A new era of high-throughput biological exploration is upon us, fueled by an ever-expanding toolkit to generate genetic and biological diversity. However, for most engineering applications, this approach necessitates an increased throughput for metabolite detection. In this regard, the primary bottleneck of most high-throughput metabolic engineering approaches is detecting the desired product, not creating genetic diversification. Indeed, this presents an interesting deviation from the traditional paradigms of metabolic engineering espoused several decades ago when genetic modifications were the bottleneck. To this end, detection of metabolites, especially those that are central and active intermediates in metabolism such as malonyl-CoA, can be especially challenging. The field has been exploring biosensors to transduce the concentrations of such intermediates into readily detectable outputs, such as color or fluorescence. However, application of this work has been stymied by malonyl-CoA biosensors that invoke multistep transduction and are self-limited to Escherichia coli and Saccharomyces cerevisiae. In their recent report, Yang et al. (1) repurpose a red pigment-producing enzyme to create a single-step malonyl-CoA biosensor for rapid detection that can function in several different industrially relevant bacteria. This versatile, direct approach facilitated rapid library screening, resulting in improved production of malonyl-CoA–derived polyketide products.
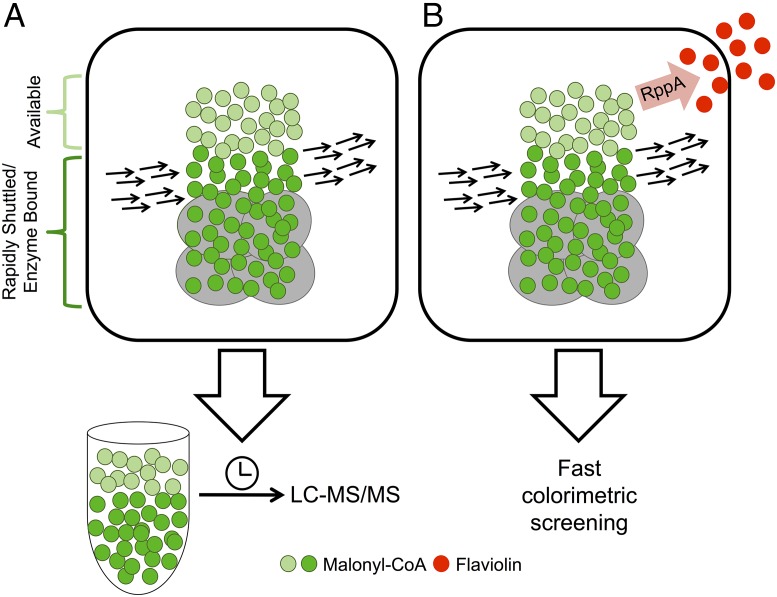
Detection of active intermediates, such as the central metabolite malonyl-CoA, poses unique challenges for the field. Unlike a downstream product that accumulates to ease detection, CoA species are highly labile and, thus, obtaining an accurate cellular concentration can be difficult due to rapid conversion to various downstream products. Moreover, although determining precise measurements via analytical techniques such as LC-MS/MS are possible, they are time-consuming and particularly challenging due to the complex chemical mixture found within cells (2). However, even measurements of total concentrations alone (either through binding-based biosensors or chromatography) do not measure what is of most importance to a metabolic engineer: metabolite availability. In this regard, determining how much of an intermediate metabolite is available to be diverted and feed a downstream product or pathway is the essence of pathway rewiring. Central metabolites such as malonyl-CoA participate in many different pathways, complicating this question of availability. Functionally, the cell controls such labile products through the use of enzyme complexes, and in eukaryotes through compartmentalization. These mechanisms can contribute to the rapid shuttling of metabolites of interest into context-specific pathways. Thus, the measurement of concentration and availability can diverge, requiring metabolite detection strategies that can parse this difference (Fig. 1).
Fig. 1.
A new RppA-based biosensor provides a method for rapidly detecting malonyl-CoA availability. The malonyl-CoA pool within a cell contains a portion that is bound by enzyme complexes and readily shuttled to downstream reactions. This rapidly shuttled portion is shown in dark green, whereas the portion available to be diverted into an alternative pathway is represented by light green. (A) Sample preparation extracts the entire pool of malonyl-CoA for time-consuming LC-MS/MS analysis. (B) The RppA biosensor converts available malonyl-CoA into the red pigment, flaviolin, which can be rapidly assessed through colorimetric screening.
