Significance
Erectile dysfunction is a common condition of men in middle and older ages. Twin studies suggest that about one-third of the risk is due to genetic factors, independent of other known erectile dysfunction risk factors. However, studies that have searched for specific genetic contributors have been limited due to small sample sizes, candidate gene approaches, and weak phenotyping. As a result, there are no confirmed genetic risk factors for erectile dysfunction. This study finds a specific genetic cause for erectile dysfunction.
Keywords: genome-wide association, erectile dysfunction, SIM1, genetic, melanocortin
Abstract
Erectile dysfunction affects millions of men worldwide. Twin studies support the role of genetic risk factors underlying erectile dysfunction, but no specific genetic variants have been identified. We conducted a large-scale genome-wide association study of erectile dysfunction in 36,649 men in the multiethnic Kaiser Permanente Northern California Genetic Epidemiology Research in Adult Health and Aging cohort. We also undertook replication analyses in 222,358 men from the UK Biobank. In the discovery cohort, we identified a single locus (rs17185536-T) on chromosome 6 near the single-minded family basic helix-loop-helix transcription factor 1 (SIM1) gene that was significantly associated with the risk of erectile dysfunction (odds ratio = 1.26, P = 3.4 × 10−25). The association replicated in the UK Biobank sample (odds ratio = 1.25, P = 6.8 × 10−14), and the effect is independent of known erectile dysfunction risk factors, including body mass index (BMI). The risk locus resides on the same topologically associating domain as SIM1 and interacts with the SIM1 promoter, and the rs17185536-T risk allele showed differential enhancer activity. SIM1 is part of the leptin–melanocortin system, which has an established role in body weight homeostasis and sexual function. Because the variants associated with erectile dysfunction are not associated with differences in BMI, our findings suggest a mechanism that is specific to sexual function.
Erectile dysfunction is a common and costly disease of men in middle and older ages (1, 2). Its pathophysiology is tied to psychosocial, neurological, hormonal, and vascular factors (3). Epidemiological studies have shown that age, obesity, diabetes, benign prostatic hyperplasia (BPH), lower urinary tract symptoms, hyperlipidemia, cardiovascular disease, and smoking are important risk factors in erectile dysfunction susceptibility (4). There is also substantial evidence that genetics influence the risk of erectile dysfunction. A twin study in middle-aged male veterans found that about one-third of the risk is heritable, independent of known erectile dysfunction risk factors (5). However, subsequent association studies searching for specific genetic contributors have been limited by small sample sizes, a reliance on limited candidate-gene approaches, and weak phenotyping. As a result, there are no confirmed genetic risk factors for erectile dysfunction (6). Understanding the genetic basis of erectile dysfunction can provide insight into its etiology and lead to the development of new therapies.
Here, we undertook a genome-wide association study (GWAS) of erectile dysfunction in the large and ethnically diverse Genetic Epidemiology Research in Adult Health and Aging (GERA) cohort, which includes 36,649 men from four race/ethnicity groups (non-Hispanic whites, Hispanic/Latinos, East Asians, and African Americans). We then validated genome-wide significant associations in an external independent cohort of 222,358 men from the UK Biobank. We further examined the effect of the validated risk locus in race/ethnicity and phenotype subgroups. Finally, through in silico and in vitro functional investigations, we linked our risk locus to gene function.
Results
The GERA Cohort.
We conducted the primary-discovery erectile dysfunction GWAS using the survey phenotype definition in 36,349 men from four race/ethnicity groups (non-Hispanic whites, 81.4%; Hispanic/Latinos, 8.1%; East Asians, 7.5%; and African Americans, 3.0%) in the GERA cohort (Table 1). Cases were older than controls (68.9 ± 10.8 vs. 56.1 ± 11.4 y), had slightly higher body mass indices (BMIs) (27.7 ± 4.7 vs. 26.9 ± 4.3), were more likely to have diabetes (29.8% vs. 14.6%), and were more likely to be current smokers (6.2% vs. 5.5%) or former smokers (53.1% vs. 37.2%). Cases were also more likely than controls to have a clinical diagnosis recorded in the electronic health record (EHR) (39.3% vs. 23.3%) and were more likely to have filled a phosphodiesterase type 5 inhibitor (PDE5i) prescription to treat erectile dysfunction (59.2% vs. 29.0%).
Table 1.
Characteristics of GERA men
| Characteristic | Control, n = 22,434 | Case, n = 14,215 | Total, n = 36,649 |
| Race/ethnicity, no. participants (%) | |||
| Non-Hispanic whites | 17,995 (80.2) | 11,864 (83.5) | 29,859 (81.5) |
| Hispanic/Latinos | 1,954 (8.7) | 1,016 (7.1) | 2,970 (8.1) |
| East Asian | 1,799 (8.0) | 938 (6.6) | 2,737 (7.5) |
| African American | 686 (3.1) | 397 (2.8) | 1,083 (3.0) |
| Age, y* | 56.1 ± 11.4 | 68.9 ± 10.8 | 61.1 ± 12.8 |
| BMI* | 26.9 ± 4.3 | 27.7 ± 4.7 | 27.2 ± 4.5 |
| Diabetes, no. participants (%) | |||
| Yes | 3,265 (14.6) | 4,240 (29.8) | 7,505 (20.5) |
| No | 19,169 (85.4) | 9,975 (70.2) | 29,144 (79.5) |
| Smoking history, no. participants (%)† | |||
| Never | 12,586 (57.2) | 5,545 (40.7) | 18,131 (50.9) |
| Former | 8,181 (37.2) | 7,235 (53.1) | 15,416 (43.3) |
| Current | 1,220 (5.5) | 841 (6.2) | 2,061 (5.8) |
| EHR diagnosis, no. participants (%) | |||
| Yes | 5,237 (23.3) | 5,591 (39.3) | 10,828 (29.5) |
| No | 17,197 (76.7) | 8,624 (60.7) | 25,821 (70.5) |
| PDE5i prescription, no. participants (%) | |||
| Yes | 6,513 (29.0) | 8,422 (59.2) | 14,935 (40.8) |
| No | 15,921 (71.0) | 5,793 (40.8) | 21,714 (59.2) |
Age and BMI are presented as means ± SD.
