Abstract
The recent successes of cryo-electron microscopy fostered great expectation of solving many new and previously recalcitrant biomolecular structures. However, it also brings with it the danger of compromising the validity of the outcomes if not done properly. The Map Challenge is a first step in assessing the state of the art and to shape future developments in data processing. The organizers presented seven cases for single particle reconstruction, and 27 members of the community responded with 66 submissions. Seven groups analyzed these submissions, resulting in several assessment reports, summarized here. We devised a range of analyses to evaluate the submitted maps, including visual impressions, Fourier shell correlation, pairwise similarity and interpretation through modeling. Unfortunately, we did not find strong trends. We ascribe this to the complexity of the challenge, dealing with multiple cases, software packages and processing approaches. This puts the user in the spotlight, where his/her choices becomes the determinant of map quality. The future focus should therefore be on promulgating best practices and encapsulating these in the software. Such practices include adherence to validation principles, most notably the processing of independent sets, proper resolution-limited alignment, appropriate masking and map sharpening. We consider the Map Challenge to be a highly valuable exercise that should be repeated frequently or on an ongoing basis.
Keywords: Single particle analysis, Fourier shell correlation, power spectra, resolution, reconstruction, cryo-electron microscopy, validation
Introduction
Cryo-electron microscopy (cryoEM) is undergoing an enormous expansion due to the recent introduction of direct electron detectors (McMullan et al., 2016; Vinothkumar and Henderson, 2016). These detectors have a significantly enhanced the signal-to-noise (SNR) ratio combined with fast image acquisition, enabling solving biomolecular structures to atomic resolution (Bartesaghi et al., 2018; Merk et al., 2016; Tan et al., 2018). This newfound popularity comes with a price: The methodologies need to be robust enough to avoid pitfalls (Henderson, 2013; Subramaniam, 2013; van Heel, 2013) and ensure valid outcomes (Heymann, 2015; Rosenthal, 2016). The Map Challenge was conceived to start addressing these issues. The stated goals are to: “Develop benchmark datasets, encourage development of best practices, evolve criteria for evaluation and validation, compare and contrast different approaches” (http://challenges.emdatabank.org/?q=2015_map_challenge).
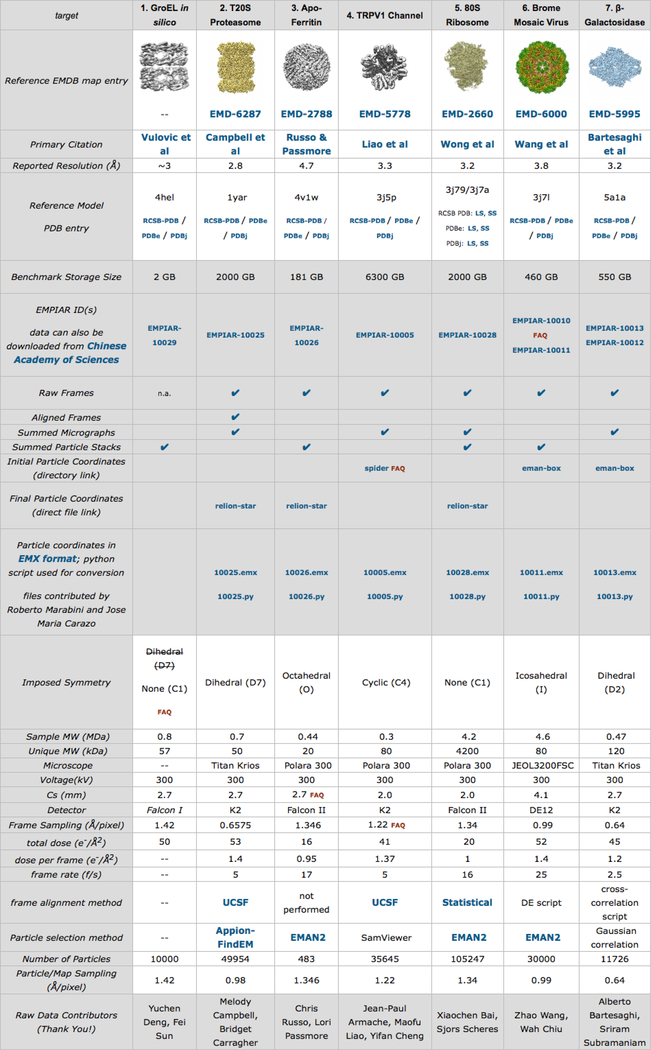
The Map Challenge organizers selected seven cases covering different symmetries and sizes during the development phase (January - June 2015) (Figure 1). The challenge phase started in August 2015 and submissions closed in April 2016. Twenty-seven map challengers submitted 66 reconstructions distributed over all cases. Assessors were asked to analyze the submissions, first with limited metadata (blind assessment: November 2016 – April 2017), and then with full metadata (June – September 2017, Supplemental Material). An initial assessment by visualization and FSC was done by MH and AP, followed by six assessment reports submitted by other participants. The exercise culminated in the CryoEM Structure Challenges Workshop in October 2017 on the SLAC campus at Stanford University with presentations by both map challengers and assessors.
Figure 1:
Screen shot of the Map Challenge web page with all the donated data sets. (http://challenges.emdataresource.org/?q=2015_map_challenge)
The Map Challenge is the first of its kind, which means that many issues have not been clearly formulated. The challenge was posed as an open-ended exercise, with users allowed great freedom in processing the data, and assessors asked to come up with their own analyses. Here we present a summary of the map challenge assessments ((Heymann, 2018b; Jonic, 2018; Marabini et al., 2018; Pintilie and Chiu, 2018; Stagg and Mendez, 2018) , Appendix). We devised several different approaches to assess the submitted maps. Some of them are traditional (visual inspection and FSC calculation), while others are newly proposed (pairwise comparisons and model fitting). The organizers formulated the challenge with an aim to identif the best software and approaches in SPA. However, the issues are subtler and we concluded that the current major determinant of reconstruction quality is the user. Given good data and an appropriate workflow, most of the software packages are capable of producing high quality reconstructions. We attempt here to understand why some of the submissions are of lesser quality, and why some seems to be overfitted.