To address the detection bottleneck, researchers have developed several techniques that have increased throughput. Outside of high-throughput mass spectrometry methods, the most contemporary approach for sampling metabolic concentration is the use of biosensors. In such a scheme, enzymes, transcription factors, and aptamers are repurposed and optimized to transduce metabolite concentration into a more readily detectable attribute such as fluorescence or color (3). This approach replaces the often-limited scale of analytical techniques such as liquid or gas chromatography and mass spectrometry with more rapid, liquid-handling–based methods such as FACS or plate-reader assays. The resulting increase in scale has allowed researchers to improve production of a vast array of products (3). In the case of malonyl-CoA [the intermediate of interest in the Yang et al. (1) paper], biosensor detection has been previously developed and applied to improve the production of malonyl-CoA–derived products for both E. coli (4–6) and S. cerevisiae (7, 8). In this scheme, the transcription factor system FapR (adapted from Bacillus subtilis) binds the associated fapO operator region, inducing transcriptional repression. In the presence of malonyl-CoA, FapR undergoes a conformational change, which releases the operator and thus relieves repression, enabling expression of a downstream fluorescent reporter gene (4). However, this system requires a multistep, heterologous signal transduction that is not amenable to all host organisms. Moreover, the use of fluorescence-based methods is also incompatible with some host organisms due to high levels of natural autofluorescence that can obfuscate the biosensor signal (9). Although biosensors have provided valuable improvements to metabolic engineering, several issues remain to be addressed to fully realize the potential of high-throughput strain engineering.
In contrast to previous reliance on a transcription factor-based fluorescent sensor, Yang et al. (1) take the approach of opting to detect malonyl-CoA availability—rather than absolute concentration—directly in a colorimetric assay. Previously in the field, a related strategy was used to screen for improved isoprenoid production by detecting lycopene (10). Such an approach relies on the hypothesis that improved intermediate production can translate to improved yields in a different (but related) heterologous pathway. We see this hypothesis at work in Yang et al. (1) as well; however, their study is unique in the focus on a central metabolite for detection and the more direct measurement of precursor availability. The authors utilize a heterologous enzyme, RppA, to directly transduce the availability of malonyl-CoA into a red pigment. RppA (found in several Streptomyces spp.) has previously been described to produce the secondary metabolite 1,3,6,8-tetrahydroxynaphthalene (THN), an intermediate that can spontaneously oxidize into the red pigment flaviolin (11, 12). When Yang et al. (1) expressed rppA in E. coli, they were able to readily detect flaviolin by eye and quantify it using absorbance. Moreover, this signal was found to exhibit a dose–response behavior to malonyl-CoA, as determined by experiments using the chemical inhibitor of fatty acid synthesis, cerulenin (13). Thus, this work bypasses the need for a multistep, transcription factor-based process to detect malonyl-CoA. In converting malonyl-CoA directly and permanently into flaviolin, this approach also provides a more specific measure of intermediate availability in comparison with techniques that rely upon the transient interactions required for transcription factor activation. In other words, malonyl-CoA must be fully available to be converted into flaviolin, rather than just present to activate a transcription factor. Furthermore, Yang et al. (1) demonstrate the generalizability of this approach by importing this sensor into the industrially relevant organisms Pseudomonas putida and Corynebacterium glutamicum, in which cellular autofluorescence can be an issue. In this regard, Yang et al. successfully address limitations to previous malonyl-CoA detection schemes.
Armed with this way to measure malonyl-CoA, Yang et al. (1) embarked on a series of experiments using this enzyme-based biosensor to improve the throughput of metabolic engineering for malonyl-CoA–derived products. To this end, an E. coli sRNA
Yang et al. repurpose a red pigment-producing enzyme to create a single-step malonyl-CoA biosensor for rapid detection that can function in several different industrially relevant bacteria.
knockdown library was screened to identify 14 top gene targets that led to increased production of flaviolin. These identified targets were then applied to the production of four malonyl-CoA–derived polyketides (6-methylsalicylic acid, aloesone, naringenin, and resveratrol) that possess a variety of different potential health applications (14–16). Through the efforts in this work, Yang et al. (1) were able to improve production significantly across all products, with the highest increase of fourfold observed for resveratrol production. Moreover, this work also boosts microbial production of the polyketide aloesone. As described above, one of the hallmarks of this scheme is direct detection of the central intermediate availability, rather than a surrogate downstream molecule as explored by previous approaches. To this end, the same identified sRNA-based knockdown was found to yield the greatest production improvement in all four cases, clearly demonstrating the malonyl-CoA–specific nature of their sensor. This exciting result suggests that the RppA biosensor could be used to create platform strains preoptimized for malonyl-CoA availability. Such strains could be applied to malonyl-CoA products beyond even those tested by Yang et al. (1), further reducing the overall strain development timeline. Therefore, in addition to addressing previous malonyl-CoA biosensor limitations, this work also opens up exciting future applications.
