Significance
Neuronal functions of long noncoding RNAs (lncRNAs) are poorly understood. Here we describe identification and function of lncRNA GM12371 in regulating synaptic transmission, synapse density, and dendritic arborization in primary hippocampal neurons. GM12371 expression is regulated by cAMP signaling and is critical for the activity regulated synaptic transmission. Importantly, GM12371 is associated with transcriptionally active chromatin and regulates expression of several genes involved in neuronal growth and development. Taken together, these results suggest that GM12371 acts as a transcriptional regulator of synapse function.
Keywords: long noncoding RNA, synaptic plasticity, synapse morphology, synapse function, gene expression
Abstract
Despite the growing evidence suggesting that long noncoding RNAs (lncRNAs) are critical regulators of several biological processes, their functions in the nervous system remain elusive. We have identified an lncRNA, GM12371, in hippocampal neurons that is enriched in the nucleus and necessary for synaptic communication, synapse density, synapse morphology, and dendritic tree complexity. Mechanistically, GM12371 regulates the expression of several genes involved in neuronal development and differentiation, as well as expression of specific lncRNAs and their cognate mRNA targets. Furthermore, we find that cAMP-PKA signaling up-regulates the expression of GM12371 and that its expression is essential for the activity-dependent changes in synaptic transmission in hippocampal neurons. Taken together, our data establish a key role for GM12371 in regulating synapse function.
One of the most important challenges in modern molecular neurobiology is to understand the dialogue between genes and synapses. Decades of studies have led to the identification of key molecular players that govern synapse function. For example, neurotransmitter receptors at postsynaptic compartments (1–3), synaptic vesicle release machinery at presynaptic compartments (4, 5), various pre- and postsynaptic scaffolding proteins (1–3, 6), transsynaptic signaling (7), and translation machinery at the synapse (8–10) have been identified. In the nucleus, expression of specific genes and remodeling of chromatin (11–16) are known to regulate synapse function. However, the molecular underpinnings of how changes in gene expression results in regulating synapse function remain to be understood in detail. Particularly, we know little about the contribution of the noncoding transcriptome in regulating synapse function. The noncoding transcriptome is highly diverse and includes ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), microRNAs (miRNAs), and long noncoding RNAs (lncRNAs) (17–22). Among these, the neurobiology of lncRNAs remains the least understood. A large number of lncRNAs have now been discovered as a consequence of unbiased sequencing of genomes and transcriptomes (23–27).
lncRNAs are transcripts that are more than 200 nucleotides long and are important regulators of gene expression (23–28). An increasing number of functional studies have established that lncRNAs regulate almost every stage of gene expression, from epigenetic modifications in the nucleus (29–31) to messenger RNA (mRNA) stability (32–34) and translation (35) in the cytoplasm. Recent studies have also profiled lncRNA metabolism profiles relative to mRNAs, demonstrating that lncRNAs differ in their half-lives, posttranscriptional modifications, subcellular localization patterns, and sequence conservation (19, 36). Notably, while the promoters for lncRNAs show high sequence conservation, the gene products show less conservation, although the presence of lncRNA orthologs across species (37, 38) have been documented. Despite this trend, lncRNAs have been shown to hold important regulatory functions in mammalian neurons. Previously, it has been shown that Malat1—a 7-kb single exon lincRNA—is highly enriched in nuclear speckles in the mouse hippocampus. This transcript, while showing low sequence conservation between rodents and humans, regulates alternative splicing of genes involved in nuclear organization and neuronal function (39). It has also been shown that a natural antisense transcript of BDNF can regulate BDNF expression in the mouse hippocampus, and shows only partial conservation within the BDNF overlapping region (40). Furthermore, the noncoding RNA BC1 regulates fragile X mental retardation protein-mediated protein synthesis in dendrites (41). While these discoveries suggest the significance of lncRNAs in the brain, fundamental gaps remain in our understanding of whether and how lncRNAs regulate synaptic transmission, synaptic architecture, and synaptically relevant protein-coding gene expression.
Here we describe the identification and characterization of an lncRNA, GM12371. We find that GM12371 is critical for synapse function in hippocampal neurons. Inhibition of its function resulted in a decrease in synaptic transmission, total synapse density, number of mushroom spines, and dendritic arborization. We also identified molecular targets of this lncRNA; among them are two lncRNAs that regulate expression of synaptically pertinent mRNAs in cis. Furthermore, we find that expression of GM12371 is regulated by cAMP-PKA signaling and that expression of GM12371 is necessary for the activity-dependent changes in synaptic transmission.
Results
Discovery of GM12371.
Our previously published work has uncovered differentially expressed lncRNAs in the hippocampus, as well as hippocampal subregions (23). Using lncRNAs identified in this study, we employed a candidate approach by quantitative real-time PCR (qPCR) to identify lncRNAs expressed in mouse primary hippocampal neurons. We identified lncRNA GM12371, transcribed from chromosome 4 (Fig. 1A), as expressed in primary hippocampal neurons. We next confirmed its neuronal expression by FISH using a digoxigenin (DIG)-labeled 300-nt-long riboprobe (SI Appendix, Fig. S1A) and found that GM12371 is mostly localized in the nucleus.
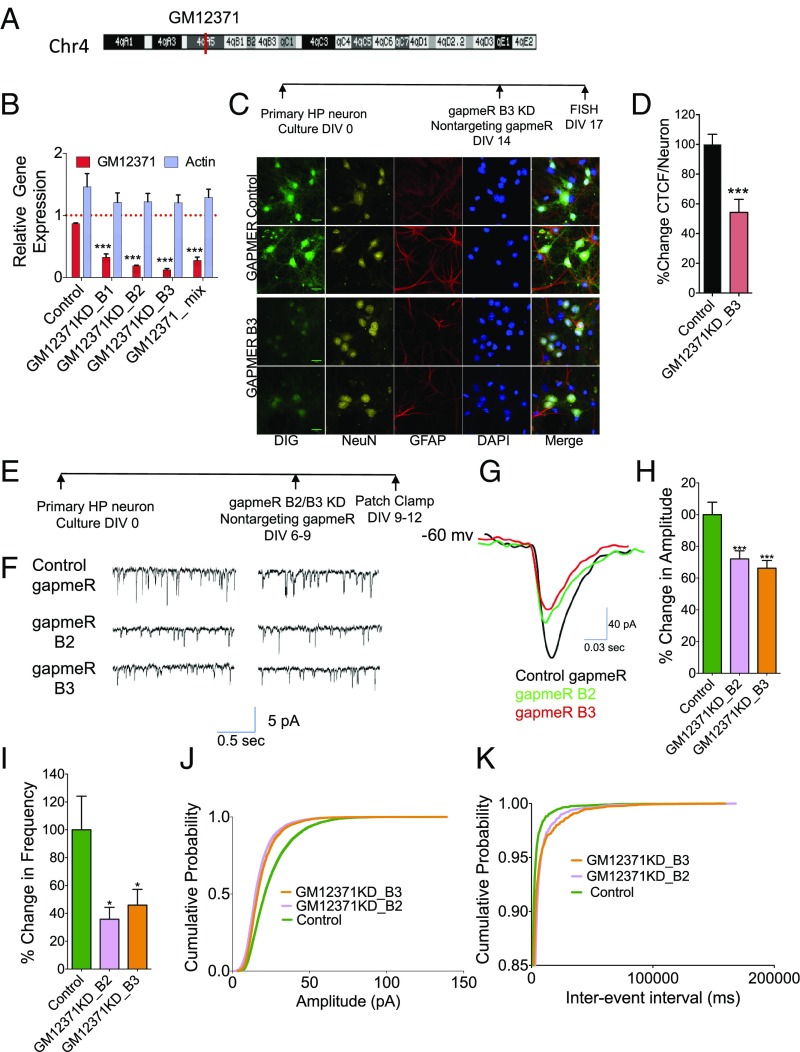
Fig. 1.
