Significance
There is a strong need for new analgesic and antiinflammatory medicines that are both effective and safe. Animal studies have shown that inhibition of N-acylethanolamine acid amidase (NAAA)—an intracellular enzyme that degrades the lipid mediator palmitoylethanolamide—causes profound analgesic and antiinflammatory effects. To facilitate the discovery of drugs targeting this protein and to better understand its mechanism of action, we determined its 3D structure. Our results illustrate the sequential steps leading to the activation of NAAA at lipid membranes, and reveal how current inhibitors block this enzyme.
Keywords: endocannabinoid, palmitoylethanolamide, N-terminal nucleophile hydrolase, N-acylethanolamine acid amidase, interfacial activation
Abstract
Palmitoylethanolamide is a bioactive lipid that strongly alleviates pain and inflammation in animal models and in humans. Its signaling activity is terminated through degradation by N-acylethanolamine acid amidase (NAAA), a cysteine hydrolase expressed at high levels in immune cells. Pharmacological inhibitors of NAAA activity exert profound analgesic and antiinflammatory effects in rodent models, pointing to this protein as a potential target for therapeutic drug discovery. To facilitate these efforts and to better understand the molecular mechanism of action of NAAA, we determined crystal structures of this enzyme in various activation states and in complex with several ligands, including both a covalent and a reversible inhibitor. Self-proteolysis exposes the otherwise buried active site of NAAA to allow catalysis. Formation of a stable substrate- or inhibitor-binding site appears to be conformationally coupled to the interaction of a pair of hydrophobic helices in the enzyme with lipid membranes, resulting in the creation of a linear hydrophobic cavity near the active site that accommodates the ligand’s acyl chain.
Cells employ a variety of signaling lipids to communicate with their surroundings or to propagate signals intracellularly. One class of bioactive lipids, the N-acylethanolamines (NAEs, or fatty acid ethanolamides), acts through several receptors to exert a diversity of biological effects in the nervous and immune systems. The best-characterized NAE is arachidonoylethanolamide (anandamide), which together with the related compound 2-arachidonoylglycerol (2-AG), form the classical endocannabinoids that bind to CB1 and CB2 cannabinoid receptors (1). NAEs bearing a shorter and more saturated acyl chain, such as palmitoylethanolamide (PEA), do not activate cannabinoid receptors (2), but act mainly through the peroxisome proliferator-activated receptor-α (PPAR-α) (3), a ligand-operated transcription factor. PEA is a well-recognized analgesic, antiinflammatory, and neuroprotective mediator (4) whose production occurs constitutively but is diminished following various proinflammatory stimuli (5–7). This decrease contributes to the inflammatory response via downstream gene targets of PPAR-α (6).
NAEs are primarily inactivated by hydrolysis to ethanolamine and fatty acid (8, 9) carried out by two enzymes: fatty acid amide hydrolase (FAAH) (10) and N-acylethanolamine acid amidase (NAAA) (9). FAAH is widely distributed (11) although it is most abundant in the brain and liver (12), whereas NAAA is almost exclusively expressed in immune cells (13, 14) such as monocytes and tissue macrophages (15, 16) but is also found in prostate epithelium (13, 17). In light of the analgesic and antiinflammatory actions of PEA, multiple NAAA inhibitors have been developed over the last decade (18, 19) and shown to exhibit beneficial effects in a range of rodent models of human disease, including inflammation (20–25), allergic contact dermatitis (26), spinal cord trauma (27), neuropathic pain (28, 29), chronic pain (30), inflammatory bowel disease (31), lung inflammation (25, 32), arthritis (14), and multiple sclerosis (33).
No NAAA-targeting compound has yet reached clinical trials. On the other hand, FAAH inhibitors are currently being tested in humans for the treatment of anxiety and depression (18). These properties are mediated by the activity of the FAAH substrate anandamide on cannabinoid receptors (18). Whereas FAAH hydrolyzes long-chain polyunsaturated NAEs, including anandamide, more efficiently than short unsaturated ones such as PEA (34), NAAA demonstrates an opposite substrate preference in vivo (27, 28, 31, 33) and in vitro (35, 36), with PEA being its optimal substrate (15, 37). Another major difference between the two enzymes, besides tissue distribution, is their intracellular localization: FAAH is found in the cytosol tethered to the outer face of mitochondria and the endoplasmic reticulum (38), whereas NAAA is localized to acidic organelles such as the lysosome (16, 37, 39). The two proteins are structurally unrelated: FAAH belongs to the amidase signature superfamily (40), while NAAA is part of the N-terminal nucleophile (NTN) hydrolases that undergo proteolytic self-activation (41, 42). NTN hydrolases cleave amide bonds in their mostly soluble substrates, and as NAAA acts on the amide bonds in the lipid PEA, it is both an amidase and a lipase. The crystal structure of FAAH has been determined, revealing a dimeric protein that stably embeds into membranes via a hydrophobic helix-turn-helix segment, enabling substrate access to its active site (40). On the other hand, NAAA is a soluble enzyme (35) with no sequence homology to FAAH and its lipid association and substrate-binding modes are uncharacterized. Its in vitro activity is increased by certain detergents and nonsubstrate lipids (15, 36) through an unknown mechanism. Although NAAA was discovered (35), cloned (39), and purified (15) almost 20 years ago, questions regarding its mechanism of action remain unanswered due to a lack of structural information. Homology models have been built (27, 43) and employed for inhibitor development (20, 44, 45). Such models are, however, of little heuristic value because they rely on a bacterial homolog sharing only 13% sequence identity with NAAA. Structural information about the only mammalian homolog of NAAA, acid ceramidase (ASAH1), was recently obtained (46), but there are significant functional differences between these two enzymes. Here, crystal structures of NAAA in distinct activation states are presented, revealing how proteolytic self-activation uncovers the enzyme’s active site, and how exposure to a hydrophobic environment induces a conformational rearrangement that generates the substrate-binding cavity. This putative interfacial activation mechanism is reminiscent of those seen with certain lipid hydrolases such as monoacylglycerol lipase (47), but has not been previously reported for any other member of the NTN superfamily. Complexes of the protein with a covalent and a noncovalent inhibitor are also described. These results further our understanding of the function of NAAA and will facilitate its targeting for therapeutic purposes.
Results
NAAA Adopts an αββα-Fold with a Helical α-Subunit.
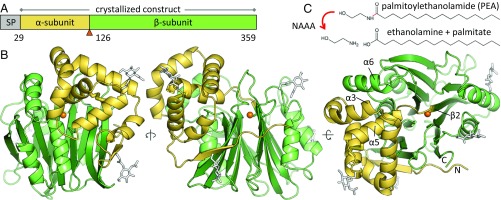
NAAA is part of the NTN hydrolase superfamily, a diverse group of amidases that carry out catalysis via their N-terminal cysteine, serine, or threonine residue (41, 42). These proteins are produced as precursors and undergo activation by removal of the initiator methionine or by self-cleavage of an internal peptide bond. This results either in the separation of a short propeptide or, in the case of other family members including NAAA (15, 37, 48, 49), formation of the mature enzyme composed of an α- and a β-subunit that remain associated. We determined the crystal structure of human NAAA (hNAAA) precursor produced recombinantly in insect cells, bearing the Cys126Ala mutation that prevents self-cleavage (48), as well as of the cleaved and active form of the murine enzyme (mNAAA) (SI Appendix, Table S1). NAAA comprises an N-terminal signal peptide that is removed cotranslationally, a small α-subunit spanning residues 29–125 (49), and a large catalytic β-subunit (126–359) (Fig. 1A).
Fig. 1.
