Significance
Populations often show “islands of divergence” in the genome. Analysis of divergence between subspecies of Antirrhinum that differ in flower color patterns shows that sharp peaks in relative divergence occur at two causal loci. The island is shaped by a combination of gene flow and multiple selective sweeps, showing how divergence and barriers between populations can arise and be maintained.
Keywords: hybrid zone, Antirrhinum, genomic island, selective sweep, speciation
Abstract
Genomes of closely-related species or populations often display localized regions of enhanced relative sequence divergence, termed genomic islands. It has been proposed that these islands arise through selective sweeps and/or barriers to gene flow. Here, we genetically dissect a genomic island that controls flower color pattern differences between two subspecies of Antirrhinum majus, A.m.striatum and A.m.pseudomajus, and relate it to clinal variation across a natural hybrid zone. We show that selective sweeps likely raised relative divergence at two tightly-linked MYB-like transcription factors, leading to distinct flower patterns in the two subspecies. The two patterns provide alternate floral guides and create a strong barrier to gene flow where populations come into contact. This barrier affects the selected flower color genes and tightly-linked loci, but does not extend outside of this domain, allowing gene flow to lower relative divergence for the rest of the chromosome. Thus, both selective sweeps and barriers to gene flow play a role in shaping genomic islands: sweeps cause elevation in relative divergence, while heterogeneous gene flow flattens the surrounding “sea,” making the island of divergence stand out. By showing how selective sweeps establish alternative adaptive phenotypes that lead to barriers to gene flow, our study sheds light on possible mechanisms leading to reproductive isolation and speciation.
Genome scans of closely-related species or populations have revealed “genomic islands” as peaks of high relative sequence divergence (Fst) that stand out against a lower “sea” of divergence (1–5). The causes of genomic islands remain unclear, but they have been suggested to contain key loci involved in local adaptation and/or reproductive isolation (6). However, their significance for speciation with or without gene flow between populations is a matter of debate (6–9). One hypothesis is that gene flow is unimpeded across most of the genome, reducing between-population diversity, except for loci under divergent selection and loci in close physical linkage to selected loci (8). Another hypothesis is that genomic islands reflect selective sweeps, where specific alleles are driven to high frequency, thus reducing within-population diversity (7, 9, 10). These two hypotheses are typically presented as alternatives, although they are not mutually exclusive: both barriers to gene flow and selective sweeps may play a role. Here, we determine how these processes contribute to a genomic island that controls floral differences between two subspecies of Antirrhinum majus: A.m.striatum and A.m.pseudomajus. This system has the advantage of being genetically tractable and having a hybrid zone that allows selection and gene flow to be analyzed in nature (11, 12).
Antirrhinum has closed flowers that are prised open by pollinating bees. A.m.striatum and A.m.pseudomajus exhibit two different floral patterns that signpost the bee entry point (Fig. 1 A and B). A.m.striatum flowers have restricted veins of magenta anthocyanin on upper petals, which contrast against a yellow aurone background (Fig. 1A). A.m.pseudomajus exhibits a complementary pattern, with a patch of yellow at the bee entry point on lower petals contrasted against magenta (Fig. 1B). Yellow patterning is controlled by SULF (12). Here we focus on control of magenta by the ROSEA (ROS) and ELUTA (EL) loci (13–15). The advantage of studying these loci is that they are tightly linked, allowing variation in intervening regions to provide insights into evolutionary forces. A further locus influencing magenta pigmentation pattern is VENOSA, which promotes magenta in dorsal veins (14). Many natural accessions carry VEN alleles, while the cultivated species A. majus used for genetic analysis typically carries ven, allowing its effects to be seen in genetic crosses.
Fig. 1.
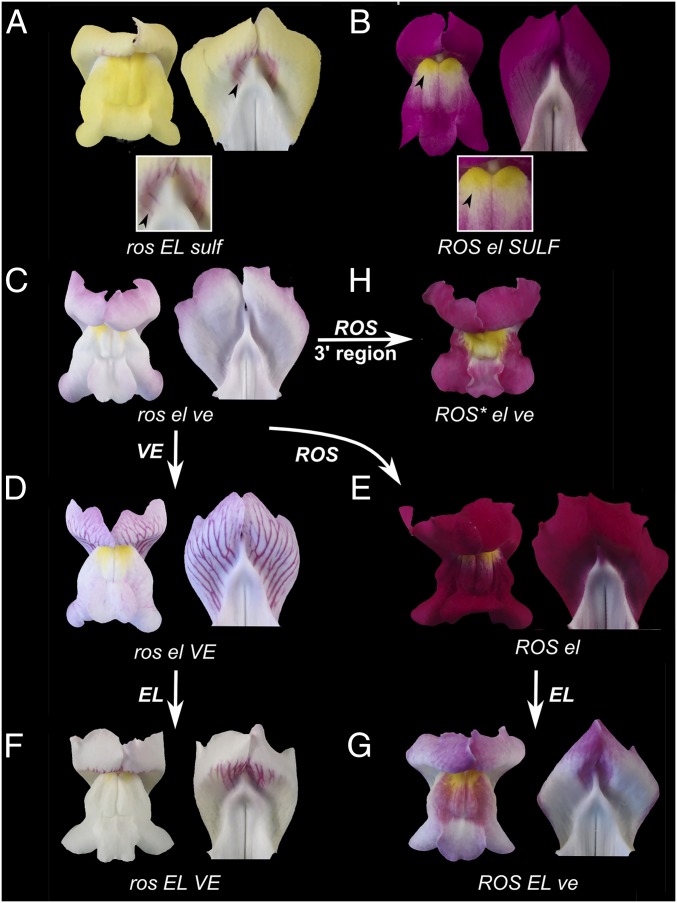
Genetics of flower color. Flowers of A.m.striatum (A, ross/ross ELs/ELs sulfs/sulfs) and A.m.pseudomajus (B, ROSp/ROSp elp/elp SULFp/SULFp). Each panel shows face view (Left), inside of dorsal petals (Right), and closeup (Bottom). Arrowheads highlight dorsal (A) and ventral (B) patterns. (C–G) Progeny of crosses between plants from the hybrid zone and lines of A. majus, illustrating phenotype of various allele combinations. All are SULFm/- or SULFp/-. (C) ross/rosd elp/elm ve/ve gives a flower with pale magenta color on petal periphery. (D) ross/ross elp/elp VE/- has flowers with magenta veins because of VE. (E) ROSp/ROSp elp/elp gives strong magenta throughout the flower due to ROS allele (venosa genotype unknown). (F) ross/ross ELs/ELs VE/- has vein pigment restricted to a central region. (G) ROSp/ROSp ELs/ELs ve/ve giving a restricted pattern of pigmentation compared with E. (H) ROS*/ROS* elp/elp ve/ve have spread magenta but of weaker intensity than conferred by ROS (compare with E). Allele superscripts and abbreviations used in figure legend: *, recombinant; d, dorsea (mutant in A. majus background); m, majus; p, A.m.pseudomajus; s, A.m.striatum; X/-, unknown whether homozygous or heterozygous for dominant allele X.