Summaries of individual assessments
We adopted several different approaches to assess the submitted maps: visual inspection, the FSC curve (Harauz and van Heel, 1986) between the so-called “even” and “odd” maps (FSCeo), an FSC curve against a more or less external reference derived from the published atomic structure (FSCref), and judging how well the maps can be interpreted through modeling. We ranked and classified the maps, highlighting those that show signs of poor quality or overfitting.
Holmdahl and Patwardhan
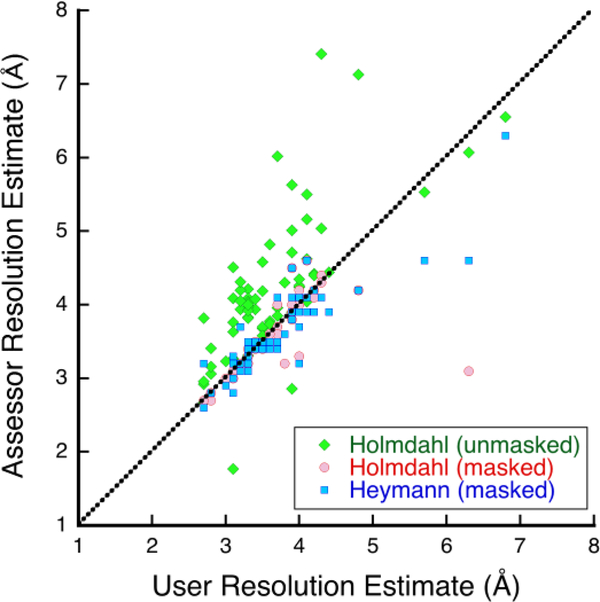
MH and AP did an initial assessment (http://challenges.emdataresource.org/?q=map-submission-overview), providing visual impressions of the submissions as well as FSC analysis based on the submitted even-odd or halfmaps and masks using EMAN2 (Tang et al., 2007). The reported and re-analyzed resolution estimations differ considerably in several instances (Figure 2), indicating some uncertainty in what it means and how to compare the submissions.
Figure 2:
Resolution estimates from even-odd FSC curves at a threshold of 0.143 done by the submitters (users) compared to those redone in various ways by assessors. Holmdahl did the analysis without masks (green diamonds) and with user-submitted masks (red discs), while Heymann used new, case-specific masks (blue squares). The dotted line indicates equality.
Heymann
JBH inspected the maps and their power spectra for signs of problems (Heymann, 2018b) . The maps varied in many properties, with some indicating issues that affect their quality, such as artifacts and inappropriate masking or sharpening. For a more quantitative assessment, JBH compared the even-odd maps by FSC (FSCeo), as well as the full unfiltered maps against a reference calculated from the atomic structure (FSCref), using the Bsoft package (Heymann, 2018a). However, the submitted maps have different sizes, samplings and orientations, complicating comparison. JBH posed the maps in the same configuration (scale and orientation) with minimal interpolation, and calculated shaped masks with proper low-pass filtering to avoid influence on the FSC calculation (other than removing extraneous noise). He then calculated FSCeo and FSCref curves for the unfiltered maps. FSCref gives an indication of how closely the map represents the structure. For some submissions, the FSCeo curves are much better than expected compared to the FSCref curves, suggesting overfitting. The FSC results show that in each case, there is a cluster of submissions that are comparable – a consensus. JBH concluded that some reconstructions simply suffer from lack of data, while others have issues that point to problems with data processing.
Jonic
SJ analyzed the unfiltered maps by visual inspection in Chimera, as well as by quantitative evaluation of pairwise similarities among the maps and among Gaussian-based map approximations (Jonic and Sorzano, 2016). The pairwise similarities are based on the Pearson correlation coefficient (CC). To be able to compare the maps, she first aligned and resized the maps to a common reference in each case (Jonic, 2018). The Gaussian-based map approximation and CC were calculated within the area determined by a mask created from the common reference. A distance matrix was constructed from the pairwise dissimilarities (1-CC) and projected onto a low-dimensional (3D) space using non-metric multidimensional scaling (Sanchez Sorzano et al., 2016). SJ identified clusters of the most similar maps in each case that show a consensus structure. She noted that the assessment is complicated by ambiguous information about processing, and the dominance of one software package. She therefore did not observe any clear trends with regard to users or software packages.
Marabini, Kazemi, Sorzano and Carazo
RM and co-workers devised a pair comparison method, where they modified the FSC curve calculated between each two unfiltered maps to produce a weighted integrated similarity value 5 (Marabini et al., 2018). They then used the pairwise similarity matrix to identify dissimilar submissions, and conversely, clusters of similar maps. They also aggregated all similarity values for each submission with respect to other maps, and used it to do a hierarchical classification of the maps. They find that all the algorithms in the software packages have the potential to work properly if the data is good. When looking at the initial frame alignment methods, they observe a small improvement in cases where optical-flow (Abrishami et al., 2015) was used over motioncorr (version 1) (Li et al., 2013), potentially due to local translational refinement. It is also not clear if dose-weighting is beneficial, but that could be hidden behind stronger influences.
Stagg and Mendez
SMS and JHM examined the submissions by how well a model can be built into them (Stagg and Mendez, 2018). Their assumption is that the user-sharpened map represents the most interpretable representation of the structure. They extracted small parts of the maps and built multiple (thousands) of de novo models using Rosetta (Bradley et al., 2005; Raman et al., 2009). They then assessed the quality of the models by calculating the RMSD with respect to the reference model, and the average RMSD of 100 randomly selected pairs of generated models. They were able to produce models for all cases except GroEL. One of their conclusions is that the user-reported FSC did not show good agreement with their modeling results. Another is that there are subsets of particles contributing most to the map quality, but at least in one case the largest number of particles used gave the best map (ribosome submission 123). Finally, they find that the familiarity of the user with SPA influences the map quality.
Pintilie and Chiu
GP and WC calculated Z-scores for secondary structure elements (SSEs) and side chains in the submitted fitted models to assess to what degree these features are resolved in the density (Pintilie and Chiu, 2018). They point out that the reported resolution for a given map, calculated by FSC, is a global parameter which does not reveal the variation of resolvability within the map. The proposed Z scores quantify the match between the local features in the model and the observed densities. In their analysis, GP and WC show that Z scores correlate moderately to reported resolution, and provide a ranking of the submitted maps which more closely corresponds to visual analysis.
