Significance
Retroviruses have invaded vertebrate hosts during millions of years by occasional germline infections, which were transmitted to the host offspring through generations as inherited endogenous retroviruses (ERVs). Thus, ERVs provide a remarkable record of virus–host interactions. State-of-the-art sequencing methods facilitate ERV identification to decipher their long-term associations with host species. By expanding the scope beyond presence or absence in a single reference genome assembly, it is possible to estimate ERV frequencies and spread in related wild and domestic host populations. Here we demonstrate a considerable ERV makeup variance in wild and domestic rabbit populations, which offers insights into retrovirus–host coevolution and an extension to common strategies in population comparative genomics.
Keywords: endogenous retrovirus, host population, segregation, comparative genomics, evolution
Abstract
Although recent advances in sequencing and computational analyses have facilitated use of endogenous retroviruses (ERVs) for deciphering coevolution among retroviruses and their hosts, sampling effects from different host populations present major challenges. Here we utilize available whole-genome data from wild and domesticated European rabbit (Oryctolagus cuniculus sp.) populations, sequenced as DNA pools by paired-end Illumina technology, for identifying segregating reference as well as nonreference ERV loci, to reveal their variation along the host phylogeny and domestication history. To produce new viruses, retroviruses must insert a proviral DNA copy into the host nuclear DNA. Occasional proviral insertions into the host germline have been passed down through generations as inherited ERVs during millions of years. These ERVs represent retroviruses that were active at the time of infection and thus present a remarkable record of historical virus–host associations. To examine segregating ERVs in host populations, we apply a reference library search strategy for anchoring ERV-associated short-sequence read pairs from pooled whole-genome sequences to reference genome assembly positions. We show that most ERVs segregate along host phylogeny but also uncover radiation of some ERVs, identified as segregating loci among wild and domestic rabbits. The study targets pertinent issues regarding genome sampling when examining virus–host evolution from the genomic ERV record and offers improved scope regarding common strategies for single-nucleotide variant analyses in host population comparative genomics.
Retroviruses have colonized vertebrate host genomes for millions of years by integrating proviral DNA copies as permanent parts in the host germline, which have been passed down to the host offspring through generations as inherited endogenous retroviruses (ERVs) (1). The genomic ERV record represents retroviruses across all currently known retroviral genera at the time of integration and constitutes large fractions of vertebrate genomes today (2–5).
ERVs are identified in host genomes from their genetic structures and sequence motif similarities to exogenous retroviruses (6). Although ERVs do not experience the considerably faster evolution rates of exogenous viruses, they may further be eventually rendered undetectable following their long-term residence within the host genome. ERV contributions to host genome structure and function include shuffling of genomic sequences into new contexts by mediating genomic recombination (1), which also generates ERV isoforms known as solitary long terminal repeats (solo-LTRs) that vastly outnumber persisting full-length ERV loci (7, 8). Overall, the genomic ERV record provides a remarkable source for an evolutionary perspective on virus–host interactions.
Research on ERVs by means of paleovirology has benefited from advancements in sequencing technologies and computational analyses (9). Taking advantage of the growing vertebrate genome assembly catalog, recent studies have focused on comparing ERVs across different host species (4, 5). Population-based analyses of whole genomes should offer deeper insights into ERV–host genome variation.
Identification of genomic structural variation and polymorphisms involving ERVs from unassembled short-sequence reads requires independent ERV libraries, which allow anchoring of reference as well as nonreference ERVs to host genome assembly positions. Along these lines, a recent study could successfully identify specific ERV variants in the human population (10), given large sampling of genomes for recovering ERV loci and limiting overestimation of segregating ERV frequencies. However, the novelty status for each locus depends on the detectable ERV record in the genome assembly of a given host species, which, due to the severe sampling effect introduced by using a single reference individual, does not accurately represent ERV frequencies in the host population. Also, utilizing a single host population could introduce sampling error compared with the entire host species. However, the sampling effect can be mitigated with supporting information from orthologous common variant ERV insertions detected in related host populations. This approach provides additional means for frequency estimates even from lower sampling from each population, under the assumption that shared ERV loci in related host populations are inherited from retroviral genomic invasion of a common host ancestor.
Although a host species, or a population thereof, can be examined by comparative genomics to determine its relevant host–pathogen interactions, genetic disorders, and inherited traits, it is meaningful to extend the scope of analysis to include support from closely related host species and more than one population. To this end, domestic animals offer advantages including sampling from many related populations, often with documented breeding history and known phenotypic changes or other adaptations. Domestic animals also share much of human environment and therefore have been studied extensively in the scope of human diseases (11).
As a means for population-based analysis of whole genome sequences, we apply a computational strategy for ERV identification utilizing unassembled paired-end short-read sequences. The European rabbit (Oryctolagus cuniculus sp.) genome shows high nucleotide diversity and was shaped by multiple evolutionary events, including domestication, hybridization, and subspeciation (12–14). The divergence between rabbit species covers a time frame of about 1 My, and our dataset includes samples from populations across the Iberian Peninsula and southern France. By contrasting ERV loci among wild subspecies populations of Oryctolagus cuniculus cuniculus and Oryctolagus cuniculus algirus to domestic rabbit populations (O. c. cuniculus) as well as the domestic rabbit individual from which the reference assembly version oryCun2.0 was generated (13), we show that although ERV diversity tends to follow the host species’ divergence, a subset of phylogenetically distinct retroviruses expanded along the rabbit host phylogeny. The results indicate standing variation across ERV loci in different host populations that may be utilized to complement conventional comparative genomics strategies currently focusing on single-nucleotide variant analyses for genome-wide association studies.
