavior of a proteinogenic amino. At all positions in the PUMA and CP2 peptides, mutations to 3Th are energetically equivalent to mutations to the structurally similar Phe; likewise for tBu and Leu (Fig. 5 and SI Appendix, Fig. S13).
Fig. 5.
Certain nonproteinogenic amino acids closely mimic those in the universal genetic code. Graph shows ∆∆G mutant scans for proteinogenic (black) and nonproteinogenic amino acids (rainbow). MeB shows a similar ∆∆G profile to Pro in the folding and binding of α-helical PUMA (Top), whereas the ∆∆G profile of 3Th mimics Phe (Middle) and tBu mimics Leu (Bottom), in both PUMA and β-sheet CP2. Errors in ∆∆G propagated from SDs in log2E.
Discussion
By coupling the synthetic abilities of genetic code reprogramming to the massively parallel analysis of deep mutational scanning, we describe a method to exhaustively trial multiple, diverse nonproteinogenic amino acids in the thermodynamics of protein–protein interactions. This approach permits the study of large numbers of nonproteinogenic amino acid mutants in a single experiment, on a scale that would not be possible using classical cycles of synthesis and biophysical analysis.
Understanding the binding of peptides and improving their affinity is a recurring problem in biomedical science. Here, we show how deep mutational scanning can provide exhaustive peptide structure–activity relationships, down to single-atom resolution. One use of this data is to assess the extent that peptide affinity can be improved and whether there is any benefit to using nonproteinogenic chemistry. In the example of PUMA binding MCL1, many proteinogenic mutations stabilized the interaction with MCL1, suggesting that PUMA had not evolved solely for optimal affinity. Nonetheless, a nonproteinogenic amino acid substitution was the most favorable at many positions in the PUMA sequence, highlighting the use of chemistry beyond the universal genetic code to improve interaction interfaces. We used this information to redesign a PUMA peptide for greater affinity. Importantly, the bound structure was not required for this optimization, and some of the nonproteinogenic mutations (e.g., A139tBu) would have been difficult to predict, even with such structural information.
In contrast to PUMA, few mutations to the macrocyclic CP2 stabilized the interaction with its binding partner. This suggests that the trillion-member cyclic peptide library of the RaPID system, while insufficient to cover sequence space entirely, is large enough to discover molecules highly optimized for binding. However, even when affinity is difficult to improve upon, the deep mutational scanning data are still a valuable resource. Many substitutions were energetically neutral, and many of these were to amino acids with physical properties that increase protease resistance and membrane permeability (13). This approach applied to the CP2 cyclic peptide was significantly quicker and suggested more suitable modifications than previous structure-based design (26).
A longstanding basic biological question asks whether the collection of proteinogenic amino acids is optimal for making folded, functional proteins. Here, in the context of the two main protein-folding secondary structures (the α-helix of PUMA and the β-sheet CP2), our deep mutational scanning identified and validated nonproteinogenic bioisosteric replacements: nonproteinogenic amino acids that closely mimicked the behavior of a proteinogenic amino acid. This finding suggests that alternative genetic codes are possible that are as “fit” as the universal genetic code (15) with regard to producing folding and binding polypeptides.
Here, we examined 1,360 mutants of PUMA (714 nonproteinogenic) and 468 mutants of CP2 (240 nonproteinogenic). This is has not taken advantage of the maximum mRNA display library size (>1013) or the full throughput of next-generation DNA sequencing. Therefore, nonproteinogenic deep mutational scanning can be expanded to study multiple mutants or longer protein sequences. This method can be applied to other natural, de novo discovered or designed peptides or proteins that function through binding or through binding and folding.
The ability to rapidly generate high-resolution structure–activity maps will be particularly useful in the development of drug-candidate peptides, as these information-rich datasets can be used to guide modifications for greater binding and potency. In essence, this approach is a highly parallelized version of hit-to-lead exploration in small molecule drug development. Lastly, this method can be extended to include any number of nonproteinogenic amino acids tolerated by the ribosome, a set that contains hundreds of structures and continues to expand (23, 35, 36). Deep mutational scanning can be used to comprehensively assess these nonproteinogenic amino acids for use in binding, folding, and protein engineering.