Zhao, Palovcak, Armache and Cheng
JZ and coworkers only analyzed the TRPV1 case (see Appendix). They visually inspected the maps, did an analysis with EMRinger (Barad et al., 2015), calculated an FSC curve with respect to a model (PDB 3J5P) using a spherical (isotropic) mask, and refined a model for each map with Phenix (Adams et al., 2010). Several of the maps cluster with FSC0.5 ≈ 7 Å (7/8 maps) and FSC0.143 ≈ 4.1 Å (5/8 maps). They report a Molprobity score (Chen et al., 2010) for each refined model, indicating much better quality of all the fitted structures than expected from the estimated resolutions. Submission 156 is clearly overfit as indicated by much better apparent resolution than the others. Submissions 133 and 135 achieved low scores with EMRinger and correlation after refinement with Phenix. They conclude that the EMRinger results agree best with visual inspection, while the resolution estimates by FSC and the refinement in Phenix are not reliable quality indicators.
Comparison of results
The assessment approaches were all different, making comparisons complicated. Here we attempt to relate results with some overlap, aiming at a consistent interpretation. We also want to highlight those maps that show signs of problems with an eye to improved practices. Several of us calculated FSC curves between the even and odd maps (Harauz and van Heel, 1986). Figure 2 shows the correspondence of the resolutions reported by the submitters to the re-estimates. The users presumably calculated their estimates with their own masks, also used by MH and AP. There is general agreement (barring a few outliers), indicating consistency of the relatively straightforward FSC calculations between different software packages. MH and AP also calculated FSC curves without masks, that produced estimates that are in general worse (as expected). JBH estimated resolutions on rescaled maps with consistent case-specific masks (Heymann, 2018b), which for the most part correspond to user-reported estimates.
Three of the assessments used a form of modeling as a measure of map quality ((Pintilie and Chiu, 2018; Stagg and Mendez, 2018), Appendix). We assume that the most representative basis for modeling is the map as filtered and sharpened by the user. While the different ways to prepare the map can potentially affect the results, we did not investigate how this would influence the ranking results.
We compared the maps either through a measure such as the resolution or model fitting score, or by pairwise similarity. With each type of measure we were able to identify clusters of similar maps and rank these as shown in Table 1. We calculated an optimal ranking using the RankAggreg method (Pihur et al., 2009) with the “BruteForce” option that considers all possible combinations. For the ribosome and ß-galactosidase cases with more than 10 contributions, this approach is too time consuming and we used the “RankAggreg” option with the Cross-Entropy Monte Carlo algorithm. We assessed the similarity between rankings with the Spearman distance. The algorithm does not allow several maps with the same rank, so in those cases all possible permutations were tested with appropriate weights (e.g., for a case with clusters of 3, 3, 2, and 1, there are 6×6×2×1 = 72 equally probable rankings and the weight for each is thus 1/72). The results are shown in the “Optimal” columns in Table 1 (also see the last column in the Supplemental spreadsheet).
Table 1:
Case 1: GroEL. We ranked the maps based on various analyses and impressions†. Where the maps were similar enough as judged by the assessor, they received the same rank.
| Rank | JBH | RM | GP | SJ | Optimal |
|---|---|---|---|---|---|
| 1 | 132,143, 165 |
104,120, 132,143, 165,169 |
132 | 104,120, 132,143, 153,165 |
132 |
| 2 | 169 | 143 | |||
| 3 | 143 | 165 | |||
| 4 | 169 | 120 | 169 | ||
| 5 | 104,120 | 165 | 120 | ||
| 6 | 168 | 104 | |||
| 7 | 158,168 | 153,158, 168 |
158 | 158 | 158 |
| 8 | 153 | 168 | 168 | ||
| 9 | 153 | 104 | 169 | 153 |
The bases for the different rankings used by the assessors: JBH: FSCref (Heymann, 2018b)
RM: FSCi (Marabini et al., 2018)
GP: Side chain Z-score (Pintilie and Chiu, 2018)
JHM(1): Combined score (Stagg and Mendez, 2018)
JHM(2): Internal RMSD (Stagg and Mendez, 2018)
SJ: CC-based distances between maps approximated with Gaussians (Jonic, 2018)
JZ: Visual inspection (Zhao and coworkers, Appendix)
Optimal: An optimal ranking calculated using RankAggreg (Pihur et al., 2009)
The clustering and ranking varies with assessment as expected from the different approaches that address different aspects of map quality. However, there is some consistency to the cluster patterns for each of the cases. What we learn from these rankings is not only which are the best maps, but even more importantly, which maps have problems that we can address through better software development or practices. In the following discussion we focus on the best and worst submissions and the possible reasons for this outcome.
The ranking results for GroEL (Table 1) is a mixed bag, with some disagreement between the assessors. This could result from the issues with the generation of the original images. Unfortunately, the images were produced without considering that the GroEL molecule is asymmetric at high resolution. The projections were calculated with orientations within only one asymmetric unit, omitting the necessary views from other angles. Some submitted maps have D7 symmetry imposed, while others were done asymmetrically. The best map seems to be 132, and the worst 158 and 168. One person submitted 168, done without symmetry, and 169, done with D7 symmetry. The latter is consistently better, likely because of better noise suppression due to symmetrization. Unfortunately, this dataset does not produce much insight into how SPA was done by the different submitters.
For the 20S proteasome case (Table 2), there is some difference of opinion between the assessors. For example, 103 and 108 score high in half of the assessments, but low in the others. On the other side, 130 and 131 are mostly ranked at the bottom. RM and co-workers (Marabini et al., 2018) found that most of the maps are similar, and the variable ranking might indicate that the differences are very small and not of much significance.