Flowers homozygous for recessive alleles at all three loci (ros el ven) have very weak magenta pigmentation (Fig. 1C). Introduction of VEN leads to magenta overlying the veins of dorsal petals (Fig. 1D), whereas introduction of ROS leads to strong magenta throughout the corolla (Fig. 1E). The semidominant EL allele restricts the magenta conferred by VEN and ROS to lie over the bee entry point (Fig. 1 F and G). The ROS locus contains three MYB-like transcription factors, ROS1, ROS2, and ROS3, with ∼90% protein sequence identity in the MYB domain. So far, only ROS1 and ROS2 have been functionally characterized, with ROS1 exerting the major control on anthocyanin levels and pattern (14). EL is tightly linked to ROS but has not been previously isolated (11, 14). Selection at ROS has been inferred from analysis of a hybrid zone between A.m.striatum and A.m.pseudomajus: both magenta pigmentation and ROS allele frequencies show sharp clines, ∼1 km wide, whereas markers >5 cM from ROS show more uniform allele frequency distributions (11).
Flower color differences between A.m.striatum and A.m.pseudomajus are unlikely to be maintained by adaptation to local conditions, as there are no clear differences in environment or pollinators across the hybrid zone (16). Rather, hybrids and recombinants may be selected against because their flower patterns are less effective as signposts for bee entry than the parental patterns (12, 17) and possibly because bees favor the commonest local phenotype (18–20). This situation is similar to how wing color pattern differences are maintained in Heliconius butterflies (21–23). Heliconius genes interact to generate distinct color patterns, which signal distastefulness to predators (24). Several patterns can deter Heliconius predators, just as several can highlight Antirrhinum flower entry. Sharp clines in Heliconius are maintained because hybrid phenotypes are less effective (23) and because the commonest pattern is fitter (22). Genomic islands are observed at the wing pattern loci and are particularly striking near hybrid zones (2, 21, 25).
Here we combine analysis of pooled DNA sequences and SNP frequencies from across the hybrid zone between A.m.striatum and A.m.pseudomajus, with genetic and gene expression analysis of parental and recombinant genotypes. We pinpoint the loci responsible for differences in anthocyanin flower color pattern and show that they underlie genomic islands of high Fst. Through examination of sequence variation around and between the islands, combined with simulations, we show that the islands reflect multiple selective sweeps, which raise relative divergence locally. The sweeps create a barrier to gene flow, which leads to the islands standing out from the genomic sea. Thus, both selective sweeps and barriers to gene flow play key roles in the creation and shaping of genomic islands.
Results and Discussion
Patterns of Differentiation and Diversity.
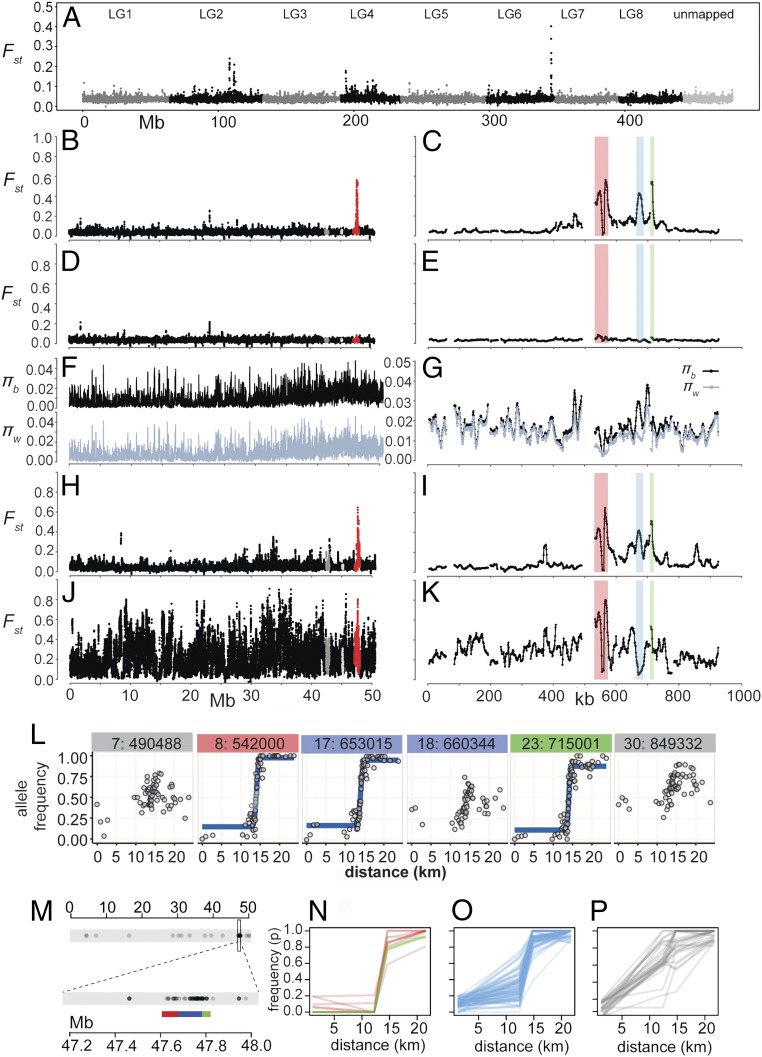
To determine the pattern of sequence diversity around the ROS locus, we estimated relative sequence divergence, Fst, between A.m.striatum and A.m.pseudomajus by sequencing pools of ∼50 individuals sampled from either side of the hybrid zone, with the centers of the pools separated by ∼2.5 km (SI Appendix, Fig. S1 and Table S1) (26). SNP analysis of individuals showed that these pools provided good estimates of allele frequencies (SI Appendix, Fig. S2). Low Fst was observed throughout the genome except for regions with elevated Fst on chromosomes 2, 4, and 6 (Fig. 2A). We focused our analysis on the peak on chromosome 6 as this is where the ROS locus maps (Fig. 2B). At a finer scale, three sharp peaks were found in the ROS region superimposed on a broader region of increased Fst (Fig. 2C). The left peak included ROS1 and ROS2 (ROS3 is in a region of lower Fst). These Fst peaks were not observed between pools from the same side of the hybrid zone (Fig. 2 D and E and SI Appendix, Fig. S3). Thus, the Fst peaks in the ROS region represent genomic islands of divergence between A.m.striatum and A.m.pseudomajus.