Some participants (n = 1,041) did not have a smoking history.
Discovery of an Erectile Dysfunction Risk Locus in the GERA Cohort and Replication in the UK Biobank Cohort.
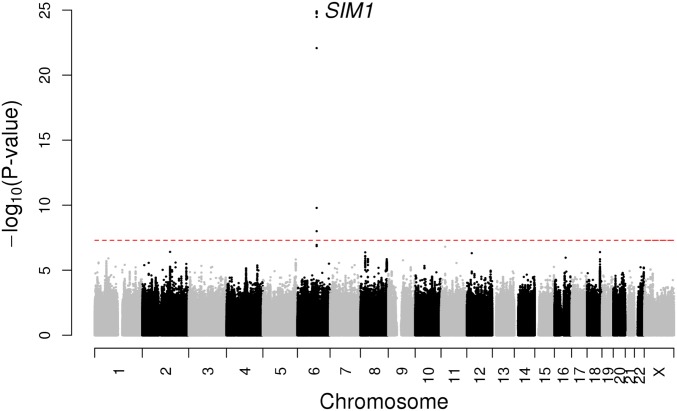
In our discovery multiethnic GWAS analysis, we identified a single locus on chromosome 6 with multiple noncoding SNPs that were associated at a genome-wide level of significance with erectile dysfunction (P < 5 × 10−8) (Fig. 1). To prioritize associated SNPs for follow up analyses, we used a Bayesian approach to derive the smallest set of variants that included the causal variant with 95% probability (95% credible set) (7). Five SNPs were included in this 95% credible set (SI Appendix, Table S1). We then conducted a replication association analysis of these five SNPs in an independent cohort of 222,358 men (2,957 cases and 219,401 controls) from the UK Biobank (SI Appendix, Table S2). All five credible set SNPs were significantly associated with erectile dysfunction in the replication analysis (P < 0.01 required for multiple testing; all SNPs were associated P < 10−13) in the same direction as the GERA cohort results (SI Appendix, Table S3). As evolutionary conservation is a strong marker of functional genomic sequences, we focused our follow-up analyses on one of the five SNPs, rs17185536, which was the only SNP located in an evolutionarily conserved region (8).
Fig. 1.
Manhattan plot of the GERA discovery cohort multiethnic genome-wide association meta-analysis of erectile dysfunction. A GWAS of erectile dysfunction was conducted in 36,649 men (14,215 cases and 22,434 controls) from four race/ethnicity groups (non-Hispanic white, Latino, East Asian, and African American). Association results (−log10 P values) are plotted for each chromosome. The SIM1 gene name at the locus associated with erectile dysfunction is indicated.
Subgroup and Sensitivity Analyses Show That the Association of rs17185536 Is Independent of Known Erectile Dysfunction Risk Factors.
To investigate whether the effect of the replicated erectile dysfunction risk locus was influenced by race/ethnicity, we examined the association of rs17185536 separately by GERA race/ethnicity group. The T allele of rs17185536 (rs17185536-T) was associated with an increase in the risk of erectile dysfunction in non-Hispanic whites (odds ratio 1.25, 95% CI 1.19–1.31), Hispanic/Latinos (odds ratio 1.35, 95% CI 1.16–1.57), East Asians (odds ratio 1.05, 95% CI 0.65–1.71), and African Americans (odds ratio 1.36, 95% CI 1.07–1.71) (SI Appendix, Fig. S1A). While the association was not significant (P > 0.05) in the East Asian group, this appears to be due to the lower frequency of the T allele in that group (2%) than in the other race/ethnicity groups (26% in non-Hispanic whites, 19% in Hispanic/Latinos, and 21% in African Americans). We also examined the association of rs17185536 by decade of age. We observed significant associations across each decade, with the strongest effect in men aged 50–59 y (odds ratio 1.32, 95% CI 1.24–1.41).
Because the risk of erectile dysfunction has been associated with a number of other risk factors, including higher BMI, diabetes, benign prostatic hyperplasia, lower urinary tract symptoms, hyperlipidemia, cardiovascular disease, and smoking status, we conducted analyses adjusting for each of these risk factors individually and combined in GERA to determine whether the risk locus imparted its effect via one of these risk factors. After adjusting for BMI, the effect of rs17185536-T remained similar to the overall GERA result (odds ratio 1.26, 95% CI 1.21–1.32) (SI Appendix, Fig. S1B), which is consistent with a lack of association between the SNP and BMI (P = 0.51). Similarly, the association between rs17185536 and erectile dysfunction was similar after adjusting for the other risk factors individually and in a model including all risk factors as covariates at the same time (odds ratio 1.27, 95% CI 1.21–1.33), suggesting that these risk factors do not explain the observed association. In the fully adjusted model, rs17185536 explained 1.6% of the heritability of the risk of ED. Finally, we used LD Hub to conduct a genetic correlation analysis with 177 traits with available GWAS summary statistics (9). After correcting for multiple testing, there were no significant associations with other traits.