Table 2:
Table 2: Case 2: 20S proteasome. See Table 1 for how the ranks were done
| Rank | JBH | RM | GP | JHM(1) | JHM(2) | SJ | Optimal |
|---|---|---|---|---|---|---|---|
| 1 | 103,108 | 103,107, 108,131, 141,144, 145,162 |
103 | 141 | 144,145 | 107,141, 144,145, 162 |
141 |
| 2 | 108 | 144 | 144 | ||||
| 3 | 141 | 141 | 145,162 | 107,141 | 145 | ||
| 4 | 130,131 | 162 | 162 | ||||
| 5 | 145 | 107,108 | 162 | 107 | |||
| 6 | 144,145, 162 |
144 | 108 | 130,131 | 108 | ||
| 7 | 107 | 103 | 130 | 103 | |||
| 8 | 131 | 130 | 103 | 108 | 131 | ||
| 9 | 107 | 130 | 130 | 131 | 131 | 103 | 130 |
In the apo-ferritin case (Table 3) the maps 112 and 121 are deemed the best by most. Several of the maps were calculated from small numbers of images, with mixed results. Map 124 was calculated from only 1371 images, but is of better quality than the worst maps calculated from 243 (map 122), 1370 (map 147) and 7304 (map 155) images. SJ found a small conformational difference in map 124 compared to the better ones (Jonic, 2018), potentially due to the limited data included. Nevertheless, the demonstration that a good map can be reconstructed from few particle images, suggests that there is a problem with the processing of the other maps.
Table 3:
Case 3: Apo-ferritin. See Table 1 for how the ranks were done.
| Rank | JBH | RM | GP | JHM(1) | JHM(2) | SJ | Optimal |
|---|---|---|---|---|---|---|---|
| 1 | 121 | 112,118, 121,124, 166 |
121 | 112,121 | 112 | 112,118, 166 |
121 |
| 2 | 112 | 112 | 121 | 112 | |||
| 3 | 166 | 166 | 166 | 118 | 166 | ||
| 4 | 118 | 118 | 118 | 166 | 155 | 118 | |
| 5 | 124 | 124 | 124 | 124 | 121 | 124 | |
| 6 | 122 | 155 | 155 | 155 | 155 | 122 | 155 |
| 7 | 155 | 122,147 | 147 | 122,147 | 122 | 124 | 122 |
| 8 | 147 | 122 | 147 | 147 | 147 |
For TRPV1 (Table 4), several maps are ranked mostly in the top half (115, 133, 135, 161). The consistently poor quality maps are 146, 156 and 163. The problems in this case might be due to flexibility in some of the structure, as reflected in relatively low correlation at low frequencies (see FSCeo curves by (Heymann, 2018b)).
Table 4:
Case 4: TRPV1. See Table 1 for how the ranks were done.
| Rank | JBH | RM | GP | JHM(1) | JHM(2) | SJ | JZ | Optimal |
|---|---|---|---|---|---|---|---|---|
| 1 | 133,135 | 101,115, 133,135, 161 |
161 | 101 | 101 | 101,115, 133,135 |
115,161 | 101 |
| 2 | 115 | 115 | 115 | 115 | ||||
| 3 | 115 | 135 | 161 | 133,161 | 101 | 161 | ||
| 4 | 163 | 133 | 133,135 | 133,135 | 133 | |||
| 5 | 146 | 101 | 135 | 161 | 135 | |||
| 6 | 161 | 156,163 | 156 | 163 | 156,163 | 163 | ||
| 7 | 101,156 | 146 | 156 | 156 | ||||
| 8 | 146 | 163 | 146 | 146 | 146 |
The best map for the ribosome case (Table 5) is 123, and the worst cases are 111 and 129. Here we have the interesting situation that a best (123) and a worst (129) were done by the same person, the difference being the number of particle images (~8 fold difference in number). For BMV (Table 6) the best maps are 102 and 140, while the worst ones are 110 and 152. Both the latter maps were reconstructed from fewer particle images than the better maps. Submission 152 was done with a new algorithm (SAF-FPM) that potentially needs refinement.
Table 5:
Case 5: Ribosome. See Table 1 for how the ranks were done.
| Rank | JBH | RM | GP | JHM(1) | JHM(2) | SJ | Optimal |
|---|---|---|---|---|---|---|---|
| 1 | 123, 125 | 123 | 123 | 123 | 123 | 114,123, 125,126, 127,128, 148,149, 150,151 |
123 |
| 2 | 151 | 126 | 129,148, 151,119, 127 |
125 | 125 | ||
| 3 | 114, 126, 151 |
114,125, 126,149, 150 |
151 | 114 | 151 | ||
| 4 | 114 | 149 | 114 | ||||
| 5 | 125 | 127 | 126 | ||||
| 6 | 149,150 | 149 | 148 | 149 | |||
| 7 | 150 | 114,126 | 126 | 150 | |||
| 8 | 127 | 119,127, 128,148 |
127 | 128,151 | 127 | ||
| 9 | 111,128 | 128 | 125 | 128 | |||
| 10 | 148 | 128,149 | 150 | 148 | |||
| 11 | 119 | 129 | 129 | 111 | 129 | ||
| 12 | 148 | 111 | 111 | 150 | 119 | 119 | 119 |
| 13 | 129 | 129 | 119 | 129 | 111 |
Table 6:
Case 6: BMV. See Table 1 for how the ranks were done.
| RM | GP | JHM(1) | JHM(2) | SJ | Optimal | ||
|---|---|---|---|---|---|---|---|
| 1 | 102 | 102,136, 137,140, 142 |
102 | 140 | 110 | 102,136, 137,140, 142 |
102 |
| 2 | 140 | 140 | 102,136, 137 |
102,137, 140 |
140 | ||
| 3 | 142 | 142 | 142 | ||||
| 4 | 137 | 137 | 137 | ||||
| 5 | 136 | 136 | 142 | 136,142 | 136 | ||
| 6 | 110,152 | 110 | 110 | 110 | 110 | 110 | |
| 7 | 152 | 152 | 152 | 152 | 152 | 152 |
The ß-galactosidase case (Table 7) is perhaps the most straightforward case, and one assessor (RM and co-workers (Marabini et al., 2018)) could not find significant differences between the maps. The others of us could distinguish differences in quality that seem to be important. The maps consistently deemed good are 138 and 139, while the bad maps are 159 and 164 (these maps are identical) and 167. These were produced by a new reconstruction algorithm generating artifacts, to the extent that one assessor (SMS and JHM, (Stagg and Mendez, 2018)) using modeling approaches could not analyze them.