Fig. 2.
Divergence between A.m.striatum and A.m.pseudomajus. (A) Fst comparisons between pools of A.m.striatum and A.m.pseudomajus populations either side of a hybrid zone (YP1 vs. MP2) and ∼2.5 km apart across the whole genome summarized in 50-kb windows with a 25-kb step size. (B) Same pools as A at 10-kb window resolution with 1-kb step size for chromosome 6. A region of high Fst is within a ∼930-kb scaffold containing the ROS gene (red). Linked scaffolds contain DICHOTOMA (dark gray) and PALLIDA (light gray). (C) Closeup of region of high Fst at ROS comprising three peaks: left (red, 530–575 kb), middle (blue, 663–687 kb), and right (green, 707–720 kb on the ROS scaffold). The ∼930-kb scaffold corresponds to positions 47.088–48.015 Mb on chromosome 6. (D and E) Pools from the same side of the hybrid zone (YP1 vs. YP2, both A.m.striatum, 0.2 km apart). (F and G) πb and mean πw for the same sequence data as used in B and C. (H and I) Pools sampled from populations either side of the hybrid zone (YP4 vs. MP11), ∼20 km apart. (J and K) Pools sampled from remote populations (∼100 km apart, ML vs. CIN). (L) Clines for selected SNPs genotyped across the hybrid zone population. Headings denote the SNP identifier and position within the ROS 930-kb scaffold. (M) Distribution of 115 differential SNPs showing allele frequency differences >0.8 between the outer pools (YP4 and MP11) and coverage of 20–200× in all pools. Enlarged Inset shows regions corresponding to ROS peak (red), intervening region (blue), and EL peak (green). (N) SNP allele frequencies in the pools for eight differential SNPs within the ROS peak (red) and six within the EL peak (green) exhibit clines centered at the hybrid zone. (O) Most of the 74 SNPs located within the interval between the ROS and EL peaks, plotted in blue, exhibit clines centered at the hybrid zone. (P) SNP frequencies outside the ROS and EL peaks derive from flanking regions on the ROS superscaffold (n = 13) or elsewhere on LG6 (n = 14).
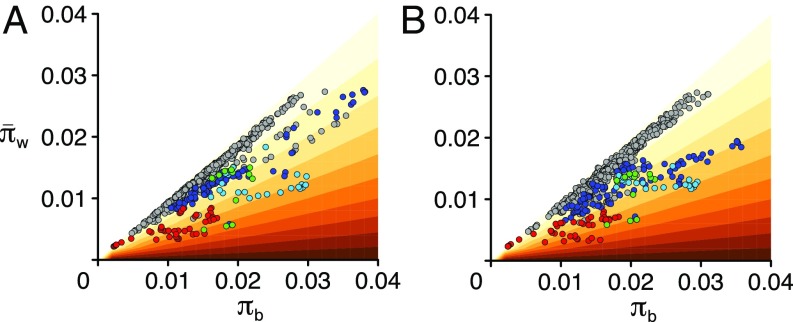
Fst is defined as (πb − πw)/(πb + πw), where πb (also known as dxy) and πw are the absolute pairwise divergence between and within populations, respectively (7). An increase in Fst can therefore be due to an increase in πb, a decrease in πw, or a combination of the two. Plotting πb against πw revealed that for the Fst peak lying over the ROS locus (left peak), πw is low, whereas πb is similar to that across the rest of the genome (Fig. 2 F and G; red points, Fig. 3A). The ROS/EL region does not fall in a region of reduced recombination (SI Appendix, Fig. S4), so low recombination cannot explain the observed reduced diversity, unlike in other cases (27). Instead, reduced diversity at ROS is likely due to fixation of one or more favorable mutations (selective sweeps). The right Fst peak, ∼150 kb downstream of ROS, is also primarily due to a decrease in πw (lower green points, Fig. 3A). πw is reduced in both populations, for both the left and right peaks, implying at least four sweeps (i.e., at two loci for each of the two populations). By contrast, the middle peak does not have low πw but, rather, relatively high πb (light blue points, Fig. 3A). The middle peak is absent or reduced in some population comparisons (detailed below), suggesting that selective sweeps were not involved in generating it. The above results thus indicate that only the left and right Fst peaks arose through selective sweeps.
Fig. 3.
Comparison of within- and between-population divergence in the ROS/EL region. Relationship between πb and πw for pools sampled either side of the hybrid zone, separated by ∼2.5 km (A, YP1 and MP2, corresponding to Fig. 2 B and C) or ∼20 km (B, YP4 and MP11, corresponding to Fig. 2 H and I), summarized in 10-kb windows, with a color gradient indicating the respective Fst (light colors, low; dark colors, high). The left, middle, and right Fst peaks indicated in Fig. 2C are shown as red, light blue, and green points, respectively. The dark blue points indicate windows between those Fst peaks. Other windows from around the ROS region are shown in gray.
Mapping the Causal Loci.