Collectively, this recent work by Yang et al. (1) tackles many challenges associated with the rapid detection of energetic metabolites, as exemplified by malonyl-CoA. By detecting the red pigment of flaviolin, the authors, in essence, devised a direct screen for metabolite availability. Moreover, this screen is applicable to other host organisms and generalizable for improving other malonyl-CoA–derived products. This work highly complements the emerging high-throughput biology approach. In this regard, this RppA biosensor scheme is compatible with the field’s ever-increasing capacity to diversify strains. Additionally, the underlying principle of establishing a platform strain (for malonyl-CoA through flaviolin production) and applying this strain to a multitude of other alternative products is rather appealing. Such approaches have started (and will continue) to speed engineering and science goals. Lastly, this approach should inspire similar detection schemes for other key labile intermediates in cellular metabolism.
Footnotes
The authors declare no conflict of interest.
See companion article on page 9835 in issue 40 of volume 115.
References
- 1.Yang D, et al. Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc Natl Acad Sci USA. 2018;115:9835–9844. doi: 10.1073/pnas.1808567115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao L, et al. Simultaneous quantification of malonyl-CoA and several other short-chain acyl-CoAs in animal tissues by ion-pairing reversed-phase HPLC/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;853:303–313. doi: 10.1016/j.jchromb.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 3.Lin JL, Wagner JM, Alper HS. Enabling tools for high-throughput detection of metabolites: Metabolic engineering and directed evolution applications. Biotechnol Adv. 2017;35:950–970. doi: 10.1016/j.biotechadv.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Xu P, et al. Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli. ACS Chem Biol. 2014;9:451–458. doi: 10.1021/cb400623m. [DOI] [PubMed] [Google Scholar]
- 5.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci USA. 2014;111:11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu D, Xiao Y, Evans BS, Zhang F. Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator. ACS Synth Biol. 2015;4:132–140. doi: 10.1021/sb400158w. [DOI] [PubMed] [Google Scholar]
- 7.Li S, Grüschow S, Dordick JS, Sherman DH. Molecular analysis of the role of tyrosine 224 in the active site of Streptomyces coelicolor RppA, a bacterial type III polyketide synthase. J Biol Chem. 2007;282:12765–12772. doi: 10.1074/jbc.M700393200. [DOI] [PubMed] [Google Scholar]
- 8.David F, Nielsen J, Siewers V. Flux control at the malonyl-CoA node through hierarchical dynamic pathway regulation in Saccharomyces cerevisiae. ACS Synth Biol. 2016;5:224–233. doi: 10.1021/acssynbio.5b00161. [DOI] [PubMed] [Google Scholar]
- 9.Loeschcke A, Thies S. Pseudomonas putida—A versatile host for the production of natural products. Appl Microbiol Biotechnol. 2015;99:6197–6214. doi: 10.1007/s00253-015-6745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Özaydın B, Burd H, Lee TS, Keasling JD. Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production. Metab Eng. 2013;15:174–183. doi: 10.1016/j.ymben.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Funa N, et al. A new pathway for polyketide synthesis in microorganisms. Nature. 1999;400:897–899. doi: 10.1038/23748. [DOI] [PubMed] [Google Scholar]
- 12.Funa N, Ohnishi Y, Ebizuka Y, Horinouchi S. Properties and substrate specificity of RppA, a chalcone synthase-related polyketide synthase in Streptomyces griseus. J Biol Chem. 2002;277:4628–4635. doi: 10.1074/jbc.M110357200. [DOI] [PubMed] [Google Scholar]
- 13.Omura S. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol Rev. 1976;40:681–697. doi: 10.1128/br.40.3.681-697.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson JA. Polyketide synthase complexes: Their structure and function in antibiotic biosynthesis. Philos Trans R Soc Lond B Biol Sci. 1991;332:107–114. doi: 10.1098/rstb.1991.0038. [DOI] [PubMed] [Google Scholar]
- 15.Abe I, Utsumi Y, Oguro S, Noguchi H. The first plant type III polyketide synthase that catalyzes formation of aromatic heptaketide. FEBS Lett. 2004;562:171–176. doi: 10.1016/S0014-5793(04)00230-3. [DOI] [PubMed] [Google Scholar]
- 16.Abe I, Morita H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat Prod Rep. 2010;27:809–838. doi: 10.1039/b909988n. [DOI] [PubMed] [Google Scholar]