Results
ERV Identification in Diverse Host Populations.
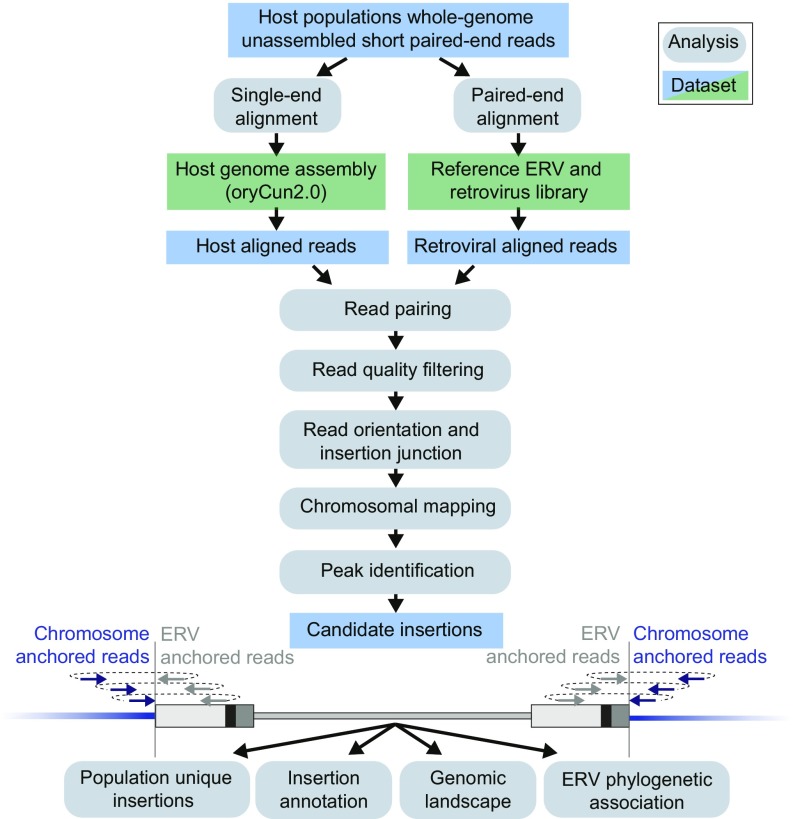
The reference rabbit genome assembly, O. c. cuniculus version oryCun2.0 (13), was analyzed using the RetroTector software (6) to identify 945 ERVs, which were curated for autosomal loci including one or both LTRs flanking the internal proviral sequence, resulting in a reference library containing 567 ERVs (Dataset S1). Additional proviral reference sequences (4, 5) were appended to the rabbit ERV set for use in our computational strategy for identification of candidate ERV insertions in rabbits (Fig. 1). Briefly, unassembled short-read sequences derived from pooled whole genomes sampled from 14 rabbit populations across the Iberian Peninsula, southern France, and the Porto Santo Island, as well as samples from six domestic rabbit breeds and the rabbit individual used to generate the reference assembly (13) were analyzed for reference as well as nonreference ERV insertions (Fig. 2). The paired sequence read coordinates were curated according to the computational strategy (Fig. 1) to reduce signal to noise ratios, after which window read counts were detected as signal peaks indicating candidate ERV and nuclear DNA junctions. Mapping results were assessed by applying false discovery rate-corrected thresholds (FDR = 1:1,000) along autosomal DNA coordinates. These FDR thresholds were calculated for each population based on its read depth and were used for candidate ERV insertion junction identification in that population. If available, presence of ERV-associated sequence reads at orthologous loci in related rabbit populations were then used to rescue candidate ERV identification, at lower thresholds facilitated by cross-population support, for these loci (Datasets S2 and S3).
Fig. 1.
ERV identification in unassembled paired-end short-read sequence libraries. Computational strategy for detecting reference and nonreference assembly ERVs utilizing unassembled paired-end short-read sequence libraries. The output presents a list of candidate insertions (blue data box) anchored to chromosomal positions along host DNA to which downstream analyses can be applied.
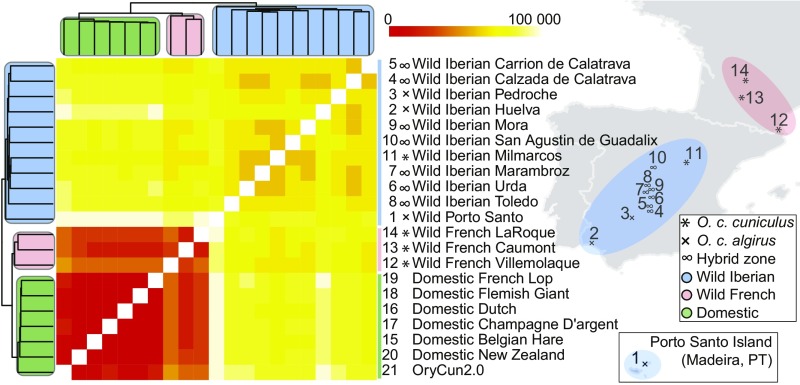
Fig. 2.
Oryctulagus cuniculus sp. geographical distribution and ERV segregation profiles. Hierarchical clustering and heatmap (Dataset S4) illustrating presence or absence of ERV insertion junctions among O. cuniculus sp., sequenced as pooled DNA sampled from wild populations across the Iberian Peninsula, southern France, and the Porto Santo Island, as well as from domestic rabbits (13). The heatmap and cladogram show pairwise differences among individual groups: domestic (green), wild French (red), and wild Iberian (blue). The two subspecies O. c. cuniculus and O. c. algirus are indicated, as well as rabbit populations sampled from a hybrid zone on the Iberian Peninsula (12, 13).
