Significance
R loops form during transcription when the mRNA hybridizes back to the template DNA forming a stable DNA–RNA hybrid. Stable R loops can block replication and transcription machineries, leading to genome instability and human diseases. We report that the human mitochondrial degradosome (mtEXO), formed by SUV3 and PNPase, plays an important role in R-loop metabolism in the mitochondria apart from preventing dsRNA accumulation, with implications in replication and preventing instability of the mitochondrial genome. Our findings help establish a connection between the RNA processing activities of the mtEXO and mitochondrial genome stability, affecting mitochondria homeostasis, cellular metabolism, and human diseases.
Keywords: genome instability, mitochondrial degradosome, R loops, RNA metabolism, transcription–replication conflicts
Abstract
R loops are nucleic acid structures comprising an DNA–RNA hybrid and a displaced single-stranded DNA. These structures may occur transiently during transcription, playing essential biological functions. However, persistent R loops may become pathological as they are important drivers of genome instability and have been associated with human diseases. The mitochondrial degradosome is a functionally conserved complex from bacteria to human mitochondria. It is composed of the ATP-dependent RNA and DNA helicase SUV3 and the PNPase ribonuclease, playing a central role in mitochondrial RNA surveillance and degradation. Here we describe a new role for the mitochondrial degradosome in preventing the accumulation of pathological R loops in the mitochondrial DNA, in addition to preventing dsRNA accumulation. Our data indicate that, similar to the molecular mechanisms acting in the nucleus, RNA surveillance mechanisms in the mitochondria are crucial to maintain its genome integrity by counteracting pathological R-loop accumulation.
Very-short DNA–RNA hybrids occur during both replication and transcription, serving specific purposes in DNA or RNA metabolism, such as priming lagging-strand DNA synthesis or extending the nascent RNA inside the active pocket of RNA polymerases. R loops, on the other hand, are a distinct class of nucleic acid structures, comprising an DNA–RNA hybrid and a displaced single-stranded DNA (1). These structures also occur naturally, mostly in a cotranscriptional manner and have been shown to play essential biological functions in different organisms. Namely, R loops are essential for Escherichia coli plasmid replication, mitochondrial DNA replication, and human Ig class switch recombination, and evidence accumulates supporting their involvement in DNA damage repair, transcription regulation, and chromatin states in human cells and other organisms (2–5). Indeed, the presence of R loops in genomes has proven to be more frequently detected than initially anticipated, as shown by the growing number of whole-genome sequencing studies mapping the genomic locations where these structures arise. These studies dissecting the nature and distribution of R loops have helped establish common features aiding R-loop formation and stabilization, showing that negative supercoiling, GC content, repetitive sequences, and the occurrence of nicks or breaks on the DNA molecule are major contributing factors (6–10). Despite the specific cellular functions of certain R loops, these can alter the surrounding chromatin landscape and become an obstacle on the DNA molecule that can stall the replication machinery and, subsequently, lead to DNA breaks, representing an important source of DNA damage and a driver of genome instability (11, 12). To regulate the accumulation of R loops, cells have developed different mechanisms that avoid or remove them. As an initial measure to avoid R-loop formation, RNA-binding proteins (RBPs) that associate with the transcription machinery, such as the THO complex, bind the nascent RNA and reduce its ability to hybridize with the negatively supercoiled DNA template behind the traveling RNA polymerase (13, 14). Indeed, other factors involved in the RNA packing into messenger ribonucleoprotein complexes (mRNPs), splicing and export of the mature mRNA can lead to the accumulation of harmful R loops, indicating that RNA biogenesis and export in general contribute to prevent R-loop accumulation (15, 16). Additionally, cells have also developed means to deal with R loops once they form. These include a growing number of RNA and DNA helicases, such as Senataxin and DDX23, which unwind the RNA hybridized from the DNA molecule, with yet unknown genomic locus specificity, if any (5, 17). The best-studied R-loop–removing factors, and likely most important, are the RNase H endonucleases, which are present in most organisms and specifically target and degrade the RNA moiety within the DNA–RNA hybrids (18). RNase H1, in particular, has been proven central for proper DNA replication in the nucleus, removing Okazaki fragments, and in the mitochondria, where replication initiation relies on a persistent R loop used as a primer for the replicative DNA polymerase (19). In addition, well-established genome guardians from the DNA damage response and repair pathways have also been implicated in reducing harmful R loops associated with replication fork stalling, such as BRCA1, BRCA2, and Fanconi anemia factors, which likely deal with persistent and unresolved R loops in cells entering the S–G2 phase of the cell cycle (20, 21). This is in favor of persistent R loops threatening genome integrity by impairing replication fork progression and/or causing DNA breaks, making the contribution of replication-associated DNA repair pathways in eliminating them of paramount importance.
The human mitochondrial DNA (mtDNA) is a small 16.6-kb circular double-stranded DNA (dsDNA) molecule, containing 37 genes encoding essential components of the mitochondrial respiratory chain and specific tRNAs and rRNAs. Within each mitochondrion, several hundreds of mtDNA copies are present and organized in mtDNA–protein complexes termed nucleoids (22). mtDNA has a C-rich light (L) strand and a complementary G-rich heavy (H) strand, with one large noncoding region (NCR) where most of the regulatory elements locate, including the H strand origin of replication (OH), and the L and H strand promoters (LSPs and HSPs, respectively). The NCR, or control region, of many mtDNA molecules presents a three-stranded complex structure that contains a D loop formed by the 7S DNA, where replication is initiated from the OH. Also at the control region, a stable R loop is found at high frequency, formed by a small RNA transcribed from the LSP by the mitochondrial RNA polymerase (POLRMT) (23). This plays a critical role in mtDNA replication serving as a primer for leading-strand synthesis by the mitochondrial DNA polymerase Polγ from OH, after processing by RNase H1 (19, 24). Both the D loop and R loop have been implicated not only in replication of the mtDNA, but also in the organization and distribution of the mitochondrial genome (25, 26). Transcription of the mtDNA by POLRMT is initiated from the HSPs and LSPs, producing long polycistronic transcripts from both strands that undergo nucleolytic processing to yield the mitochondrial mRNA, rRNA, and tRNA species (27, 28). Posttranscriptional processing of mtRNAs and assembly of mitochondrial ribosomes has been shown to occur in specialized centers adjacent to nucleoids, termed RNA granules (29, 30).