Expression of lncRNA GM12371 is required for sEPSCs of synapses of hippocampal neurons. (A) Chromosomal position of GM12371. (B) Analysis of knockdown of GM12371 by three different gapmeRs and a gapmeR mix. A nontargeting gapmeR was used as specificity control. Expression level of actin mRNAs was used to assess nonspecific targeting effect of gapmeRs. Error bars are SEM, ***P < 0.0001, one-way ANOVA followed by Tukey test. (C) FISH analysis of knockdown of GM12371 by gapmeR B3 in primary hippocampal neurons. A nontargeting gapmeR was used as control for knockdown. A DIG-labeled riboprobe (SI Appendix, Fig. S1) was used to visualize cellular localization of GM12371. Two representative confocal projection images are shown for each condition. (Scale bars, 20 µm.) (D) Quantification of FISH data shown in C. Number of neurons analyzed: nontargeting gapmeR control = 166, GM12371 knockdown = 180, Error bars are SEM, ***P < 0.0001, Student’s t test. (E) Experimental outline for patch clamp electrophysiology to measure sEPSCs. (F) Two representative traces of sEPSC measurements in hippocampal neurons following control or GM12371 knockdown (KD) using gapmeRs. (G) Representative traces of sEPSCs recording show a decrease in sEPSC amplitudes in gapmeR B2 (green) and gapmeR B3 (red) groups relative to the control gapmeR (black). (H and I) Quantitation of changes in sEPSC amplitude and frequency, respectively. Bar graphs show percent changes in amplitude/frequency of sEPSCs. Error bars are SEM, *P < 0.05, ***P < 0.0001, one-way ANOVA followed by Tukey test. (J and K) Cumulative probability of changes in sEPSC amplitude and frequency respectively following GM12371 knockdown by gapmeRs. (Control gapmeR, n = 37; gapmeR B2, n = 35; gapmeR B3, n = 35). HP, hippocampus.
To explore the potential role of GM12371, we first asked whether its expression is necessary for synaptic communication in primary hippocampal neurons. We carried out loss-of-function experiments by depletion of GM12371 using locked nucleic acid long RNA gapmeR oligonucleotide (gapmeR)-assisted knockdown (42, 43) in hippocampal neurons. To assess the efficiency of knockdown, we used three different gapmeRs against GM12371, as well as a mix that contained all three gapmeRs. A nontargeting gapmeR was used as a specificity control. qPCR analysis in Fig. 1B shows that the gapmeRs we synthesized could specifically knockdown GM12371 in hippocampal neurons [fold-change in GM12371 levels compared with control nontargeting gapmeR: 0.88 ± 0.012, gapmeR B1 0.32 ± 0.05, B2 0.19 ± 0.01, B3 0.12 ± 0.02, mix (B1 + B2 + B3) 0.27 ± 0.05; fold-change in actin mRNA levels compared with vehicle control: nontargeting gapmeR 1.5 ± 0.2, gapmeR B1 1.2 ± 0.2, B2 1.2 ± 0.0.14, B3 1.2 ± 0.0.13, mix (B1 + B2 + B3) 1.3 ± 013; n = 4 for all; P < 0.05, one-way ANOVA followed by Tukey test] (Dataset S1, Table S1). We then confirmed the gapmeR-mediated knockdown of GM12371 in hippocampal neurons using FISH analysis (Fig. 1 C and D) (percent mean fluorescence intensity of GM12371 staining in neurons following knockdown by gapmeR B3 compared with nontargeting gapmeR: 52.4 ± 8.7, P < 0.05; n = 20; unpaired two-tailed t test) (Dataset S1, Table S1).
To examine whether GM12371 has a critical role in synaptic communication, we measured the effect of knockdown of GM12371 on spontaneous excitatory postsynaptic currents (sEPSCs), using whole-cell patch-clamp recordings (Fig. 1 E and F). To knock down GM12371, we used two different gapmeRs (B2 and B3) (SI Appendix, Fig. S1B) identified from the above experiments. A nontargeting gapmeR was used as a specificity control. As shown in Fig. 1 F–K, we found that both gapmeRs produced a decrease in sEPSCs. Specifically, we observed a decrease in the amplitude and frequency of sEPSCs following GM12371 knockdown compared with the nontargeting gapmeR control (amplitude: control gapmeR, n = 37, 100 ± 7.8% vs. gapmeR B2 knockdown, n = 35, 72.12 ± 5.07%, and gapmeR B3 knockdown, n = 35, 66.24 ± 4.86%, P = 0.0003139, one-way ANOVA followed by Tukey test; frequency: control, n = 37, 100 ± 24.12%, gapmeR B2 knockdown, n = 35, 35.79.12 ± 8.52%, and gapmeR B3 knockdown, n = 35, 45.94 ± 11.29%, P = 0.01418, one-way ANOVA followed by Tukey test).
Because the amplitude of EPSCs is related to the postsynaptic strength, whereas the frequency is correlated to the number of functional synapses and to the presynaptic release machinery, these results suggest that GM12371 has a significant role in excitatory synaptic transmission in hippocampal neurons. We then calculated the multiplicative shift in sEPSCs and found that sEPSC events from neurons with gapmeR B2 knockdown and with gapmeR B3 knockdown were decreased by a multiplicative factor of 0.72 and 0.66, respectively (compared with control gapmeR). It was previously shown that a multiplicative shift in the cumulative probability fraction of EPSC amplitudes is indicative of a cell-wide change in synaptic strength (44, 45). Taken together, these results suggest that normal expression of GM12371 is necessary for maintaining excitatory synaptic transmission in hippocampal neurons.
GM12371 Expression Regulates Spine Density and Dendritic Tree Complexity.
The changes we observed in synaptic communication with GM12371 knockdown suggested specific disruptions at the synapse, such as reduction in spine density or a specific change in spine morphology. Alternatively, the reduction in synaptic transmission may be indicative of more subtle molecular defects at pre- and postsynaptic compartments. Therefore, to understand the mechanism underlying the regulation of synaptic transmission by GM12371, we first assessed whether knockdown of GM12371 can produce structural changes in hippocampal neurons. To address this, hippocampal neurons were transfected with pEGFPN1 to visualize neuronal architecture and confocal projection images were collected (Fig. 2A). Using confocal live cell imaging, we examined spine morphology and assessed spine density and spine type in GFP-labeled hippocampal neurons following knockdown of GM12371 by the two gapmeRs we used in the electrophysiology experiments. The image analyses (Fig. 2 B–D) showed that knockdown of GM12371 produced a decrease in total synapse density in hippocampal neurons, a specific decrease in mushroom and stubby spines, and an increase in thin spines (total spine density expressed as percentage of EGFP control; nontargeting gapmeR: 90.6 ± 1.6; gapmeR B2: 58.1 ± 1.7; gapmeR B3: 60.7 ± 3.1, P < 0.05, unpaired two-tailed t test; spine morphology: mushroom, stubby, thin spines: EGFP control 48.2 ± 5.5; 5.9 ± 1.8; 45.8 ± 3.9; nontargeting control gapmeR: 41.5 ± 2.8; 17.3 ± 3; 41.2 ± 3.8; gapmeR B2 25.5 3; 15.4 ± 4.6; 58.9 ± 2.1; gapmeR B3 30.6 ± 2.3; 16.5 ± 3.5; 52.9 ± 3.1 P < 0.05 for mushroom and thin spines, one-way ANOVA followed by Tukey test) (Dataset S1, Table S2) following GM12371 knockdown.