Accuracy for ERV coordinate identification, using the short paired-end read sequences for chromosomal anchoring and dynamic read count thresholds, was determined by data permutation of the observed candidate insertion junctions and compared with coordinates of ERVs identified in the rabbit reference assembly oryCun2.0 (4) using the RetroTector software (6), to about 20× that of random and recovering about 70% of RetroTector identified loci (Chi2, P < 10−15).
Although the available pooled genome sequences (13) did not permit complete reconstruction of exact ERV insertion breakpoints or confident pairing of upstream and downstream insertion junction due to short paired-read insert sizes and sequencing depth estimated below 1× for each genome, we identified 346,225 candidate junctions of which 241,068 were identified in the oryCun2.0 reference individual, demonstrating the benefit of including population data (Dataset S3). Rabbit RepeatMasker consensus sequences matched 37,223 of these 241,068 junctions, with the remaining junctions captured using our large and diverse search library. In summary, 4,061 junctions indicated strongly divergent frequencies, defined as having successful calls in 70% of host populations in one comparison group (i.e., domestic or wild French) and no successful call in the other comparison group. Of these 4,061 loci, 3,007 were more common in domestic rabbits, and 1,054 were more common in wild French rabbits. Rabbit RepeatMasker consensus sequences matched 1,308 of these 4,061 junctions (877 domestic and 431 wild French), whereas 16 of these junctions overlapped with RetroTector identified ERVs in the oryCun2.0 assembly, all in the domestic group. Read counts, calls, and classifications for candidate insertion junctions are presented in Dataset S3.
ERV Segregation in Rabbit Host Populations.
To investigate ERV segregation among rabbit hosts, we performed a hierarchical clustering for presence or absence of ERV junctions across the sampled host populations (Fig. 2 and Dataset S4). The ERVs generally follow the rabbit phylogeny (13), indicating that most ERVs predate the rabbit domestication as well as the divergence of European rabbit subspecies. All domestic rabbits cluster together showing fewer ERV insertion differences (population average 66,918; SD 2,153) compared with other groups, whereas the wild Iberian rabbit populations clade shows the largest ERV insertion diversity (wild French, average 74,697, SD 3,154; Wild Iberian, average 89,761, SD 2,556). The observed pattern indicates reduced genetic diversity among domestic rabbits, which correlates with the previously described O. cuniculus sp. nucleotide diversity (13). We also show that the clade containing wild French populations is more closely related to domestic rabbits than other wild rabbits. These results are consistent with single-nucleotide polymorphism (SNP) data (13), and the pattern is expected because domestication is estimated to have occurred in France about 1,000 y ago. The oryCun2.0 rabbit and the related domestic New Zealand White rabbits form basal branches together in the domestic rabbit population clade. Branches representing wild rabbits from the Porto Santo Island, which is expected to be the most diverged O. c. algirus due to its history (12), form a basal branch to the wild rabbit clade. The Porto Santo Island rabbit population is derived from a female founder and its litters in the mid-15th century, which could explain fixation or loss of rare ERV insertions because of the severe bottleneck.
ERV Confirmation.
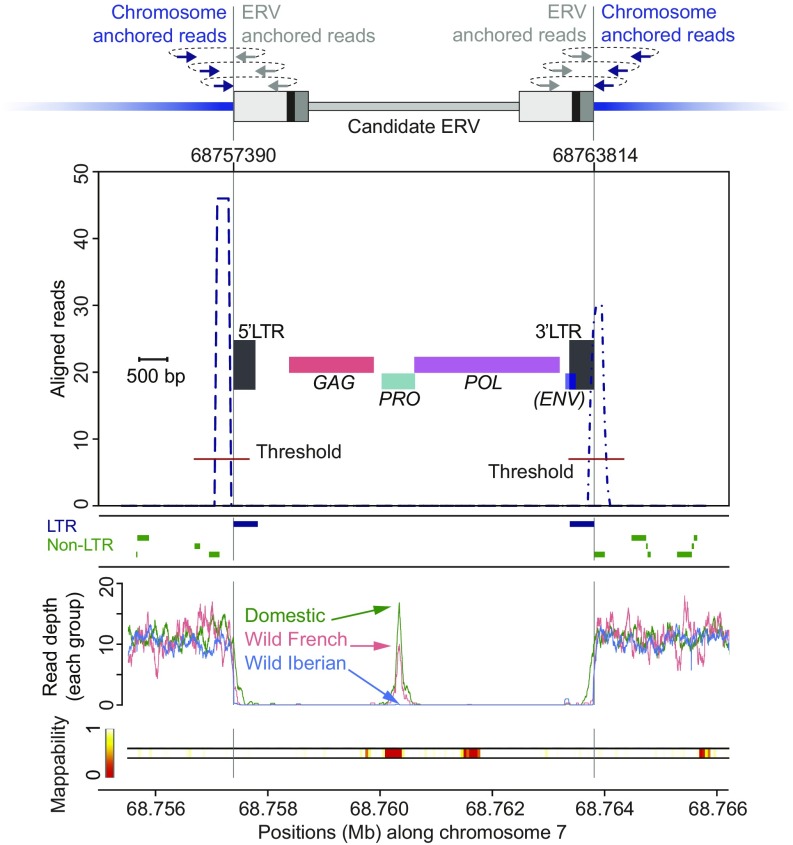
We analyzed a reference assembly ERV locus on chromosome 7 (Fig. 3 and Dataset S1; ERV-id oc1092), which was selected because it was identified in domestic rabbits and RetroTector (6) identified both LTRs as well as most of the internal provirus sequence in the reference assembly. Briefly, unassembled short reads were aligned separately to both the oryCun2.0 assembly and our ERV library for anchoring of ERV-associated read pairs to positions along the rabbit autosomal DNA (Fig. 1). Read pairs were filtered for matching ERV and autosomal DNA junctions before read counts were tallied in sliding windows to generate a graph with distinct peaks at the ERV insertion junctions (Fig. 3). Thresholds were calculated separately for each read count peak based on sample-specific false discovery rates (FDR = 1:1,000) along the host autosomes and adjusted with support from ERV-associated reads identified at the orthologous locus in the related host populations (Dataset S2). The predicted ERV gene structure and LTR coordinates from read count peaks agree with results from RetroTector and the RepeatMasker LTR track (Fig. 3).