Materials and Methods
Nonproteinogenic amino acids for mutagenesis (ester activated) were loaded onto tRNAEnGluCAU using flexizymes (21) (SI Appendix, Table S3). Nonproteinogenic amino acids for translation initiation (N-acetyl-l-phenylalanine for PUMA and N-chloroacetyl-d-tyrosine for CP2) were loaded onto tRNAfMetCAU, using the flexizyme eFx (22). Loaded tRNA were separately added to methionine-deficient in vitro translation systems, along with site-saturation mRNA libraries (with C-terminal HA tag) for PUMA or CP2, and translated into peptides according to an mRNA display protocol (25). Reverse transcription was carried out using barcoded DNA primers unique for each reprogrammed genetic code, and the products from each genetic code were pooled. Nonproteinogenic amino acids were incorporated with high fidelity (SI Appendix, Fig. S2), but to account for any differences in translation efficiencies, cDNA-linked peptide mutants (with C-terminal HA tag) were purified using anti-HA magnetic beads to remove any incompletely translated by-products.
Twenty microliters of an approximately 200 nM library of anti-HA–purified cDNA-linked peptide mutants (PUMA or CP2) was incubated with 200 nM protein binding partner biotin-streptavidin immobilized onto magnetic beads (MCL1 or KDM4A). A 200 nM concentration of partner protein was low enough to produce a broad distribution of log2E, while high enough to avoid excessive PCR cycles during DNA recovery (SI Appendix, Fig. S14). Binding was allowed to reach equilibrium (3 h at 25 °C), and the beads were twice washed with buffer (20 µL each), allowing binding to reach equilibrium after each buffer addition (three 3-h incubations in total). Samples of the anti-HA–purified cDNA libraries before and after binding were prepared for Illumina sequencing. DNA reads were analyzed using a modified version of the Enrich pipeline (2), calculating enrichment scores (E) for each proteinogenic and nonproteinogenic mutant. Repeat experiments of the incubation, washing, and PCR amplification, (starting from the same library preparation, n = 3 for PUMA, n = 4 for CP2) were averaged to give the reported log2E values, and the SDs were used to estimate the error in log2E. Depending on the function ∆∆G = f(log2E), errors were propagated appropriately to give reported errors in ∆∆G.
A collection of proteinogenic and nonproteinogenic mutants of PUMA and CP2, covering the range of E scores, was synthesized using solid-phase peptide synthesis, and binding to MCL1 and KDM4A, respectively, was tested using surface plasmon resonance. The resulting KD/∆∆G values, plus any published values (30), were used to find empirical relationships between E and ∆∆G, and these functions were used to calculate KD/∆∆G for all PUMA and CP2 mutants.
Supplementary Material
Acknowledgments
We thank A. Kawamura for the gift of biotinylated KDM4A. This study was supported by Grants-in-Aid for Japan Society for the Promotion of Science Fellows (P13766) (to J.M.R.), a Joint French National Research Agency/Japan Science and Technology Agency Grant (ANR-14-JITC-2014-003 and JST-SICORP), a Japan Science and Technology Agency Core Research for Evolutionary Science and Technology Grant (JPMJCR12L2), and a Japan Agency for Medical Research and Development Grant (JP18am0301001).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809901115/-/DCSupplemental.
References
- 1.Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci USA. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fowler DM, et al. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocklin GJ, et al. Global analysis of protein folding using massively parallel design, synthesis, and testing. Science. 2017;357:168–175. doi: 10.1126/science.aan0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araya CL, et al. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci USA. 2012;109:16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson CA, Wu NC, Sun R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr Biol. 2014;24:2643–2651. doi: 10.1016/j.cub.2014.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Rosmalen M, et al. Affinity maturation of a cyclic peptide handle for therapeutic antibodies using deep mutational scanning. J Biol Chem. 2017;292:1477–1489. doi: 10.1074/jbc.M116.764225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitehead TA, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Podgornaia AI, Laub MT. Protein evolution. Pervasive degeneracy and epistasis in a protein-protein interface. Science. 2015;347:673–677. doi: 10.1126/science.1257360. [DOI] [PubMed] [Google Scholar]
- 9.Klesmith JR, Bacik JP, Wrenbeck EE, Michalczyk R, Whitehead TA. Trade-offs between enzyme fitness and solubility illuminated by deep mutational scanning. Proc Natl Acad Sci USA. 2017;114:2265–2270. doi: 10.1073/pnas.1614437114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadam RU, et al. Potent peptidic fusion inhibitors of influenza virus. Science. 2017;358:496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilbs J, Middendorp SJ, Heinis C. Improving the binding affinity of in-vitro-evolved cyclic peptides by inserting atoms into the macrocycle backbone. ChemBioChem. 2016;17:2299–2303. doi: 10.1002/cbic.201600336. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen DS, et al. Orally absorbed cyclic peptides. Chem Rev. 2017;117:8094–8128. doi: 10.1021/acs.chemrev.6b00838. [DOI] [PubMed] [Google Scholar]
