Abstract
Bacterial infections cause acute and chronic diseases. Antimicrobial resistance and aging-related immune weakness remain challenging in therapy of infectious diseases. Vaccines are however an alternative to prevent bacterial infections. Here we report a facile method to rapidly generate bacterium-membrane-formed nanovesicles as a vaccine using nitrogen cavitation. The vaccine is comprised of double-layered membrane vesicles (DMVs) characterized by cryo-TEM, biochemistry and proteomics, showing DMVs possess the integrity of bacterial membrane and contain a wide range of membrane proteins required for vaccination. In the mouse sepsis model induced by Pseudomonas aeruginosa, we found that DMVs can improve mouse survival after mice were immunized with DMVs. The increased adaptive immunity and unique biodistribution of DMVs were responsible for enhanced protection of bacterial infection. Our studies demonstrate that this simple and innovative approach using nitrogen cavitation would be a promising technology for vaccine developments.
Keywords: Vaccines, OMVs, DMVs, Pseudomonas aeruginosa, Infections, Nanotechnology
1. Introduction:
Bacterial infections cause severe acute and chronic inflammation resulting in tissue damage and multi-organ failure, such as acute lung injury and sepsis (1-3). The incidence of infectious diseases dramatically increases due to antimicrobial resistance (4,5) and aging-related weakness of immune systems (6), therefore, it is needed to develop novel approaches and technologies to tackle infectious diseases (7,8).
One of the strategies is vaccination, a process of administration of antigens that activate the host response to eliminate bacteria from the body. Antigen subunits of a bacterium were exploited to generate vaccines, for example, lipopolysaccharide (LPS), O-polysaccharides (9-11) and multivalent LPS-based vaccines have been evaluated in several disease models(12-15). The vaccines act by targeting specified toxins (16,17), but the multiple cellular pathways regulate the host responses to bacterial infections (18-20), therefore, this type of vaccination is unlikely effective for a wide range of pathogens.
Nanotechnology is a powerful tool to generate and assemble materials to form nanoscale structures with bio-functionality (21-26). Nanoparticles have been exploited in vaccine developments (27-29), for instance, nanoparticle-based vaccines were formulated by nanoparticles carrying antigens and adjuvants to mimic microbes (30-33). Nanoparticles are easily taken up by antigen-presenting cells (APCs) that induce T cell activation (34). However, nanoparticle-based vaccines are inadequate to replicate multiple pathways induced by a bacterium because a wide array of antigens expressed on a bacterium is required for the effectiveness of vaccines (35,36).
Outer membrane vesicles (OMVs) are derived from bacterial membrane and have gained much attention in the applications of vaccination and targeted drug delivery (37-45). For example, a gram-negative bacterium is comprised of outer and inner membranes, peptidoglycan and antigens across both membranes(46). It has been shown that bacteria constantly liberate OMVs to extracellular space(47,48), and OMVs show a single-layered spherical membrane with diameters of 20-250 nm (49). Despite their promising applications, OMVs have several limitations (45). Since OMVs are secreted from culture media, their composition, size and reproducibility vary batch by batch(50). In addition, scalability of OMVs is challenging using current approaches (51). Another issue is related to the stability of OMVs in vivo due to their single-layer cell membrane. To resolve this problem, gold nanoparticles were inserted in OMVs to increase their stability and long circulation time in vivo resulting in enhanced vaccination efficacy (50).
Herein, we report a facile approach to generate bacterial membrane nanovesicles using nitrogen cavitation. We show that nanovesicles are comprised of a whole bacterial membrane with inner and outer membrane components, and they act as a vaccine to prevent severe infections from bacteria. To demonstrate the principle of concept, we used Pseudomonas aeruginosa (P. aeruginosa) as a model, a major pathogen in infectious diseases (52,53). Vaccines against P. aeruginosa have been widely explored (54), however, there still remaining challenges, such as poor efficacy (55) and clinical safety (56,57). Our studies reveal that the nitrogen cavitation approach may be a simple technology to rapidly generate bacterial membrane nanovesicles for vaccine developments to overcome the safety issues of whole bacterial vaccines.