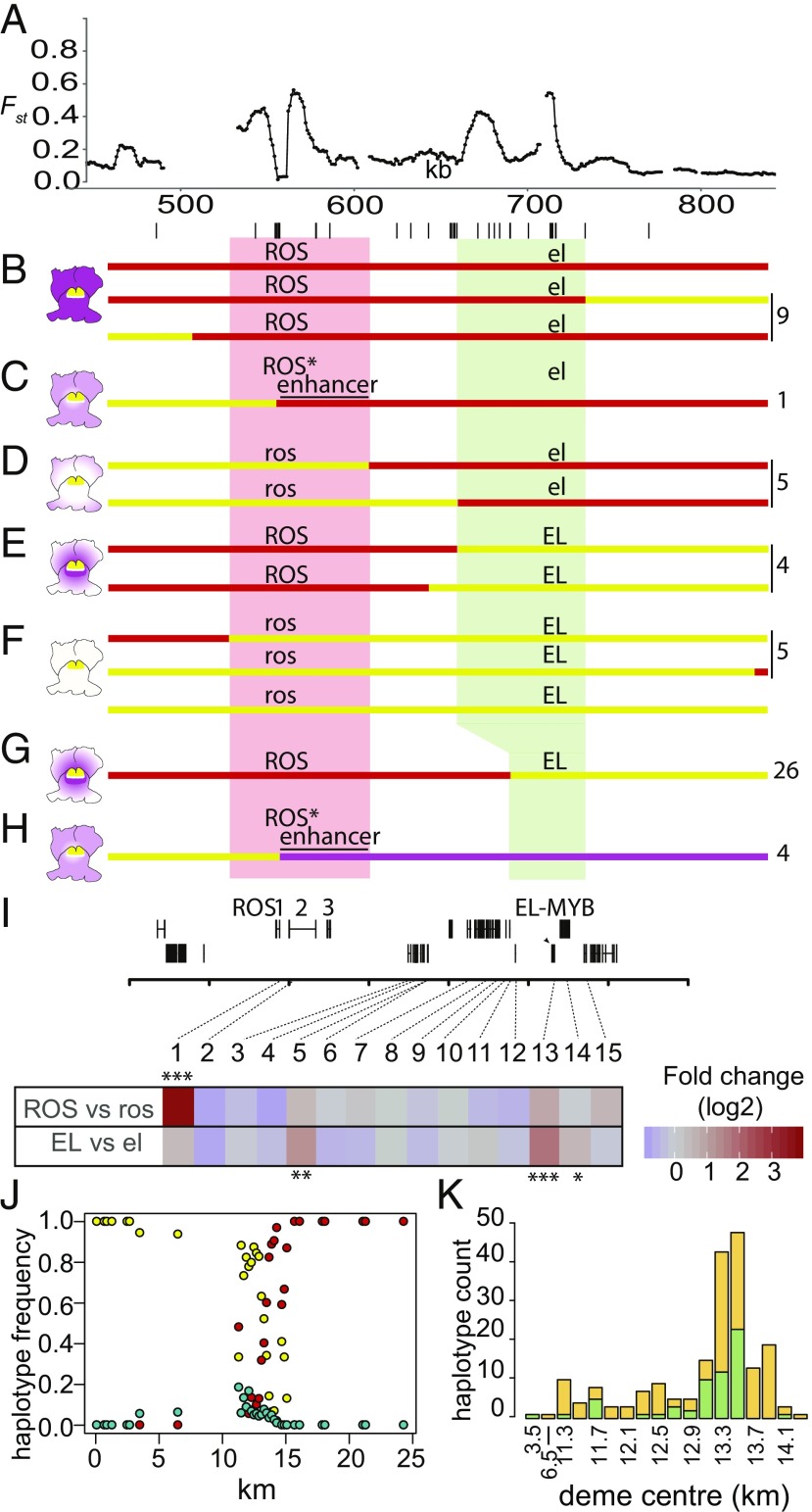
To determine whether the regions subject to selective sweeps had phenotypic effects, we introgressed ros EL from A.m.striatum into A. majus (ROS el) and genotyped F2 populations. Recombinants were backcrossed or self-pollinated to determine their homozygous phenotypes (Fig. 4 B–F). Regions causing the ROS phenotype mapped to the left Fst peak, while the EL phenotype mapped to the middle and/or right Fst peaks. The limits of ROS and EL were further refined by crossing plants heterozygous for ros EL (from A.m.striatum) and ROS el (from A.m.pseudomajus or A. majus) to a ros el/ros el line. Screening 10,261 progeny yielded 26 ROS EL recombinants, mapping EL to an interval of ∼50 kb (Fig. 4G), below the right Fst peak. The map distance between ROS and EL was 0.5 cM, corresponding to ∼3 cM/Mbp, which is of the same order as the genome-wide average of 1.8 cM/Mbp. No phenotypic effect mapped to the middle Fst peak.
Fig. 4.
Mapping loci in relation to Fst peaks. (A) Fst profile for pools in Fig. 2B (YP1 vs. MP2) showing location of genes and markers (lines below) used for mapping. (B–H) Mapping ROS and EL. Pale red and pale green boxes indicate mapping intervals for ROS and EL, respectively. Parental haplotypes shown as lines in red (A. majus JI7), magenta (A.m.pseudomajus), or yellow (A.m.striatum). Recombination to the left and right of the Fst peak gives parental phenotypes (B and F); recombination 3′ of ROS1 gives pale magenta (C and H); recombination between ROS and EL gives very pale (D) or restricted (E) patterns. Numbers of each class recovered shown, Right. (I) Floral bud expression of 15 genes found in or between the ROS and EL mapping intervals. Significant differential expression for ROS vs. ros or EL vs. el comparisons at q (false discovery rate) < 0.05, q < 0.01, and q < 0.001 is indicated by one, two, or three asterisks, respectively. Only genes with a mean expression of >5 transcripts per million are shown. The sole gene in the region with significant differential expression in ROS vs. ros comparisons was ROS1 (q < 5.6e−29). EL-MYB showed the most significant differential expression in the EL vs. el comparison (q < 2.3e−9) with two further genes (Gene 5, which is outside the mapped EL interval) and Gene 14, which is immediately adjacent to EL-MYB) reporting differential expression at lower significance thresholds. (J) Frequency of A.m.pseudomajus (magenta), A.m.striatum (yellow), and recombinant (turquoise) haplotypes in demes with ≥8 individuals along the hybrid zone transect. (K) Barplot showing counts of recombinant haplotypes for all demes with ≥8 individuals (ross elp in green; ROSp ELs in orange). Deme center locations between 11.3 and 14.3 km are at 0.2-km intervals. For details of genotyping, see SI Appendix, Supplementary Text S3.