The Association of rs17185536 Is Robust to Changes in Phenotype Definition.
We also conducted sensitivity analyses to determine whether the effect of this locus was influenced by phenotype definition. Since the self-reported questionnaire included four severity levels, we compared the different response levels, using men who answered that they are “Always” able to get an erection as the reference group (SI Appendix, Table S4). We observed a greater effect of rs17185536-T with each increase in severity level, with odds ratios of 1.15 (1.09–1.21) for the “Usually” group, 1.30 (1.23–1.38) for the “Sometimes” group, and 1.41 (1.31–1.51) for the “Never” group (SI Appendix, Fig. S1C). We also observed genome-wide significant associations between rs17185536-T and an EHR-based clinical diagnosis of erectile dysfunction (odds ratio 1.12, 95% CI 1.08–1.17) as well as with the use of PDE5i drugs or other erectile dysfunction treatments (odds ratio 1.16, 95% CI 1.12–1.19). Finally, because of the incomplete concordance across these different phenotype definitions, we conducted an analysis using a strict definition of case and control, requiring cases to meet case criteria for our survey and clinical and treatment definitions and controls to meet control criteria in all three definitions. We observed an even stronger association between the rs17185536-T allele and erectile dysfunction (odds ratio 1.37, 95% CI 1.31–1.43).
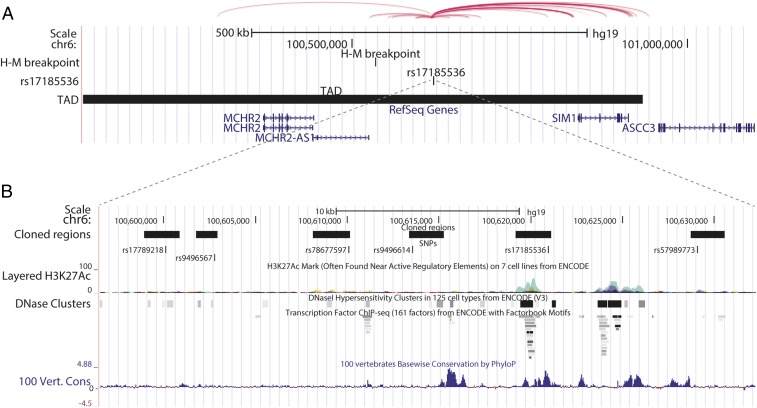
rs17185536 Resides in an Enhancer That Interacts with the Promoter of the Single-Minded Family Basic Helix-Loop-Helix Transcription Factor 1 (SIM1) Gene.
We then undertook chromatin interaction and evolutionary conservation analyses to determine whether the region around rs17185536 interacts with nearby genes. Chromosomes are organized into topologically associating domains (TADs); enhancers interact with genes in the same TAD more frequently than with genes located in other parts of the genome (10). rs17185536 resides within a TAD that includes the genes SIM1, MCHR2, PRDM13, CCNC, and USP45, indicating that the erectile dysfunction risk locus could interact with one of these genes (10). However, of those genes, only SIM1 is located within a human–mouse synteny block that contains rs17185536. This synteny block is defined at one end by a mouse chromosomal breakpoint ∼93 kb distal to MCHR2 (11), the next closest gene to rs17185536, which suggests that the physical proximity of the erectile dysfunction risk locus and SIM1 has been preserved over evolutionary time (Fig. 2A). Analyses of various chromatin conformation capture assays using the 3D Genome Browser (12) show that the region around rs17185536 interacts with the SIM1 promoter (Fig. 2A). Consistent with this interaction, rs17185536 is located within an evolutionarily conserved sequence that has an H3K27ac ChIP-seq peak in human skeletal muscle cells and myoblasts from ENCODE (13) data, suggesting that this region may act as an enhancer (Fig. 2B).
Fig. 2.
Genomic and epigenetic annotations of the SIM1 locus. (A) A University of California, Santa Cruz genome browser snapshot of the SIM1 locus showing rs17185536, the TAD in this region, the human–mouse (H–M) synteny breakpoint, and the virtual 4C (circularized chromosome conformation capture) interactions from human GM12878 cells adapted from the 3D Genome Browser (54). (B) A zoomed-in view of the regions that were cloned for enhancer assays showing the cloned regions, the SNPs, the ENCODE human skeletal muscle cells and myoblasts ChIP-seq peaks (green), ENCODE DNaseI hypersensitivity sites, ENCODE transcription factor ChIP-seq sites, and evolutionary conservation peaks (blue peaks) (B).