2. Materials and Methods:
2.1. Materials:
Gram-negative bacteria of P. aeruginosa (PA-103) were purchased from ATCC (29260™). HBSS (without Ca2+, Mg2+ and Phenol red) was purchased from Corning (Inc, NY). A nitrogen cavitation vessel was purchased from Parr instrument (Moline, IL). Sarkosyl and RIPA buffer were purchased from Sigma-Aldrich (St. Louis, MO, USA). DiD (3H-Indolium, 2-(5-(1,3-dihydro-3,3-dimethyl-1-octadecyl-2H-indol-2-ylidene)-1,3-pentadienyl)-3,3-dimethy l-1-octadecyl-, perchlorate) [Ex(640nm)\Em(670nm)], DAPI, Penicillin streptomycin (pen strep) and glutamine (100x) were purchased from Life Technologies (Grand Island, NY). A QuantiFluor dsDNA detection kit was purchased from Promega. Pierce™ BCA protein assay kit was purchased from Thermo Fisher Scientific. rmGM-CSF, anti-CD11c Abs, anti-CD40 Abs, anti-CD80 Abs, anti-CD86 Abs, murine IL-6 ELISA kit, murine IL-1β ELISA kit and murine TNF-α ELISA kit, murine IL-12p70 ELISA kit and murine IL-2 ELISA kit were purchased from Biolegend.
2.2. Bacteria culture and Generation of Bacterial Outer Membrane Vesicles
Gram-negative bacteria of P. aeruginosa were cultured in Luria broth agar overnight at 37 °C. A single colony was incubated in LB medium. Following incubation, the medium was cultured in a shaker at 37 °C until OD600 of the medium reached mid log phase approximately 1.0, indicating the logarithmic growth phase. P. aeruginosa OMVs were produced following the published protocol (58). Briefly, bacterial culture media were pelleted at 6,000 g for 30 min, the supernatant was filtered through a 0.45 μm filter followed by 0.22 μm filtration. The resultant suspension was concentrated via a 100 kDa tangential filtration concentration unit (Pall-Gellman). OMVs were pelleted at 200,000 g for 4 h and resuspended in HBSS buffer (1 ml) for future experiments.
2.3. Preparation of double-layered membrane vesicles (DMVs) from bacteria.
P. aeruginosa cells (1 L) were cultured and 100 mL cells were collected and washed with HBSS (without Ca2+, Mg2+ and Phenol red, Corning, Inc, NY). Then, these 100 mL cells were collected and were resuspended in HBSS at a concentration of 1-1.5×109 ml−1. The cell suspension (10-20 ml) was placed in a nitrogen cavitation vessel (Parr instrument, Moline, IL) under a pressure of 1500 psi for 20 min and the pressure was quickly released to disrupt cells. To completely disrupt cells, nitrogen cavitation was repeated twice. The resulting suspension was centrifuged at 6,000 g at 4 °C for 30 min. The resulting supernatant was centrifuged at 100,000 g at 4 °C for 30 min (Ultra TLX Beckman). After the supernatant was removed, a pellet was suspended in HBSS (1 ml). After cells were disrupted via nitrogen cavitation, the product after each centrifugation was quantitatively analyzed. Protein concentrations were determined via the BCA assay. The final vesicles after lyophilization were weighted for in vitro and in vivo experiments.
2.4. Characterization of DMVs
DMVs were characterized using dynamic light scattering (DLS) and cryo-TEM. The particle sizes and zeta potentials were measured by Malvern Zetasizer Nano ZS90 (Westborough, MA). Measurements were usually repeated five times. For cryo-TEM, a drop of vesicle solution (1 mg ml−1) was deposited on a carbon-coated grid discharged by PELCO easiGlow. After soaked by a piece of filter paper, the grid was quickly dropped in liquid nitrogen and stored overnight. The vesicles were imaged using TF20 TEM with a liquid nitrogen stage. Stability of nanovesicles was measured via monitoring nanovesicle size over time using DLS. The vesicles were labeled for imaging and biodistribution studies. Briefly, a pellet after ultracentrifuge was resuspended and the suspension was quickly mixed with DiD dye (1 μl, 1 mM) at 37 °C for 30 min in water bath. The suspension was centrifuged at 100,000 g twice to remove free dye molecules. To investigate whether DMVs contain specific antigens from their source of bacteria, a 96-well plate was coated with purified DMVs or P. aeruginosa, respectively, at a concentration of 100 ng per well in HBSS, then blocked with BSA/HBSS (1%), and loaded with anti-OmpA antibody (Gift). OmpA concentration in OMVs was measured as control. OmpA protein concentration were detected using secondary peroxidase-conjugated anti-mouse IgG Abs followed by adding a chemiluminescent substrate.