- 13.Walport LJ, Obexer R, Suga H. Strategies for transitioning macrocyclic peptides to cell-permeable drug leads. Curr Opin Biotechnol. 2017;48:242–250. doi: 10.1016/j.copbio.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Ahlbach CL, et al. Beyond cyclosporine A: Conformation-dependent passive membrane permeabilities of cyclic peptide natural products. Future Med Chem. 2015;7:2121–2130. doi: 10.4155/fmc.15.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber AL, Miller SL. Reasons for the occurrence of the twenty coded protein amino acids. J Mol Evol. 1981;17:273–284. doi: 10.1007/BF01795749. [DOI] [PubMed] [Google Scholar]
- 16.Checco JW, et al. α/β-Peptide foldamers targeting intracellular protein-protein interactions with activity in living cells. J Am Chem Soc. 2015;137:11365–11375. doi: 10.1021/jacs.5b05896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinert ZE, Lengyel GA, Horne WS. Protein-like tertiary folding behavior from heterogeneous backbones. J Am Chem Soc. 2013;135:12528–12531. doi: 10.1021/ja405422v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon MD, et al. d-amino acid scan of two small proteins. J Am Chem Soc. 2016;138:12099–12111. doi: 10.1021/jacs.6b03765. [DOI] [PubMed] [Google Scholar]
- 19.Lyamichev VI, et al. Stepwise evolution improves identification of diverse peptides binding to a protein target. Sci Rep. 2017;7:12116. doi: 10.1038/s41598-017-12440-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin JW. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 21.Murakami H, Ohta A, Ashigai H, Suga H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods. 2006;3:357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- 22.Goto Y, et al. Reprogramming the translation initiation for the synthesis of physiologically stable cyclic peptides. ACS Chem Biol. 2008;3:120–129. doi: 10.1021/cb700233t. [DOI] [PubMed] [Google Scholar]
- 23.Passioura T, Suga H. A RaPID way to discover nonstandard macrocyclic peptide modulators of drug targets. Chem Commun (Camb) 2017;53:1931–1940. doi: 10.1039/c6cc06951g. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka Y, et al. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature. 2013;496:247–251. doi: 10.1038/nature12014. [DOI] [PubMed] [Google Scholar]
- 25.Yamagishi Y, et al. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem Biol. 2011;18:1562–1570. doi: 10.1016/j.chembiol.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Kawamura A, et al. Highly selective inhibition of histone demethylases by de novo macrocyclic peptides. Nat Commun. 2017;8:14773. doi: 10.1038/ncomms14773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reich LL, Dutta S, Keating AE. SORTCERY-A high-throughput method to affinity rank peptide ligands. J Mol Biol. 2015;427:2135–2150. doi: 10.1016/j.jmb.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams JM, Cory S. The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 2018;25:27–36. doi: 10.1038/cdd.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers JM, Wong CT, Clarke J. Coupled folding and binding of the disordered protein PUMA does not require particular residual structure. J Am Chem Soc. 2014;136:5197–5200. doi: 10.1021/ja4125065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers JM, et al. Interplay between partner and ligand facilitates the folding and binding of an intrinsically disordered protein. Proc Natl Acad Sci USA. 2014;111:15420–15425. doi: 10.1073/pnas.1409122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rezaei Araghi R, Ryan JA, Letai A, Keating AE. Rapid optimization of Mcl-1 inhibitors using stapled peptide libraries including non-natural side chains. ACS Chem Biol. 2016;11:1238–1244. doi: 10.1021/acschembio.5b01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tjhung KF, et al. Silent encoding of chemical post-translational modifications in phage-displayed libraries. J Am Chem Soc. 2016;138:32–35. doi: 10.1021/jacs.5b10390. [DOI] [PubMed] [Google Scholar]
- 33.Passioura T, Bhushan B, Tumber A, Kawamura A, Suga H. Structure-activity studies of a macrocyclic peptide inhibitor of histone lysine demethylase 4A. Bioorg Med Chem. 2018;26:1225–1231. doi: 10.1016/j.bmc.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Gray VE, Hause RJ, Fowler DM. Analysis of large-scale mutagenesis data to assess the impact of single amino acid substitutions. Genetics. 2017;207:53–61. doi: 10.1534/genetics.117.300064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 36.Hartman MC, Josephson K, Lin CW, Szostak JW. An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS One. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.