To determine whether the flower color phenotypes reflect variation in gene expression levels, we performed RNAseq on flower buds from homozygous progeny of individuals used in the genetic mapping experiments. Two of fifteen genes detected in the ROS-EL region showed highly significant expression differences (Fig. 4I, q < 0.001; SI Appendix, Fig. S6). One transcript derived from ROS1 and was about 10 times more abundant for samples with a dominant ROS allele compared with those with recessive ros, consistent with ROS conferring strong magenta. The second differential transcript encoded a MYB-like transcription factor with 57% protein identity to ROS1 in the MYB domain and mapped to the EL region (SI Appendix, Figs. S5 and S6). This EL-MYB was expressed about threefold more in samples with a dominant EL allele compared with those with recessive el, consistent with it being a repressor of magenta pigmentation (SI Appendix, Fig. S6C). These results indicate that EL encodes a MYB-like transcription factor and show that at least some of the differences in gene activity are transcriptional. The EL-MYB gene maps to the rightmost Fst peak (Fig. 4A). Two other transcripts showed differences in expression between el and EL genotypes (genes 5 and 14, Fig. 4I, q < 0.01, q < 0.05, respectively) but showed a much weaker correlation with genotype than the EL-MYB gene (SI Appendix, Fig. S6 B and C).
We also analyzed recombinants, termed ROS1*, with breakpoints just downstream of the ROS1 gene (Fig. 4H). ROS1* is expressed at a similar level to A.m.pseudomajus ROS1, although it carries the ROS1 coding and upstream region of A.m.striatum (SI Appendix, Fig. S6C). Thus, variation in ROS1 transcript levels largely maps to a downstream enhancer. The paler flowers of ROS1* compared with A.m.pseudomajus ROS1 (Fig. 1E vs. Fig. 1H) suggests that variation in the coding region also contributes to the phenotype. Taken together with the observation of low πw for only the left and right Fst peaks, these findings suggest that selective sweeps at ROS and EL caused these Fst peaks.
Gene Flow Lowers Fst Outside the ROS/EL Region.
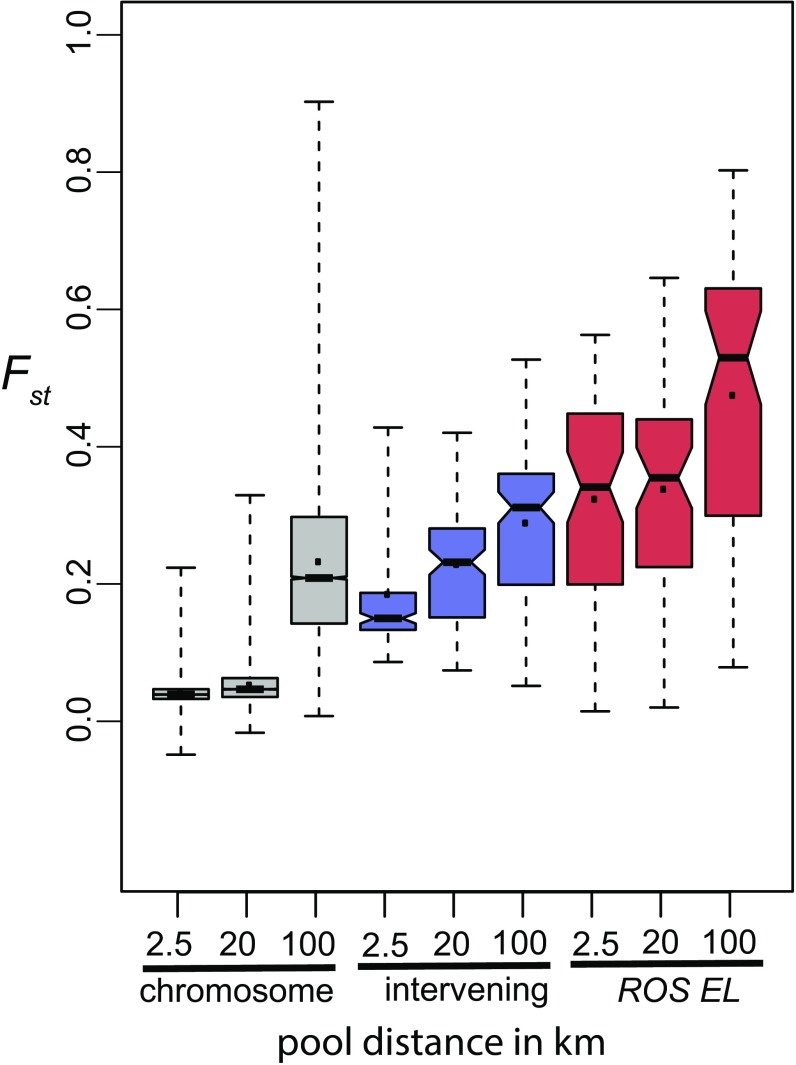
Sequence pools for populations of A.m.pseudomajus and A.m.striatum away from the center of the hybrid zone (∼20 km apart instead of ∼2.5 km) showed a higher median Fst (0.048 ± 0.0008 compared with 0.040 ± 0.0004) and more variable profile for chromosome 6 than for nearby populations (Figs. 2 H and I, 3B, and 5). By contrast, Fst values at ROS, EL, and the intervening region were similar to those for the nearby populations (Figs. 2 H and I and 5). More remote populations showed a further increase in Fst for chromosome 6, with some comparisons yielding numerous Fst peaks, so that those at ROS and EL no longer stood out (Figs. 2 J and K and 5 and SI Appendix, Fig. S3 A and D and Table S9). Such a pattern of “isolation by distance” is often seen and indicates that gene flow reduces local divergence. In contrast, Fst is elevated across the whole ROS/EL region (Fig. 5), as expected from a strong barrier to gene flow generated by selection on ROS and EL (28). The statistical significance of these patterns is considered in SI Appendix, Supplementary Text S1.3.
Fig. 5.