2.5. Proteomics and analysis of DMVs
P. aeruginosa outer membrane protein extracts were prepared based on papers published (49,59). Briefly, P. aeruginosa were pelleted as previously described and resuspended in Tris-HCl (1 ml, 20 mM, PH 8.0) containing 20% sucrose. Lysozyme (5 μl, 15 mg ml−1) and EDTA (10 μl, 0.5 M) were added to the suspension cells, followed by 40 min incubation on ice. Then MgCl2 (20 μl, 0.5 M) was added and the spheroplasts were pelleted at 9,500 g for 20 min. The supernatant containing periplasmic components was stored at −80 °C. The pellet was resuspended in ice-cold Tris-HCl (1 ml, 10 mM, PH 8.0) and then sonicated and centrifuged at 8,000 g for 5 min. The whole membrane then was pelleted from the supernatant at 40,000 g for 1 h, washed with Tris-HCl (10 mM, PH 8.0), resuspended in distilled water, and freeze-thawed. The membranes were then incubated with sarkosyl (0.5%, Sigma-Aldrich, St. Louis, MO, USA) at 25 °C for 20 min, and then the outer membrane was pelleted at 40,000 g for 90 min. Finally, these pellets were resuspended in ice-cold Tris-HCl (10 mM, PH 8.0) and stored in −80 °C. Proteins (5 μg) from outer membrane and purified DMVs fractions were electrophoresed on 10% SDS-PAGE. After electrophoresis, the gel was stained with CBB R-250 (Bio-Rad). Three bands from DMVs were dissected from SDS-PAGE gel for proteomics analysis.
For P. aeruginosa total proteins and P. aeruginosa-derived DMV protein extract preparations, the samples were collected and delipidation was performed by preparing an aqueous solution of protein (1-2 mg) in sterile milli-Q water (10 ml). An equal volume of Tris-buffered phenol was added and the mixture vortexed followed by heating at 70 °C for 10 min. The samples were placed on ice and centrifuged at 5,000 g for 10 min at 4 °C. The aqueous phase was discarded, and phenol extraction was repeated using equal volume of milli-Q water. Then, ice-cold acetone (2-fold volumes) was added to the phenol phase and centrifuged at 5,000 g for 10 min at 4 °C and this step was repeated twice. The pellet was left dry and dissolved in milli-Q water and stored at −20 °C.
Proteomics of P. aeruginosa and P. aeruginosa-derived DMVs were conducted in Tissue imaging & Proteomics Lab in Washington State University, Pullman. P. aeruginosa protein and genome database were obtained from Uniprot (release 15.12) and Pseudomonas Genome Database (60). Thermo Scientific™ Orbitrap FusionTM Tribrid™ Mass Spectrometer was used. Proteome Discoverer™ software was used to identify proteins. SEQUEST was utilized during peptide and protein identification processes.
2.6. P. aeruginosa -derived DMVs evoke innate immunity
(0.8 mg/kg) DMVs were i.p injected into CD1 mice and sera were collected at 12 h,24 h post injections. LPS (10 mg kg−1) injection was conducted as a positive control. Pro-inflammatory cytokines (IL-6, IL-1 β and TNF- α ) were measured via ELISA according to the manufacturer’s instructions (Biolegend).
2.7. Ex vivo studies of DCs activation
2.7.1. Bone marrow-derived DCs isolation
In bone marrow-derived DCs (BMDCs) isolation, CD1 mice (20-25 g) were euthanized and femur were dissected and trimmed at both ends. Marrow contents were flushed with HBSS (2 ml) using 1-ml insulin syringe with a 29G × ½ needle. The contents were collected into a sterile 50-ml centrifuge tube and washed twice. Bone marrow cells were seeded at a density of 2×106 cells per 90 mm petri dish and cultured with culture medium (RPMI-1640 +10% FBS+20 ng ml−1 rmGM-CSF +20 mM penicillin/streptomycin). Bone marrow cells were incubated at 37 °C, 5% CO2, and 95% humidity in CO2 incubator. A fresh culture medium (10 ml) was added into each petri dish at day 3. Primary BMDCs in each petri dish were harvested by collecting non-adherent cells by gently pipetting them with culture medium. BMDCs were washed twice with HBSS and collected for the future ex vivo studies of isolated DCs (61).