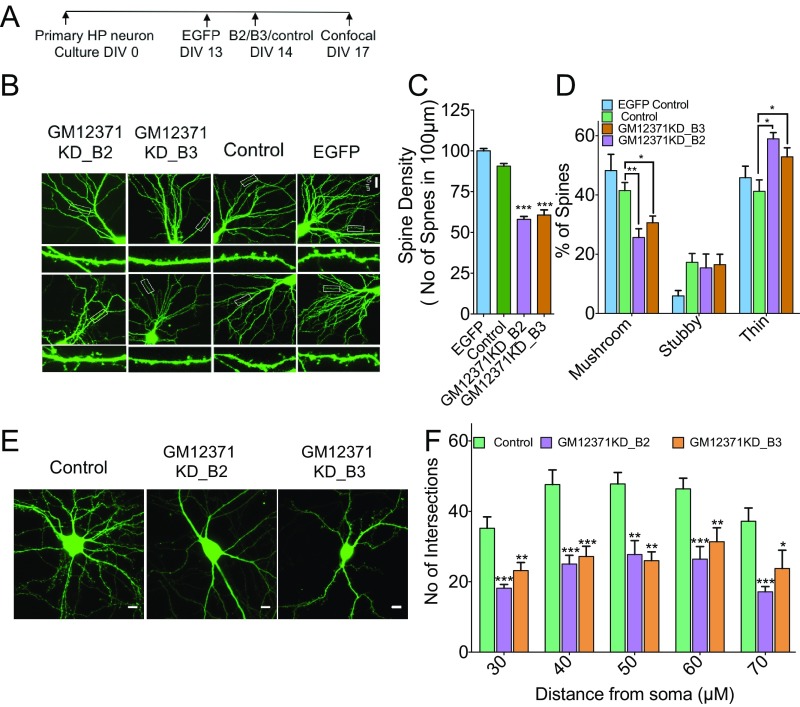
Fig. 2.
GM12371 regulates spine density, spine morphology, and dendritic tree complexity in hippocampal neurons. (A) Experimental outline. (B) Two representative confocal projection images of spines collected at different conditions are shown. (Scale bar, 20 µm.) Two different gapmeRs were used to knock down GM12371. A nontargeting gapmeR was used as control for knockdown. (C) Quantification of total spine density. Bar graphs show number of spines per 100 μm of distal dendrites quantified in EGFP control, control gapmeR, and gapmeR B2 and B3 knockdown neurons. (D) Quantitation of specific changes in spine morphology. Number of neurons analyzed for EGFP control = 20, gapmeR control = 18, gapmeR B2 = 17, gapmeR B3 = 24. (E) Sholl analysis to assess the effect of GM12371 knockdown on dendritic tree complexity. Shown are confocal projection images of EGFP expressing hippocampal neurons following transfection by nontargeting gapmeR (neg control), gapmeR B2 (GM12371 KD_B2), and B3 (GM12371 KD_B3) to specifically knock down GM12371. (Scale bars, 20 μm.) (F) Bar graphs show number of intersections at varying distances from soma. Number of neurons analyzed for gapmeR control = 18, gapmeR B2 = 18, gapmeR B3 = 25. Error bars are SEM, *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA followed by Tukey test.
Because we observed a major change in spine morphology with GM12371 knockdown, we asked whether GM12371 expression might be critical for dendritic tree complexity. As described in the spine analysis experiments above, hippocampal neurons were transfected with pEGFPN1 to visualize neuronal architecture and confocal projection images of dendritic arbors were collected for Sholl analysis (46). Intriguingly, GM12371 knockdown produced a significant reduction in dendritic tree complexity (Fig. 2 E and F) (distance from soma 30, 40, 50, 60, and 70 µm; number of intersections EGFP control: 35.2 ± 3,2; 47.6 ± 4.2; 47.8 ± 3.2; 46.4 ± 3; 37.2 ± 3.7; gapmeR B2: 18.1 ± 1.1; 25 ± 2.5; 27.7 ± 4; 26.4 ± 3.6; 17.1 ± 1.5; gapmeR B3: 23.2 ± 2.3; 27.2 ± 2.9; 26 ± 2.5; 31.4 ± 3.9; 23.75 ± 5.2, n = 12, P < 0.05, one-way ANOVA followed by Tukey test) (Dataset S1, Table S2). To assess whether the morphology changes that we observed in day in vitro (DIV) 17 neurons also occur in younger neurons that we used in the electrophysiology experiments, we carried out imaging analysis as described in Fig. 2. Data shown in SI Appendix, Fig. S2 suggest that expression of GM12371 is also required for dendritic arborization and spine density in DIV 10–12 neurons. Taken together, these results suggested that normal expression of GM12371 is essential for spine density and morphology and dendritic tree complexity.
Molecular Targets of GM12371 Reveal a trans Mechanism of Action.
We next investigated the molecular mechanism underlying GM12371 function in neurons. Because of its nuclear localization, we considered its role as a regulator of transcription. As described earlier, several studies have shown that lncRNAs impact cell function by modulating transcription of their RNA targets that may lie within close proximity of the lncRNA transcription site (cis regulation) or elsewhere in the genome (trans regulation) (25, 27, 28). Therefore, to understand the molecular basis of the neuronal phenotypes we observed due to GM12371 knockdown, using the University of California, Santa Cruz (UCSC) Genome Browser (mm10) we first searched for transcripts that are within 200 kb of the GM12371 locus, on chromosome 4. Changes in the levels of specific transcripts within the 200-kb region of GM12371 following its knockdown would suggest cis regulation of targets by GM12371. The only such gene we identified within its genomic locus was the protein-coding gene lingo2. However, our qPCR results showed that GM12371 knockdown did not produce any significant change in the lingo2 mRNA levels (SI Appendix, Fig. S3 and Dataset S1, Table S3), suggesting that GM12371 likely modulates the expression of its molecular targets by a trans mechanism. To address the possibility of trans regulation of gene expression, we analyzed global changes in the neuronal transcriptome using next-generation RNA sequencing (RNAseq) following GM12371 knockdown. Differential expression analysis between GM12371 knockdown and control yielded 908 mRNAs and 7 lncRNAs (n = 3 biological replicates; adjusted P < 0.05) (Dataset S1, Tables S4–S7) in hippocampal neuronal cultures. Circos plot analysis of the RNAseq data show that these mRNAs and lncRNAs are transcribed from different chromosomes (Fig. 3A), suggesting a genome-wide effect, and possibly a trans mechanism underlying GM12371 function.