Relative divergence between populations at different geographic locations. Notched boxplots of Fst for three genomic regions: chromosome 6 (gray, from position >35 Mb excluding the ROS/EL region), interval between ROS and EL (blue), and the ROS and EL loci (pink). For each boxplot: the horizontal waistline indicates the median, the point indicates the mean, the length of the waist indicates the 95% confidence interval of the median, the box indicates the interquartile range, and the whiskers extend to the data minima and maxima. For each genomic region, three A.m.striatum/A.m.pseudomajus comparisons are shown, separated by 2.5 km (YP1 and MP2), 20 km (YP4 and MP11), or 100 km (ML-CIN). Distributions are based on values calculated for 10-kb windows, 1-kb step size. Windows overlying ROS and EL: midpoints 530–575 kb and 707–720 kb on ROS scaffold. Windows between ROS and EL: midpoints 576–706 kb on ROS scaffold.
A barrier to gene flow is also expected to cause sharp clines at any loci within it, regardless of whether they are selected. Indeed, we observe sharp clines at all divergent SNPs within or near the genomic islands, including those that lie outside ROS or EL (Fig. 2L and SI Appendix, Supplementary Text S2 and Fig. S7). Of the ∼6 × 105 biallelic SNPs on chromosome 6, 115 showed frequency differences greater than 0.8 between the outer pools (∼20 km apart). One hundred and one of these differential SNPs were within an ∼0.5 Mbp ROS/EL region (Fig. 2M and SI Appendix, Fig. S3C), 14 of which were within the ROS and EL Fst peaks, 74 were between these peaks, and 13 were in flanking regions. Comparing SNP allele frequencies in the pools showed that the 14 differential SNPs within the ROS and EL Fst peaks, together with most of the 74 SNPs from the intervening region, exhibited clines centered at the hybrid zone (Fig. 2 N and O), confirmed and further refined by individual genotyping (Fig. 2L and SI Appendix, Fig. S7). The remaining differential SNPs, including 14 that were distributed sparsely along the chromosome (Fig. 2M), mainly showed a frequency change over a geographic region where the population density is low (Fig. 2P and SI Appendix, Fig. S7C). The change in frequency for these SNPs likely reflects fluctuations caused by the reduced gene flow created by the population density gap.
These findings support the hypothesis of a selective barrier at the ROS/EL region. The yellow flower patterning gene SULF exhibits steep SNP clines centered at the same geographical location as ROS-EL clines (12), supporting the idea that selection on flower color is the basis of the barrier.
Based on the 0.5-cM distance between ROS and EL, recombinants should be generated at hybrid zones, at a rate of 0.5% per heterozygote. Genotyping 2,393 individuals at the hybrid zone, using haplotype-specific markers in ROS1 and EL, identified 201 recombinant haplotypes, which reached ∼10% frequency at the center of the hybrid zone (Fig. 4 J and K). Genotyping and testcrossing of progeny grown from 27 recombinants confirmed that most gave the expected phenotypes (SI Appendix, Supplementary Text S3). Assuming a neutral model with no selection against recombinants, we estimated a lower bound of ∼85 generations for the age of this hybrid zone (SI Appendix, Supplementary Text S4). If the hybrid zone is older than this, then selection must have acted to eliminate recombinants. A note attached to a herbarium specimen of A.m.pseudomajus from 1928 (London Natural History Museum) describes extensive color polymorphism at the geographic location of the hybrid zone, further suggesting that the hybrid zone is at least 90 y old.
The barrier to gene flow observed at ROS/EL raises the question of whether this alone could be responsible for the Fst peaks. According to this view, the drop in Fst in the intervening region between the peaks would be due to gene flow. However, selection at two linked loci (ROS and EL) generates a strong barrier to gene flow throughout the intervening region because two recombination events are required to transfer a neutral allele onto the opposite genetic background (SI Appendix, Supplementary Text S5 and Figs. S15 and S16). A barrier of this form would therefore not be expected to generate two separate sharp peaks in Fst, as is observed. Thus, the barrier to gene flow alone cannot be responsible for the two sharp Fst peaks. This argument illustrates the value of having two linked loci for distinguishing hypotheses. A further advantage of having two linked loci is that it allows a region of elevated Fst to be readily picked out because the barrier extends over 0.5 cM and >200 kb. Single selected loci would generate a barrier over a narrow region, which would be harder to detect.
The observation that flower color variation under selection derives from two closely-linked loci (ROS and EL) seems to lend support to the idea that divergent loci tend to cluster because linkage hinders swamping of locally adapted alleles (5, 29). However, other pigment loci under selection (e.g., SULF) are unlinked to ROS and EL, showing that tight linkage is not essential. Moreover, ROS and EL are both MYB-like transcription factors and so may be clustered due to gene duplication. Thus, clustering may not be due to selection for linkage (SI Appendix, Supplementary Text S1.6).
Role of Selective Sweeps and Barriers to Gene Flow in Generating Genomic Islands.
Taken together, the clines, genetic analysis, transcriptional differences, and analysis of Fst peaks indicate that the ROS/EL genomic island and its surround have been shaped by two processes: (i) historic selective sweeps that led to different ROS and EL alleles becoming fixed in A.m.pseudomajus and A.m.striatum populations and (ii) selection against hybrid genotypes generated where A.m.pseudomajus and A.m.striatum populations meet, creating a local barrier to gene flow (28). We performed simulations to explore scenarios consistent with the data and modes of selection.
To provide constraints on simulations, we first estimated the age of the selective sweeps. Based on the residual diversity within the sharp peaks at ROS and EL, we estimated the date of the most recent sweeps to be ∼90,000 generations ago (SI Appendix, Supplementary Text S1); this is an upper bound, since “soft sweeps” might not have eliminated all diversity. We also estimated the age of the barrier to gene flow. As detailed in SI Appendix, Supplementary Text S1, the time required for Fst in the ROS/EL interval to accumulate to the observed value of 0.125 is T ∼ 0.54 Ne ∼ 45,000 generations (where Ne = effective population size). Thus, both estimates suggest that selective sweeps and a barrier to gene flow were established roughly Ne ∼ 105 generations ago.