The mitochondrial degradosome (mtEXO) is a functionally conserved complex from bacteria to humans, specifically involved in maintaining mitochondrial integrity in eukaryotes mostly by its function in RNA surveillance and degradation (31). It is composed of the ATP-dependent RNA and DNA helicase SUV3 (SUV3 or SUPV3L1) and PNPase (PNPT1), which has poly(A) polymerase (PAP) and exoribonuclease activities. Both are encoded in the nuclear genome and imported to the mitochondria to exert their functions (32, 33). It has been shown in vitro that purified SUV3 has the ability to unwind double-stranded RNA (dsRNA), DNA–RNA, or dsDNA, and its helicase activity is enhanced when in complex with PNPase, which then promotes 3′–5′ degradation of the RNA molecules (34–36). In yeast, the mtEXO complex also shows severe mitochondrial RNA turnover defects and leads to loss of mitochondrial genome, rendering the cells “petite” (37, 38). In mammalian cells, in addition to their well-established role in RNA turnover as part of the mtEXO, SUV3 and PNPase have been implicated in other processes related to mitochondrial integrity and correct cellular function. In particular, SUV3 and PNPT1 homozygous KO mice are embryonic lethal (39, 40), and SUV3 heterozygous mice show increased tumor development and premature aging phenotypes (40), serving the idea that it might act as a tumor suppressor gene. Still, the causes of these phenotypes and their relation with the known mtEXO function are unclear.
Here we show that the loss of SUV3–PNPase activity, in addition to disrupting mtRNA turnover, favors the accumulation of dsRNA and leads to the accumulation of R loops in the mitochondrial genome in HeLa and U2OS cells, generating problems in replication and mitochondrial genome integrity. Our findings suggest a role for the mitochondrial degradosome in counteracting deleterious R loops at specific hybrid-prone regions in the mitochondrial genome. In addition to a role in preventing R loops at mitochondrial genes, which can impair replication progression, we propose that the mtEXO might have a particularly relevant role at the control region, modulating the abundance of the RNA species used for priming replication from OH. Ultimately, our results suggest that RNA surveillance mechanisms in the mitochondria, similar to the molecular mechanisms acting in the nucleus, are crucial in avoiding genomic instability by counteracting pathological R-loop accumulation and preventing replication–transcription conflicts, supporting the notion that this is a general mechanism for genome integrity.
Results
Loss of mtEXO Leads to Accumulation of DNA–RNA Hybrids and dsRNA in Mitochondria.
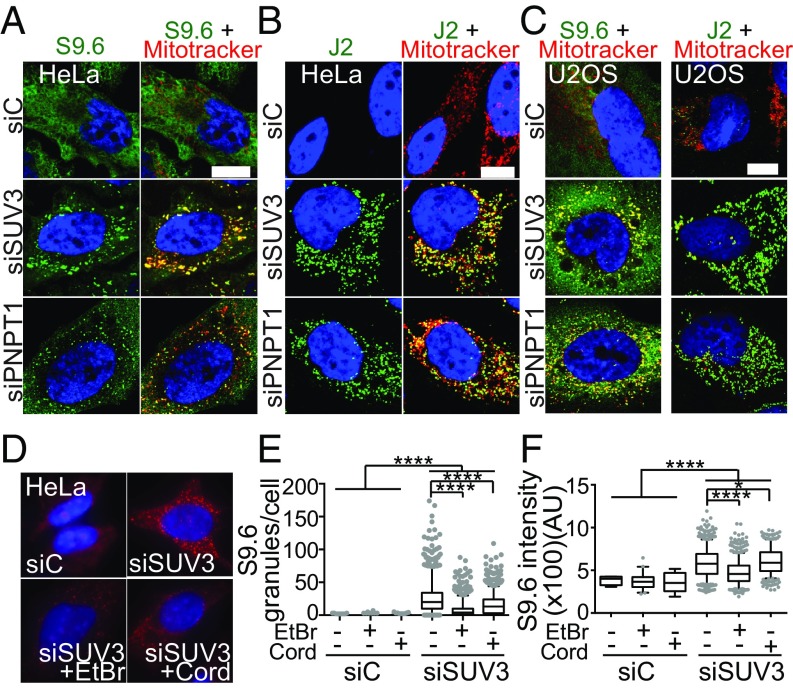
A number of nuclear-encoded proteins involved in various aspects of DNA and RNA metabolism including nuclear RNA surveillance, have been implicated in nuclear R-loop prevention and R-loop–associated genome instability (11). To address whether these processes are conserved in nonnuclear genomes, we analyzed whether mitochondrial RNA surveillance, in particular the mitochondrial degradosome (mtEXO), has any role in mitochondrial R-loop homeostasis and genome integrity. To visualize and establish R-loop detection we relied on immunofluorescence (IF) with the S9.6 anti-DNA–RNA hybrid antibody and RNase H1 overexpression and/or in vitro treatment, previously demonstrated to be bona fide readouts of R-loop presence (6, 14, 15, 41). We found that silencing the mtEXO exoribonuclease-encoding gene PNPT1 using siRNA (siPNPT1) produced an unexpected punctate S9.6 staining pattern in the cytoplasm. Importantly, depletion of the other mtEXO subunit, the SUV3 helicase by siRNA (siSUV3), conferred the same punctate S9.6 staining pattern. To test whether the S9.6 signal was confined or in the soluble cytoplasmic fraction, we assessed colocalization with the mitochondria using Mitotracker. Confocal microscopy analysis of SUV3- and PNPase-depleted cells showed that the S9.6 signal colocalized within the mitochondria in aggregates (henceforth referred to as granules), compared with the generally diffuse S9.6 signal observed in control cells (Fig. 1A). Since mtEXO is involved in mtRNA degradation and turnover, and loss of mtEXO function leads to the accumulation of processing intermediates and decay RNA species (32, 33, 42), we also tested the levels of dsRNA in both mtEXO-depleted cells using the J2 antibody, which recognizes dsRNA molecules (43). mtEXO-depleted cells showed a J2 granular pattern similar to the S9.6 staining, localizing within the mitochondrial network, evidencing the defective RNA turnover and degradation caused by mtEXO loss (Fig. 1B). The same S9.6 and J2 staining phenotypes were confirmed in U2OS cells (Fig. 1C).