Structure of the NAAA precursor. (A) Domain organization of human NAAA. The enzyme is composed of a cleavable signal peptide (SP), a small α-subunit, and a large β-subunit generated from a single polypeptide chain by proteolytic self-activation. The active site residue Cys126 is marked by an orange triangle. (B) Self-cleavage is disabled in the Cys126Ala mutant whose crystal structure is displayed. The active site residue 126 is modeled here as cysteine (sulfur atom as orange sphere). A disulfide bond within the α-subunit is represented by thick yellow sticks, and N-linked glycans (white sticks) are simplified for clarity. The N terminus and C terminus are labeled, as are several key helices and strands described later. (C) Reaction catalyzed by NAAA.
NTN hydrolases share a αββα-sandwich fold with a core of two antiparallel β-sheets surrounded by α-helices (41, 42). In the NAAA β-subunit, these sheets consist of five and eight β-strands, flanked by two α-helices on either side (Fig. 1B). The α-subunit begins with a strand associated with the central β-sheets and forms a bundle of five helices positioned at the corner of the “top” face of the β-subunit (Fig. 1B), with a buried interface area of about 2,200 Å2. NAAA is part of a subfamily of NTN hydrolases whose best-characterized members are bacterial enzymes involved in antibiotic biosynthesis (SI Appendix, Fig. S1B). In mammals, this group is composed of phospholipase B-like proteins 1 and 2 (PLBD1 and -2)—two lysosomal amidases of known structure but unknown function (SI Appendix, Fig. S1C) (50, 51)—and the acid ceramidase (ASAH1), which degrades lysosomal ceramide into sphingosine and fatty acid, whose crystal structure was recently determined (SI Appendix, Fig. S1A) (46). NAAA shares 33% sequence identity with ASAH1, whereas its identity to all other NTN hydrolases is below 20%. In this subfamily, the α-subunit exhibits the highest structural variability, being completely absent in some members (SI Appendix, Fig. S1B).
NAAA bears two N-linked glycans on each subunit (Fig. 1B) (48, 49). A fifth potential site (Asn315) is also surface exposed but not glycosylated. hNAAA contains one disulfide bond between helices α4 and α5 within the α-subunit (Fig. 1B). This feature is conserved in about 70% of mammals but not in the other rodent structures described here and is also absent from nonmammalian vertebrates. Unlike NAAA (37, 49), a disulfide bridge links both subunits of ASAH1 (46).
Self-Proteolysis Exposes the NAAA Active Site.
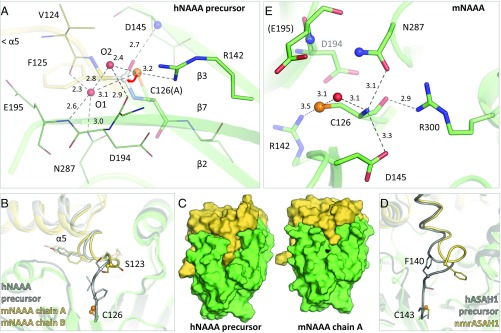
Most NTN hydrolases, including NAAA (48), are converted from a precursor to an active form by self-proteolysis, a reaction mediated by the same catalytic residue that subsequently carries out substrate hydrolysis (41, 42). NAAA undergoes self-cleavage at acidic pH but not in neutral conditions (37, 48, 52). To capture the precursor form, the catalytic Cys126 residue was mutated to alanine. Cys126 (Ala126) is located at the start of strand β2 of the β-subunit and is connected to the last helix α5 of the α-subunit by a short linker in hNAAA (Fig. 1B). Two water molecules are found in the precursor active site within hydrogen-bonding distance of the Cys126 side chain (modeled as cysteine in Fig. 2A), which also interacts with Arg142. The self-proteolytic mechanism of NTN hydrolases begins with deprotonation of the catalytic cysteine, serine, or threonine side chain that then carries out a nucleophilic attack on the carbonyl carbon of the preceding peptide bond (Phe125). This is facilitated by the backbone nitrogen of Asp145 serving as the oxyanion hole for the carbonyl oxygen (Fig. 2A). Several different means of initiating this reaction were proposed in these enzymes, including deprotonation of the nucleophile by a nearby water molecule (SI Appendix, Fig. S2A) (53–55) or side chain (56). It has also been suggested that the intrinsic nucleophilicity of cysteine is sufficient to initiate the attack (57, 58). Distortion and strain of the scissile peptide bond (50) or a peptide flip (59) were observed in other NTN hydrolase precursors, but not in the NAAA Cys126Ala structure. Regardless of the precise mechanism, the attack is followed by dissociation of the Cys126 backbone amine from Phe125 (N-S acyl shift). The resulting thioester intermediate is subsequently hydrolyzed to yield Cys126 at the N terminus of the β-subunit. In the structure of wild-type activated mNAAA, the end of helix α5 and the linker leading to the β-subunit, which is now the C terminus of the α-subunit, shifts relative to the hNAAA precursor (Fig. 2B); its last two residues (Val124 and Phe125) also become disordered and not discernible in the electron density. This rearrangement results in exposure of the active site to the environment, which was previously buried under the linker (Fig. 2C). A similar conformational change and disordering occurs in ASAH1 (Fig. 2D) (46).
Fig. 2.
Active site and proteolytic self-activation of NAAA. (A) Active site of the human NAAA precursor (Cys126Ala mutant) with the catalytic residue modeled here as cysteine. The nucleophilic attack by the Cys126 sulfur atom (orange sphere) on the carbonyl carbon of the preceding amino acid (white sphere) is marked by a red arrow. Residues forming part of the active site are labeled, including two water molecules (O1 and O2, red spheres), as are secondary structure elements. Hydrogen bonds are represented by dashed lines, with interatomic distances (in angstroms). (B) Comparison of the human precursor (gray) and murine active enzyme (yellow) illustrating the rearrangement of helix α5, with slightly different orientations in both protein copies in the crystal. The last visible residue in the electron density of the active structure is Ser123 (sphere); the subsequent two C-terminal amino acids of the α-subunit are disordered. (C) Proteolytic self-activation results in the exposure of the catalytic residue (black) on the protein’s surface. (D) Similarly, self-cleavage of acid ceramidase (ASAH1, PDB ID codes 5U7Z and 5U81) leads to a larger rearrangement and partial disordering of the C-terminal end of the α-subunit. (E) Active site of murine NAAA (human numbering). Nitrogen atoms forming the oxyanion hole are displayed as blue spheres, and a water molecule, as a red sphere. Although Cys126 was partially oxidized in the murine structure, the active site arrangement remained unchanged relative to the other structures described later.
In the active site of mature mNAAA, Cys126 hydrogen-bonds to Arg300 via its backbone oxygen and to Asn287 and Asp145 via its N-terminal amine (Fig. 2E). Most of these stabilizing interactions are conserved in this NTN hydrolase subfamily (SI Appendix, Fig. S2B). Their mechanism of substrate amide bond hydrolysis is well characterized (42) and begins with deprotonation of the catalytic residue’s side chain by its own N-terminal amine. In ASAH1 (46) and NAAA, this moiety may form an acid-base pair with Asp145, facilitating catalysis at acidic pH which is optimal for NAAA (15, 35, 37, 52). The pKa of the sulfhydryl group is probably lowered by the nearby cationic Arg142 (50). Although a water molecule bridges the Cys126 side chain and N-terminal amine (Fig. 2E), direct proton transfer between the two was proposed (60). The side chain then carries out a nucleophilic attack on the substrate’s carbonyl carbon, enabled by stabilization of the carbonyl oxygen at the oxyanion hole composed of the Glu195 backbone nitrogen and Asn287 side chain amine. Dissociation of the substrate’s amine moiety (ethanolamine), protonated by the Cys126 backbone amine, results in an acyl-enzyme intermediate that is subsequently hydrolyzed. All of the residues involved are conserved in ASAH1, with the exception of Asp194 (Asn), which is peripheral to the active site (Fig. 2E and SI Appendix, Fig. S2B).