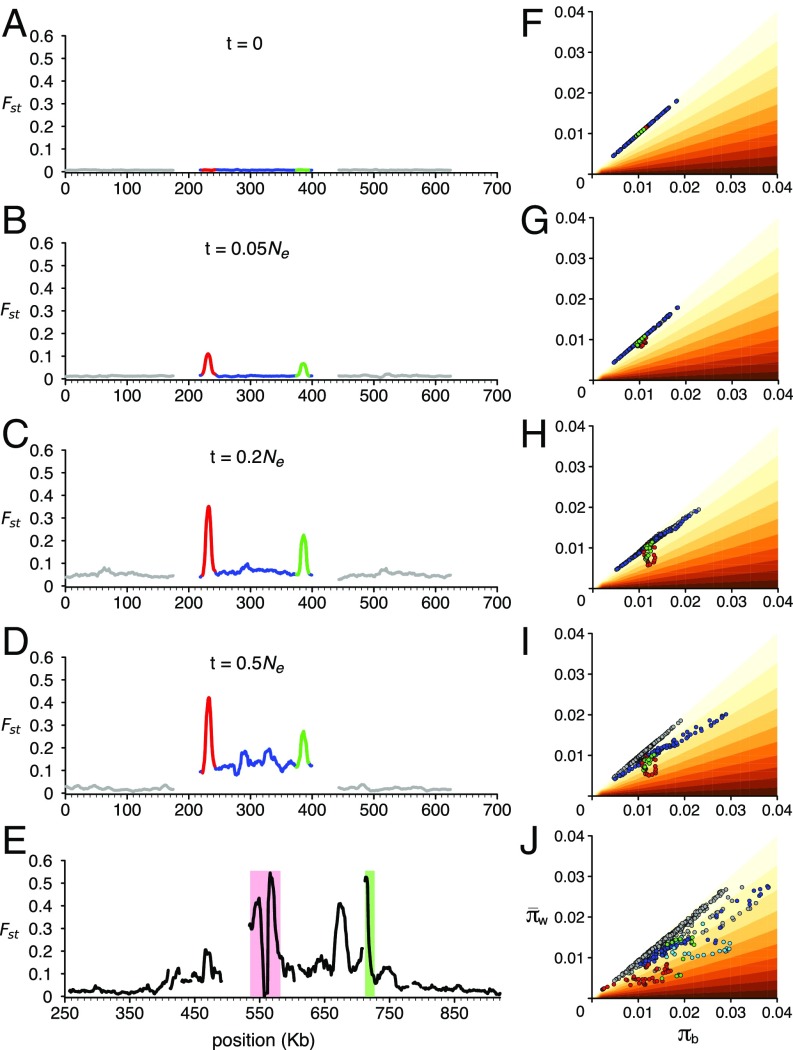
We assume that a homogeneous ancestral population is first split by a geographic barrier, allowing sweeps to occur independently in each population (Fig. 6 A and F, for simplicity assuming an initial Fst 0.0). Geographic separation is a simple way of ensuring that alleles swept in one population do not sweep into the other, although other scenarios such as environmental heterogeneity are possible; the sequence data are also compatible with divergence in primary contact. Sweeps at ROS and EL (red, green in Fig. 6 B and G) reduce diversity, πw, generating peaks in Fst. These sweeps presumably reflect the selective advantage of a change in flower color, compared with the ancestral phenotype in each population. Given that both populations underwent sweeps, the ancestral flower phenotype would have been different from both of the current phenotypes in A.m.pseudomajus or A.m.striatum. Further sweeps at ROS and EL strengthen the Fst peaks (Fig. 6 C and H). Unlike the simulations, in real populations, it is possible that global and/or local sweeps occur at many other genetic loci and spatial locations, in addition to ROS and EL, creating a more rugged Fst profile across the genome.
Fig. 6.
Simulations of gene flow and selective sweeps. Combined effects of a barrier to gene flow and selective sweeps on Fst (Left) and on πb and πw (Right). (A and F) A homogeneous population is split by a geographic barrier. (B and G) Alleles at ROS and EL (red, green) sweep through the separate populations, reducing diversity, πw, generating peaks in Fst. (C and H) Further sweeps occur at ROS and EL, strengthening the Fst peaks. By t = 0.2 Ne generations, divergence has increased genome-wide, with Fst 0.05. At this time, the divergent populations meet and exchange genes everywhere except between ROS and EL. (D and I) By time 0.5 Ne, Fst outside ROS/EL has decreased due to mixing (Left, black), but has increased between ROS and EL (Left, blue). Although in this scenario, population contact was established at 0.2 Ne, similar final profiles for Fst, πb, and πw would be generated, with contact being made earlier or later than this. (E and J) The πb, πw observed in pools YP1, MP2, 2.5 km apart, with the maximum Fst observed at ROS indicated by pale red (E) or red (J), and at EL indicated by green. Note that Ne is estimated as roughly 8.3 × 104 (SI Appendix, Supplementary Text S1.3). For further details, see SI Appendix, Supplementary Text S1.5.
After a period of time (0.2 Ne generations in the simulation shown in Fig. 6), the divergent populations come into contact. Gene flow leads to a lowering of Fst from the chromosome-wide average, except at loci where a barrier has been established. We propose that a barrier to gene flow occurs for only a subset of swept loci: those for which epistatic interactions or frequency dependence maintain divergence. ROS and EL represent one such case, as their interactions, together with loci controlling yellow, lead to alternative floral guides. Other loci that underwent sweeps, but led to no incompatibility (presumably the majority of sweeps) would undergo gene flow, with the allele conferring higher overall fitness going to fixation in both populations. By time 0.5 Ne, Fst outside ROS/EL has decreased due to gene flow (gray), but has further increased between ROS and EL (blue) because of the local barrier to gene flow (Fig. 6 D and I). The resulting Fst, πb, and πw values are comparable to those observed (compare Fig. 6 D and I with Fig. 6 E and J). According to the above scenario, selective sweeps led to fixation of different alleles in each population, and selection maintains a local barrier to gene flow. Multiple changes in alleles are involved, a reasonable assumption given these events occurred over a period of ∼105 generations, extending over glacial periods, during which populations and the environment were in a state of flux.
Our analysis indicates that both selective sweeps and barriers to gene flow combine to shape genomic islands of differentiation. The barrier to gene flow at ROS/EL is insufficient to prevent exchange for much of the genome. However, if the barrier were more severe and applied to additional loci, it could prevent gene flow more completely, leading to speciation. The mechanisms that created the genomic islands may therefore represent partial steps toward reproductive isolation and speciation.