Fig. 1.
Accumulation of transcription-dependent DNA–RNA hybrids and dsRNA in mitochondria of HeLa and U2OS cells depleted of SUV3 and PNPase. (A) IF of DNA–RNA hybrids using the S9.6 antibody and (B) dsRNA using the J2 antibody in HeLa cells transfected with siC (control), siSUV3, and siPNPT1. Mitochondria are stained with MitoTracker. (C) IF with MitoTracker and S9.6 and J2 antibodies in siC, siSUV3, and siPNPT1 in U2OS cells. (D) Effect of transcription-blocking agents in the formation of S9.6 granules in HeLa cells seen by IF. Cells were treated with 50 ng/mL EtBr for 5 h or 100 μM cordycepin for 2 h before fixation. Nuclei were stained with DAPI. (Scale bar, 10 μm.) (E) Box plots represent the median and quartile levels of total S9.6 granules per cell or (F) granule intensity obtained from three independent experiments. ****P < 0.0001; *P < 0.05 according to Mann–Whitney U test. AU, arbitrary units.
We next analyzed the dependency of these granules on active transcription. For this, we treated cells with low doses of ethidium bromide (EtBr), which have been shown to effectively inhibit mtDNA transcription (44, 45), or cordycepin (3′ deoxyadenosine), an RNA chain elongation inhibitor shown to effectively inhibit both nuclear (46) and mitochondrial RNA synthesis without affecting RNA stability (47). A marked decrease in the number and intensity of S9.6 granules was observed in SUV3-depleted cells when transcription was inhibited with either drug (Fig. 1 D–F), confirming that the signals detected were cotranscriptional.
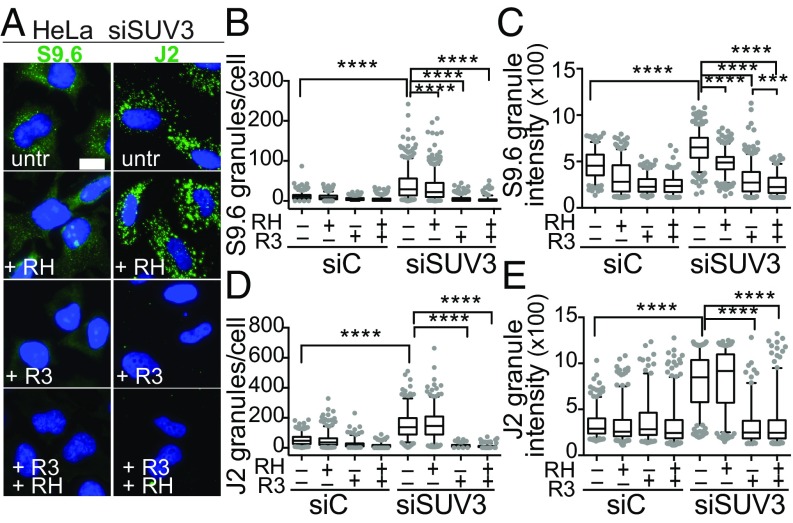
The IF data indicated that mtEXO-depleted cells accumulate both dsRNA and DNA–RNA hybrid molecules in mitochondria. However, the high dsRNA signal observed in mtEXO-depleted cells led us question whether high dsRNA levels could interfere with the S9.6 antibody, despite the high specificity of S9.6 for DNA–RNA hybrids (48, 49). To shed light on the nature of the structures lighted up by the antibodies, we performed IF with the S9.6 and J2 antibodies in samples treated in vitro with RNases H and III, (Fig. 2A). First, we confirmed that the two enzymes had substrate specificity for DNA–RNA hybrids and dsRNA in vitro, respectively (SI Appendix, Fig. S1A), as expected (45, 50). Importantly, we found mtEXO-depleted HeLa cells treated with RNase H showed a small reduction of S9.6 granules and a significant reduction in their intensity (Fig. 2 B and C and SI Appendix, Fig. S1B), which was further reduced when treated with RNase III or both ribonucleases simultaneously. In contrast, RNase H did not affect the number or intensity of J2 granules (Fig. 2 D and E and SI Appendix, Fig. S1C). However, RNase III removed the granular staining of both S9.6 and J2 (Fig. 2 B and D). These results indicate that the granular structures observed by IF demark local accumulation of both dsRNA and DNA–RNA hybrids, DNA–RNA hybrids being a smaller but significant fraction of the retrieved IF signal.
Fig. 2.
IF analysis of DNA–RNA hybrids and dsRNA in mitochondria of mtEXO-depleted HeLa cells. (A) Representative IF of S9.6 (hybrids) and J2 (dsRNA) granules in siSUV3 HeLa cells in control conditions [buffer only, untreated (utr)] or treated in vitro with RNase H (RH), RNase III (R3), or both. Nuclei were stained with DAPI. (B) Quantification of S9.6 granules per cell and (C) intensity. (D) Quantification of J2 granules per cell and (E) intensity. ****P < 0.0001 according to Mann–Whitney U test.