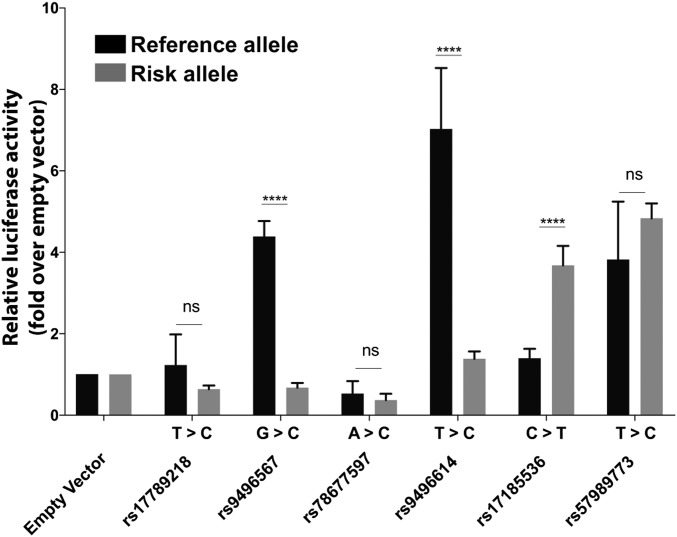
We next set out to determine whether the region encompassing rs17185536 or other regions nearby that have SNPs in strong linkage disequilibrium (r2 >0.8) with rs17185536 function as enhancers and whether the erectile dysfunction-associated variant(s) might lead to differential enhancer activity. We cloned six different regions (SI Appendix, Table S5) containing both the risk and reference alleles into an enhancer assay vector and tested them for enhancer activity in HEK 293T cells. We chose this cell line as SIM1 is known to be expressed in the kidney. We observed differential enhancer activity between the risk and reference alleles for three of the constructs, including the rs17185536-T (risk) allele, which showed significant enhancer activity compared with empty vector, and the rs17185536-C (reference), which did not have significant enhancer activity (Fig. 3). Combined, our results suggest that the rs17185536-T (risk) allele or other erectile dysfunction-associated alleles in this region lead to differential enhancer activity and that this region may regulate the expression of SIM1.
Fig. 3.
Enhancer assay results in HEK 293T cells. Results for each allele compared with the empty vector are presented as mean ± SD. ****P < 0.0001; ns, not significant; t test.
Discussion
We identified a single locus near the SIM1 gene that was significantly associated with the risk of erectile dysfunction and confirmed that association in a large, independent cohort. The association was robust to changes in phenotype definition and was independent of known erectile dysfunction risk factors. Through a series of analyses, we showed that the region containing the lead variant likely interacts with the promoter of the SIM1 gene and that the risk allele of the lead variant alters an enhancer.
Several different lines of evidence suggest a biologically plausible role for SIM1 in erectile dysfunction susceptibility. SIM1 encodes a transcription factor that is active in the leptin–melanocortin pathway, a system that plays a central role in body weight homeostasis and sexual function (14). The melanocortin peptides alpha melanocortin-stimulating hormone (α-MSH) and adrenocorticotrophic hormone (ACTH) have long been known to stimulate penile erection in male animals (15, 16); MT-II, a synthetic analog of α-MSH, has been shown to induce penile erection in men (17).
While both MT-II and α-MSH are nonselective melanocortin agonists, it is believed that their effect on sexual function is mediated by the melanocortin 4 receptor (MC4R). Mice lacking Mc4r display impaired copulatory behavior (18), an effect that is reversed when MC4Rs are reexpressed only on SIM1-expressing neurons (19). In the latter study, SIM1-dependent expression of MC4R was observed in the paraventricular nucleus of the hypothalamus and medial amygdala. Rare mutations in the coding sequences of both MC4R and SIM1 cause severe forms of human obesity (20), and neurons coexpressing MC4R and SIM1 in the paraventricular nucleus of the hypothalamus have been shown to be both necessary and sufficient for the regulation of feeding and body weight in mice (21).
In our study, the SNPs in the erectile dysfunction risk locus were not associated with variation in BMI, nor was the effect of this locus on the risk of erectile dysfunction changed after adjusting for BMI. We hypothesize that the SIM1 enhancer harboring rs17185536 or the other erectile dysfunction-associated alleles that show differential enhancer activity are active in neurons that control erectile function but not in those controlling feeding and body weight homeostasis. Melanocortin agonists have been shown to initiate penile erection when administered in both the brain and the spinal cord (22). Determining whether the neurons that are sufficient for erectile function are located in the brain, the spinal cord, or both will be essential to understand the specificity of the erectile dysfunction risk locus identified in this study. Further in vivo analyses of enhancers in this region have the potential to address these questions.
An important limitation of the current investigation is the potential for phenotype misclassification when using a self-reported, survey-based phenotype. We addressed this limitation by conducting a number of sensitivity analyses using alternative phenotype definitions based on EHRs and replication in an independent cohort with a different phenotype definition. While the difference in phenotype definition can reduce power to confirm associations, we observed strong confirmation of the discovery association, which provides further support for the robustness of the observed association to changes in phenotype definition. The incomplete concordance between the survey, clinical, and treatment-based phenotype definitions is expected and has been described previously in the literature (2). Many men with erectile dysfunction do not seek medical care for the condition. For this reason, the absence of a clinical diagnosis or PDE5i prescription is not on its own an indicator of an absence of erectile dysfunction. To limit the potential presence of cases among our control group, and vice versa, we conducted an analysis using strict case and controls definitions in which each strict case met the case criteria for the survey and clinical and treatment-based phenotype definitions, and each strict control met the control criteria for all three definitions. We again observed a significant association with the same locus, with a modestly stronger effect size.
While MC4R and SIM1 loss of function are associated with erectile dysfunction, we observed increased enhancer activity for the rs17185536 erectile dysfunction-associated allele. This could be due to this SNP not being the causative variant, to interaction between this variant and other SNPs, or to this region having an additional, as yet uncharacterized function. Further analyses of this region will be needed to characterize its functional role.
Another limitation is that we did not have a comparable phenotype available in women. Based on both human and animal studies of the effects of melanocortin agonists on sexual function in females, it is possible that our erectile dysfunction risk locus may also affect female sexual function, including sexual desire and sexual arousal (23, 24).