Table 7:
Case 7: ʰ-galactosidase. See Table 1 for how the ranks were done.
| Rank | JBH | RM | GP | JHM(1) | JHM(2) | SJ | Optimal |
|---|---|---|---|---|---|---|---|
| 1 | 134 | 106,113, 116,134, 138,139, 154,157, 159,160, 164,167 |
138 | 138 | 138 | 106,113, 134,138, 139,154 |
138 |
| 2 | 116,138, 139 |
139 | 106,139 | 106,139 | 139 | ||
| 3 | 134 | 106 | |||||
| 4 | 113 | 154 | 154 | 154 | |||
| 5 | 160 | 106 | 113 | 113 | 113 | ||
| 6 | 167 | 154 | 116,157, 160 |
116 | 134 | ||
| 7 | 154 | 116 | 134 | 157,160 | 160 | ||
| 8 | 106 | 160 | 160 | 116 | |||
| 9 | 157 | 157 | 134 | 157 | 116 | 157 | |
| 10 | 113 | 159/164* | 159/164* | 159/164* | |||
| 11 | 159/164* | ||||||
| 12 | 167 | 167 | 167 |
159 and 164 are identical.
Discussion
The value of the Map Challenge is in giving the 3DEM community an opportunity to ascertain the state of SPA. One of the main questions is whether we are doing the best image processing we can, and what needs to improve. The availability of high quality data sets is crucial to obtain a relevant answer to these questions. The generously provided data sets cover typical sizes and symmetries of particles being analyzed (Figure 1). For a first challenge, the selection focused on rigid particles that should be easy to reconstruct.
Doing SPA today requires a significant investment in time and computational resources. The number of cases and the open-ended nature of the analysis were issues that both the users and assessors struggled with. In the end, 66 maps were submitted, spread over 7 cases, with about half done with only one software package. After the initial assessment by MH and AP, only six further individual assessments were submitted ((Heymann, 2018b; Jonic, 2018; Marabini et al., 2018; Pintilie and Chiu, 2018; Stagg and Mendez, 2018) , Appendix) , using several different approaches to analyze the submitted maps. The associated statistics is therefore insufficient to draw hard conclusions, and the assessment here at best provides some guidelines for the further evolution of SPA and future challenges.
The inherent nature of SPA makes assessing its results non-trivial. The two traditional ways of judging the quality of a map are visual inspection and calculating FSC between independently reconstructed maps. The first is subjective, relying heavily on the experience of the observer. The second is subject to subtle influences arising from the processing workflow, or otherwise overt manipulation. The more recent way of assessing the interpretability of a map through modeling implicitly assumes that the map was properly reconstructed and filtered. So, we are faced with the dilemma that there is no absolute “gold standard”. What we can look for is consistency.
An important observation made by several assessors is that for each case, the submitted maps could be clustered based on quality measures (Tables 1–7). In each case, one cluster with high quality maps then represent a form of validation through consensus, as these were processed by different people and with different software packages. The ranking patterns also show some consistency across the various assessments. These allow us to identify those maps with poor quality, likely resulting from either lack of data, issues with particle image alignment, or overfitting.
One of the aims of the Map Challenge was to understand influential aspects of SPA. We could not find a strong relationship between the map quality and a software package or workflow used. We conclude that with good data and an appropriate workflow, the highest possible quality can be achieved. Instead, the success of the outcome seems to correspond best to the user, suggesting that familiarity with the software is a key determinant.
Our consensus is therefore that a good SPA result is achieved through taking meticulous care of every aspect of the workflow. RM and co-workers (Marabini et al., 2018) concluded that it is beneficial to do frame alignment with local refinement. The issues of CTF fitting and particle selection remain important elements in reconstruction. JBH (Heymann, 2018b) emphasized the correct generation and use of masks for reference modification and FSC calculation. Proper validation principles must be followed, making sure that they are not compromised in some way. Many of these aspects are further discussed in the next section.
One issue that emerged from the rankings is that new algorithms for alignment or reconstruction fared relatively poorly. However, this does not mean these approaches are less useful than the more established ones. Most likely, the well-used methods have been refined over many years to more carefully deal with statistical realities. With perseverance, the authors of newer methods may very well develop better ways of processing the data. Indeed, after the assessment reported here was done, several software developers improved their SPA algorithms and produced better reconstructions (Bell et al., 2018; Sorzano et al., 2018). We encourage future participants of map challenges to contribute novel methods of processing.
Best practices
The Map Challenge exposed the extent to which the outcome of SPA is still determined by the ability of the user. The familiarity of the user and the choices he/she makes impacts the quality of reconstructions (Heymann, 2018b; Stagg and Mendez, 2018). This means that the current understanding of best practices is either not sufficient, or not adequately taught. It also indicates that the typical software lacks safeguards and warnings that should guide users towards valid reconstructions. The latter is particularly important as we move towards full automation. The following list includes practices that are already common in SPA, and some that need to become the norm.
Validation
All of the Map Challenge submitters processed two separate data subsets for each submission. While processing such separated subsets is certainly a best practice, it is not sufficient to ensure validity. Several reconstructions show signs of overfitting, and thus a violation of the independence of the individual workflows. Potential solutions to avoid this are: resolution-limited particle alignment, appropriate masking (see below), and low-pass filtering reference maps (see below).