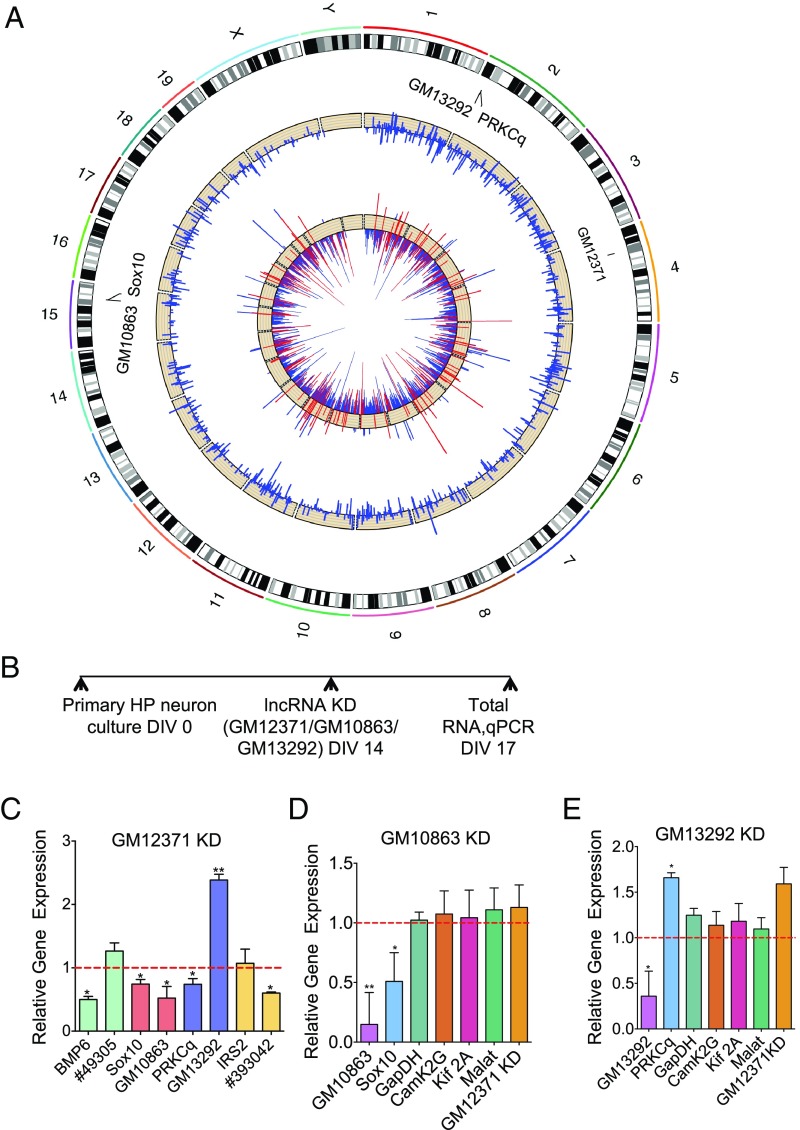
Fig. 3.
Mechanism of action of GM12371. (A) Circos plot showing genome-wide differentially expressed lncRNAs and mRNAs from RNAseq analysis (n = 3) of hippocampal neurons transfected with gapmeR B2 to knock down GM12371 and a control nontargeting gapmeR. The outer ring displays the chromosomes annotated with GM12371 and two lncRNA:mRNA pairs. The middle track shows log2 relative changes of lncRNAs, and the inner track shows log2 relative change of protein-coding genes belonging to three selected functions (cellular development, growth, and differentiation in red) and all other protein-coding genes (blue). (B) Experimental outline for assessing regulation of lncRNA:mRNA pairs by qPCR. (C–E) qPCR analysis of lncRNAs and mRNAs following GM12371 knockdown n = 5 (C), lncRNA GM10863 knockdown, n = 6 (D), and GM13292 knockdown n = 6 (E). Expression of GAPDH, Kif2A, and CamK2G were used as controls for nonspecific effects of gapmeRs in D and E. Data normalized to 18S rRNA levels. Error bars are SEM, *P < 0.05, **P < 0.01, unpaired two-tailed t test.
We then validated the RNAseq data by analyzing 19 differentially expressed mRNAs (10 down-regulated and 9 up-regulated) by qPCR following knockdown of GM12371. Consistent with the RNAseq data, qPCR analyses of these mRNAs also showed a differential expression pattern following GM12371 knockdown (SI Appendix, Fig. S4 and Dataset S1, Table S8) (n = 6, P < 0.05, unpaired two-tailed t test).
Indicative of the lack of lncRNA characterization in neurons, none of the differentially expressed lncRNAs we identified are known to have any specific neuronal function. To understand the biological significance of the differentially expressed mRNAs, we looked to see if any pathways were either down- or up-regulated with GM12371 knockdown. Ingenuity pathway analysis of differentially expressed mRNAs (Dataset S1, Tables S9–S12) have identified three canonical pathways: glutamate receptor signaling, bone morphogenetic protein (BMP) signaling, and synaptic long-term depression, which are significantly down-regulated (P < 0.05) following GM12371 knockdown. We next analyzed RNAseq data by gene ontology (GO) analyses and identified molecules involved in cell–cell signaling, cellular development, maintenance, nervous system development and function, and tissue morphology that are down-regulated (SI Appendix, Figs. S5 and S6) following GM12371 knockdown. These pathways are indicative of the structural and morphological phenotypes we observed upon knockdown of GM12371, suggesting that GM12371 regulates expression of specific genes that are critical for synapse function.
To better understand the trans regulation mechanism of gene expression by GM12371, we next studied RNA targets of differentially expressed lncRNAs we identified from RNAseq data. Using the UCSC Genome Browser we searched for lncRNA:mRNA pairs (mRNAs located within 200 kb of the lncRNA gene body). We then examined whether these mRNAs are also differentially expressed due to knockdown of GM12371 (Dataset S1, Tables S4–S7). This analysis identified four lncRNA:mRNA pairs of potential interest (49305:BMP6, GM10863:Sox10, GM13292:PRKCq, 393042:IRS2).
To assess whether these lncRNA:mRNA pairs are indeed targets of GM12371, we examined GM12371 knockdown-induced changes in gene expression using qPCR (Fig. 3B and Dataset S1, Table S13). Fig. 3C shows that the expression levels of BMP6, Sox10, and PRKCq (protein kinase C θ) are down-regulated upon GM12371 knockdown (normalized fold-change compared with control gapmeR: BMP6 0.5 ± 0.05; Sox10: 0.5 ± 0.07; PRKCq 0.74 ± 0.09, n = 5, P < 0.05, unpaired two-tailed t test), whereas expression of IRS2 did not change. Among the four lncRNA candidates, the expression levels of three (GM10863, GM13292, 393042) changed significantly (normalized change compared with control gapmeR: GM 10863 0.52 ± 0.18; GM13292 2.4 ± 0.09; 393042 0.6 393042 0.01, n = 5, P < 0.05, unpaired two-tailed t test) with GM12371 knockdown. Importantly, these results show that expression of Sox10 mRNA is positively correlated with the expression of its cognate lncRNA GM10863, whereas expression of PRKCq is negatively correlated with the expression of its cognate lncRNA GM13292. The expression of lncRNA (#49305) for BMP6 did not change significantly with GM12371 knockdown (normalized change compared with control gapmeR 1.2 ± 0.13). Similarly, IRS2 mRNA did not change (normalized change compared with control gapmeR 1.07 ± 0.22), although levels of its putative lncRNA changed with GM12371 knockdown.
We thus focused our efforts on the two lncRNA:mRNA pairs validated by qPCR. Sox10 is a transcription factor known to be involved in neuronal crest differentiation (47–50) and PRKCq is involved in modulating synapse elimination (51). Recently, the Sox family member, Sox5, has been implicated in autism spectrum disorder (52). However, the regulation of Sox10 and PRKCq expression by lncRNAs in neurons remains unknown.
We first examined whether gapmeR-mediated knockdown of GM10863 and GM13292 lncRNAs would result in the predicted corresponding expression of the respective mRNA targets (Sox10 and PRKCq) by qPCR (Fig. 3B). GAPDH, Camk2G, Kif2A, lncRNA MALAT1, and GM12371 levels were used as controls (Dataset S1, Table S13). Consistent with our hypothesis that GM10863:Sox10 and GM13292:PRKCq are indeed lncRNA:mRNA pairs, we found that knockdown of GM10863 specifically reduced the expression of Sox10 mRNA and that knockdown of GM13292 enhanced the expression of PRKCq mRNA (Fig. 3 D and E) (n = 6; P < 0.05, unpaired two-tailed t test). Taken together, our results indicate that these two lncRNA:mRNA pairs are regulated by GM12371 by a trans mechanism.
GM12371 Is Associated with Transcriptionally Active Chromatin.