Our study is a large-scale investigation of the genetics of erectile dysfunction. We anticipate that future studies involving even larger samples will uncover additional risk loci, providing further insights into the etiology of erectile dysfunction. Our functional analyses, along with previous studies in the literature, point toward a previously unknown mechanism underlying erectile dysfunction, which opens the possibility of developing drug therapies with a more specific target. Those treatments may have the potential to improve sexual function in both men and women.
Methods
GERA Study Population.
We performed a GWAS of erectile dysfunction in 36,349 men from the GERA cohort (Table 1). The full GERA cohort includes 110,266 adult men and women who have provided informed consent as participants in the Research Program on Genes, Environment, and Health (RPGEH) drawn from adult (at least 18-y-old) members of the Kaiser Permanente Medical Care Plan, Northern California Region (KPNC) (25, 26). The cohort is diverse, with representation from non-Hispanic white, Hispanic/Latino, East Asian, and African American race/ethnicity groups. All study procedures were approved by the Institutional Review Board of the Kaiser Foundation Research Institute.
Phenotype Definition.
Upon enrollment in the study, RPGEH participants answered a health questionnaire. On this survey, male participants were asked the following question: “Many men have difficulty getting and keeping an erection (or hard-on) that is rigid enough for satisfactory sexual activity. How would you describe your experience during the past year? (Without the use of a medication such as Viagra, Cialis, Levitra, injectable drugs, or penis implant or pump device.)” Respondents were given a choice of four possible answers: (i) Always able to get and keep an erection good enough for sexual activity, (ii) Usually able to get and keep an erection good enough for sexual activity, (iii) Sometimes able to get and keep an erection good enough for sexual activity, (iv) Never able to get and keep an erection good enough for sexual activity. For the purpose of the discovery GWAS, we defined erectile dysfunction cases as those subjects answering “Sometimes” or “Never” (iii or iv) on the survey and controls as those subjects answering “Always” or “Usually” (i or ii) on the survey. This survey definition of erectile dysfunction has been used in other large-scale studies (27) and demonstrated reasonable accuracy in detecting erectile dysfunction in men undergoing a detailed clinical examination (28). Men with a history of prostate cancer at the time of survey were excluded from the analyses.
Genotyping and Imputation.
DNA samples from GERA individuals were extracted from Oragene kits (DNA Genotek, Inc.) at KPNC and were genotyped at the Genomics Core Facility of the University of California, San Francisco (UCSF). DNA samples were genotyped at over 665,000 SNPs on four race/ethnicity-specific Affymetrix Axiom arrays (Affymetrix) optimized for individuals of European, Hispanic/Latino, East Asian, and African American ancestry (29, 30). Genotype quality-control (QC) procedures for the GERA samples were performed on an arraywise basis (26). SNPs with an initial genotyping call rate ≥97%, an allele frequency difference ≤0.15 between men and women for autosomal markers (even though women were not included in this analysis), and genotype concordance rate >0.75 across duplicate samples were included. About 94% of samples and more than 98% of genetic markers assayed passed QC procedures. In addition to those QC criteria, SNPs with genotype call rates <90% were removed.
Imputation was also conducted on an arraywise basis. Following the prephasing of genotypes with Shape-IT v2.r72719 (31), variants were imputed from the cosmopolitan 1000 Genomes Project reference panel (phase I integrated release; www.internationalgenome.org/) using IMPUTE2 v2.3.0 (32–34). As a QC metric, we used the info r2 from IMPUTE2, which is an estimate of the correlation of the imputed genotype to the true genotype (35). We excluded variants with an imputation r2 <0.3 or a minor allele frequency (MAF) <1%.
For SNPs that were associated at a genome-wide level of significance in the genome-wide association analyses, we conducted further QC checks. For directly genotyped SNPs, we examined missing rates and Hardy–Weinberg tests in the control group and the overall analysis group. All SNPs in the associated region had low missing rates (<0.005) and were found to be in Hardy–Weinberg equilibrium (P > 0.05).
GWAS Analysis and Covariate Adjustment.
We first analyzed each of the four race/ethnicity groups (non-Hispanic whites, Hispanic/Latinos, East Asians, and African Americans) separately. We performed a logistic regression of erectile dysfunction case/control status with the covariates age at the time of the health survey and ancestry as principal components (PCs). The Eigenstrat method (36) v4.2 was used to calculate the PCs on each of the four race/ethnicity groups (26). The top 10 ancestry PCs were included as covariates for the non-Hispanic whites, while the top six ancestry PCs were included for the other three race/ethnicity groups. We then performed a logistic regression of the residuals on each SNP using PLINK (37) v1.9 (www.cog-genomics.org/plink/1.9/) to assess genetic associations. Data from each SNP were modeled using additive dosages to account for the uncertainty of imputation (38).
We then undertook a GERA meta-analysis of erectile dysfunction to combine the results of the four race/ethnicity groups using the R (39) package meta. We calculated fixed-effects summary estimates under an additive model, and we assessed the heterogeneity index, I2, (0–100%) among groups as well as Cochrane’s Q heterogeneity statistic.
To identify SNPs for follow-up analyses, we used a Bayesian approach (CAVIARBF) (7). This approach uses SNP association test statistics and information on the correlation of individual SNPs to estimate the posterior inclusion probability of each individual SNP. These probabilities can be used to derive the smallest set of SNPs that includes the causal variant with 95% probability. This set of SNPs is called the “95% credible set.” For the significantly associated SNPs in this study, we computed each variant’s ability to explain the observed signal within a 2-Mb window (±1.0 Mb with respect to the original lead SNP) to derive the 95% credible set. Previous studies (40, 41) have used similar approaches to prioritize variants near index SNPs for follow-up.