Fig. 3.
Candidate ERV. (Top) A schematic shows identification strategy and chromosomal anchoring of ERV-associated sequence read pairs, which demonstrates marked peaks in sliding window counts with their independent dynamic thresholds immediately adjacent to the ERV insertion junctions. The present locus was also identified in the reference rabbit genome assembly (oryCun2.0) using RetroTector, which shows the predicted gene structure and confirms a truncated ERV. The RepeatMasker track for LTRs (blue) and non-LTRs (green) lines up with our identification as well as RetroTector prediction. (Bottom) Average read depth along the host DNA as well as the mappability are summarized. Decreased sequence read depth across the locus is because of poor locus-specific mapping due to similar ERVs elsewhere in the genome, whereas the peak in the middle of the locus indicates a short segment permitting locus-specific sequence read pair mapping, which in this case also allow estimation of segregation at this locus among the domestic, wild French, and wild Iberian rabbit populations (indicated by arrows).
Sequence read depth analysis for the domestic, wild French, and wild Iberian rabbit populations show that this ERV locus could be identified across all domestic rabbits but only at low frequency in the wild French rabbit populations and was completely missing in wild Iberian rabbit populations (Fig. 3). Because the whole-genome sequence data were generated from pooled DNA samples rather than individually sequenced rabbits (13), limited sequencing depth (<1×) for each genome prevents reconstruction of exact ERV insertion breakpoints and hallmarks such as target site duplications (TSDs). However, we infer that the read support for each ERV insertion might serve as an approximation for its allele frequency in a given host population. In the available data, the correlation between read counts and insertion frequency is not complete, because other factors including mappability for sequence reads and read depth fluctuations present confounding factors.
To evaluate the present locus, we collected read depths with the same mapping quality threshold used in our computational strategy (MAPQ > 20 corresponding to about 1:100 error rate for short-read alignments to chromosomal positions) and plotted read count graphs across the entire ERV and flanking DNA as averages for each domestic, wild French, and wild Iberian group next to the short reads mappability track derived from all rabbit populations (Fig. 3). As expected, given the large numbers of similar ERV insertions elsewhere in the host genomes, there is decreased locus-specific mapping inside the ERV, except for a small segment in the middle of this locus (about 2× paired-end read insert length), where locus-specific read mapping was possible. Similar patterns, also showing locus-specific mapping across the entire ERVs at levels comparable to flanking DNA, are common, and mapping specificity depends on presence of similar ERVs elsewhere in the host genome. In agreement with the ERV locus frequency estimations among rabbit populations (above), the graphs show sequence read depth at baseline values for this locus in domestic breeds, whereas sequence read depths were approximately half of the genomic average for wild French and absent among wild Iberian rabbit populations. We interpret these three graph tiers as an indication for incomplete allelic sorting at this locus resulting from standing ERV variation that existed before divergence of the rabbit comparison groups.
Standing ERV Variation Among Host Populations.