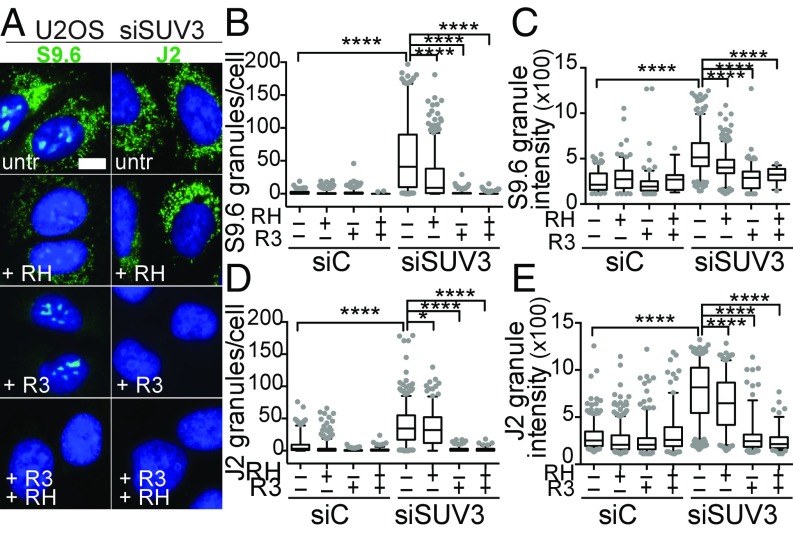
We also analyzed the effect of transient in vivo overexpression of human RNase H1 on the S9.6 staining of mtEXO-depleted cells and observed a decrease in the number of S9.6 granules (SI Appendix, Fig. S1 D and E), further confirming the accumulation of hybrids in the mitochondria of SUV3-deficient cells. The same sensitivity of the S9.6 and J2 signals to both RNase H and RNase III treatment was observed in mtEXO-depleted U2OS cells (Fig. 3 and SI Appendix, Fig. S2). Since the same results were obtained with siSUV3 and siPNPT1, further analyses were mostly focused on SUV3-depleted cells, since depletion of the SUV3 protein was more efficient than PNPase (SI Appendix, Fig. S3 A–D).
Fig. 3.
IF analysis of DNA–RNA hybrids and dsRNA in mitochondria of mtEXO-depleted U2OS cells. (A) Representative IF of S9.6 (hybrids) and J2 (dsRNA) granules in siSUV3 U2OS cells treated in vitro as in Fig. 2. (B) Quantification of S9.6 granules per cell and (C) intensity. (D) Quantification of J2 granules per cell and (E) intensity. ****P < 0.0001; *P < 0.05 according to Mann–Whitney U test.
Finally, we could also observe changes in mitochondrial morphology of mtEXO-deficient cells with respect to the control. Using the MiNa script for mitochondrial network analysis in mammalian cells in Fiji software (51, 52), we could see a shift from the normal tubular mitochondrial network to a bulkier granular morphology (SI Appendix, Fig. S4), confirming previous observations in SUV3-depleted cells (53).
Loss of mtEXO Leads to Increased R Loops in the Mitochondrial Genome.
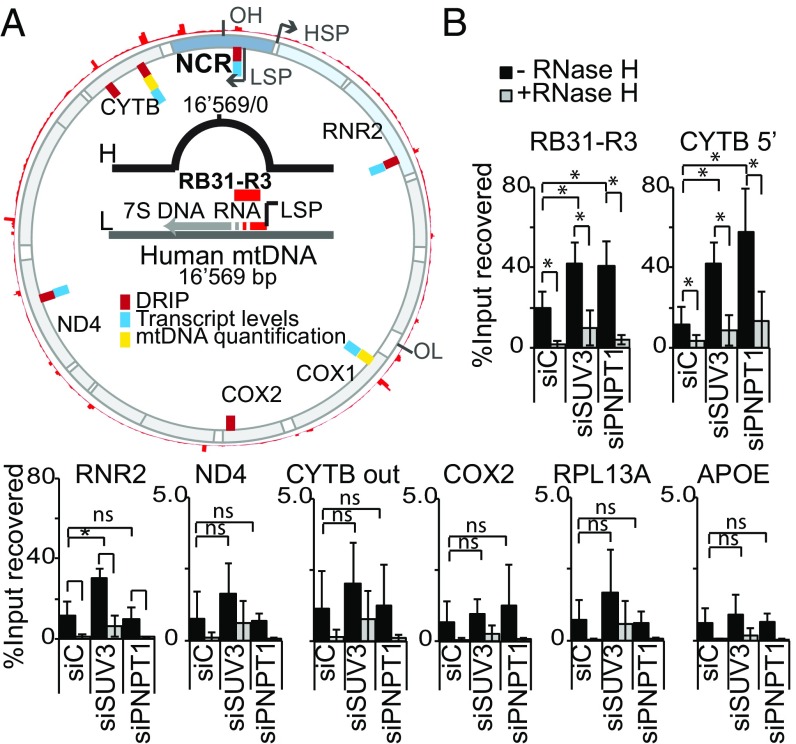
To confirm at the molecular level that mtEXO-deficient cells accumulate DNA–RNA hybrids in the mitochondrial genome, we performed DNA–RNA immunoprecipitation (DRIP). Apart from the stable R loop formed at the control region between the LSP and OH, which can be readily detected by DRIP analysis using the primer pair RB31-3B (54), R loops have been identified to occur naturally and consistently in several discrete regions around the mitochondrial genome (8). Based on the available DRIP-seq data (accession no. GSE68953), we designed different sets of primers coinciding either with R-loop peak regions or not, to analyze the accumulation of hybrids at different mitochondrial loci and allowing us to distinguish between previously described (native) or new (nonnative) hybrid-accumulating regions (Fig. 4A). Loss of mtEXO function by depletion of either SUV3 or PNPase led to a significant increase in hybrids found at the control region (RB31-3B) and at the native R-loop–forming sites that we studied (RNR2 and CYTB), which could no longer be detected when samples were treated with RNase H in vitro before immunoprecipitation (Fig. 4B). Conversely, we did not observe, at levels detectable by this technique, a significant increase in hybrids either at nuclear control genes (RPL13A and APOE) or at sites in the mitochondrial genome where no R loops tend to occur naturally (ND4, CYTB out, COX2) (Fig. 4B). This result is consistent with an increase in the number of mitochondrial DNA molecules with hybrids detected in mtEXO-depleted cells, in support of the IF data.