Visual cues
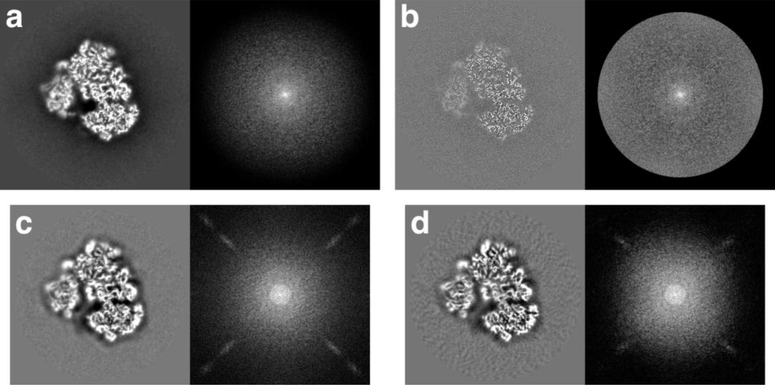
The most important assessment of a map is still visual inspection. We trust our eyes to tell us if a map is reasonable at the reported level of detail. We expect a well-constructed map to have clear density for the particle with detailed features. Ideally, unfiltered maps should not be masked, bandpass-limited or amplitude-modified (such as sharpened). Figure 3a shows the central slice and central section of the best ribosome map, unfiltered. It has significant low-pass character that is modified during the filtering that produced the map in Figure 3b. The noise in the background of this map has a good texture that is muted so that it is not amplified by filtering. The power spectrum has an even distribution of intensities up to the cutoff frequency in Figure 3b. In contrast, the worst ribosome maps (111 and 119, Figure 3c,d) have artifacts (streaks) in their power spectra. Furthermore, the central slice in Figure 3d has a background with radial streaks. A significant number of the submissions have such undesirable issues (see Heymann 2018 supplement for more detail (Heymann, 2018b)). The potential causes include inadequate particle picking, inappropriate alignment, the reconstruction algorithm and unsuitable post-processing. Identifying such a cause for a particular case requires a deeper, quantitative analysis.
Figure 3:
Examples of submitted maps for the ribosome case to highlight some issues that can be identified by visual inspection. In each panel the left side shows a central slice and the right side shows the logarithm of the power spectrum. (a-b) The best ribosome map (123) unfiltered (a) and filtered (b). (c-d) Two of the worst ribosome maps (c – 111, d – 119). For both these submissions the unfiltered and filtered maps are identical
Calculating FSC curves
The FSC analysis (Harauz and van Heel, 1986) remains the most important quantitative assessment of map quality. Nevertheless, it is still beset by uncertainty and controversy (Rosenthal and Rubinstein, 2015; Sorzano et al., 2017; van Heel and Schatz, 2005). The maps are usually masked prior to analysis to remove noise outside the particle boundaries. The nature of the masks are however important to ensure a valid FSC curve, as any high frequency features may introduce artificial correlations and lead to erroneous resolution estimates (Rosenthal and Rubinstein, 2015). Such an over-optimistic impression of the reliable detail in the maps is commonly referred to as “overfitting”. The most appropriate way is to remove background with a fuzzy mask low-pass filtered with a hard cutoff well below the expected resolution of the map. When reporting a masked FSC curve, the mask used should also be provided.
Reference map handling
The reference map used to align the particle images should be appropriately handled to avoid problematic issues. Most important is to have some form of low-pass filtering of the map itself, or limiting projection-matching to some high resolution limit (i.e., resolution-limited alignment). Where the reference map is masked, the mask itself should not introduce high frequency elements. This is best achieved by low-pass filtering the mask to a resolution well below the estimate for the map.
A second issue is how the map should be weighted. Sharpening may or may not influence alignment, depending on the particular algorithm used in the software package. The user should be aware whether sharpening has an influence on particle alignment. This should be stated more explicitly in the documentation of each software package.
Final map processing
For the Map Challenge, access to the original unfiltered maps is important to understand aspects of the image processing workflow that produced them. These maps should be reconstructed to the Nyquist frequency with no masking or amplitude modification (sharpening). We acknowledge that some reconstruction algorithms already impose such modifications, but the ideal would be to omit any form of filtering during reconstruction.
The purpose of the filtered map after reconstruction is to aid interpretation, most often modeling the structure. It should be masked to remove background noise and low-pass filtered to just beyond the estimated resolution. It further should be sharpened to allow the best chance for model building. Current practice is to either filter it with a so-called “negative B-factor” (for the effects of different B values on maps, see (DeLaBarre and Brunger, 2006)), or with a more sophisticated algorithm that attempts to weigh structure factors more according to theoretical expectation (Rosenthal and Henderson, 2003). Finally, the map appearance should conform to the guidelines stated in the section “Visual cues”.
Minimum metrics report
The X-ray crystallography community has a common practice of providing experimental details for data collection and processing in a summary table. The 3DEM community can benefit from a similar common or standard table to incorporate in manuscripts and aid in future challenges. We used the spreadsheet with all the submission details (see Supplemental Material) as a source of elements that should go into such a table. Table 8 shows a suggested layout for a minimum metrics report for a single particle reconstruction.
Table 8.
Minimum metrics report based on the submission 123 for the 80S ribosome, including information from the original data collection (Wong et al., 2014).
| Parameter | Example values |
|---|---|
| Data collection | |
| Microscope | FEI Polara |
| Detector | FEI Falcon II |
| Acceleration voltage (kV) | 300 |
| Number of micrographs | 1081 |
| Frames per micrograph | 16 |
| Frame rate (/s) | 16 |
| Dose per frame (e-/pixel) | 2.24 |
| Accumulated dose (e-/Å2) | 20 |
| Defocus range (ʼm) | 0.8 – 3.8 |
| Frames: | |
| Alignment software | motion_corr |
| Frames used in final reconstruction | 1 – 16 |
| Dose weighting | yes |
| CTF: | |
| Fitting software | CTFFIND4 |
| Correction | Full |
| Particles: | |
| Picking software | Relion 1.4 |
| Picked | 123232 |
| Used in final reconstruction | 123232 |
| Alignment: | |
| Alignment software | Relion 1.4 |
| Initial reference map | EMDB 2275 |
| Low-pass filter limit (Å) | 60 |
| Number of iterations | 25 |
| Local frame drift correction | yes |
| Reconstruction: | |
| Reconstruction software | Relion 1.4 |
| Size | 380×380×380 |
| Voxel size (Å) | 1.34 |
| Symmetry | C1 |
| Resolution limit (Å) | 2.68 |
| Resolution estimate (Å, FSC0.143) | 3.1 |
| Masking | no |
| Sharpening | B-factor: −62.4 |
Future map challenges
The goal of the Map Challenge is to understand the current state of SPA and what to improve. An ideal outcome would have included an even distribution of software packages and workflows. As assessors, we struggled with the dominance of one software package, combined with the numerous ways in which workflows were constructed. This likely influenced the assessment results in ways that are not obvious. For future challenges, it would be useful to have a more targeted approach. For one, the number of cases should be reduced to two or three to allow more direct comparisons between submissions. An effort should be made to have a better representation of different software packages. Specific goals could be aimed at particular issues, such as frame alignment, the effect of masking on particle alignment and FSC calculation, and the differences in reconstruction algorithms.