Detergent-Induced Rearrangement Generates the NAAA Substrate-Binding Site.
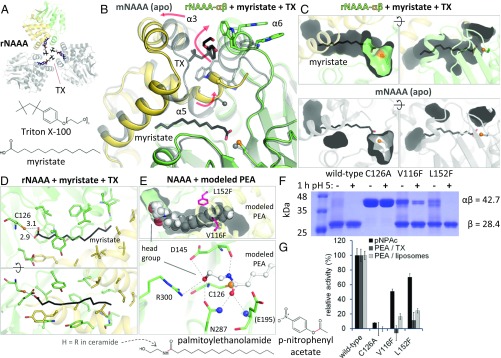
NAAA hydrolyzes its optimal substrate PEA (15, 35–37) into ethanolamine and palmitate, a 16-carbon saturated fatty acid (Fig. 1C). To identify the substrate-binding site, we crystallized the protein with fatty acids; the detergent Triton X-100 (TX) was included to solubilize these ligands. The structure of activated rabbit NAAA (rNAAA) in complex with myristate (14 carbons, saturated) was obtained. Electron density revealed the presence of detergent molecules at a crystallographic interface formed by helices α3 and α6 from three protein copies (Figs. 1B and 3A and SI Appendix, Fig. S3E), including one distinct TX molecule bound deeply between the two helices (Fig. 3B). To accommodate it, helix α3 from the α-subunit is displaced relative to its position in apo mNAAA, and helix α6 from the β-subunit is shifted and rotated by one residue. The latter rearrangement results in the surface exposure of two tryptophan side chains from a “WWW” segment (residues 200–202) that were buried in apo-activated mNAAA, but now contact interfacial detergent molecules. The space previously occupied by these Trp residues is now filled by the end of helix α5 and the C-terminal portion of the α-subunit: the helix becomes bent and the C terminus becomes fully ordered (Fig. 3B). Importantly, this last conformational change generates a pocket between both subunits, in which a myristate molecule is bound (Fig. 3 B and C and SI Appendix, Fig. S3E). This pocket is fragmented in apo-activated mNAAA, preventing accommodation of the fatty acid (Fig. 3C).
Fig. 3.
Substrate binding by NAAA. (A) Crystal arrangement of rNAAA, with crystallographic symmetry copies in gray and TX molecules as colored sticks. (B) Comparison of mNAAA apo form (transparent gray) with rNAAA (yellow and green) bound to myristate and TX (black sticks). Secondary structure elements are labeled. Rearrangements that occur in presence of detergent are illustrated by red arrows. The last residue visible in the mNAAA α-subunit electron density is marked by a sphere in both structures. (C) As in B, these conformational changes produce a cavity where the fatty acid is inserted (Top), whereas this ligand would not fit into the apo protein (Bottom). (D) Residues lining the cavity are displayed as sticks, with interatomic distances (in angstroms). (E) PEA was manually placed into the rNAAA substrate-binding site and is shown as full-scale spheres (Top). Potential hydrogen bonds between PEA and active site residues (Bottom, human numbering) are represented by dashed lines, including to the oxyanion hole atoms (blue spheres). The location of ethanolamine or bulky artificial head groups is labeled. (F) Proteolytic self-activation upon incubation at acidic pH of two hNAAA point mutants designed to partially block substrate binding; these residues are displayed as pink sticks in E. Calculated molecular weights of the glycosylated precursor and of the active enzyme’s β-subunit are provided. (G) Relative in vitro hydrolysis rates of p-nitrophenyl acetate (pNPAc), PEA in TX micelles, and PEA in liposomes. Full activity corresponds to: 0.05 μM pNPAc/nM NAAA per hour (means and SDs of a representative of two experiments performed in quadruplicates); 2.2 μM PEA in TX/nM NAAA per hour (means and SDs of three experiments performed in quadruplicates); 0.7 μM PEA in liposomes/nM NAAA per hour (means and SDs of a representative of three experiments performed with n = 8). In the substrate chemical structures, the scissile bond is in red.
Hydrophobic residues surround the myristate lipid tail, whereas its carboxyl group hydrogen-bonds to the Cys126 side chain (Fig. 3D). This does not reflect the productive substrate-binding mode in which the carbonyl carbon should be positioned closer to the sulfur atom. However, the location of the lipid tail facilitated manual modeling of a PEA molecule into the NAAA structure (Fig. 3E), further guided by the expected position of the carbonyl carbon, and of the carbonyl oxygen at the oxyanion hole. In this manner, the PEA amide nitrogen hydrogen-bonds with the backbone of Asp145. Although the orientation of the ethanolamine head group is less certain, its hydroxyl moiety can hydrogen-bond with any of four surrounding residues. To validate this pocket as the substrate-binding site, two side chains lining the cavity were individually mutated to bulkier phenylalanine residues which should partially impede substrate entry (Fig. 3E). The purified mutants underwent (almost) full self-activation (Fig. 3F) but were somewhat less stable than the wild-type protein, based on their yield and lower enzymatic activity on the small nonlipid substrate p-nitrophenyl acetate (50–70% of wild type) (Fig. 3G). However, hydrolysis of PEA in TX micelles and also in liposomes decreased to 20% or lower. These results, together with the structural data, suggest that this pocket is not a detergent-induced artifact, but represents the N-acylethanolamine–binding site of NAAA.
The shape and size of the cavity offer an explanation for the substrate specificity of this enzyme. The endocannabinoid hydrolases FAAH and monoacylglycerol lipase (MGL) act most readily on anandamide and 2-AG, respectively (18); these compounds contain a 20-carbon acyl chain with four cis double bonds. On the other hand, NAAA displays low to no activity on these molecules, preferring shorter saturated lipids (15, 35–37). The substrate-binding site proposed here is narrow and relatively straight, better suited for accommodating saturated acyl chains. In addition, although the cavity extends beyond the end of the manually modeled PEA (16 carbons) (Fig. 3E), it becomes too narrow to allow longer ligands without further expansion. This cannot be excluded, as this region of the enzyme demonstrates significant flexibility, and additional rearrangements may occur upon contact with a lipid bilayer. The modeled ethanolamine portion is solvent exposed, explaining the enzyme’s ability to hydrolyze a substrate bearing a bulky fluorescent head group (49) or react with probes containing a large head group substituent (61). These moieties would take the place of ethanolamine, projecting outside of the cavity into the solvent (Fig. 3E).
NAAA Inhibitors Block the Substrate-Binding Site.
The functions of endocannabinoids and other N-acylethanolamines in the nervous and immune systems established their degrading enzymes—FAAH, MGL, and NAAA—as therapeutic targets in a variety of disorders (18). NAAA has been the focus of intense inhibitor discovery in the last decade. Most compounds against this hydrolase consist of a polar group linked to a hydrophobic moiety. Polar groups include amines (62), esters, retroesters, amides, and retroamides (44, 63–66), and oxazoline (22). Whereas these likely associate noncovalently and reversibly with the enzyme, other classes of molecules contain reactive groups that bind covalently to the active site cysteine: oxazolidones (29, 30, 67), β-lactones (20, 27, 45, 68–70), β-lactams (23, 32, 71, 72), isothiocyanates (43), and a tetrazole-carboxamide (43). To facilitate structure-guided design of NAAA inhibitors, we crystallized the enzyme in complex with ARN726 (IC50 27 nM) (32), a β-lactam that irreversibly reacts with Cys126, and with ARN19702 (IC50 230 nM) (33), a noncovalent benzothiazole-piperazine derivative dissimilar to the other compounds listed above. The detergent-dependent conformations of the protein are discussed later; the inhibitor-binding modes are first described.