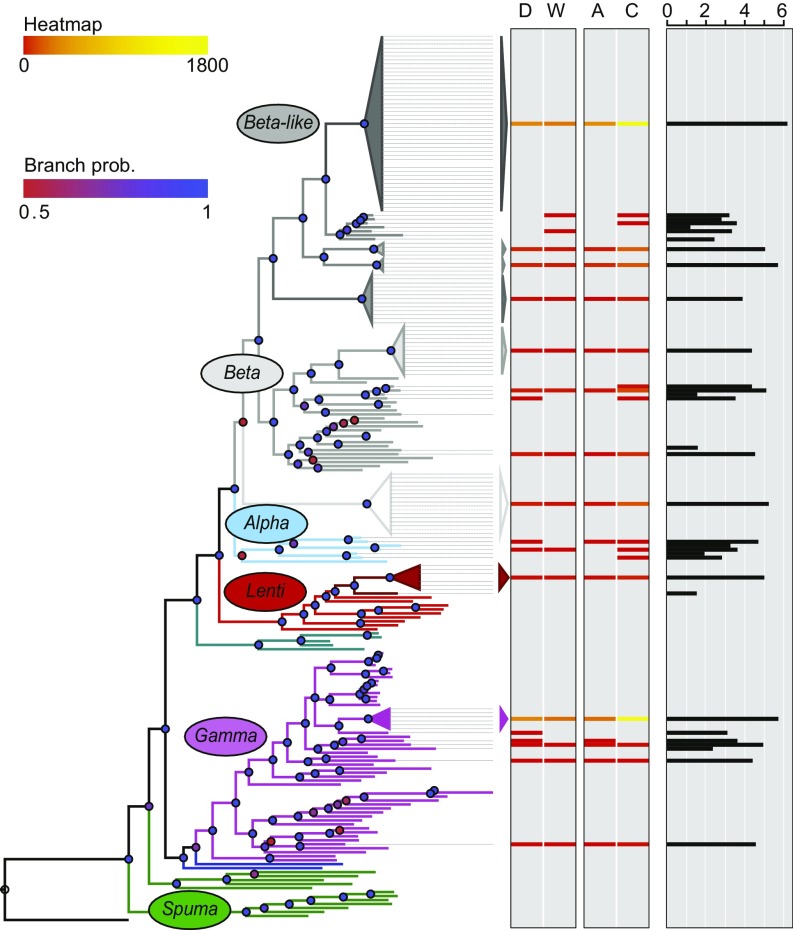
Because paired-end short reads and their insert sizes limit sequence annotation to the immediate ERV–host DNA junctions, we constructed a phylogenetic tree for associating candidate ERV loci by similarities to retroviral reference sequences and ERVs previously identified in the rabbit reference assembly (oryCun2.0) as described by Hayward et al. (4) (Fig. 4). The phylogeny recovers the main tree topology observed in previous studies (4, 5) and shows several rabbit ERV clades associated mainly with Betaretroviruses and Gammaretroviruses, as well as previously described mammalian endogenous Lentiviruses, including the rabbit endogenous lentivirus type K (RELIK) reference sequence (15–17). A radiation of ERVs displaying short terminal branches are associated with the RERVH reference sequence (18) and form a large Betaretrovirus-like crown group (Fig. 4 and Dataset S5), which recently was suggested to have colonized Leporidae host ancestors about 9 Mya (19).
Fig. 4.
ERV phylogenetic association and spread among O. cuniculus sp. Retroviral gag- and pol-based tree derived from reference retroviruses and ERVs where colors indicate retrovirus-like nomenclature as previously described (4, 5). Dashed lines indicate ERVs identified in the rabbit reference genome assembly, oryCun2.0 (Dataset S1). The heatmap shows number of ERV insertions junctions in pairwise comparisons between domestic rabbit populations (D) and wild French rabbit populations (W), as well as between wild O. c. algirus populations (A) and wild O. c. cuniculus populations (C). The overall presence of candidate ERV insertion junctions across all O. cuniculus sp. populations is represented by the black histogram using log10 scale.
The overall ERV insertion junction counts (Fig. 4, black histogram) from the rabbit populations in this study broadly reflect expected number of insertions extrapolated from the oryCun2.0 assembly ERV sequences used to generate the phylogenetic tree (Dataset S1), such that numerous candidate ERVs among the host populations are associated with clades showing large radiations in the phylogenetic analysis. Of ∼200,000 candidate ERV insertion junctions in each of the 20 rabbit populations as well as the single rabbit used to generate the oryCun2.0 assembly (Dataset S3; about 4 million junctions among all rabbit populations), we could associate 3,748,740 insertion junctions by their short paired-end read sequence similarities to proviral taxa represented in the phylogenetic tree (Fig. 4 and Dataset S5). The Betaretrovirus-like clade dominates in the reference genome analysis as well as among all rabbit populations (1,713,446 or about 45% of the ERV insertion junctions), and the Gammaretrovirus-like ERVs could be more common among rabbits than suggested by the phylogenetic analysis based on the single oryCun2.0 assembly. However, because the phylogeny is based mainly on gag and pol motifs (4, 5) and was generated from relatively complete ERVs compared with the candidates insertions identified here, we recognize scope for improvements in future studies involving whole-genome sequences derived from individual hosts to, for example, pair upstream and downstream ERV insertion junctions better in combination with identification of proviral integration hallmarks such as TSDs flanking the proviral insertion, which was unfortunately not feasible using the current dataset.
To investigate potential recent retroviral expansion or coevolution among specific ERVs and rabbit hosts, we identified pairwise ERV insertion differences along the phylogenetic clades across grouped rabbit populations: the domestic and wild rabbit populations as well as the wild O. c. algirus and the wild O. c. cuniculus populations—D vs. W and A vs. C, respectively (Fig. 4, colored heatmap). Although intergroup differences are observed (for example, wild O. c. cuniculus show candidate ERVs absent among wild O. c. algirus, and candidate ERV insertions show differences between domestic O. c. cuniculus and wild O. c. cuniculus), the current data do not permit specific ERV taxa expansions to be distinguished from distantly related ERVs along the phylogeny (Fig. 4). Pairwise rabbit population group differences are detected in clades that have undergone radiation as well as in clades with fewer taxa and longer branch lengths, representing inactivated and truncated ERVs identified from the oryCun2.0 assembly. Intersecting candidate ERV locations with previously identified domestication sweeps (13) support the standing ERV variation pattern because an increased proportion of candidate ERV loci showing divergent frequencies between wild and domestic rabbits (Chi2, P < 10−3) were found inside the domestication sweep regions relative to the genomic average. Locations for divergent ERVs and genes were not correlated, which is also consistent with most ERV insertions differing due to standing genomic variation involving the identified ERV loci in the ancestral host populations rather than recently active replication of specific retroviruses in rabbit hosts.