In this challenge, several submitters expressed an interest in resubmitting improved maps after the initial assessment. One possibility is to incorporate such a process into challenges. The first assessment is communicated to the submitters, who are then expected to either keep their original submissions, or resubmit new ones. The second assessment is then on the final submissions, and forms the basis of an evaluation of the state of SPA. The focus of the reporting is therefore on what was done to improve the reconstructions.
Conclusion
The Map Challenge is a valuable experience that exposed the areas in which SPA needs attention. It is also the first time that multiple assessors analyzed the submitted maps and devised ways to examine the quality of reconstructions. Many of these approaches will need to be refined to have consistent measures for future challenges. Nevertheless, we showed that in each case we could identify a consensus structure supported by the combined analysis. We believe this provides a sound basis for assessments in future challenges. The outcome of this challenge strongly indicated that the choices made by the users dictate the quality of the processing. The short-term solution is therefore to promote best practices when using current software packages. In the long term, these practices should be encoded in the algorithms used in SPA. Future challenges should preferably be focused on specific aspects of SPA to simplify assessment, increasing its value.
Supplementary Material
Acknowledgements
We thank the Map Challenge organizing committee for their efforts to advance single particle analysis: Bridget Carragher, Wah Chiu, Cathy Lawson, Jose-Maria Carazo, Wen Jiang, John Rubinstein, Peter Rosenthal, Fei Sun, Janet Vonck, and Ardan Patwardhan. We also thank all the participants in the Map Challenge. This work was supported by:
The Intramural Research Program of the National Institute for Arthritis, Musculoskeletal and Skin Diseases, NIH (JBH).
The Spanish Ministry of Economy and Competitiveness through Grants BIO2013–44647-R, BIO2016–76400-R(AEI/FEDER, UE) and AEI/FEDER BFU 2016 74868P, the Comunidad Autónoma de Madrid through Grant: S2017/BMD- 3817, European Union (EU) and Horizon 2020 through grant CORBEL (INFRADEV-1–2014-1, Proposal: 654248). This work used the EGI Infrastructure and is co-funded by the EGI-Engage project (Horizon 2020) under Grant number 654142. European Union (EU) and Horizon 2020 through grant West-Life (EINFRA-2015–1, Proposal: 675858) European Union (EU) and Horizon 2020 through grant Elixir - EXCELERATE (INFRADEV-3– 2015, Proposal: 676559) European Union (EU) and Horizon 2020 through grant iNEXT (INFRAIA-1–2014-2015, Proposal: 653706). The authors acknowledge the support and the use of resources of Instruct, a Landmark ESFRI project (RM, MK, COSS and JMC).
Partially by NIH grant R01GM108753 (JHM and SMS).
NIH grants: P41GM103832 and R01GM079429 (GP and WC).
Abbreviations
- SPA
Single particle analysis
- EM
electron microscopy
- EMDB
Electron microscopy data bank
- SNR
Signal-to-noise ratio
- FSC
Fourier shell correlation
- FSCeo
Even-odd FSC
- FSCref
FSC relative to a reference map
Appendix: Assessment of TRPV1 submissions to the Map Challenge
Jianhua Zhao1, Eugene Palovcak1, Jean-Paul Armache1 and Yifan Cheng1
1Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94143, USA
Eight TRPV1 maps were evaluated by the Cheng laboratory. We conducted four different tests to evaluate map quality: 1) visual inspection of the maps by four separate experts, 2) EMRinger analysis (Barad et al., 2015), 3) map-to-model FSC calculation, 4) Phenix refinement and resulting statistics (Adams et al., 2010). Criteria for ranking of maps by visual inspection include visualization of features in the transmembrane region, identification of bound lipids, and minimal noise and artefacts in the map. EMRinger analysis was conducted with the PDB model 3J5P. The map-to-model FSCs were calculated using a soft spherical mask. A single round of Phenix refinement was completed with PDB model 5IRZ. Results of the analyses are summarized in Tables A1 and A2.
Through visual inspection, we consider maps 161 and 115 to be of the highest quality, followed by map 101. Maps 133 and 135 used the super-resolution pixel size and therefore appears suspiciously smooth. Maps 156 and 163 had fewer visible side-chain densities and were ranked lower. Map 146 appears to have strong artefacts and was ranked lowest. The only metric that appeared to corroborate the visual ranking is the EMRinger score (Table A1). EMRinger is a method that uses side-chain density to evaluate the placement of backbone atoms in EM maps. Higher EMRinger scores indicate better models and, with the exception of maps 133 and 135, higher ranked maps also had higher EMRinger scores. One possible reason maps 133 and 135 had low EMRinger scores could be errors with the header information of the maps, distorting the alignment between map and model. Maps 133 and 135 also had unreasonably low map cross-correlation scores (Table A2, Map CC). The FSC analysis and Phenix refinement statistics were not found to be good indicators of map quality. It is worth noting that map 156 had a very sharp drop-off in the FSC, with the FSC falling below 0.5 and 0.143 at inverse spatial frequencies of 3.4 and 3.3 Å, respectively.
In conclusion, EMRinger works well in helping to assess map quality when side-chain densities are present. However, in general, the best way to assess the quality of an EM map remains visual inspection by experts. Further methods development could provide more quantitative and accurate means of assessing map quality.
Table A1.
EMRinger and FSC analysis
| Rank | Map # | EMRinger | FSC=0.5 (Å) | FSC=0.143 (Å) |
|---|---|---|---|---|
| 1* | 161 | 4.03 | 6.9 | 4.1 |
| 1* | 115 | 3.92 | 6.9 | 4.1 |
| 2 | 101 | 2.58 | 7.1 | 4.2 |
| 3** | 133 | 0.22 | 6.9 | 4.1 |
| 3** | 135 | 0.65 | 7.2 | 4.2 |
| 4*** | 156 | 2.37 | 3.4 | 3.3 |
| 4*** | 163 | 2.29 | 7.4 | 5.7 |
| 5 | 146 | 0.43 | 7.0 | 4.7 |
identical rankings
identical rankings
identical rankings
Table A2.