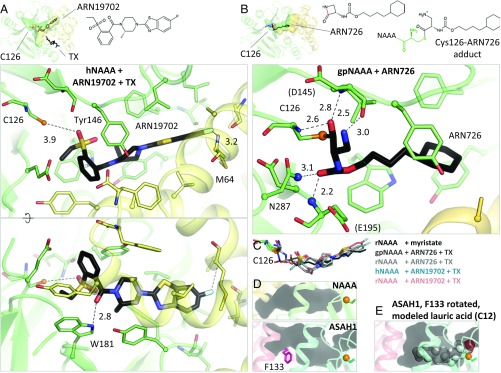
The structures of hNAAA and rNAAA with ARN19702 were determined and are essentially identical in terms of enzyme–ligand arrangement (Fig. 4C and SI Appendix, Fig. S3 A and B). The compound occupies the same cavity as myristate (Fig. 4 A and C), but its closest contact to Cys126 is 3.9 Å away. It forms only two hydrogen bonds with NAAA: one between its carbonyl oxygen and the side chain of Trp181, and the second between its terminal fluorine atom and the backbone nitrogen of Met64. The inhibitor also establishes a T-shaped π-stacking interaction with Tyr146 and van der Waals contacts with several surrounding hydrophobic residues.
Fig. 4.
NAAA inhibitor complexes. (A) Crystal structure of hNAAA with ARN19702 in presence of TX. Residues forming hydrogen bonds (dashed lines) with the inhibitor are labeled, with interatomic distances (in angstroms). (B) Crystal structure of gpNAAA covalently bound to ARN726. The scissile bond is colored in red in the free inhibitor chemical structure. Nitrogen atoms forming the oxyanion hole are displayed as blue spheres. (C) Comparison of binding orientations of inhibitors and product. (D) In acid ceramidase (ASAH1, PDB ID code 5U7Z), the corresponding cavity is shorter and partially blocked by a phenylalanine side chain (pink). (E) This side chain was reoriented and a lauric acid molecule was manually placed into the ASAH1 cavity.
The structures of rNAAA and of the guinea pig enzyme (gpNAAA) covalently bound to ARN726 were also determined (SI Appendix, Fig. S3 C and D). This complex is the product of a nucleophilic attack on the carbonyl carbon of the strained β-lactam moiety by the Cys126 side chain (Fig. 4B) and was stable for at least 4 weeks at acidic pH during crystallization. The two homologs exhibit a slightly different inhibitor orientation, possibly due to the moderate resolution of the structures (2.7 Å and 3.0 Å) (Fig. 4C). Again, the compound is found in the same pocket as myristate, and its relatively short hydrocarbon tail forms van der Waals interactions with the protein (Fig. 4B). The carbonyl oxygen of its carbamate group is positioned at the oxyanion hole, establishing two hydrogen bonds, whereas its carbonyl oxygen derived from the β-lactam ring contacts the N-terminal amine of Cys126 and the backbone nitrogen of Asp145. Another hydrogen bond is present between the β-lactam–derived primary amine and the backbone of Asp145. These results will facilitate the design of more potent compounds targeting the enzyme.
Proposed Membrane Interactions via Hydrophobic Helices α3 and α6.
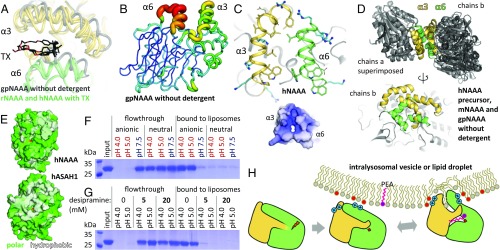
Due to their lipid characteristics, N-acylethanolamines are not likely to occur as freely soluble molecules in the cell, but are transported via carrier proteins, membrane vesicles, or lipid droplets (73). Consequently, their degrading enzymes must be able to access them within these hydrophobic settings. Crystal structures of FAAH and MGL revealed the ways these hydrolases interact with lipids (40, 74, 75). FAAH contains a transmembrane helix but binds stably to bilayers even without this segment, as it possesses a hydrophobic helix-turn-helix motif for monotopic (nonbilayer-spanning) membrane insertion (40). MGL is both cytosolic and membrane associated; it contains a hydrophobic helix as part of its cap domain that is the proposed site of membrane anchoring (74). Conversely, NAAA is a soluble protein (35) and its lipid access mode has not been characterized. Lysosomal lipid degradation occurs on intraluminal anionic vesicles (76), and liposome-binding assays confirmed that NAAA only associates stably with vesicles via electrostatic interactions (Fig. 5F). However, the enzyme must still disrupt the membrane to reach its embedded substrate, and helices α3 and α6 are likely candidates for this function. They bear almost exclusively hydrophobic and cationic side chains, resulting in an overall positive charge for this face of the hydrolase (Fig. 5C); such a lipophilic and cationic plateau is a common feature of monotopic membrane proteins (77, 78). When crystallized in the absence of detergent, NAAA from three different species displayed dimerization with varying orientations but always mediated by the α3 and α6 helices (Fig. 5D), and partial dimerization was also observed on size exclusion chromatography. These observations suggest this is not a specific, functional dimerization interface, but occurs to prevent the unfavorable solvent exposure of this hydrophobic patch. In presence of TX, the crystal forms of the rat and human enzyme contained detergent molecules covering this face (Fig. 3A), with a recurring TX ligand present between the two helices (Fig. 5A), further supporting this region as the membrane-associating element.
Fig. 5.
NAAA membrane interaction. (A) Conformational differences in helices α3 and α6 between gpNAAA bound to ARN726 in absence of detergent (gray), and other NAAA structures (each in yellow and green) in complex with inhibitors or product in presence of TX. (B) In gpNAAA, conformational flexibility of these helices is depicted by a plot of crystallographic B factors; a redder and thicker trace indicates higher disorder. (C) Helices α3 and α6 are mainly composed of hydrophobic residues (yellow and green sticks) and cationic side chains (blue sticks), resulting in an overall positive electrostatic surface potential at pH 4.5 (−5 kT/e, red to +5 kT/e, blue). (D) Superposition of crystallographic dimers (via chains a) from all crystal forms of NAAA obtained without detergent (Top), and alternate view of chains b, illustrating their variable orientations (Bottom). (E) In acid ceramidase (ASAH1, PDB ID code 5U7Z), a hydrophobic (white) bowl-shaped surface spans the α- and β-subunits. Residues were colored according to the Eisenberg hydrophobicity scale. (F) hNAAA was incubated with anionic or uncharged liposomes at various pH values. The vesicles were washed and pelleted, and bound protein was visualized by SDS/PAGE. (G) As in F, with the cationic amphiphilic drug desipramine added during incubation. (H) Proposed model of NAAA activation. After self-proteolysis in the lysosome, the enzyme electrostatically associates with intraluminal membranes. Contacts between lipids and its hydrophobic helices lead to a conformational change that exposes the substrate-binding site.
As noted above, binding of TX was accommodated by displacement of both helices and a rotation of helix α6 with the exposure of tryptophan side chains from a WWW segment to the protein’s surface (Fig. 3B). This was accompanied by a bending motion and ordering of helix α5, generating the substrate- and inhibitor-binding sites (Fig. 3C). To clarify whether the detergent induces these conformational changes allowing substrate binding, or vice versa, ligand-free rNAAA was crystallized in the presence of TX in the same crystal form, which relies on this rearrangement, but it diffracted X rays poorly (as was also the case for the majority of tested ligand-bound crystals). On the other hand, the gpNAAA–ARN726 complex was produced without detergent (Fig. 4B). The covalently bound inhibitor prevents the substrate-binding site from collapsing and reverting to the conformation observed for apo mNAAA (Fig. 3B). However, helix α6 is shifted toward the cavity otherwise occupied by TX (Fig. 5A) and becomes strikingly disordered (Fig. 5B), with poor electron density for the WWW segment (SI Appendix, Fig. S3C) whose side chains are flexible in this forced unstable configuration. These observations suggest that a hydrophobic environment is required for maintaining the substrate-binding site.