2.7.2. Ex vivo studies of DCs activation
For ex vivo study of DMV uptake by DCs, bone marrow-derived DCs were seeded on a cover slip at a concentration of 1.5 × 105 in a 12-well plate. Labeled vesicles (2 mg ml−1) were added into each well and incubated at 37°C for 25 min under continuous agitation. Cells were washed twice with HBSS and fixed with 4% PFA for 10 min on ice. Cells were then incubated with mouse anti-CD11c Abs (Dendritic cells marker, Biolegend). After incubated with anti-CD11c Abs, cells were washed twice with HBSS and were mounted on a slide with a mounting reagent containing DAPI (Life technologies Grand Island, NY). Cells were imaged using a confocal microscope (Nikon A1R+ laser scanning confocal microscope).
To test activation and maturation of dendritic cells, bone marrow-derived dendritic cells were cultured as described and plated at 50% confluency in a tissue culture-treated 6-well plate. For the study of DCs cytokine release, various doses (0, 10, 100 and 1000 ng ml−1) of DMVs were added into DCs at a density of 104 cells /well in 96-well plate and co-cultured for 48h. The supernatant was collected for ELISA assay of IL-12p70, IL-1β and IL-6. For maturation studies of DCs, DMVs (0, 5 and 10 μg ml−1) were incubated with dendritic cells for 48 h before the cells were collected by scraping and washed 3 times using HBSS with bovine serum albumin (2%, BSA, Sigma Aldrich). The cells were then immunostained with Alexa Fluor 488 anti-mouse CD11c Abs (N418, Biolegend), Alexa Fluor 647 anti-mouse CD40 Abs (HM40-3, Biolegend), PE anti-mouse CD86 Abs (GL-1, Biolegend) and PE/Cy7 anti-mouse CD80 Abs (16-10A1, Biolegend) for 20 min at room temperature in the dark followed by washing with BSA HBSS (3 ml, 0.1%) under centrifugation for three times. Cells were then resuspended in BSA HBSS (400 μl, 0.1%) and analyzed by Accuri C6 flow cytometer (BD Biosciences, San Jose, CA).
2.8. P. aeruginosa-derived DMVs evokes adaptive immunity
2.8.1. Immunization procedure
In the immunization experiments, mice were i.p. administered with DMVs every week for three times. Vaccines were prepared with DMVs dissolved in HBSS and an equal volume of either Freund’s complete adjuvant(62,63) (1st) or Freund’s incomplete adjuvant (62,63) (2nd, 3rd). The control group was HBSS with the same volume of Freund’s adjuvants.
2.8.2. DMVs -protein specific B cell and T cell responses
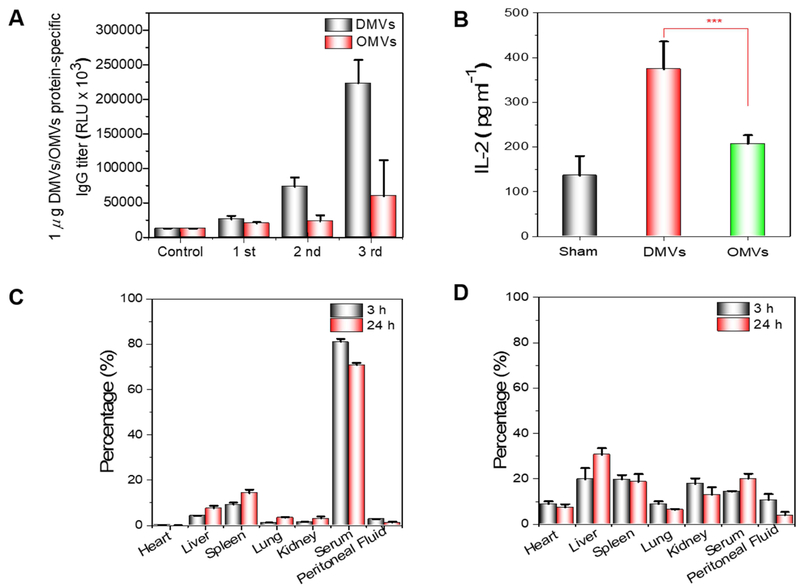
For evaluating the DMVs -protein specific IgG, mice were immunized with P. aeruginosa derived -DMVs (0 μg, 1 μg and 5 μg/mouse) as described in Section 2.8.1 and sera were harvested via facial vein 7 days after each immunization from DMVs and sham-immunized mice. The 96-well plates were coated with purified DMVs at the concentration of 100 ng per well in HBSS, and then were blocked with BSA/HBSS (1%) and were loaded with serum samples (1:5000 dilutions in 1% BSA/HBSS). DMVs-protein specific IgG were detected by secondary peroxidase-conjugated anti-mouse IgG Abs followed by adding a chemiluminescent substrate. The antibody IgG titer was measured by a plate reader at the greatest dilution (1:5000).