Phenix refinement
| Rank | Map # | Rama. Favored |
MolProbity score† |
Clashscore | RMSD bonds |
RMSD angles |
Map CC |
|---|---|---|---|---|---|---|---|
| 1* | 115 | 0.926 | 1.74 | 5.18 | 0.006 | 1.198 | 0.81 |
| 2 | 101 | 0.896 | 1.78 | 4.44 | 0.012 | 1.379 | 0.811 |
| 3** | 133 | 0.9311 | 1.73 | 5.4 | 0.005 | 1.145 | 0.034 |
| 3** | 135 | 0.919 | 1.88 | 7.06 | 0.007 | 1.368 | 0.052 |
| 4*** | 156 | 0.9082 | 1.79 | 5.01 | 0.008 | 1.284 | 0.722 |
| 4*** | 163 | 0.9158 | 1.84 | 6.18 | 0.01 | 1.32 | 0.788 |
| 5 | 146 | 0.9247 | 1.67 | 4.22 | 0.004 | 1.105 | 0.0 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrishami V, Vargas J, Li X, Cheng Y, Marabini R, et al. , 2015. Alignment of direct detection device micrographs using a robust Optical Flow approach. J Struct Biol 189, 163–176. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, et al. , 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, et al. , 2015. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12, 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Aguerrebere C, Falconieri V, Banerjee S, Earl LA, et al. , 2018. Atomic Resolution Cryo-EM Structure of beta-Galactosidase. Structure 26, 848–856 e843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JM, Fluty AC, Durmaz T, Chen M, Ludtke SJ, 2018. New software tools in EMAN2 inspired by EMDatabank map challenge. J Struct Biol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P, Misura KM, Baker D, 2005. Toward high-resolution de novo structure prediction for small proteins. Science 309, 1868–1871. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, et al. , 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT, 2006. Considerations for the refinement of low-resolution crystal structures. Acta Crystallogr D Biol Crystallogr 62, 923–932. [DOI] [PubMed] [Google Scholar]
- Harauz G, van Heel M, 1986. Exact filters for general geometry three dimensional reconstruction. Optik 73, 146–156. [Google Scholar]
- Henderson R, 2013. Avoiding the pitfalls of single particle cryo-electron microscopy: Einstein from noise. Proc Natl Acad Sci U S A 110, 18037–18041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, 2015. Validation of 3D EM Reconstructions: The Phantom in the Noise. AIMS Biophys 2, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, 2018a. Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci 27, 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, 2018b. Map Challenge Assessment: Fair comparison of single particle cryoEM reconstructions. J Struct Biol, in press. [DOI] [PubMed] [Google Scholar]
- Jonic S, 2018. A methodology using Gaussian-based density map approximation to assess sets of cryo-electron microscopy density maps. J Struct Biol, in press. [DOI] [PubMed] [Google Scholar]
- Jonic S, Sorzano CO, 2016. Versatility of Approximating Single-Particle Electron Microscopy Density Maps Using Pseudoatoms and Approximation-Accuracy Control. Biomed Res Int 2016, 7060348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, et al. , 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10, 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabini R, Kazemi M, Sorzano COS, Carazo J-M, 2018. Map Challenge: Analysis using Pair Comparison Method. J Struct Biol, in press. [DOI] [PubMed] [Google Scholar]
- McMullan G, Faruqi AR, Henderson R, 2016. Direct Electron Detectors. Methods Enzymol 579, 1–17. [DOI] [PubMed] [Google Scholar]
- Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, et al. , 2016. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell 165, 1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihur V, Datta S, Datta S, 2009. RankAggreg, an R package for weighted rank aggregation. BMC Bioinformatics 10, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintilie G, Chiu W, 2018. Assessment of Structural Features in Cryo-EM Density Maps using SSE and Side Chain Z-Scores. J Struct Biol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, et al. , 2009. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 77 Suppl 9, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, 2016. Testing the Validity of Single-Particle Maps at Low and High Resolution. Methods Enzymol 579, 227–253. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R, 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Rubinstein JL, 2015. Validating maps from single particle electron cryomicroscopy. Curr Opin Struct Biol 34, 135–144. [DOI] [PubMed] [Google Scholar]
- Sanchez Sorzano CO, Alvarez-Cabrera AL, Kazemi M, Carazo JM, Jonic S, 2016. StructMap: Elastic Distance Analysis of Electron Microscopy Maps for Studying Conformational Changes. Biophys J 110, 1753–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorzano CO, Vargas J, Oton J, Abrishami V, de la Rosa-Trevin JM, et al. , 2017. A review of resolution measures and related aspects in 3D Electron Microscopy. Prog Biophys Mol Biol 124, 1–30. [DOI] [PubMed] [Google Scholar]
- Sorzano COS, Vargas J, Trevin JMR, Jiménez A, Melero R, et al. , 2018. High-resolution reconstruction of Single Particles by Electron Microscopy. J Struct Biol, in press. [DOI] [PubMed] [Google Scholar]
- Stagg SM, Mendez JH, 2018. Assessing the quality of single particle reconstructions by atomic model building. J Struct Biol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, 2013. Structure of trimeric HIV-1 envelope glycoproteins. Proceedings of the National Academy of Sciences 110, E4172–E4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YZ, Aiyer S, Mietzsch M, Hull JA, McKenna R, et al. , 2018. Sub-2 Å Ewald Curvature Corrected Single-Particle Cryo-EM. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, et al. , 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38–46. [DOI] [PubMed] [Google Scholar]
- van Heel M, 2013. Finding trimeric HIV-1 envelope glycoproteins in random noise. Proc Natl Acad Sci U S A 110, E4175–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Schatz M, 2005. Fourier shell correlation threshold criteria. J Struct Biol 151, 250–262. [DOI] [PubMed] [Google Scholar]
- Vinothkumar KR, Henderson R, 2016. Single particle electron cryomicroscopy: trends, issues and future perspective. Q Rev Biophys 49, e13. [DOI] [PubMed] [Google Scholar]
- Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, et al. , 2014. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.