Given its nuclear localization, we hypothesized that GM12371 regulates transcription by direct association with chromatin. We asked whether neuronal activation could lead to recruitment of GM12371 RNA to the transcriptionally active chromatin. Therefore, we carried out native chromatin immunoprecipitation (ChIP) (53) experiments and isolated both DNA and RNA from immunoprecipitated complexes (Fig. 4 A and B). Specifically, we analyzed the H3K27ac modification, which has been previously studied in activity-dependent hippocampal function (54). We immunoprecipitated H3K27ac complexes from hippocampal neurons exposed to forskolin that increased cAMP levels leading to transcriptional changes. Forskolin has been used to elicit chemically induced long-term potentiation in neuronal cultures (55) as well as in hippocampal slices (56). To assess the promoter of GM12371, we utilized publicly available ENCODE data from mouse early-postnatal forebrain H3K27ac ChIP (Gene Expression Omnibus accession no. GSE82428) and designed primers spanning the region upstream of the transcriptional start site with the most sequencing coverage. As a positive control, we also measured enrichment of the cFos promoter. IgG alone in ChIP assays were used to normalize data. We find that the GM12371 promoter was enriched in H3K27ac complexes immunoprecipitated from forskolin-treated neurons (Fig. 4C) (fold-change compared with DMSO, cFos: 1.9 ± 0.26; GM12371: 1.7 ± 0.14, n = 5; unpaired two-tailed t test, P < 0. 01). We then purified chromatin-associated RNAs from H3k27ac ChIP and assessed the abundance of GM12371 lncRNA using cFos RNA as a negative control. Consistent with its role in transcription, we find that GM12371 is enriched in transcriptionally active chromatin (Fig. 4D) (fold-enrichment in forskolin compared with DMSO 2.9 ± 0.39, n = 5, unpaired two-tailed t test, P < 0.01). Despite its being an immediate early gene, cFos RNA was not detected, indicating that the RNAs associated with the complexes are not nascent transcripts, but are likely regulatory.
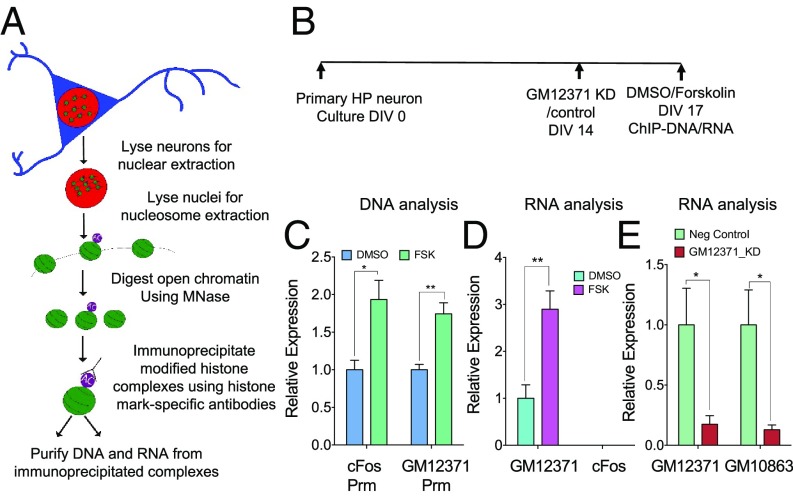
Fig. 4.
Forskolin regulates the association of GM12371 to transcriptionally active chromatin. (A) Schematics of the native ChIP to assess transcriptionally active chromatin and associated RNAs. A histone marker of transcriptionally active chromatin, H3K27ac, was used in the ChIP analysis. (B) Experimental timeline for GM12371 knockdown and forskolin treatment for ChIP experiments. (C) qPCR analysis of GM12371 and cFOS promoter in ChIP. (D) qPCR analysis of GM12371 and cFOS RNA in ChIP. (E) qPCR analysis of GM12371 and GM10863 lncRNAs in the ChIP following the knockdown of GM12371 by gapmeR B3. A nontargeting gapmeR was used as a specificity control. All groups are normalized to IgG control, before indicated control group normalization. Bar graphs show fold-enrichment in the ChIP following forskolin stimulation. Error bars are SEM, n = 5, unpaired two-tailed t test, *P < 0.05, **P < 0.01. FSK, forskolin; Prm, promoter.
We next asked whether knockdown of GM12371 by gapmeR would reduce the levels of GM12371 associated with transcriptionally active chromatin and whether its targets are also subsequently affected. As suggested earlier, the GM10863:Sox10 lncRNA:mRNA pair is a trans target of GM12371 (Fig. 3). Therefore, we assessed the association of GM10863 lncRNA (Fig. 3 C–E) to transcriptionally active chromatin after knockdown of GM12371. We found that knockdown of GM12371 resulted in a decrease in the association of both GM12371 and its target GM10863 lncRNA to transcriptionally active chromatin (Fig. 4E) (fold-change compared with knockdown using nontargeting control gapmeR, GM12371: 0.17 ± 0.07; GM10863: 0.13 ± 0.04; n = 5, unpaired two-tailed t test, P < 0.05). Taken together, these results suggest that GM12371 lncRNA associates with active chromatin to regulate transcription of its targets.
GM12371 Expression Is Necessary for the Activity-Dependent Changes in Synaptic Transmission.
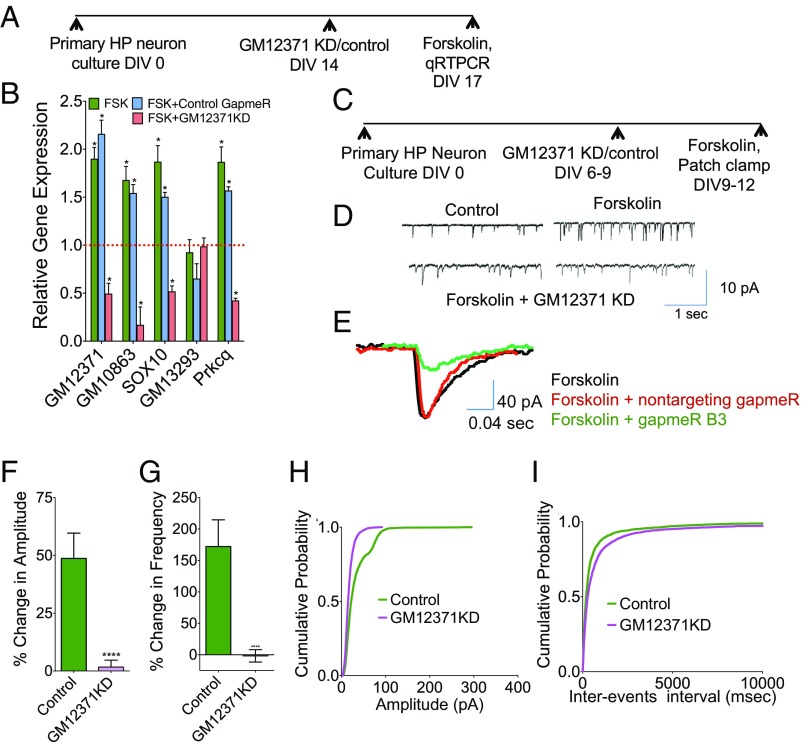
We next asked whether expression of GM12371 RNA is regulated by forskolin. qPCR analysis of the expression of GM12371 and two other lncRNAs randomly selected from our previous study (23) (GM11549, A43008) in SI Appendix, Fig. S7 shows that lncRNA GM12371 is up-regulated in response to exposure of 25 μM forskolin (fold-change 2.2 ± 0.3; n = 12; P < 0.05, one-way ANOVA followed by Tukey test) (Dataset S1, Table S14), whereas PKA inhibitor 14-22 amide blocked this up-regulation (fold-change 1.2 ± 0.03; n = 12; P < 0.05, one-way ANOVA followed by Tukey test) (Dataset S1, Table S14), suggesting cAMP-PKA signaling can regulate GM12371 levels in hippocampal neurons. Expression levels of two other lncRNAs were unchanged in primary hippocampal neurons with forskolin exposure (n = 12; P > 0.05) (Dataset S1, Table S14). We then investigated the temporal regulation of forskolin-induced GM12371 expression by carrying out a time course analysis (SI Appendix, Fig. S7B). We observed a significant change in GM12371 expression within 10 min of forskolin exposure; this then increased and peaked at 20 min, but persisted for 3 h after forskolin treatment [fold-change compared with DMSO (vehicle) 10 min, 1.9 ± 0.09, n = 3; 20 min, 3.6 ± 0.78, n = 3; 30 min, 3.11 ± 0.58, n = 6; 1 h, 1.96 ± 0.29, n = 4; 3 h, 1.86 ± 0.21, n = 3; 6 h, 1.23 ± 0.18, n = 3; P < 0.05 except for 6 h, unpaired two-tailed t test]. This activity-dependent expression pattern suggests that GM12371 acts as an immediate response gene in the cAMP-PKA signaling pathway.