Fig. 4.
Analysis of R-loop accumulation in the mitochondrial genome by DRIP-qPCR in HeLa cells depleted of SUV3 and PNPase. (A) Diagram of the human mtDNA showing features relevant for this study. Outermost red peak profile represents the alignment of R-loop accumulating sequences along the mitochondrial genome. Inner red dashes represent the location of primers used for DRIP experiments; blue dashes, primers used for mtRNA expression levels; and yellow dashes, primers used for quantifying mtDNA relative to nuclear DNA content. Diagram in the center details the position of the primers targeting the stable R loop present at the NCR, downstream of the LSP (RB31-R3). (B) DRIP analysis showing the percentage of recovered input from S9.6 IP as mean value ± SD from at least four independent experiments for each region of interest. Nuclear genes included as controls (RPL13A and APOE). Differences were analyzed using t test (*P < 0.05).
To investigate whether we could see any striking difference in transcript levels in mtEXO-deficient cells, we analyzed the accumulation of different RNA species by RT-qPCR. cDNA was synthesized from whole RNA extracts without any denaturation step, allowing us to analyze directly nascent, processed, or unprocessed single-stranded mtRNA molecules without accounting for signal deriving from dsRNA molecules. The only readily noticeable change we found was in the abundance of the RNA species forming the R loop at the control region (RB31-R3), which was consistently increased when silencing SUV3 or PNPT1 in all experiments (SI Appendix, Fig. S5 A and B). Our analysis could not discriminate, however, whether this highly abundant species corresponded to the short RNA species of the natural primer occurring between the LSP and CBSII sites or to alternative longer RNA species such as those formed at high frequency in mouse mtDNA (26). This result opens the possibility that the mtEXO might be involved in regulating DNA–RNA hybrids at the control region, whether by assisting RNase H1 or by recognizing and degrading an excess of nascent RNA species, thus maintaining the steady-state levels available for R-loop formation.
Loss of mtEXO Impairs mtDNA Replication Causing Mitochondrial Genomic Instability.
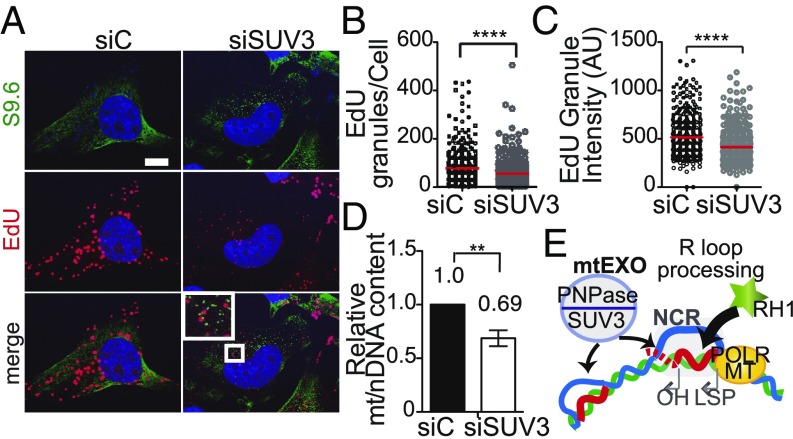
Next, we investigated how the increase of R loops in the mitochondrial genome was affecting mtDNA maintenance. To assay possible defects in mitochondrial replication, we measured 5-ethynyl-2′-deoxyuridine (EdU) incorporation into newly synthesized mtDNA and visualized replicating nucleoids by microscopy analysis (Fig. 5A). Cells were treated with aphidicolin before the EdU incubation, which inhibits the nuclear replicative DNA polymerases but not the mtDNA polymerase γ (55, 56), to have a clean measure of replicating mtDNA without the interfering signal from nuclei. In control cells, we could observe a high number of nucleoids engaged in new mtDNA synthesis that showed EdU incorporation colocalizing within the mitochondrial network (SI Appendix, Fig. S6A), while in SUV3-depleted cells, there was a decrease in mtDNA replication ability, with fewer and dimmer EdU granules (Fig. 5 B and C), representing impaired replication and overall loss of replication-competent nucleoids per cell. Together with mitochondrial aggregation and loss of replication-competent nucleoids, we also observed loss of mitochondrial DNA content (Fig. 5D), as expected from the failure to replicate and in agreement with previously published data for cells lacking SUV3 (40, 53). We could not, however, rescue the replication defect in SUV3-depleted cells by overexpression of RNase H1 in vivo (SI Appendix, Fig. S6 B–D). The increased levels of RNase H1 led, in the same extent in control and SUV3-depleted cells, to a major loss of replicative ability, consistent with the described role of RNase H1 in primer removal of the leading strand at OH and at other sites of the lagging strand (19, 26). Hence, RNase H1 overexpression is most likely removing the natural RNA primers impeding mtDNA replication and, therefore, masking the impact it might have in mitigating R-loop accumulation in SUV3-depleted cells.
Fig. 5.
R-loop–mediated mitochondrial genomic instability caused by mtEXO depletion. (A) Visualization of mtDNA replication by EdU incorporation. Cells were incubated with 6 μM aphidicolin for 4 h before incubation with EdU. Nuclei were stained with DAPI. (Scale bar, 10 μm.) (B) Quantification of total EdU granules per cell. (C) Quantification of EdU granule intensities. Red bar represents the mean value for at least three independent experiments for each condition. ****P < 0.0001; **P < 0.01 according to Mann–Whitney U test. AU, arbitrary units. (D) Changes in mtDNA content relative to nuclear DNA (nDNA) in total DNA extracts. Mean value ± SD obtained from at least four independent experiments are represented. (E) Proposed model for the contribution of mtEXO to R-loop homeostasis in mitochondria.