Discussion
The structural and biochemical results presented here allow us to propose a model of NAAA activation (Fig. 5H). The enzyme undergoes self-proteolysis in the lysosome, resulting in a disordering of the α-subunit C terminus and exposure of the catalytic cysteine in the β-subunit. Concomitantly, NAAA associates electrostatically with intraluminal vesicles or possibly lipid droplets. Its hydrophobic helices α3 and α6 possibly embed into the lipid membrane to some extent, causing a conformational change that generates a cavity between the two subunits, which accommodates the substrate’s acyl chain. This model, which remains to be validated by further biochemical and biophysical studies, explains the activating effect of certain detergents and lipids on PEA hydrolysis by NAAA in vitro (15, 36). An analogous mechanism occurs in a family of lipases that includes the endocannabinoid-deactivating enzyme, MGL: upon contact with a lipid layer, a helical lid from their cap domain undergoes a rearrangement, switching the enzyme to its open form with its active site exposed (47). Such a process, termed “interfacial activation” (47), has not yet been reported in NTN hydrolases, which act mostly on soluble substrates, and all share below 20% sequence identity with NAAA. The only NTN hydrolase similar to NAAA is ASAH1, with 33% identity. However, the proposed activation mode is unlikely to apply to ASAH1 for several reasons. The WWW segment is absent from that protein, which features instead a larger hydrophobic surface spanning additional portions of the β-subunit (Fig. 5E). ASAH1 does contain a cavity adjacent to the active site, partially blocked by a phenylalanine residue (Fig. 4D) (46). Modeling suggests that rotation of that side chain would allow entry of a short, 12-carbon fatty acyl moiety (Fig. 4E) linked to a substrate ceramide molecule, although ceramide contains two lipid chains that cannot be both accommodated inside at once. Considering that ASAH1 exhibits only low hydrolysis of short single-chain N-acylethanolamines such as PEA (37) as well as a different sensitivity to pharmacological inhibition compared with NAAA (33), other determinants of substrate and inhibitor specificity are likely to be found in this protein. Furthermore, ASAH1 activity is enhanced by the lipid-binding and membrane-disrupting cofactor protein saposin D (79), whereas no such stimulation has been reported for NAAA. The membrane-association mode of acid ceramidase remains to be experimentally determined. Ceramide recognition by ASAH1 has previously been explored by molecular docking (46); as the resulting acyl chain orientation is different from the NAAA–myristate complex (SI Appendix, Fig. S4), substrate binding by acid ceramidase merits further investigation.
Electrostatic attachment of NAAA to lipid vesicles is characteristic of certain lysosomal lipases (76). Many therapeutic drugs with cationic amphiphilic properties concentrate in lysosomes and result in dissociation of these lipases, which are then degraded by luminal proteases, leading to substrate accumulation (80). Interestingly, one such compound disrupted the binding of NAAA to liposomes (Fig. 5G), raising the possibility that these agents could have side effects by interfering with NAAA activity. Irrespective of this speculation, the structural and mechanistic insights provided by the present study will facilitate the discovery of NAAA inhibitors with potential therapeutic application in chronic pain, inflammation, and other pathologies.
Materials and Methods
Protein Expression and Purification.
Recombinant NAAA was expressed as a secreted protein in Sf9 insect cells infected with baculovirus. The endogenous signal peptide comprising the first 28–33 residues was replaced by the melittin signal peptide MKFLVNVALVFMVVYISYIYA followed by a hexahistidine tag DRHHHHHHKL. Constructs encompassed residues 29–359 of NAAA from human (UniProt: Q02083), 34–362 from mouse (UniProt: Q9D7V9 with variants Ala47Val, His142Arg, and Asn163Asp), 31–359 from rabbit (RefSeq: XP_008265950) and 29–355 from guinea pig (RefSeq: XP_012996906 with variants Ala75Arg, Xaa76Asp, Lys95Glu, and an alanine insertion at 194–195). Proteins were isolated from expression culture media by immobilized metal affinity chromatography (IMAC), purified by size exclusion chromatography (SEC) in buffer (15 mM Tris⋅HCl pH 7.5, 100 mM NaCl) and concentrated to 10 mg/mL. rNAAA was further applied to an anion exchange Q column in buffer (10 mM Tris⋅HCl pH 7.5, 50 mM NaCl); under these conditions, the protein did not bind to the resin.
Crystallization and Data Collection.
The hNAAA precursor (Cys113Ala, Cys126Ala) crystallized in 1 M LiCl, 0.1 M Mes pH 6 and 10% PEG 6000, and in multiple other conditions. The Cys113Ala mutation was introduced because this residue was initially suspected of forming spurious disulfide bonds. mNAAA (Asn112Ser, Asn338Ser) crystallized in 0.2 M NaCl, 0.1 M BIS-Tris pH 5 and 20% PEG 3350. The mutations abolished two N-linked glycosylation sites to generate new crystal forms. For the inhibitor complexes, the enzymes were preincubated with 10 mM DTT, 2 mM TX (but no detergent for gpNAAA), and either 0.67 mM ARN19702, 1 mM ARN726, or 1 mM myristate. hNAAA crystallized in 0.2 M NaSCN and 20% PEG 3350, whereas gpNAAA crystallized at 3 mg/mL in 0.2 M KH2PO4 and 20% PEG 3350. rNAAA crystallized in diverse conditions, including 0.2 M tripotassium citrate with 20% PEG 3350 (ARN19702); 0.8 M NaH2PO4, 0.8 M KH2PO4 with 0.1 M Hepes pH 7.5 (ARN726); and 1.1 M (NH4)2SO4 with 0.1 M sodium acetate pH 4.5 (myristate). Crystals were grown by sitting or hanging drop vapor diffusion at 22 °C and flash frozen after brief soaking in crystallization solution supplemented with 20% glycerol. X-ray diffraction data were collected at 100 K on beamlines 08B1-1 with a Rayonix MX300HE CCD detector (hNAAA with ARN19702), or 08ID-1 with a Rayonix MX300 CCD detector (hNAAA precursor) or a Pilatus3 S 6M detector (all others) at the Canadian Macromolecular Crystallography Facility, Canadian Light Source.
Structure Determination.
Data were processed by HKL2000 (81) with autocorrections enabled. This option applies several corrective procedures including ellipsoidal truncation which reduces high-resolution data completeness depending on anisotropy. This is made apparent in the number of reflections and completeness values in “data collection” versus “refinement” in SI Appendix, Table S1. The structure was solved by molecular replacement using the acid ceramidase structure (PDB ID code 5U7Z) as search model with Phaser (82) in Phenix (83), and manually rebuilt in Coot (84). Refinement was carried out by phenix.refine (85) with the following settings: noncrystallographic symmetry restraints for all hNAAA and gpNAAA structures, and translation–libration–screw parameters for the hNAAA precursor and mNAAA. A covalent bond was specified between ARN726 and Cys126. Inhibitor restraints were generated by eLBOW (86). Electron density maps indicated partial oxidation of the mNAAA active site cysteine; this residue was modeled as cysteine sulfonate (SI Appendix, Fig. S3F). However, the active site arrangement was unaffected relative to the other structures. Crystallographic data collection and structure refinement statistics are presented in SI Appendix, Table S1. Structural images were prepared with PyMOL (The PyMOL Molecular Graphics System, version 1.3; Schrödinger, LLC). Electrostatic surface potentials were generated with PDB ID code 2PQR (87) and APBS (88) as part of PyMOL APBS Tools (89). Interface surface areas were calculated with PISA (90).
Liposome-Binding Assays.