To further understand the up-regulation of GM12371 by cAMP signaling, we asked whether expression of GM12371 targets are also affected by activation of the cAMP signaling. Based on our data that gapmeR-mediated reduction in GM12371 levels resulted in corresponding changes in two lncRNA:mRNA pairs—GM10863:Sox10 and GM13292:PRKCq—we assumed that we would observe a change in the expression levels of GM10863:Sox10 and GM13292:PRKCq pairs in a GM12371-dependent manner in response to activation of cAMP-PKA signaling (SI Appendix, Fig. S7C). We assessed these possibilities by examining the expression of GM12371 and the two lncRNA:mRNA pairs following exposure to forskolin in the presence of GM12371 knockdown (Fig. 5 A and B). qPCR analysis shown in Fig. 5B suggest that consistent with the trans regulation of GM10863:Sox10 and GM13292:PRKCq pairs by GM12371, forskolin exposure produced an increase in the levels of lncRNAs as well as their cognate mRNAs (forskolin exposure alone mean fold-change ± SEM: GM12371, 1.9 ± 0.12; GM10863, 1.7 ± 0.15; SOX10, 1.9 ± 0.17; GM13293, 0.9 ± 0.14; PRKCq 1.7 ± 0.16; forskolin + nontargeting gapmeR control: GM12371, 2.2 ± 0.15; GM10863, 1.5 ± 0.0.09; SOX10, 1.5 ± 0.0.05; GM13293, 0.65 ± 0.16; PRKCq 1.6 ± 0.04; forskolin + GM12371 knockdown: GM12371, 0.49 ± 0.11; GM10863, 0.17 ± 0.19; SOX10, 0.51 ± 0.06; GM13293, 0.98 ± 0.09; PRKCq 0.4 ± 0.03, n = 6, unpaired two-tailed t test, P < 0.05 for comparison between forskolin alone/forskolin + control gapmeR vs. GM12371 gapmeR, expression changes were not significant for GM13293). Taken together, these results suggest that forskolin-induced changes in the GM10863:Sox10 pair depends on GM12371 levels, whereas regulation of the GM13292:PRKCq pair by forskolin might involve multiple mechanisms. However, in the basal condition, expression of the GM13292:PRKCq pair is dependent on GM12371 levels.
Fig. 5.
Expression of GM12371 is necessary for the forskolin-induced enhancements in synaptic transmission. (A) Experimental outline for assessing forskolin (FSK)-induced changes in GM12371 and its lncRNA:mRNA targets. (B) qPCR analysis of forskolin-induced changes in the expression of lncRNA:mRNA targets following GM12371 knockdown. Bar graphs show fold-changes. Data normalized to 18s rRNA levels. *P < 0.05, Student’s t test. (C) Experimental outline for assessing the necessity of GM12371 expression for forskolin-induced changes in synaptic transmission. (D) One representative sEPSC traces of control, forskolin, and two representative traces of forskolin + GM12371 knockdown (KD) are shown. (E) Representative sEPSC traces showing changes in amplitude in the presence of forskolin, forskolin + control KD, and forskolin + GM12371 KD. (F and G) Bar graphs showing percent changes in the amplitude and frequency following GM12371 KD in the presence of FSK. ****P < 0.0001, one-way ANOVA followed by Tukey test. (H and I) Cumulative probability analysis showing changes in the amplitudes and frequencies of sEPSCs.
Because activation of cAMP-PKA signaling produced by forskolin exposure results in enhanced synaptic transmission in hippocampal neurons, we next asked whether expression of GM12371 is necessary for forskolin-induced changes in synaptic transmission. Following GM12371 knockdown, we measured sEPSCs before and after the treatment of forskolin (25 μM for 5 min) (Fig. 5 C–I). Quantitative analysis of sEPSC traces (Fig. 5 D and E) showed a decrease in sEPSC amplitudes (Fig. 5F) (mean ± SEM, nontargeting gapmeR control: 48.7 ± 10.93; GM12371 knockdown using gapmeR B3: 1.67± 3.02; P < 0.01, one-way ANOVA followed by Tukey test) and frequencies (Fig. 5G) (mean ± SEM: control knockdown: 172.12 ± 42.51; GM12371 knockdown: −1.71 ± 9.9; P < 0.01, one-way ANOVA followed by Tukey test). Consistent with these results, cumulative frequency analysis of electrophysiology data (Fig. 5 H and I) showed differences in both amplitude and frequencies, indicating that expression of GM12371 is necessary for the forskolin-induced changes in synaptic transmission in hippocampal neurons.
trans Regulation of GM10863:Sox10 Pairs by GM12371.