Discussion
Here we examined wild and domestic rabbit hosts populations (13), using them as a model to identify fixed and segregating ERV loci to decode evolutionary associations among retroviruses and rabbit hosts. Although recent studies have focused on identification of ERVs across diverse reference genome assemblies (4, 5), by expanding this scope to analyze a host population, Wildschutte et al. (10) could recently demonstrate specific ERV variants in the human population. However, sampling from a single host population limits the species-wide estimation of ERV frequencies because of lack of diversity within the sample set. This bias is better addressed by including related host populations in the sampling design, as in this present study, rather than increasing the total number of samples. This strategy minimizes the population-level sampling error at a given total sample size, while preserving power because identification of orthologous ERV loci in one of the populations can rescue the corresponding loci in populations with low sequencing support, assuming that these loci were inherited from a common host ancestor.
ERV analyses utilizing unassembled sequence libraries allow identification of nonreference ERV insertions, which is not possible when comparing reference genome assemblies. However, limited sequence coverage into the candidate ERV hampers detailed locus annotation of insertions missing from the reference assembly. Replacing population pools with individually sequenced host genomes using the same sampling design across populations will provide a better estimate of the ERV allelic variation within and across host populations. More precise ERV frequencies could then be used to determine segregation in the host population and thus present novel insights into potential selection during evolution.
The ERV loci heatmap (Fig. 2) shows that although ERVs tended to segregate along the O. cuniculus sp. phylogeny, wild rabbit populations show higher ERV loci diversity, which could be explained by potential new insertions after rabbit divergence, selection of ERV loci (for or against) in host nuclear DNA from a standing genetic variance of ERV insertions across different breeds, or a combination of these possibilities. The present data do not permit excluding any of these possibilities. However, because our results indicate that ERV insertions are older than the divergence times of the sampled populations (12), it is reasonable to assume that most of the observed differences are due to standing ERV variation in the ancestral host population germline.
Until long-read sequencing technologies for extended ERV coverage using individually sequenced host genomes are readily available, we find a working strategy to associate short-read sequences with reference ERV libraries. The strategy supports extended annotation of reference and nonreference ERVs by, for example, phylogenetic analyses (4, 5), and estimation of retrovirus diversity from identified insertion frequencies in host populations. Individually sequenced genomes using longer reads will also allow better evaluation of allelic variation and discriminating between full-length ERVs and solo LTRs, compared with utilizing insufficient sequencing depth, <1× in the present data, generated from pooled DNA sequencing.
Here we observe more differences between wild O. c. cuniculus and O. c. algirus than between domestic and wild rabbit populations, which could be expected as the evolutionary divergences correspond to about 1.8 My among wild rabbit populations and about 1,000 y for the domesticated rabbits (12–14). Remarkably, we observe host population insertion frequency differences among sequences associated with the Betaretrovirus-like crown group (Fig. 4 and Dataset S5) related to the previously described RERVH (18), recently suggested to have colonized Leporidae host ancestors about 9 Mya (19). These findings support our impression that observed ERV insertion differences broadly reflect standing genomic variation in host populations, and therefore, single population sampling must be sufficient to recover rare and segregating ERV variants. We also observe lentiviral ERV insertion differences, albeit fewer than for other taxa in our phylogenetic analysis, between wild O. c. cuniculus and O. c. algirus, supporting previous estimates that these ERVs represent past retroviral colonization of a Leporidae host ancestor because genomic fragments from these insertions have been dated as old as 12 My (15–17), which appear to segregate in rabbit host populations today. As discussed above, we favor standing ERV variation in the host population genomic makeup for explaining these insertion differences over population-specific retroviral activities because ERV loci segregate within the two wild subspecies but not between domestic and wild rabbit populations. Neither could we identify any candidate replication competent ERV loci in the domestic reference genome assembly, oryCun2.0.
Transposable elements are important for genome architecture and as regulators of diverse biological functions for the host (20), and additional population data could be applied to ERVs for investigating their role in structural variation and effects on host genome function across diverse populations. ERVs further provide improved means for accurate dissection of host genomic associations with diseases and inherited traits by presenting new options for deep comparative analyses within and across related host populations.
An advantage from studying ERVs as markers in comparative genomics in addition to conventional SNP analyses commonly used in genome-wide associations, clinical and population genetics studies (11), is that the ERV variant allele can always be considered the derived allele, whereas determining the ancestral states of single nucleotide differences in a population is complicated. As we demonstrate for O. cuniculus sp., ERVs mainly segregate along host phylogeny, and it is therefore conceivable that ERVs could serve as alternative markers for identifying genetic variants as well as improved dating because ERV fixation is unidirectional with the possibility to estimate time of insertion.
In summary, we demonstrate the advantage of studying multiple related genomes sampled across populations to better understand the genomic variation in a host species. Because comparative genomic studies often rely on SNP analyses for genome-wide associations, we suggest the use of additional genomic markers, such as ERVs along this study, to sort out derived and ancestral allelic variants in diverse host species evolution.
Materials and Methods
Data Analyzed.
Whole-genome Illumina paired-end sequence libraries (76 nucleotides reads with average 200 nucleotides insert sizes) were previously produced for the rabbit used to generate the reference genome assembly (oryCun2.0) as well as 20 wild and domestic rabbit populations using pooled DNA samples for domestic rabbits (OryCun2.0, n = 1; Belgian hare, n = 17; Champagne D’argent, n = 16; Dutch, n = 13; Flemish Giant, n = 18; French Lop, n = 20; New Zealand, n = 16) and wild rabbits (French Caumont, n = 10; French LaRoque, n = 10; French Villemolaque, n = 10; Iberian Pedroche, n = 11; Iberian Calzada de Calatrava, n = 16; Iberian Carrion de Calatrava, n = 17; Iberian Marambroz, n = 16; Iberian Milmarcos, n = 11; Iberian Huelva, n = 16; Iberian San Agustin de Guadalix, n = 20; Iberian Mora, n = 13; Iberian Urda, n = 16; Iberian Toledo, n = 14; Porto Santo, n = 17) to generate ∼10× sequencing coverage for each population as previously described (13).