Liposomes were prepared by extrusion through 100-nm polycarbonate filters. Anionic liposomes were composed of 50 mol% dimyristoyl phosphatidylcholine, 20% cholesterol, 20% bis(monooleoylglycero)phosphate, and 10% C12 ceramide. Neutral liposomes consisted of 70% dimyristoyl phosphatidylcholine, 20% cholesterol, and 10% PEA. Untagged hNAAA was produced from a construct that contained the tobacco etch virus (TEV) protease cleavage sequence ENLYFQ-SG after the hexahistidine tag, cleaved by TEV protease, and repurified by IMAC and SEC. The tag was removed to prevent its contribution to electrostatic interactions with the charged vesicles. Liposomes at 15 mM total lipid concentration were incubated with untagged hNAAA at 0.25 mg/mL in binding buffer (50 mM sodium acetate pH 4 or sodium acetate pH 5 or Tris⋅HCl pH 7.5, and 50 mM NaCl) for 1 h at 22 °C, washed once in 10 volumes of binding buffer, pelleted, and analyzed by SDS/PAGE. Optionally, desipramine-HCl at 5 or 20 mM was included in the binding buffer.
Enzymatic Activity Assays.
Wild-type and mutant hNAAA were preincubated in assay buffer (50 mM sodium acetate pH 4.5, 100 mM NaCl) with 5 mM DTT for 1 h at 37 °C to allow for full proteolytic self-activation and reduction of the active site cysteine. For small molecule hydrolysis, the enzyme at 10 μM was incubated with 2 mM p-nitrophenyl acetate in assay buffer with 1 mM DTT for 30 min at 37 °C. The p-nitrophenol product was quantified by absorbance measurement at 347 nm. The ester substrate was used instead of its amide counterpart p-nitroacetanilide because no hydrolysis of that compound by NAAA was detected. In the detergent-based PEA hydrolysis assay, the protein at 100–200 nM was incubated with 0.5 mM PEA and 5 mM TX in assay buffer with 1 mM DTT for 1 h at 37 °C. The reaction was stopped at 95 °C for 5 min and the palmitate product was quantified by the free fatty acid fluorometric assay kit (Cayman Chemical). For liposomal PEA hydrolysis, the enzyme at 300 nM was incubated with neutral liposomes at 5 mM total lipid concentration containing 0.5 mM PEA in assay buffer with 1 mM DTT for 1 h at 37 °C. The reaction was stopped at 95 °C for 5 min and TX was added to 20 mM to solubilize lipids before quantification by the free fatty acid fluorometric assay kit. Background fluorescence from control samples containing substrate but no enzyme was subtracted from the experimental samples, for both PEA hydrolysis assays.
Data Availability.
Atomic coordinates and structure factors were deposited into the Protein Data Bank under ID codes 6DXW–6DXZ and 6DY0–6DY3.
Supplementary Material
Acknowledgments
Research described in this paper was performed using beamlines 08ID-1 and 08B1-1 at the Canadian Light Source, which is supported by the Canada Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, the University of Saskatchewan, the Government of Saskatchewan, Western Economic Diversification Canada, the National Research Council Canada, and the Canadian Institutes of Health Research (CIHR). B.N. is supported by an operating grant from the CIHR (Grant MOP-133535).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6DXW–6DXZ and 6DY0–6DY3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811759115/-/DCSupplemental.
References
- 1.Wang J, Ueda N. Biology of endocannabinoid synthesis system. Prostaglandins Other Lipid Mediat. 2009;89:112–119. doi: 10.1016/j.prostaglandins.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Griffin G, Tao Q, Abood ME. Cloning and pharmacological characterization of the rat CB(2) cannabinoid receptor. J Pharmacol Exp Ther. 2000;292:886–894. [PubMed] [Google Scholar]
- 3.Lo Verme J, et al. The nuclear receptor peroxisome proliferator-activated receptor-alpha mediates the anti-inflammatory actions of palmitoylethanolamide. Mol Pharmacol. 2005;67:15–19. doi: 10.1124/mol.104.006353. [DOI] [PubMed] [Google Scholar]
- 4.Petrosino S, Iuvone T, Di Marzo V. N-palmitoyl-ethanolamine: Biochemistry and new therapeutic opportunities. Biochimie. 2010;92:724–727. doi: 10.1016/j.biochi.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Piomelli D, Sasso O. Peripheral gating of pain signals by endogenous lipid mediators. Nat Neurosci. 2014;17:164–174. doi: 10.1038/nn.3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alhouayek M, Muccioli GG. Harnessing the anti-inflammatory potential of palmitoylethanolamide. Drug Discov Today. 2014;19:1632–1639. doi: 10.1016/j.drudis.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Pontis S, Ribeiro A, Sasso O, Piomelli D. Macrophage-derived lipid agonists of PPAR-α as intrinsic controllers of inflammation. Crit Rev Biochem Mol Biol. 2016;51:7–14. doi: 10.3109/10409238.2015.1092944. [DOI] [PubMed] [Google Scholar]
- 8.Sun YX, et al. Involvement of N-acylethanolamine-hydrolyzing acid amidase in the degradation of anandamide and other N-acylethanolamines in macrophages. Biochim Biophys Acta. 2005;1736:211–220. doi: 10.1016/j.bbalip.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Tsuboi K, Takezaki N, Ueda N. The N-acylethanolamine-hydrolyzing acid amidase (NAAA) Chem Biodivers. 2007;4:1914–1925. doi: 10.1002/cbdv.200790159. [DOI] [PubMed] [Google Scholar]
- 10.Ueda N, Puffenbarger RA, Yamamoto S, Deutsch DG. The fatty acid amide hydrolase (FAAH) Chem Phys Lipids. 2000;108:107–121. doi: 10.1016/s0009-3084(00)00190-0. [DOI] [PubMed] [Google Scholar]
- 11.Giang DK, Cravatt BF. Molecular characterization of human and mouse fatty acid amide hydrolases. Proc Natl Acad Sci USA. 1997;94:2238–2242. doi: 10.1073/pnas.94.6.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desarnaud F, Cadas H, Piomelli D. Anandamide amidohydrolase activity in rat brain microsomes. Identification and partial characterization. J Biol Chem. 1995;270:6030–6035. doi: 10.1074/jbc.270.11.6030. [DOI] [PubMed] [Google Scholar]
- 13.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonezzi FT, et al. An important role for N-acylethanolamine acid amidase in the complete Freund’s adjuvant rat model of arthritis. J Pharmacol Exp Ther. 2016;356:656–663. doi: 10.1124/jpet.115.230516. [DOI] [PubMed] [Google Scholar]
- 15.Ueda N, Yamanaka K, Yamamoto S. Purification and characterization of an acid amidase selective for N-palmitoylethanolamine, a putative endogenous anti-inflammatory substance. J Biol Chem. 2001;276:35552–35557. doi: 10.1074/jbc.M106261200. [DOI] [PubMed] [Google Scholar]
- 16.Tsuboi K, et al. Predominant expression of lysosomal N-acylethanolamine-hydrolyzing acid amidase in macrophages revealed by immunochemical studies. Biochim Biophys Acta. 2007;1771:623–632. doi: 10.1016/j.bbalip.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, et al. Expression and secretion of N-acylethanolamine-hydrolysing acid amidase in human prostate cancer cells. J Biochem. 2008;144:685–690. doi: 10.1093/jb/mvn122. [DOI] [PubMed] [Google Scholar]
- 18.Tuo W, et al. Therapeutic potential of fatty acid amide hydrolase, monoacylglycerol lipase, and N-acylethanolamine acid amidase inhibitors. J Med Chem. 2017;60:4–46. doi: 10.1021/acs.jmedchem.6b00538. [DOI] [PubMed] [Google Scholar]