Sensitivity Analyses.
Because BMI, diabetes, benign prostatic hyperplasia, lower urinary tract symptoms, hyperlipidemia, cardiovascular disease, and cigarette smoking have previously been reported to be associated with the risk of erectile dysfunction (27), we conducted additional analyses of our top associated SNPs using these risk factors as covariates. We first assessed each risk factor individually by including a covariate representing its presence or absence, or in the case of BMI, the value itself, in the logistic regression model. We also included all risk factors together in a single model. For diabetes status at the time of the health survey, we used the KPNC diabetes registry to identify diabetic subjects. Registry eligibility is determined by pharmacy prescription for diabetes medications, abnormal HbA1c or glucose values, and outpatient, emergency room, and hospitalization diagnoses of diabetes (42, 43). A validation study found that the registry was 99.5% sensitive for diagnosed diabetes (44). Men with BPH and lower urinary tract symptoms were defined by an American Urological Association Score Index of 8 or more, or a clinical diagnosis of BPH [International Classification of Diseases, Ninth Revision (ICD9) codes of 600.0, 600.2, or 600.9] (45–47). Hyperlipidemia was defined using ICD9 codes 272.0–272.4 or the prescription of a lipid-lowering drug, and cardiovascular disease was defined by either revascularization procedures (coronary artery bypass grafting or vascular stents) or ICD9 codes 410, 411, and 413 (ischemic heart disease), 431–434 and 436 (stroke), and 402.x1 and 428 (congestive heart failure) (48–50). For smoking status, we used subjects’ responses to the GERA health survey to create two variables: Ever/Never, and, within the Ever category, Current/Former smokers. Finally, we used LD Hub to conduct a genetic correlation analysis with 177 traits with available GWAS summary statistics (9).
In addition to the survey-based phenotype definition, we also examined the association of SNPs identified in the discovery GWAS with alternative phenotype definitions as sensitivity analyses. The KPNC EHRs contain information on patient clinical diagnoses as a well as treatment records, including prescriptions of PDE5i, alprostadil, Caverject, vacuum erectile devices, and penile implants used to treat erectile dysfunction. We used this information to create two additional erectile dysfunction phenotype definitions. For the clinical EHR-based definition, we included individuals with at least one record of organic erectile dysfunction (ICD9 607.84/ICD10 N529) as cases and those without such a diagnosis as controls. Using the treatment records, we defined cases as men with one or more recorded prescription for a PDE5i or any of the other forms of treatments and those with no recorded prescription or other treatment as controls. Finally, we created a strict case and control definition, in which each case had to meet criteria for all three survey, clinical, and treatment-based definitions and controls also had to meet the definition of control by all three definitions.
Replication of Significant SNP Associations in the Independent UK Biobank Cohort.
To test genome-wide significant SNPs from the GERA analyses for replication, we evaluated associations in the multiethnic UK Biobank cohort (51) which included 222,358 men between the ages of 40 and 69 y from five race/ethnicity groups (European/white, South Asian, mixed, African British, and East Asian) (SI Appendix, Table S2). These data have been imputed to the Haplotype Reference Consortium (www.ukbiobank.ac.uk/). We note that the GERA data were imputed to the 1,000 Genomes reference panel. Given that the Haplotype Reference Consortium panel is larger, we would expect that some SNPs might be captured at a higher imputation r2 than in the 1,000 Genomes imputed SNPs. Variation in coverage can affect statistical power; for this reason we examined the imputation r2 of the lead SNPs at the significantly associated locus (SI Appendix, Table S6) (52). Coverage is very good for all SNPs across all subsets (r2 ≥ 0.88), which indicates similar statistical power to resolve associations among these SNPs.
Cases were defined as men with a record of erectile dysfunction (“impotence of organic origin”; UK Biobank record 41202) or self-reporting of any of the following medications on the medications list (data field 20003): alprostadil, Caverject, sildenafil, Viagra (25, 50, or 100 mg), tadalafil, Cialis (10 mg or 20 mg), vardenafil, or Levitra (5, 10, or 20 mg). The control group included men who did not report any of these conditions or medications. As in the GERA analyses, we performed a logistic regression of erectile dysfunction case/control status using age and ancestry principal components as covariates.
In Silico Analyses.
We investigated the functional effects of the associated variants using three in silico tools. To test SNPs for expression of quantitative trait locus effects, we queried the Genotype-Tissue Expression (GTEx) Portal (53). None of the SNPs was contained in the GTEx database. We then queried HaploReg (8) to identify sequence conservation, enhancer marks, and changes to regulatory motifs. Finally, we used the 3D Genome Browser (54) to explore long-range chromatin interactions.
Luciferase Assays.
We used a well-established in vitro model system to assess enhancer function of the five SNPs identified in the credible set analysis and a sixth SNP (rs57989773) in strong linkage disequilibrium with these five SNPs. Selected sequences (SI Appendix, Table S5) were PCR amplified from human genomic DNA, cloned into a pGL4.23 enhancer assay vector (Promega), and sequence-verified for having either allele. We used an empty pGL4.23 as a negative control. HEK 293T cells were grown in DMEM (Invitrogen) supplemented with FBS 10% (UCSF cell-culture facility), 2 mM of L-glutamine (UCSF cell-culture facility), and 1% penicillin/streptomycin (UCSF cell-culture facility). Twenty-four hours before transfection, 50,000 HEK 293T cells were plated out in 24-well plates and were grown up to 70% confluency. Cells were transfected with 0.75 μg of the assayed plasmid and 0.1 μg of pGL4.73[hRluc/SV40] (Promega) containing Renilla luciferase to correct for transfection efficiency, using X-tremeGENE (Roche) according to the manufacturer’s protocol. Transfections were performed in quadruplets. Twenty-four hours after transfection, cells were lysed, and luciferase activity was measured using the Dual-Luciferase Reporter Assay Kit (Promega). Measurements were performed on a GloMax 96-microplate luminometer (Promega). Each experiment was repeated on two different days using four technical triplicates and three independent readings per each technical replicate.