The reference rabbit genome assembly (oryCun2.0) was analyzed using the RetroTector software (6). High-quality ERVs (Dataset S1) were assembled into a reference ERV library to which additional proviral reference sequences (4, 5) were appended for identification of ERV-associated reads in the unassembled short-read libraries. We also downloaded the oryCun2.0 RepeatMasker track (hgdownload.soe.ucsc.edu/goldenPath/oryCun2/database/) and rabbit RepeatMasker consensus ERVs (https://www.girinst.org/repbase/) for confirmation.
Alignment to ERVs and Host DNA.
Unassembled Illumina read sequences were aligned separately to the reference proviral library (as paired-end reads) and to the rabbit reference assembly, oryCun2.0 (as single-end reads), using multithreading and default settings of the BWA aligner with the MEM option (21) to recover all ERV aligned reads.
Results from single-end read alignments to the proviral library were filtered for matching identified LTRs, within 200 nucleotides from the insertion junction coordinates. Additional filtering was performed to select ERV-associated chimeric read pairs for which the paired reads did not match the proviral library and therefore must align elsewhere in the host DNA.
To identify chromosomal locations for ERV-associated read pairs, mapping quality (MAPQ) > 20, indicating about 1:100 risk of false alignment locations along host DNA for any given read, was applied to the independent sequence read alignments to the oryCun2.0 assembly. The resultant sequence reads were then paired to their corresponding proviral associated reads (in the chimeric read pairs) to identify chromosomal locations of the read pairs covering candidate ERV insertion junctions—one read in nuclear DNA and the corresponding read in the candidate ERV.
Autosomal Locations.
To identify locations for candidate ERV insertions along host autosomes, sequence read pairs were divided depending on their mapping direction in proviral and chromosomal alignments, which indicate orientation of the candidate ERV insertion. Next, coordinates derived from chromosomally anchored sequence reads were used to group ERV-associated sequence read pairs in 250 nucleotide windows determined by the sequence read lengths and insert sizes for maximum covered distance from the ERV insertion into the autosomal DNA.
ERV–host junction FDR was estimated from number of ERV-associated read pairs detected in each population and chance of those reads adding up to a threshold under the assumption that they do not represent locations for ERVs but instead include genomic reads and false chimeric ERV–host read pairs. This procedure aims to adjust candidate calling stringency to the power of the read datasets. Reconstructing complete insertions from the current pooled data (13) is not possible, because there is on average less than 1× coverage of any individual chromosome (see above). More specifically, FDR = 1:1,000 was estimated by tallying the number of reads for each autosomal strand and orientation multiplied by the bin size (250 nucleotides) over the effective autosomal length (considering complete contigs to exclude gaps) to generate an expected count in each bin for random distribution of reads. The resulting expected average was used as λ in a Poisson distribution to estimate the likelihood of occurrence for each possible sequence read count. This likelihood multiplied by the effective autosome length measured in bins resulted in the expected number of detected peaks at a certain threshold. The expected number was compared with the observed number by calculating the ratio of expected/observed.
Thresholds for independent peaks representing candidate ERV insertion junctions in each host population were selected based on the Poisson distribution (above) such that FDR = 1:1,000. Additionally, because random peaks are expected to occur independently in different populations, we could rescue subthreshold candidate ERV insertions based on support across the collection of host populations. ERV loci rescue was achieved by calculating probabilities of finding the same locus twice by chance under the host population Poisson distributions (above), while penalizing for the number of possible pairs. The rescue generated a reduced threshold for each locus, in a population, where support from related populations could be established (Dataset S2).
ERV Identification Accuracy.
To evaluate identification accuracy, we used the number of identified candidate ERVs in each host population (about 200,000; Dataset S3) to perform a permutation test to estimate random recovery of ERV insertions identified by RetroTector in the oryCun2.0 assembly and compare these findings with the observed number of recovered loci from the RetroTector results. Enrichment of recovery over the random expected was evaluated against the Chi2 distribution.
Host Population Pairwise Comparisons.
To compare candidate ERV insertions across host populations, we performed hierarchical clustering and heatmap analysis based on pairwise comparisons for presence or absence of candidate ERV insertion junctions in subsets including all rabbit populations (excluding oryCun2.0 and wild Porto Santo), domestic rabbit populations (domestic Belgian hare, domestic Champagne D’argent, domestic Dutch, domestic Flemish Giant, domestic French Lop, and domestic New Zealand), wild French populations (wild French Caumont, wild French LaRoque, and wild French Villemolaque), O. c. algirus populations (wild Iberian Huelva, wild Iberian Pedroche, and wild Porto Santo), and O. c. cuniculus populations (wild French Caumont, wild French LaRoque, wild French Villemolaque, and wild Iberian Milmarcos).
Pairwise comparisons between grouped rabbit populations were performed where at least 70% of the query population group shared a candidate ERV insertion, whereas different ranges of the corresponding ∂ were applied for the comparison group, ranging from 0 to 0.5 (0.05 increments). Phylogenetic associations of ERV loci segregation were illustrated at ∂ = 0, i.e., presence or absence of ERV-associated reads (22).