To study the stimulation of T cell response, mice (n=4 per group) were immunized via i.p. injection with DMVs (1 μg) as described in Section 2.8.1. On day 21, mice were sacrificed, and their spleens were collected to make splenocyte single cell suspensions. Briefly, the collected spleens were minced and passed through a 40 μm cell strainer and red blood cells were removed by RBC lysis Solution (5 PRIME). The collected cells were washed and resuspended with HBSS. The cells in each suspension were then counted and seeded onto a 12-well plate at a density of 2 × 105 cells per well in RPMI1640 containing fetal bovine serum (10%), β-mercaptoenthanol (50 μM) and antibiotics. In each well, heat-killed P. aeruginosa (1 × 107CFU) of were added and the cells were incubated with heat-killed P. aeruginosa for 72 h at 37 °C and 5% CO2. Following the incubation, the supernatants from each well were collected and IL-2 was measured by ELISA MAX™ Deluxe assay kits (BIOLEGEND).
2.9. P. aeruginosa-derived DMVs immunization and bacterium-induced sepsis
In the bacteria induced sepsis model, mice were immunized with DMVs or OMVs (1 μg) following the immunization protocol as described in Section 2.8.1. Sera were collected via facial vein for IgG titer measurements and mice were alive for the further challenge of P. aeruginosa (1×1010CFU). Sera were collected from DMVs or OMVs-immunized mice and sham-mice at 6 h after the bacteria challenge. In the survival study, P. aeruginosa-dedved DMVs or OMVs (1 μg) were i.p. administrated in CD1 mice following the immunization routine as described in Section 2.8.1. 7 days after the final immunization, the mice were challenged with a lethal dose (1×1010 CFU) of live P. aeruginosa. The mice were monitored every 12 h for 5 days.
2.10. DMVs or OMVs-protein specific B cell and T cell responses
For evaluating the DMVs or OMVs -protein specific IgG, mice were immunized with P.aeruginosa derived -DMVs or OMVs (1 μg) as described in Section 2.8.1 and sera were collected via facial vein 7 days after each immunization from DMVs or OMVs and sham-immunized mice. DMVs or OMVs -protein specific IgG were measured via the method described in section 2.7.2. The antibody IgG titer was determined at the greatest dilution (1:5000).
To study the stimulation of DMVs or OMVs - specific T cell responses, mice (n=4 per group) were immunized via i.p. injection with DMVs or OMVs (1 μg) as described in Section 2.8.1. On day 21, mice were sacrificed, and their spleens were collected to make splenocyte single cell suspensions. DMVs or OMVs - specific T cell responses were measured via the method described in section 2.7.2.
2.11. In vivo bio-distribution of DMVs and OMVs
DiD-labeled DMVs or OMVs (15 μg) were intraperitoneally injected into CD1 mice, and tissues (Heart, liver, spleen, lung and kidney) were dissected at 3 h and 24 h. Tissues (100 mg) were weighted and homogenized in RIPA buffer (1 ml, Sigma-Aldrich, R0278-50ML) and centrifuged at 20,000 g for 10 min. The supernatant was collected and diluted for measurement. Sera and peritoneal fluid were collected and diluted for the further measurement as well. DiD standard curve was performed with a series of dilutions. DiD signals were measured at wavelength of 640 nm(Ex)/670 nm(Em).