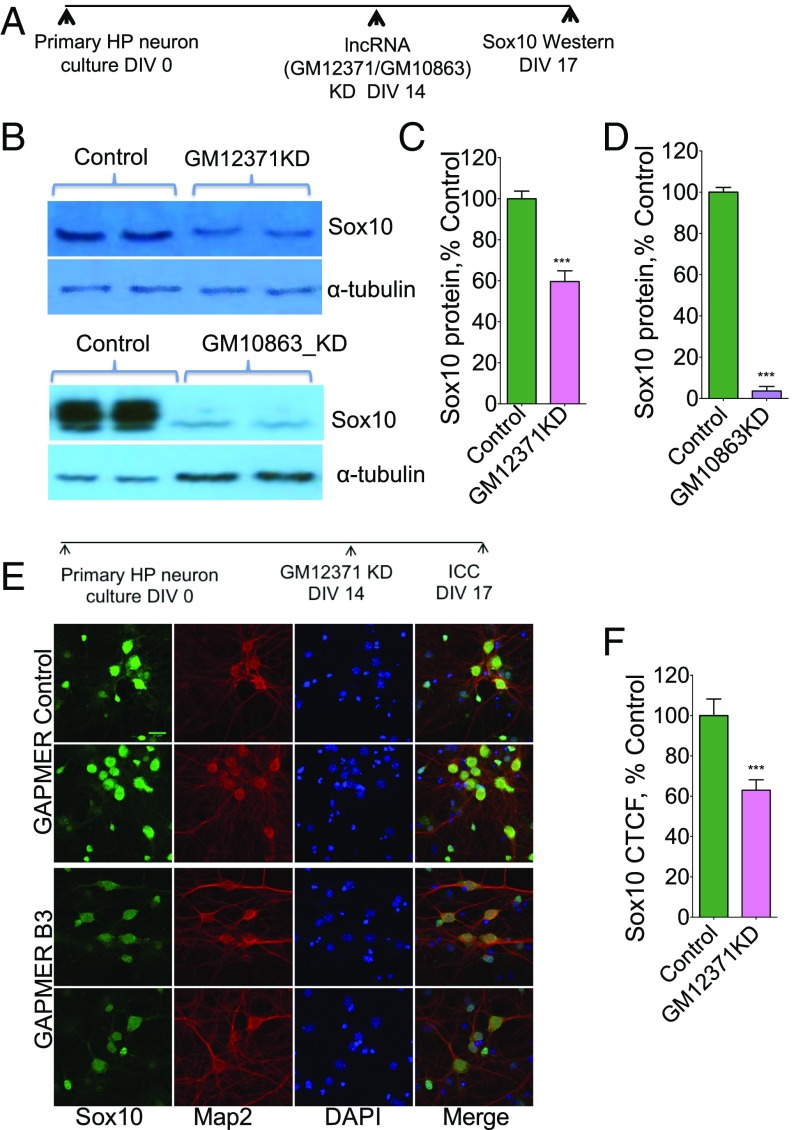
To further understand the regulation of the specific lncRNA:mRNA pairs, we next investigated the expression of Sox10 protein levels by Western blotting and immunocytochemistry following GM12371 knockdown. Western analysis showed a significant reduction in protein levels (Fig. 6 A–D) (n = 5, normalized intensity percentage of control gapmeR 59.6 ± 5.25, P < 0.05, unpaired two-tailed t test) (Dataset S1, Table S15) in the total protein extracts prepared from hippocampal neurons following 72 h of GM12371 knockdown. We then examined the effect of knockdown of the lncRNA GM10863 on Sox10 protein levels following knockdown of GM10863 by gapmeRs. Consistent with our qPCR results (Fig. 6 D–G), our Western analyses (Fig. 6 A–D) (n = 3, normalized intensity, percentage of control nontargeting gapmeR 3.6 ± 2.2, P < 0.05, unpaired two-tailed t test) suggest that Sox10 protein levels were reduced with GM10863 knockdown. In agreement with the Western data, immunolabeling of the Sox10 protein in hippocampal neuron cultures following GM12371 knockdown showed a reduced level of this protein (Fig. 6 E and F) (normalized mean intensity percent control gapmeR 63 ± 5.2, n = 149, P < 0.05, unpaired two-tailed t test) (Dataset S1, Table S15). Collectively, these experiments suggest that GM12371 exerts its effects on mRNA expression levels through a trans mechanism whereby GM12371 expression on chromosome 4 regulates the expression of lncRNA:mRNA pairs on chromosome 15 (GM10863:Sox10) and on chromosome 2 (GM13292:PRKCq). Alternately, this regulation can occur indirectly, through activation of downstream cellular pathways might eventually result in changes in Sox10 and PRKCq expression.
Fig. 6.
Regulation of GM10863-Sox10 pair by GM12371. (A) Experimental outline for assessing regulation of GM10863-Sox10 by GM12371. (B) Western blot analysis of Sox10 protein levels following GM12371 knockdown, n = 5, and GM10863 knockdown, n = 3. Sox10 protein levels in protein extracts from hippocampal neurons following lncRNA knockdown using a specific targeting gapmeR and a nontargeting gapmeR are estimated using Western blots. (C and D) Quantification of Western blots in B. Data were normalized to α-tubulin levels. Error bars are SEM, ***P < 0.001, Student’s t test. (E and F) Immunocytochemistry (ICC) analysis of expression of Sox10, a putative cognate mRNA target of lncRNA GM10863 following GM12371 knockdown. (E) Two representative confocal projection images are shown for each condition (gapmeR B3 to knock down GM12371 and control nontargeting gapmeR). (Scale bar, 20 µm.) (F) Quantitation of immunocytochemistry data. Number of neurons analyzed for control gapmeR = 149, gapmeR B3 = 149. CTCF, corrected total cell fluorescence. ***P < 0.001, Student’s t test.
Discussion
Proper development and maintenance of neuronal architecture and neuronal connections are critical for functioning of neural circuits. Formation and maintenance of synaptic connections is a well-orchestrated process that is subjected to transcriptional control. However, the specific roles of the noncoding transcriptome, especially that of recently discovered lncRNAs in synapse function, have been poorly understood.
The results presented in this study describe an lncRNA, GM12371, that plays a key role in synapse function. FISH analysis suggests that GM12371 is localized in the nucleus and is expressed in neurons as well as glia, suggesting broad functions in the nervous system. Using loss-of-function studies, we have gained critical insights into the biological functions of GM12371. Measurements of sEPSCs in hippocampal neurons suggested its key role in synaptic transmission likely through both pre- and postsynaptic mechanisms (Fig. 1). Consistent with our electrophysiology results, we found a decrease in total spine density as well as a decrease in mushroom spines (Fig. 2), often described as learning spines (57, 58). Further characterization of neuronal morphology following GM12371 knockdown showed a decrease in dendritic tree complexity (Fig. 2), accentuating the role of GM12371 in synapse function.
To understand the molecular mechanism of action of GM12371, we first searched for cis-regulated mRNAs or miRNAs. The only transcript we identified from was lingo2 mRNA; however, we found that the expression of lingo2 mRNA does not depend on GM12371 levels. It is possible that GM12371 regulate expression of other isoforms of lingo2 by a cis mechanism. We next carried out unbiased transcriptome profiling experiments in hippocampal neurons following GM12371 knockdown to assess whether a trans mechanism might explain GM12371 function. Analysis of RNAseq data revealed differential expression of coding and noncoding RNAs from different chromosomes affected by GM12371 knockdown indicative of a genome-wide effect (Fig. 3). This analysis further suggested that GM12371 selectively regulates the expression of genes involved in nervous system development, differentiation, and function (SI Appendix, Figs. S5 and S6 and Dataset S1, Tables S9–S12).
Notably, we confirmed two lncRNA:mRNA pairs among the molecular targets of GM12371. While we cannot be certain that all of the effects on transcription of genes are exclusively due to the GM12371 knockdown in hippocampal neurons, our native ChIP experiments and characterization of lncRNA:mRNA pairs suggest transcriptional control by GM12371 consistent with its nuclear localization. Collectively, these studies provide additional mechanistic insights into GM12371 function. In support of a trans mechanism, loss-of-function experiments suggested that GM12371 (transcribed from chromosome 4) regulates expression of lncRNA GM10863 and its cis target Sox 10 (on chromosome 15) and lncRNA GM13292 and its cis target PRKCQ (on chromosome 2).
Intriguingly, the expression of GM12371 and its target lncRNA:mRNA pairs (GM10863:Sox 10 pair and GM13292:PRKCQ pair) are regulated by cAMP-PKA signaling (SI Appendix, Fig. S7C). Exposure to forskolin rapidly up-regulated the expression of GM12371 as early as 10 min and its expression persisted 3 h after forskolin exposure, suggesting the potential for GM12371 as an immediate response gene in the cAMP-PKA signaling pathway. Furthermore, the regulation of GM12371 expression by forskolin suggested its possible role in activity-dependent changes in synaptic communication. Consistent with this assumption, we found that loss-of-function of GM12371 inhibited forskolin-induced enhancements in synaptic communication.
In summary, our electrophysiological, imaging, and molecular experiments demonstrate that the lncRNA GM12371 is essential for synapse function in hippocampal neurons. Importantly, GM12371 regulates expression of several genes known to be critical for synapse function. Therefore, lncRNA GM12371 acts as an important component of the transcriptional program that ensures proper synapse function. These results will further advance the molecular biology of synapse function and the physiological roles of lncRNAs in the nervous system.