To confirm candidate ERV insertion junctions that were identified in only one host population group, we searched in silico to verify that reads were completely missing from 500 nucleotides surrounding the locus and that the candidate insertion located on a single reference genome assembly contig.
Phylogenetic Association of Candidate ERVs.
Because limited sequence information covering the ends of candidate ERV insertions prevents annotation compared with related ERVs and retrovirus sequences, we performed association of best reference ERV hit among sequence reads to taxa present in phylogenetic analysis (Dataset S5). ERVs were extracted from the reference rabbit genome assembly, oryCun2.0, using RetroTector (6), and high-quality ERVs (Dataset S1), which were suitable for phylogenetic reconstruction, were aligned together with reference proviral sequences as previously described by Hayward et al. (4, 5). A retroviral gag- and pol-based phylogenetic tree was then constructed using MrBayes 3.2.4-mpi (23), which initiated from an ExaML (24) tree and ran using eight chains (four swaps) for 22 million generations (sampled every 2,000; relative burn in at 25%) at Gamma substitution rates. The phylogenetic tree (Fig. 4 and Dataset S5) was illustrated using FigTree1.4.3 (tree.bio.ed.ac.uk/software/figtree/).
Candidate ERV insertions were compared with taxa in the phylogenetic tree by sequence read similarity searches using BLAT (25) from which the best result for each query was selected.
Supplementary Material
Acknowledgments
We thank Marcin Kierczak, Nima Rafati, and Miguel Carneiro for discussions. Analyses were performed using the Uppsala Multidisciplinary Center for Advanced Computational Science computer cluster (www.uppmax.uu.se). This work was funded by the Swedish Research Council, Grant VR-M 2015-02429 (to P.J.), and the medical faculty at Uppsala University (P.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815056115/-/DCSupplemental.
References
- 1.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 2.Aiewsakun P, Katzourakis A. Endogenous viruses: Connecting recent and ancient viral evolution. Virology. 2015;479-480:26–37. doi: 10.1016/j.virol.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Farkašová H, et al. Discovery of an endogenous Deltaretrovirus in the genome of long-fingered bats (Chiroptera: Miniopteridae) Proc Natl Acad Sci USA. 2017;114:3145–3150. doi: 10.1073/pnas.1621224114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayward A, Cornwallis CK, Jern P. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc Natl Acad Sci USA. 2015;112:464–469. doi: 10.1073/pnas.1414980112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayward A, Grabherr M, Jern P. Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc Natl Acad Sci USA. 2013;110:20146–20151. doi: 10.1073/pnas.1315419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sperber GO, Airola T, Jern P, Blomberg J. Automated recognition of retroviral sequences in genomic data–RetroTector. Nucleic Acids Res. 2007;35:4964–4976. doi: 10.1093/nar/gkm515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belshaw R, et al. Rate of recombinational deletion among human endogenous retroviruses. J Virol. 2007;81:9437–9442. doi: 10.1128/JVI.02216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gemmell P, Hein J, Katzourakis A. Phylogenetic analysis reveals that ERVs “die young” but HERV-H is unusually conserved. PLOS Comput Biol. 2016;12:e1004964. doi: 10.1371/journal.pcbi.1004964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerman M, Malik HS. Paleovirology–Modern consequences of ancient viruses. PLoS Biol. 2010;8:e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wildschutte JH, et al. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci USA. 2016;113:E2326–E2334. doi: 10.1073/pnas.1602336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meadows JRS, Lindblad-Toh K. Dissecting evolution and disease using comparative vertebrate genomics. Nat Rev Genet. 2017;18:624–636. doi: 10.1038/nrg.2017.51. [DOI] [PubMed] [Google Scholar]
- 12.Rafati N, et al. A genomic map of clinal variation across the European rabbit hybrid zone. Mol Ecol. 2018;27:1457–1478. doi: 10.1111/mec.14494. [DOI] [PubMed] [Google Scholar]
- 13.Carneiro M, et al. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. 2014;345:1074–1079. doi: 10.1126/science.1253714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carneiro M, et al. The genomic architecture of population divergence between subspecies of the European rabbit. PLoS Genet. 2014;10:e1003519. doi: 10.1371/journal.pgen.1003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gifford RJ. Viral evolution in deep time: Lentiviruses and mammals. Trends Genet. 2012;28:89–100. doi: 10.1016/j.tig.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci USA. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keckesova Z, Ylinen LM, Towers GJ, Gifford RJ, Katzourakis A. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology. 2009;384:7–11. doi: 10.1016/j.virol.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths DJ, Voisset C, Venables PJ, Weiss RA. Novel endogenous retrovirus in rabbits previously reported as human retrovirus 5. J Virol. 2002;76:7094–7102. doi: 10.1128/JVI.76.14.7094-7102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Sousa-Pereira P, Abrantes J, Baldauf HM, Esteves PJ. Evolutionary studies on the betaretrovirus RERV-H in the Leporidae family reveal an endogenization in the ancestor of Oryctolagus, Bunolagus and Pentalagus at 9 million years ago. Virus Res. December 5, 2017 doi: 10.1016/j.virusres.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: From conflicts to benefits. Nat Rev Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozlov AM, Aberer AJ, Stamatakis A. ExaML version 3: A tool for phylogenomic analyses on supercomputers. Bioinformatics. 2015;31:2577–2579. doi: 10.1093/bioinformatics/btv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.