Materials and Methods
Full details of plant material, DNA extraction, genome sequence analysis, population genomics, genotyping, SNP analysis for geographic, and RNAseq analysis are given in SI Appendix, Materials and Methods. Details on inferences from pairwise diversity and divergence, geographic cline analysis, and genotypic screens are given in SI Appendix. Genomic sequence datasets are available at European Nucleotide Archive (ENA) with accession number PRJEB28287, and RNAseq datasets are deposited in National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) with accession number GSE118621. Associated scripts are provided at linked public data repositories as detailed in SI Appendix, Materials and Methods, and further information on the hybrid zone is available at www.antspec.org.
Supplementary Material
Acknowledgments
Many thanks to Christophe Thébaud for sharing his finding of the herbarium specimen referenced in the text. This work was supported by Biotechnology and Biological Sciences Research Council Grants BBS/E/J/000PR9773 and BB/G009325/1 (to E.C.), ERC Grant 201252 (to N.H.B.), and a PhD scholarship (to H.T.) from the Portuguese Foundation for Science and Technology (FCT), through the Human Potential Operating Programme (POPH) of the National Strategic Reference Framework (QREN), within the European Social Fund (Scholarship SFRH/BD/60982/2009). This research was supported in part by the Norwich BioScience Institutes Computing infrastructure for Science (CiS) group.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genomic sequence data reported in this paper are available at European Nucleotide Archive (ENA), https://www.ebi.ac.uk/ena (accession no. ENA PRJEB28287), and the RNAseq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE118621).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801832115/-/DCSupplemental.
References
- 1.Hohenlohe PA, et al. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin SH, et al. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23:1817–1828. doi: 10.1101/gr.159426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poelstra JW, et al. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 2014;344:1410–1414. doi: 10.1126/science.1253226. [DOI] [PubMed] [Google Scholar]
- 4.Clarkson CS, et al. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun. 2014;5:4248. doi: 10.1038/ncomms5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 6.Pennisi E. Disputed islands. Science. 2014;345:611–613. doi: 10.1126/science.345.6197.611. [DOI] [PubMed] [Google Scholar]
- 7.Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23:3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- 8.Ma T, et al. Ancient polymorphisms and divergence hitchhiking contribute to genomic islands of divergence within a poplar species complex. Proc Natl Acad Sci USA. 2018;115:E236–E243. doi: 10.1073/pnas.1713288114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf JB, Ellegren H. Making sense of genomic islands of differentiation in light of speciation. Nat Rev Genet. 2017;18:87–100. doi: 10.1038/nrg.2016.133. [DOI] [PubMed] [Google Scholar]
- 10.Charlesworth B, Nordborg M, Charlesworth D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet Res. 1997;70:155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- 11.Whibley AC, et al. Evolutionary paths underlying flower color variation in Antirrhinum. Science. 2006;313:963–966. doi: 10.1126/science.1129161. [DOI] [PubMed] [Google Scholar]
- 12.Bradley D, et al. Evolution of flower color pattern through selection on regulatory small RNAs. Science. 2017;358:925–928. doi: 10.1126/science.aao3526. [DOI] [PubMed] [Google Scholar]
- 13.Hackbarth J, Michaelis P, Scheller G. Untersuchungen an dem Antirrhinum-Wildsippen-Sortiment von E. Baur. Z Indukt Abstamm Vererbungsl. 1942;80:1–102. [Google Scholar]
- 14.Schwinn K, et al. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell. 2006;18:831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stubbe H. Genetik und Zytologie von Antirrhinum L. sect. Antirrhinum. Veb Gustav Fischer Verlag; Jena, Germany: 1966. [Google Scholar]
- 16.Khimoun A, et al. Ecology predicts parapatric distributions in two closely-related Antirrhinum majus subspecies. Evol Ecol. 2012;27:51–64. [Google Scholar]
- 17.Shang Y, et al. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011;189:602–615. doi: 10.1111/j.1469-8137.2010.03498.x. [DOI] [PubMed] [Google Scholar]
- 18.Gegear RJ, Laverty TM. Flower constancy in bumblebees: A test of the trait variability hypothesis. Anim Behav. 2005;69:939–949. [Google Scholar]
- 19.Smithson A, Macnair MR. Frequency-dependent selection by pollinators: Mechanisms and consequences with regard to behaviour of bumblebees Bombus terrestris (L.) (Hymenoptera: Apidae) J Evol Biol. 1996;9:571–588. [Google Scholar]
- 20.Oyama RK, Jones KN, Baum DA. Sympatric sister species of Californian Antirrhinum and their transiently specialized pollinators. Am Midl Nat. 2010;164:337–347. [Google Scholar]
- 21.Counterman BA, et al. Genomic hotspots for adaptation: The population genetics of Müllerian mimicry in Heliconius erato. PLoS Genet. 2010;6:e1000796. doi: 10.1371/journal.pgen.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallet J, Barton NH. Strong natural selection in a warning-color hybrid zone. Evolution. 1989;43:421–431. doi: 10.1111/j.1558-5646.1989.tb04237.x. [DOI] [PubMed] [Google Scholar]
- 23.Joron M, Mallet JL. Diversity in mimicry: Paradox or paradigm? Trends Ecol Evol. 1998;13:461–466. doi: 10.1016/s0169-5347(98)01483-9. [DOI] [PubMed] [Google Scholar]
- 24.Jiggins CD. The Ecology and Evolution of Heliconius Butterflies. Oxford Univ Press; Oxford: 2017. [Google Scholar]
- 25.Nadeau NJ, et al. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos Trans R Soc Lond B Biol Sci. 2012;367:343–353. doi: 10.1098/rstb.2011.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlötterer C, Tobler R, Kofler R, Nolte V. Sequencing pools of individuals–Mining genome-wide polymorphism data without big funding. Nat Rev Genet. 2014;15:749–763. doi: 10.1038/nrg3803. [DOI] [PubMed] [Google Scholar]
- 27.Payseur BA, Rieseberg LH. A genomic perspective on hybridization and speciation. Mol Ecol. 2016;25:2337–2360. doi: 10.1111/mec.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barton N, Bengtsson BO. The barrier to genetic exchange between hybridising populations. Heredity (Edinb) 1986;57:357–376. doi: 10.1038/hdy.1986.135. [DOI] [PubMed] [Google Scholar]
- 29.Yeaman S, Aeschbacher S, Bürger R. The evolution of genomic islands by increased establishment probability of linked alleles. Mol Ecol. 2016;25:2542–2558. doi: 10.1111/mec.13611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.