Finally, we assessed the levels of nuclear DNA damage that could result from mitochondrial dysfunction derived from mtEXO deficiency. By measuring the levels of 53BP1 foci in the nucleus, we found a significant increase in damage in SUV3-depleted cells (SI Appendix, Fig. S7), in agreement with previous data showing increased levels of γH2AX in stable SUV3-knockdown cells (53).
Discussion
Mitochondria are essential as the powerhouses of eukaryotic cells and there is growing evidence of their contribution to numerous human pathologies, such as neurodegenerative disorders and cancer. The mitochondrial degradosome formed by SUV3 and PNPase, plays an essential role in mitochondrial RNA turnover, contributing to mitochondrial homeostasis and overall cellular function. Here we provide evidence that the human mtEXO has an additional function in preventing harmful R-loop accumulation in the mitochondrial genome as a way to warrant mtDNA integrity. Our results show that HeLa and U2OS cells lacking a functional mtEXO, apart from accumulating dsRNA, a result consistent with a recent concomitant report (57), accumulate high levels of cotranscriptional R loops (Fig. 1), detected by immunofluorescence using the S9.6 antibody. The results were validated by showing that RNase H treatment in vitro and overexpression in vivo significantly reduced the S9.6 signal (Figs. 2 and 3). The presence of RNase H-sensitive R loops in the mitochondrial genome of SUV3- and PNPase-depleted cells was also confirmed by DRIP analysis (Fig. 4). The accumulation of hybrids seems to be mostly restricted to naturally hybrid-forming regions of the human mitochondrial genome. We could see a significant DNA–RNA hybrid increase in mtDNA regions previously identified to form R loops in undisturbed cycling cells (8), but not at other regions of the mitochondrial genome. This R-loop enrichment at specific mtDNA sites suggests a nonarbitrary occurrence of these structures, made even more striking when the general RNA metabolism is disrupted, even though we cannot discard that R loops might also form at other regions at levels undetectable by the available methodology. Optimization of whole genome-based approaches to identify R-loop–forming sequences, such as DRIP-seq and DRIPc-seq that allows strand-specific RNA-seq, will provide relevant information about the nature of these hybrid-prone sequences (10, 58).
Importantly, loss of mtEXO function leads to defective human mtDNA replication, loss of mtDNA content, and mtDNA instability (Fig. 5 A–D). Consistent with previous observations in the eukaryotic nuclear genome and in bacteria (2), this loss of mtDNA replication ability may be a consequence of the increase of unscheduled R loops, which are known to stall replication fork progression, even though it cannot be discarded at this point that the mtEXO could also exert other functions more directly associated with replication. The changes in mitochondrial morphology, dsRNA accumulation, and mtDNA instability phenotypes generated by loss of mtEXO function are expected to lead to metabolic dysfunction and increased levels of reactive oxygen species, with concomitant increase in nuclear DNA damage (SI Appendix, Fig. S7) (53). However, human SUV3 also localizes to the nucleus where it interacts with several DNA repair factors likely playing a role in nuclear DNA metabolism (59–61). It would be interesting to investigate whether SUV3 is also involved in DNA–RNA hybrid metabolism in the nucleus, even though our IF and DRIP analysis did not detect a significant increase of nuclear R loops. Nevertheless, we cannot discard that SUV3 might play a subtle role in R-loop metabolism outside the context of mitochondria. The interactions of SUV3 with nuclear DNA repair factors may be a consequence of an encounter between replication and transcription mediated by the nascent RNA molecule that would need to be investigated further to understand its physiological meaning.
Although it is well established that abrogating mtEXO function leads to deregulation of transcription and translation processes derived from defective RNA metabolism, we could not observe major changes in the steady-state levels of transcripts that we analyzed, with exception of the RNA molecule forming the R loop at the control region (SI Appendix, Fig. S5 A and B). This is not surprising, given the multiple mitochondrial genomes per cell can buffer minor changes in transcript levels in the putatively small proportion of mitochondrial genomes affected per cell. The long polycistronic transcript from HSPs contains most of the coding ORFs of the human mitochondrial genome while LSPs produce an equally large polycistronic RNA molecule that harbors only ND6 and eight tRNAs. This means that the RNA processing responsible for yielding mature transcripts also generates large antisense intergenic transcripts, termed “mirror” RNAs. Normally, these transcripts are highly unstable and rapidly degraded by the mtEXO (32, 62). In the absence of proper RNA surveillance, these lingering mirror RNAs can hybridize to the mtRNAs, resulting in the elevated dsRNA levels observed in mtEXO-depleted cells (Figs. 1–3). Nevertheless, we could greatly reduce the occurrence of the S9.6 granules by blocking transcription in mtEXO-deficient cells using ethidium bromide and cordycepin (Fig. 1 D–F). This result together with the observation that R loops are detected at the same discrete regions around the mitochondrial genome as in cells with undisturbed RNA turnover suggests that most R loops are formed cotranscriptionally in cis, similarly to nuclear R loops (11). We cannot discard, however, that in the absence of mtEXO, antisense RNAs could hybridize cotranscriptionally in trans, taking advantage of the local negative supercoiling transiently accumulated upstream of an active RNA polymerase.
In the nucleus, loss of factors that bind and process newly formed RNA is critical to prevent the formation of harmful R loops that can pose a major blockage to the moving replication and transcription machineries, leading to stalling and collapse of replication forks and further deregulation of the transcriptional process with considerable effects in genome stability (2, 12, 13). It has been shown, both in yeast and human cells, that loss of exosome function in the nucleus leads to disturbances in RNA processing with R-loop–associated genomic instability. In particular, yeast cells with the deletion of the exosome cofactor trf4 were shown to accumulate DNA–RNA hybrids and an R-loop–dependent hyperrecombinant phenotype (63). In human cells the RNA exosome also plays critical roles in R-loop metabolism, modulating the levels of sense and antisense RNA at divergent enhancer loci and acting as a cofactor of R-loop–targeted deaminases, such as AID acting on V(D)J exons during IgH class switch recombination (64, 65).