- 19.Bottemanne P, Muccioli GG, Alhouayek M. N-acylethanolamine hydrolyzing acid amidase inhibition: Tools and potential therapeutic opportunities. Drug Discov Today. 2018;23:1520–1529. doi: 10.1016/j.drudis.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Solorzano C, et al. Synthesis and structure-activity relationships of N-(2-oxo-3-oxetanyl)amides as N-acylethanolamine-hydrolyzing acid amidase inhibitors. J Med Chem. 2010;53:5770–5781. doi: 10.1021/jm100582w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrosino S, et al. Diacerein is a potent and selective inhibitor of palmitoylethanolamide inactivation with analgesic activity in a rat model of acute inflammatory pain. Pharmacol Res. 2015;91:9–14. doi: 10.1016/j.phrs.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Impellizzeri D, et al. 2-pentadecyl-2-oxazoline: Identification in coffee, synthesis and activity in a rat model of carrageenan-induced hindpaw inflammation. Pharmacol Res. 2016;108:23–30. doi: 10.1016/j.phrs.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Petracca R, et al. Progress in the development of β-lactams as N-acylethanolamine acid amidase (NAAA) inhibitors: Synthesis and SAR study of new, potent N-O-substituted derivatives. Eur J Med Chem. 2017;126:561–575. doi: 10.1016/j.ejmech.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 24.Petrosino S, et al. 2-pentadecyl-2-oxazoline, the oxazoline of pea, modulates carrageenan-induced acute inflammation. Front Pharmacol. 2017;8:308. doi: 10.3389/fphar.2017.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, et al. Inflammation-restricted anti-inflammatory activities of a N-acylethanolamine acid amidase (NAAA) inhibitor F215. Pharmacol Res. 2018;132:7–14. doi: 10.1016/j.phrs.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Sasso O, et al. The N-acylethanolamine acid amidase inhibitor ARN077 suppresses inflammation and pruritus in a mouse model of allergic dermatitis. J Invest Dermatol. 2018;138:562–569. doi: 10.1016/j.jid.2017.07.853. [DOI] [PubMed] [Google Scholar]
- 27.Solorzano C, et al. Selective N-acylethanolamine-hydrolyzing acid amidase inhibition reveals a key role for endogenous palmitoylethanolamide in inflammation. Proc Natl Acad Sci USA. 2009;106:20966–20971. doi: 10.1073/pnas.0907417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasso O, et al. Antinociceptive effects of the N-acylethanolamine acid amidase inhibitor ARN077 in rodent pain models. Pain. 2013;154:350–360. doi: 10.1016/j.pain.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, et al. Potential analgesic effects of a novel N-acylethanolamine acid amidase inhibitor F96 through PPAR-α. Sci Rep. 2015;5:13565. doi: 10.1038/srep13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren J, et al. Design, synthesis, and biological evaluation of oxazolidone derivatives as highly potent N-acylethanolamine acid amidase (NAAA) inhibitors. RCS Adv. 2018;7:12455–12463. [Google Scholar]
- 31.Alhouayek M, et al. N-acylethanolamine-hydrolyzing acid amidase inhibition increases colon N-palmitoylethanolamine levels and counteracts murine colitis. FASEB J. 2015;29:650–661. doi: 10.1096/fj.14-255208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribeiro A, et al. A potent systemically active N-acylethanolamine acid amidase inhibitor that suppresses inflammation and human macrophage activation. ACS Chem Biol. 2015;10:1838–1846. doi: 10.1021/acschembio.5b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Migliore M, et al. Second-generation non-covalent NAAA inhibitors are protective in a model of multiple sclerosis. Angew Chem Int Ed Engl. 2016;55:11193–11197. doi: 10.1002/anie.201603746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueda N, Kurahashi Y, Yamamoto S, Tokunaga T. Partial purification and characterization of the porcine brain enzyme hydrolyzing and synthesizing anandamide. J Biol Chem. 1995;270:23823–23827. doi: 10.1074/jbc.270.40.23823. [DOI] [PubMed] [Google Scholar]
- 35.Ueda N, Yamanaka K, Terasawa Y, Yamamoto S. An acid amidase hydrolyzing anandamide as an endogenous ligand for cannabinoid receptors. FEBS Lett. 1999;454:267–270. doi: 10.1016/s0014-5793(99)00820-0. [DOI] [PubMed] [Google Scholar]
- 36.Tai T, et al. Endogenous molecules stimulating N-acylethanolamine-hydrolyzing acid amidase (NAAA) ACS Chem Neurosci. 2012;3:379–385. doi: 10.1021/cn300007s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuboi K, et al. Molecular characterization of N-acylethanolamine-hydrolyzing acid amidase, a novel member of the choloylglycine hydrolase family with structural and functional similarity to acid ceramidase. J Biol Chem. 2005;280:11082–11092. doi: 10.1074/jbc.M413473200. [DOI] [PubMed] [Google Scholar]
- 38.Gulyas AI, et al. Segregation of two endocannabinoid-hydrolyzing enzymes into pre- and postsynaptic compartments in the rat hippocampus, cerebellum and amygdala. Eur J Neurosci. 2004;20:441–458. doi: 10.1111/j.1460-9568.2004.03428.x. [DOI] [PubMed] [Google Scholar]
- 39.Hong SB, et al. Molecular cloning and characterization of a human cDNA and gene encoding a novel acid ceramidase-like protein. Genomics. 1999;62:232–241. doi: 10.1006/geno.1999.5953. [DOI] [PubMed] [Google Scholar]
- 40.Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002;298:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- 41.Brannigan JA, et al. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 42.Oinonen C, Rouvinen J. Structural comparison of Ntn-hydrolases. Protein Sci. 2000;9:2329–2337. doi: 10.1110/ps.9.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West JM, et al. Biochemical and mass spectrometric characterization of human N-acylethanolamine-hydrolyzing acid amidase inhibition. PLoS One. 2012;7:e43877. doi: 10.1371/journal.pone.0043877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, et al. Design and synthesis of potent N-acylethanolamine-hydrolyzing acid amidase (NAAA) inhibitor as anti-inflammatory compounds. PLoS One. 2012;7:e43023. doi: 10.1371/journal.pone.0043023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ponzano S, et al. Synthesis, biological evaluation, and 3D QSAR study of 2-methyl-4-oxo-3-oxetanylcarbamic acid esters as N-acylethanolamine acid amidase (NAAA) inhibitors. J Med Chem. 2014;57:10101–10111. doi: 10.1021/jm501455s. [DOI] [PubMed] [Google Scholar]
- 46.Gebai A, Gorelik A, Li Z, Illes K, Nagar B. Structural basis for the activation of acid ceramidase. Nat Commun. 2018;9:1621. doi: 10.1038/s41467-018-03844-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brzozowski AM, et al. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature. 1991;351:491–494. doi: 10.1038/351491a0. [DOI] [PubMed] [Google Scholar]
- 48.Zhao LY, Tsuboi K, Okamoto Y, Nagahata S, Ueda N. Proteolytic activation and glycosylation of N-acylethanolamine-hydrolyzing acid amidase, a lysosomal enzyme involved in the endocannabinoid metabolism. Biochim Biophys Acta. 2007;1771:1397–1405. doi: 10.1016/j.bbalip.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 49.West JM, Zvonok N, Whitten KM, Wood JT, Makriyannis A. Mass spectrometric characterization of human N-acylethanolamine-hydrolyzing acid amidase. J Proteome Res. 2012;11:972–981. doi: 10.1021/pr200735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lakomek K, et al. Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography. BMC Struct Biol. 2009;9:56. doi: 10.1186/1472-6807-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Repo H, Kuokkanen E, Oksanen E, Goldman A, Heikinheimo P. Is the bovine lysosomal phospholipase B-like protein an amidase? Proteins. 2014;82:300–311. doi: 10.1002/prot.24388. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, et al. Amino acid residues crucial in pH regulation and proteolytic activation of N-acylethanolamine-hydrolyzing acid amidase. Biochim Biophys Acta. 2008;1781:710–717. doi: 10.1016/j.bbalip.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Kim Y, Kim S, Earnest TN, Hol WG. Precursor structure of cephalosporin acylase. Insights into autoproteolytic activation in a new N-terminal hydrolase family. J Biol Chem. 2002;277:2823–2829. doi: 10.1074/jbc.M108888200. [DOI] [PubMed] [Google Scholar]