Supplementary Material
Acknowledgments
We thank the KPNC members who generously provided informed consent and agreed to participate in the Kaiser Permanente RPGEH and Dr. Andrew Paterson for his encouragement and invaluable early discussions. Support for participant enrollment, survey completion, and biospecimen collection for the RPGEH was provided by the Robert Wood Johnson Foundation, the Wayne and Gladys Valley Foundation, the Ellison Medical Foundation, and Kaiser Permanente Community Benefit Programs. Genotyping of the GERA cohort was funded by National Institute on Aging, National Institute of Mental Health (NIMH), and National Institute of Health Common Fund Grant RC2 AG036607. The erectile dysfunction project was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant R01 DK104764 (to H.W., J.M.H., and S.K.V.D.E.). Data analyses were facilitated by National Eye Institute Grant R01 EY027004 (to E.J.). This article was supported in part by NIDDK Grant 1R01DK090382 and a UCSF School of Pharmacy 2017 Mary Anne Koda-Kimble Seed Award for Innovation. N.A. is also supported by National Human Genome Research Institute Grant 1UM1HG009408, NIMH Grant 1R01MH109907, National Institute of Child and Human Development Grant 1P01HD084387, and National Heart, Lung, and Blood Institute Grant 1R01HL138424. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genotype data of Kaiser Permanente Northern California Genetic Epidemiology Research in Adult Health and Aging (GERA) participants who consented to having their data shared on the Genotypes and Phenotypes (dbGaP) database have been deposited in the dbGaP database (accession no. phs000674.v2.p2). The complete GERA data are available upon application to the Kaiser Permanente Research Bank (https://researchbank.kaiserpermanente.org/). The UK Biobank data are available upon application to the UK Biobank, www.biobank.ac.uk/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809872115/-/DCSupplemental.
References
- 1.McCabe MP, et al. Incidence and prevalence of sexual dysfunction in women and men: A consensus statement from the fourth international consultation on sexual medicine 2015. J Sex Med. 2016;13:144–152. doi: 10.1016/j.jsxm.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 2.Wessells H, Joyce GF, Wise M, Wilt TJ. Erectile dysfunction. J Urol. 2007;177:1675–1681. doi: 10.1016/j.juro.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 3.Lue TF. Erectile dysfunction. N Engl J Med. 2000;342:1802–1813. doi: 10.1056/NEJM200006153422407. [DOI] [PubMed] [Google Scholar]
- 4.Hatzimouratidis K, et al. European Association of Urology Guidelines on male sexual dysfunction: Erectile dysfunction and premature ejaculation. Eur Urol. 2010;57:804–814. doi: 10.1016/j.eururo.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Fischer ME, et al. A twin study of erectile dysfunction. Arch Intern Med. 2004;164:165–168. doi: 10.1001/archinte.164.2.165. [DOI] [PubMed] [Google Scholar]
- 6.Lopushnyan NA, Chitaley K. Genetics of erectile dysfunction. J Urol. 2012;188:1676–1683. doi: 10.1016/j.juro.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, et al. Fine mapping causal variants with an approximate Bayesian method using marginal test statistics. Genetics. 2015;200:719–736. doi: 10.1534/genetics.115.176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward LD, Kellis M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng J, et al. Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim MJ, Oksenberg N, Hoffmann TJ, Vaisse C, Ahituv N. Functional characterization of SIM1-associated enhancers. Hum Mol Genet. 2014;23:1700–1708. doi: 10.1093/hmg/ddt559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium EP. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wikberg JE, Mutulis F. Targeting melanocortin receptors: An approach to treat weight disorders and sexual dysfunction. Nat Rev Drug Discov. 2008;7:307–323. doi: 10.1038/nrd2331. [DOI] [PubMed] [Google Scholar]
- 15.Bertolini A, Vergoni W, Gessa GL, Ferrari W. Induction of sexual excitement by the action of adrenocorticotrophic hormone in brain. Nature. 1969;221:667–669. doi: 10.1038/221667a0. [DOI] [PubMed] [Google Scholar]
- 16.Feder HH, Ruf KB. Stimulation of progesterone release and estrous behavior by ACTH in ovariectomized rodents. Endocrinology. 1969;84:171–174. doi: 10.1210/endo-84-1-171. [DOI] [PubMed] [Google Scholar]
- 17.Wessells H, et al. Effect of an alpha-melanocyte stimulating hormone analog on penile erection and sexual desire in men with organic erectile dysfunction. Urology. 2000;56:641–646. doi: 10.1016/s0090-4295(00)00680-4. [DOI] [PubMed] [Google Scholar]
- 18.Van der Ploeg LH, et al. A role for the melanocortin 4 receptor in sexual function. Proc Natl Acad Sci USA. 2002;99:11381–11386. doi: 10.1073/pnas.172378699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semple E, Hill JW. Sim1 neurons are sufficient for MC4R-mediated sexual function in male mice. Endocrinology. 2018;159:439–449. doi: 10.1210/en.2017-00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahituv N, et al. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80:779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah BP, et al. MC4R-expressing glutamatergic neurons in the paraventricular hypothalamus regulate feeding and are synaptically connected to the parabrachial nucleus. Proc Natl Acad Sci USA. 2014;111:13193–13198. doi: 10.1073/pnas.1407843111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wessells H, et al. Ac-Nle-c[Asp-His-DPhe-Arg-Trp-Lys]-NH2 induces penile erection via brain and spinal melanocortin receptors. Neuroscience. 2003;118:755–762. doi: 10.1016/s0306-4522(02)00866-7. [DOI] [PubMed] [Google Scholar]