Altogether, our results evidence the mtEXO as an important factor in mitochondrial R-loop processing and regulation, acting as a guardian of mitochondrial genome integrity. This concept is supported by the fact that the normal levels of RNase H1 present in the mitochondria cannot counteract the excess accumulation of hybrids forming in mtEXO-deficient cells. One way the SUV3–PNPase complex could exert its role would be by preventing the formation of R loops, binding and processing new transcripts, thus avoiding their hybridization back to the DNA molecule. Alternatively, it could play a more active role, removing R loops as a complementary mechanism to RNase H1, in which the SUV3 helicase would unwind the hybridized RNA moiety, allowing its immediate degradation by PNPase (Fig. 5E). This model resembles the mechanisms acting in the nuclear genome, where a number of processing factors are involved in preventing and/or removing R loops that would compromise genome integrity by interfering with DNA replication (11). In support of our model, yeast suv3 mutants become petite as a consequence of mtDNA loss (38). As an alternative to the previously proposed direct function of Suv3 in yeast mtDNA replication to explain mtDNA loss in suv3∆ cells (38), our results open the possibility that excess of unscheduled R loops accumulating in mtEXO mutants is a major contributing factor impacting replication. The unique replication and expression requirements of mtDNA and the importance of their successful completion for the overall integrity of the cell, may demand a multilevel regulatory network in which factors with functions bridging replication and transcription would provide an effective way to overcome conflicts between both processes. The conservation of the functional relevance of nuclear and mitochondrial RNA processing and surveillance machineries in counteracting R loops reveals the universal threat that cotranscriptional DNA–RNA hybrids pose to genome integrity, regardless of organism and organelles, and the need to adapt the RNA metabolism machinery to a double role in RNA and DNA integrity.
Materials and Methods
Full methods are described in SI Appendix. Cell culture, siRNA, plasmid transfections, quantitative PCR, and molecular biology techniques were performed using standard procedures. Mitochondrial morphology was analyzed using the macro MiNa in Fiji software (51, 52). DRIP was performed as previously described (46). mtDNA replication was analyzed in cells incubated with EdU in the presence of aphidicolin. Changes in mtDNA and RNA levels were analyzed by quantitative PCR.
Supplementary Material
Acknowledgments
We thank members of the A.A. laboratory for valuable discussion, U. Galindo for technical assistance, P. Domínguez-Giménez for assistance with confocal microscopy, and A. G. Rondón for designing the R loop in vitro substrate. Research was funded by the European Research Council (Grant ERC2014 AdG669898 TARLOOP), the Spanish Ministry of Economy and Competitiveness (Grants BFU2013-42918-P and BFU2016-75058-P), and the European Union (Fondo Europeo de Desarrollo Regional). S.S. was awarded a Juan de la Cierva–Incorporación grant from the Spanish Ministry of Economy, Industry, and Competitiveness during part of this study.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. U.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807258115/-/DCSupplemental.
References
- 1.Aguilera A, García-Muse T. R loops: From transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 2.García-Muse T, Aguilera A. Transcription-replication conflicts: How they occur and how they are resolved. Nat Rev Mol Cell Biol. 2016;17:553–563. doi: 10.1038/nrm.2016.88. [DOI] [PubMed] [Google Scholar]
- 3.Castellano-Pozo M, et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell. 2013;52:583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Pichardo D, et al. Histone mutants separate R loop formation from genome instability induction. Mol Cell. 2017;66:597–609.e5. doi: 10.1016/j.molcel.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S, et al. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat Commun. 2018;9:533. doi: 10.1038/s41467-018-02894-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marinello J, Chillemi G, Bueno S, Manzo SG, Capranico G. Antisense transcripts enhanced by camptothecin at divergent CpG-island promoters associated with bursts of topoisomerase I-DNA cleavage complex and R-loop formation. Nucleic Acids Res. 2013;41:10110–10123. doi: 10.1093/nar/gkt778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadel J, et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin. 2015;8:46. doi: 10.1186/s13072-015-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016;30:1327–1338. doi: 10.1101/gad.280834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, et al. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol Cell. 2017;68:745–757.e5. doi: 10.1016/j.molcel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos-Pereira JM, Aguilera A. R loops: New modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 12.Aguilera A, Gómez-González B. DNA-RNA hybrids: The risks of DNA breakage during transcription. Nat Struct Mol Biol. 2017;24:439–443. doi: 10.1038/nsmb.3395. [DOI] [PubMed] [Google Scholar]
- 13.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Domínguez-Sánchez MS, Barroso S, Gómez-González B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhatia V, et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Sridhara SC, et al. Transcription dynamics prevent RNA-mediated genomic instability through SRPK2-dependent DDX23 phosphorylation. Cell Rep. 2017;18:334–343. doi: 10.1016/j.celrep.2016.12.050. [DOI] [PubMed] [Google Scholar]
- 18.Cerritelli SM, Crouch RJ. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes JB, et al. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc Natl Acad Sci USA. 2015;112:9334–9339. doi: 10.1073/pnas.1503653112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stirling PC, Hieter P. Canonical DNA repair pathways influence R-loop-driven genome instability. J Mol Biol. 2017;429:3132–3138. doi: 10.1016/j.jmb.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amon JD, Koshland D. RNase H enables efficient repair of R-loop induced DNA damage. eLife. 2016;5:e20533. doi: 10.7554/eLife.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogenhagen DF, Wang Y, Shen EL, Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol Cell Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wanrooij PH, et al. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012;40:10334–10344. doi: 10.1093/nar/gks802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akman G, et al. Pathological ribonuclease H1 causes R-loop depletion and aberrant DNA segregation in mitochondria. Proc Natl Acad Sci USA. 2016;113:E4276–E4285. doi: 10.1073/pnas.1600537113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez MI, et al. RNA processing in human mitochondria. Cell Cycle. 2011;10:2904–2916. doi: 10.4161/cc.10.17.17060. [DOI] [PubMed] [Google Scholar]