- 54.Kim JK, et al. Crystal structures of glutaryl 7-aminocephalosporanic acid acylase: Insight into autoproteolytic activation. Biochemistry. 2003;42:4084–4093. doi: 10.1021/bi027181x. [DOI] [PubMed] [Google Scholar]
- 55.Yoon J, et al. A bound water molecule is crucial in initiating autocatalytic precursor activation in an N-terminal hydrolase. J Biol Chem. 2004;279:341–347. doi: 10.1074/jbc.M309281200. [DOI] [PubMed] [Google Scholar]
- 56.Huber EM, et al. A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome. Nat Commun. 2016;7:10900. doi: 10.1038/ncomms10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hewitt L, et al. Structure of a slow processing precursor penicillin acylase from Escherichia coli reveals the linker peptide blocking the active-site cleft. J Mol Biol. 2000;302:887–898. doi: 10.1006/jmbi.2000.4105. [DOI] [PubMed] [Google Scholar]
- 58.Bokhove M, et al. Structures of an isopenicillin N converting Ntn-hydrolase reveal different catalytic roles for the active site residues of precursor and mature enzyme. Structure. 2010;18:301–308. doi: 10.1016/j.str.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 59.Buller AR, Freeman MF, Wright NT, Schildbach JF, Townsend CA. Insights into cis-autoproteolysis reveal a reactive state formed through conformational rearrangement. Proc Natl Acad Sci USA. 2012;109:2308–2313. doi: 10.1073/pnas.1113633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suresh CG, et al. Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members. Nat Struct Biol. 1999;6:414–416. doi: 10.1038/8213. [DOI] [PubMed] [Google Scholar]
- 61.Petracca R, et al. Novel activity-based probes for N-acylethanolamine acid amidase. Chem Commun (Camb) 2017;53:11810–11813. doi: 10.1039/c7cc06838g. [DOI] [PubMed] [Google Scholar]
- 62.Yamano Y, et al. Lipophilic amines as potent inhibitors of N-acylethanolamine-hydrolyzing acid amidase. Bioorg Med Chem. 2012;20:3658–3665. doi: 10.1016/j.bmc.2012.03.065. [DOI] [PubMed] [Google Scholar]
- 63.Vandevoorde S, et al. Esters, retroesters, and a retroamide of palmitic acid: Pool for the first selective inhibitors of N-palmitoylethanolamine-selective acid amidase. J Med Chem. 2003;46:4373–4376. doi: 10.1021/jm0340795. [DOI] [PubMed] [Google Scholar]
- 64.Tsuboi K, Hilligsmann C, Vandevoorde S, Lambert DM, Ueda N. N-cyclohexanecarbonylpentadecylamine: A selective inhibitor of the acid amidase hydrolysing N-acylethanolamines, as a tool to distinguish acid amidase from fatty acid amide hydrolase. Biochem J. 2004;379:99–106. doi: 10.1042/BJ20031695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saturnino C, et al. Synthesis and biological evaluation of new potential inhibitors of N-acylethanolamine hydrolyzing acid amidase. Bioorg Med Chem Lett. 2010;20:1210–1213. doi: 10.1016/j.bmcl.2009.11.134. [DOI] [PubMed] [Google Scholar]
- 66.Vago R, Bettiga A, Salonia A, Ciuffreda P, Ottria R. Development of new inhibitors for N-acylethanolamine-hydrolyzing acid amidase as promising tool against bladder cancer. Bioorg Med Chem. 2017;25:1242–1249. doi: 10.1016/j.bmc.2016.12.042. [DOI] [PubMed] [Google Scholar]
- 67.Li Y, et al. Identification of highly potent N-acylethanolamine acid amidase (NAAA) inhibitors: Optimization of the terminal phenyl moiety of oxazolidone derivatives. Eur J Med Chem. 2017;139:214–221. doi: 10.1016/j.ejmech.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 68.Duranti A, et al. N-(2-oxo-3-oxetanyl)carbamic acid esters as N-acylethanolamine acid amidase inhibitors: Synthesis and structure-activity and structure-property relationships. J Med Chem. 2012;55:4824–4836. doi: 10.1021/jm300349j. [DOI] [PubMed] [Google Scholar]
- 69.Ponzano S, et al. Synthesis and structure-activity relationship (SAR) of 2-methyl-4-oxo-3-oxetanylcarbamic acid esters, a class of potent N-acylethanolamine acid amidase (NAAA) inhibitors. J Med Chem. 2013;56:6917–6934. doi: 10.1021/jm400739u. [DOI] [PubMed] [Google Scholar]
- 70.Vitale R, et al. Synthesis, structure-activity, and structure-stability relationships of 2-substituted-N-(4-oxo-3-oxetanyl) N-acylethanolamine acid amidase (NAAA) inhibitors. ChemMedChem. 2014;9:323–336. doi: 10.1002/cmdc.201300416. [DOI] [PubMed] [Google Scholar]
- 71.Fiasella A, et al. 3-aminoazetidin-2-one derivatives as N-acylethanolamine acid amidase (NAAA) inhibitors suitable for systemic administration. ChemMedChem. 2014;9:1602–1614. doi: 10.1002/cmdc.201300546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nuzzi A, et al. Potent α-amino-β-lactam carbamic acid ester as NAAA inhibitors. Synthesis and structure-activity relationship (SAR) studies. Eur J Med Chem. 2016;111:138–159. doi: 10.1016/j.ejmech.2016.01.046. [DOI] [PubMed] [Google Scholar]
- 73.Maccarrone M. Metabolism of the endocannabinoid anandamide: Open questions after 25 years. Front Mol Neurosci. 2017;10:166. doi: 10.3389/fnmol.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Labar G, et al. Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. ChemBioChem. 2010;11:218–227. doi: 10.1002/cbic.200900621. [DOI] [PubMed] [Google Scholar]
- 75.Bertrand T, et al. Structural basis for human monoglyceride lipase inhibition. J Mol Biol. 2010;396:663–673. doi: 10.1016/j.jmb.2009.11.060. [DOI] [PubMed] [Google Scholar]
- 76.Kolter T, Sandhoff K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010;584:1700–1712. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 77.Bracey MH, Cravatt BF, Stevens RC. Structural commonalities among integral membrane enzymes. FEBS Lett. 2004;567:159–165. doi: 10.1016/j.febslet.2004.04.084. [DOI] [PubMed] [Google Scholar]
- 78.Balali-Mood K, Bond PJ, Sansom MS. Interaction of monotopic membrane enzymes with a lipid bilayer: A coarse-grained MD simulation study. Biochemistry. 2009;48:2135–2145. doi: 10.1021/bi8017398. [DOI] [PubMed] [Google Scholar]
- 79.Linke T, et al. Interfacial regulation of acid ceramidase activity. Stimulation of ceramide degradation by lysosomal lipids and sphingolipid activator proteins. J Biol Chem. 2001;276:5760–5768. doi: 10.1074/jbc.M006846200. [DOI] [PubMed] [Google Scholar]
- 80.Anderson N, Borlak J. Drug-induced phospholipidosis. FEBS Lett. 2006;580:5533–5540. doi: 10.1016/j.febslet.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 81.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 82.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lerner MG, Carlson HA. 2006. APBS plugin for PyMOL (University of Michigan, Ann Arbor, MI)
- 90.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors were deposited into the Protein Data Bank under ID codes 6DXW–6DXZ and 6DY0–6DY3.