- 23.Clayton AH, et al. Bremelanotide for female sexual dysfunctions in premenopausal women: A randomized, placebo-controlled dose-finding trial. Womens Health (Lond) 2016;12:325–337. doi: 10.2217/whe-2016-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaus JG, Shadiack A, Van Soest T, Tse M, Molinoff P. Selective facilitation of sexual solicitation in the female rat by a melanocortin receptor agonist. Proc Natl Acad Sci USA. 2004;101:10201–10204. doi: 10.1073/pnas.0400491101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvale MN, et al. Genotyping informatics and quality control for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics. 2015;200:1051–1060. doi: 10.1534/genetics.115.178905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banda Y, et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics. 2015;200:1285–1295. doi: 10.1534/genetics.115.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saigal CS, Wessells H, Pace J, Schonlau M, Wilt TJ. Urologic Diseases in America Project Predictors and prevalence of erectile dysfunction in a racially diverse population. Arch Intern Med. 2006;166:207–212. doi: 10.1001/archinte.166.2.207. [DOI] [PubMed] [Google Scholar]
- 28.O’Donnell AB, Araujo AB, Goldstein I, McKinlay JB. The validity of a single-question self-report of erectile dysfunction. Results from the Massachusetts Male Aging Study. J Gen Intern Med. 2005;20:515–519. doi: 10.1111/j.1525-1497.2005.0076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann TJ, et al. Next generation genome-wide association tool: Design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann TJ, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98:422–430. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 32.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 36.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 37.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang L, Wang C, Rosenberg NA. The relationship between imputation error and statistical power in genetic association studies in diverse populations. Am J Hum Genet. 2009;85:692–698. doi: 10.1016/j.ajhg.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Development Core Team 2008. A Language and Environment for Statistical Computing (The R Foundation for Statistical Computing Vienna), Version 3.4.1.
- 40.Fritsche LG, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maller JB, et al. Wellcome Trust Case Control Consortium Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karter AJ, et al. Self-monitoring of blood glucose levels and glycemic control: The Northern California Kaiser Permanente Diabetes registry. Am J Med. 2001;111:1–9. doi: 10.1016/s0002-9343(01)00742-2. [DOI] [PubMed] [Google Scholar]
- 43.Karter AJ, et al. Ethnic disparities in diabetic complications in an insured population. JAMA. 2002;287:2519–2527. doi: 10.1001/jama.287.19.2519. [DOI] [PubMed] [Google Scholar]
- 44.Huang ES, Karter AJ, Danielson KK, Warton EM, Ahmed AT. The association between the number of prescription medications and incident falls in a multi-ethnic population of adult type-2 diabetes patients: The diabetes and aging study. J Gen Intern Med. 2010;25:141–146. doi: 10.1007/s11606-009-1179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barry MJ, et al. Measurement Committee of the American Urological Association The American Urological Association symptom index for benign prostatic hyperplasia. J Urol. 2017;197(Suppl):S189–S197. doi: 10.1016/j.juro.2016.10.071. [DOI] [PubMed] [Google Scholar]
- 46.Sarma AV, et al. Olmsted County Study of Urinary Symptoms and Health Status; Flint Men’s Health Study Comparison of lower urinary tract symptom severity and associated bother between community-dwelling black and white men: The Olmsted County Study of Urinary Symptoms and Health Status and the Flint Men’s Health Study. Urology. 2003;61:1086–1091. doi: 10.1016/s0090-4295(03)00154-7. [DOI] [PubMed] [Google Scholar]
- 47.Van Den Eeden SK, et al. Impact of type 2 diabetes on lower urinary tract symptoms in men: A cohort study. BMC Urol. 2013;13:12. doi: 10.1186/1471-2490-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidney S, et al. Heterogeneity in national U.S. mortality trends within heart disease subgroups, 2000-2015. BMC Cardiovasc Disord. 2017;17:192. doi: 10.1186/s12872-017-0630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sidney S, et al. Recent trends in cardiovascular mortality in the United States and public health goals. JAMA Cardiol. 2016;1:594–599. doi: 10.1001/jamacardio.2016.1326. [DOI] [PubMed] [Google Scholar]
- 50.Sidney S, et al. Comparative trends in heart disease, stroke, and all-cause mortality in the United States and a large integrated healthcare delivery system. Am J Med. 2018;131:829–836.e1. doi: 10.1016/j.amjmed.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sudlow C, et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgenson E, Witte JS. Coverage and power in genomewide association studies. Am J Hum Genet. 2006;78:884–888. doi: 10.1086/503751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee PH, et al. Modifiers and subtype-specific analyses in whole-genome association studies: A likelihood framework. Hum Hered. 2011;72:10–20. doi: 10.1159/000327158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, et al. 2017. The 3D genome browser: A web-based browser for visualizing 3D genome organization and long-range chromatin interactions. bioRxiv:10.1101/112268. Preprint, posted February 27, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.