- 28.Rorbach J, Minczuk M. The post-transcriptional life of mammalian mitochondrial RNA. Biochem J. 2012;444:357–373. doi: 10.1042/BJ20112208. [DOI] [PubMed] [Google Scholar]
- 29.Antonicka H, Shoubridge EA. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015;10:920–932. doi: 10.1016/j.celrep.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 30.Antonicka H, Sasarman F, Nishimura T, Paupe V, Shoubridge EA. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013;17:386–398. doi: 10.1016/j.cmet.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Szczesny RJ, et al. RNA degradation in yeast and human mitochondria. Biochim Biophys Acta. 2012;1819:1027–1034. doi: 10.1016/j.bbagrm.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Szczesny RJ, et al. Human mitochondrial RNA turnover caught in flagranti: Involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013;41:1223–1240. doi: 10.1093/nar/gks1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minczuk M, et al. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res. 2002;30:5074–5086. doi: 10.1093/nar/gkf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J Biol Chem. 2009;284:20812–20821. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shu Z, Vijayakumar S, Chen CF, Chen PL, Lee WH. Purified human SUV3p exhibits multiple-substrate unwinding activity upon conformational change. Biochemistry. 2004;43:4781–4790. doi: 10.1021/bi0356449. [DOI] [PubMed] [Google Scholar]
- 37.Stepien PP, Margossian SP, Landsman D, Butow RA. The yeast nuclear gene suv3 affecting mitochondrial post-transcriptional processes encodes a putative ATP-dependent RNA helicase. Proc Natl Acad Sci USA. 1992;89:6813–6817. doi: 10.1073/pnas.89.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo XE, et al. Uncoupling the roles of the SUV3 helicase in maintenance of mitochondrial genome stability and RNA degradation. J Biol Chem. 2011;286:38783–38794. doi: 10.1074/jbc.M111.257956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen PL, et al. Mitochondrial genome instability resulting from SUV3 haploinsufficiency leads to tumorigenesis and shortened lifespan. Oncogene. 2013;32:1193–1201. doi: 10.1038/onc.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García-Rubio ML, et al. The Fanconi Anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015;11:e1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chujo T, et al. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012;40:8033–8047. doi: 10.1093/nar/gks506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schönborn J, et al. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H, Lima WF, Crooke ST. Properties of cloned and expressed human RNase H1. J Biol Chem. 1999;274:28270–28278. doi: 10.1074/jbc.274.40.28270. [DOI] [PubMed] [Google Scholar]
- 45.Nowotny M, et al. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 2008;27:1172–1181. doi: 10.1038/emboj.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrera-Moyano E, Mergui X, García-Rubio ML, Barroso S, Aguilera A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 2014;28:735–748. doi: 10.1101/gad.234070.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zylber EA, Perlman S, Penman S. Mitochondrial RNA turnover in the presence of cordycepin. Biochim Biophys Acta. 1971;240:588–593. doi: 10.1016/0005-2787(71)90716-7. [DOI] [PubMed] [Google Scholar]
- 48.Kinney JS, Viscidi RP, Vonderfecht SL, Eiden JJ, Yolken RH. Monoclonal antibody assay for detection of double-stranded RNA and application for detection of group A and non-group A rotaviruses. J Clin Microbiol. 1989;27:6–12. doi: 10.1128/jcm.27.1.6-12.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang ZZ, Pannunzio NR, Hsieh CL, Yu K, Lieber MR. Complexities due to single-stranded RNA during antibody detection of genomic rna:dna hybrids. BMC Res Notes. 2015;8:127. doi: 10.1186/s13104-015-1092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholson AW. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip Rev RNA. 2014;5:31–48. doi: 10.1002/wrna.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valente AJ, Maddalena LA, Robb EL, Moradi F, Stuart JA. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khidr L, et al. Role of SUV3 helicase in maintaining mitochondrial homeostasis in human cells. J Biol Chem. 2008;283:27064–27073. doi: 10.1074/jbc.M802991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marinello J, et al. Dynamic effects of topoisomerase I inhibition on R-loops and short transcripts at active promoters. PLoS One. 2016;11:e0147053. doi: 10.1371/journal.pone.0147053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sasaki T, Sato Y, Higashiyama T, Sasaki N. Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells. Sci Rep. 2017;7:11257. doi: 10.1038/s41598-017-10843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lentz SI, et al. Mitochondrial DNA (mtDNA) biogenesis: Visualization and duel incorporation of BrdU and EdU into newly synthesized mtDNA in vitro. J Histochem Cytochem. 2010;58:207–218. doi: 10.1369/jhc.2009.954701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dhir A, et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature. 2018;560:238–242. doi: 10.1038/s41586-018-0363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dumelie JG, Jaffrey SR. Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. eLife. 2017;6:e28306. doi: 10.7554/eLife.28306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pereira M, et al. Interaction of human SUV3 RNA/DNA helicase with BLM helicase; loss of the SUV3 gene results in mouse embryonic lethality. Mech Ageing Dev. 2007;128:609–617. doi: 10.1016/j.mad.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Venø ST, et al. The human Suv3 helicase interacts with replication protein A and flap endonuclease 1 in the nucleus. Biochem J. 2011;440:293–300. doi: 10.1042/BJ20100991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szewczyk M, et al. Human SUV3 helicase regulates growth rate of the HeLa cells and can localize in the nucleoli. Acta Biochim Pol. 2017;64:177–181. doi: 10.18388/abp.2016_1419. [DOI] [PubMed] [Google Scholar]
- 62.Clemente P, et al. SUV3 helicase is required for correct processing of mitochondrial transcripts. Nucleic Acids Res. 2015;43:7398–7413. doi: 10.1093/nar/gkv692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gavaldá S, Gallardo M, Luna R, Aguilera A. R-loop mediated transcription-associated recombination in trf4Δ mutants reveals new links between RNA surveillance and genome integrity. PLoS One. 2013;8:e65541. doi: 10.1371/journal.pone.0065541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Basu U, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pefanis E, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.