Materials and Methods
Animals, electrophysiology, reagents, imaging, genomics and bioinformatics, qPCR, and native ChIP are described in SI Appendix, Experimental Methods. Results of the bioinformatics analysis are given in Dataset S1. Housing, animal care, and experimental procedures were consistent with the Guide for the Care and Use of Laboratory Animals (59) and approved by the Institutional Animal Care and Use Committee of The Scripps Research Institute.
Supplementary Material
Acknowledgments
We thank Mohammad Fallahi for carrying out analysis of RNA sequencing data; and Drs. Ron Davis and Kirill Martemyanov of The Scripps Research Institute and Maria Concetta Miniaci of the University of Naples for critical reading of the manuscript. This project was supported by NIH Grants 1R21DA039417-01A1 and 5R21MH108929-02 (to S.V.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper has been deposited in the GenBank NCBI BioProject database (accession no. PRJNA356039).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722587115/-/DCSupplemental.
References
- 1.Emes RD, Grant SG. Evolution of synapse complexity and diversity. Annu Rev Neurosci. 2012;35:111–131. doi: 10.1146/annurev-neuro-062111-150433. [DOI] [PubMed] [Google Scholar]
- 2.Frank RA, Grant SG. Supramolecular organization of NMDA receptors and the postsynaptic density. Curr Opin Neurobiol. 2017;45:139–147. doi: 10.1016/j.conb.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scannevin RH, Huganir RL. Postsynaptic organization and regulation of excitatory synapses. Nat Rev Neurosci. 2000;1:133–141. doi: 10.1038/35039075. [DOI] [PubMed] [Google Scholar]
- 4.Südhof TC. The presynaptic active zone. Neuron. 2012;75:11–25. doi: 10.1016/j.neuron.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Südhof TC. The synaptic vesicle cycle: A cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 6.Kim JH, Huganir RL. Organization and regulation of proteins at synapses. Curr Opin Cell Biol. 1999;11:248–254. doi: 10.1016/s0955-0674(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 7.Südhof TC. Synaptic neurexin complexes: A molecular code for the logic of neural circuits. Cell. 2017;171:745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, et al. Alterations in 5-HT2A receptor binding in various brain regions among 6-hydroxydopamine-induced parkinsonian rats. Synapse. 2010;64:224–230. doi: 10.1002/syn.20722. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Tiedge H. Translational control at the synapse. Neuroscientist. 2004;10:456–466. doi: 10.1177/1073858404265866. [DOI] [PubMed] [Google Scholar]
- 10.Steward O, Worley P. Localization of mRNAs at synaptic sites on dendrites. Results Probl Cell Differ. 2001;34:1–26. doi: 10.1007/978-3-540-40025-7_1. [DOI] [PubMed] [Google Scholar]
- 11.Sultan FA, Day JJ. Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011;3:157–181. doi: 10.2217/epi.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman-Karlsson MC, Meadows JP, Gavin CF, Hablitz JJ, Sweatt JD. Transcriptional and epigenetic regulation of Hebbian and non-Hebbian plasticity. Neuropharmacology. 2014;80:3–17. doi: 10.1016/j.neuropharm.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azpurua J, Eaton BA. Neuronal epigenetics and the aging synapse. Front Cell Neurosci. 2015;9:208. doi: 10.3389/fncel.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Na ES, Nelson ED, Kavalali ET, Monteggia LM. The impact of MeCP2 loss- or gain-of-function on synaptic plasticity. Neuropsychopharmacology. 2013;38:212–219. doi: 10.1038/npp.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3:a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smalheiser NR, Zhang H, Dwivedi Y. Enoxacin elevates microRNA levels in rat frontal cortex and prevents learned helplessness. Front Psychiatry. 2014;5:6. doi: 10.3389/fpsyt.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hombach S, Kretz M. Non-coding RNAs: Classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. doi: 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee N, et al. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat Struct Mol Biol. 2017;24:86–96. doi: 10.1038/nsmb.3325. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S. Annotating noncoding RNA genes. Annu Rev Genomics Hum Genet. 2007;8:279–298. doi: 10.1146/annurev.genom.8.080706.092419. [DOI] [PubMed] [Google Scholar]
- 21.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stadler PF. Class-specific prediction of ncRNAs. Methods Mol Biol. 2014;1097:199–213. doi: 10.1007/978-1-62703-709-9_10. [DOI] [PubMed] [Google Scholar]
- 23.Kadakkuzha BM, et al. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front Cell Neurosci. 2015;9:63. doi: 10.3389/fncel.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carninci P, et al. FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 25.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnsson P, Morris KV. Expanding the functional role of long noncoding RNAs. Cell Res. 2014;24:1284–1285. doi: 10.1038/cr.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 30.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 31.Yoon JH, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beltran M, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krystal GW, Armstrong BC, Battey JF. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990;10:4180–4191. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huarte M. LncRNAs have a say in protein translation. Cell Res. 2013;23:449–451. doi: 10.1038/cr.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward M, McEwan C, Mills JD, Janitz M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J Hum Transcr. 2015;1:2–9. doi: 10.3109/23324015.2015.1077591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modarresi F, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muslimov IA, et al. RNA transport in dendrites: A cis-acting targeting element is contained within neuronal BC1 RNA. J Neurosci. 1997;17:4722–4733. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahlestedt C, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deleavey GF, Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol. 2012;19:937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- 45.O’Brien RJ, et al. Activity-dependent modulation of synaptic AMPA receptor accumulation. Neuron. 1998;21:1067–1078. doi: 10.1016/s0896-6273(00)80624-8. [DOI] [PubMed] [Google Scholar]
- 46.Sweet ES, Langhammer CL, Kutzing MK, Firestein BL. Semiautomated analysis of dendrite morphology in cell culture. Methods Mol Biol. 2013;1018:261–268. doi: 10.1007/978-1-62703-444-9_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 48.Liu Q, Melnikova IN, Hu M, Gardner PD. Cell type-specific activation of neuronal nicotinic acetylcholine receptor subunit genes by Sox10. J Neurosci. 1999;19:9747–9755. doi: 10.1523/JNEUROSCI.19-22-09747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng Y, Cheung M, Abu-Elmagd MM, Orme A, Scotting PJ. Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Brain Res Dev Brain Res. 2000;121:233–241. doi: 10.1016/s0165-3806(00)00049-3. [DOI] [PubMed] [Google Scholar]
- 50.Carney TJ, et al. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–4630. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- 51.Li MX, et al. The role of the theta isoform of protein kinase C (PKC) in activity-dependent synapse elimination: Evidence from the PKC theta knock-out mouse in vivo and in vitro. J Neurosci. 2004;24:3762–3769. doi: 10.1523/JNEUROSCI.3930-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parikshak NN, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasinathan S, Orsi GA, Zentner GE, Ahmad K, Henikoff S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods. 2014;11:203–209. doi: 10.1038/nmeth.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corbett BF, et al. ΔFosB regulates gene expression and cognitive dysfunction in a mouse model of Alzheimer’s disease. Cell Rep. 2017;20:344–355. doi: 10.1016/j.celrep.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otmakhov N, et al. Forskolin-induced LTP in the CA1 hippocampal region is NMDA receptor dependent. J Neurophysiol. 2004;91:1955–1962. doi: 10.1152/jn.00941.2003. [DOI] [PubMed] [Google Scholar]
- 56.Huang YY, Kandel ER. Recruitment of long-lasting and protein kinase A-dependent long-term potentiation in the CA1 region of hippocampus requires repeated tetanization. Learn Mem. 1994;1:74–82. [PubMed] [Google Scholar]
- 57.Hering H, Sheng M. Dendritic spines: Structure, dynamics and regulation. Nat Rev Neurosci. 2001;2:880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 58.Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Curr Opin Neurobiol. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 59.National Research Council of the National Academies . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.