2.12. Measurement of cytokines
Levels of cytokines in serum, cell-culture supernatants were measured by ELISA according to the manufacturer’s instructions (Biolegend Inc.): IL-6, TNF-α and IL-1β in serum; IL-6, IL-1β, IL-12p70 in supernatants of DCs, and IL-2 in supernatants of isolated splenocyte single cell suspensions.
2.13. Statistical Analysis
Data are expressed as mean ± SD. Statistical analysis was conducted using one or two-way T-test using Origin 8.5. p values< 0.05 are considered significant (*). p values< 0.001 are considered extremely significant (***).
3. Results
3.1. Double-layered Structure of Nanovaccines:
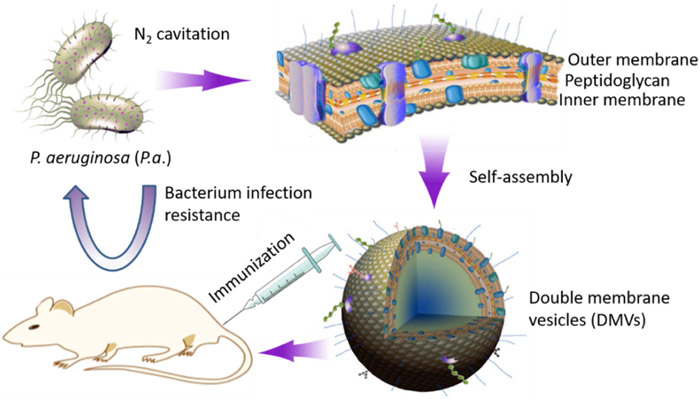
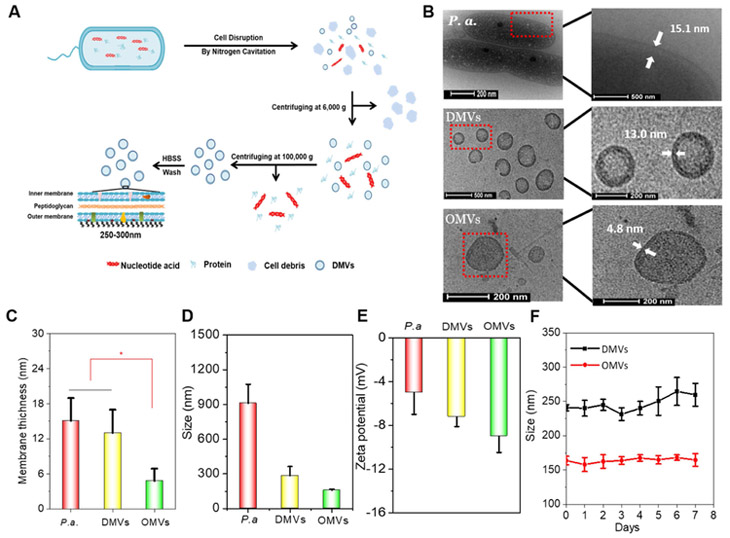
Scheme 1 shows our hypothesis that double-layered membrane vesicles (DMVs) directly derived from pathogens (such as bacteria) are made using a facile approach of nitrogen cavitation, and DMVs act as a vaccine that prevents bacterial infections. Here, we proposed a physical method to generate bacterial membrane vesicles using nitrogen cavitation. Nitrogen cavitation can generate membrane-formed nanovesicles comprised of a whole cell membrane since nitrogen cavitation is a mechanic force formed in a chamber to rapidly disrupt cell membrane, and subsequently intracellular contents are released and cellular membrane automatically forms a nanosized compartment (64,65). Fig.1A shows a detailed process of generation of bacterial membrane nanovesicles. Bacteria (here as P. aeruginosa) were collected and disrupted via nitrogen cavitation at a pressure of 1500 psi, and then we performed a series of differential centrifugations to isolate membrane-formed nanovesicles. We analyzed proteins using BCA assay after each step of centrifugation (Fig. S1). The pellet after the centrifugation at 6,000 g showed 10% proteins of the cell lysate. Subsequently, the resulting supernatant was centrifuged at 100,000 g, and it contained 98.53% ± 10.03%of proteins of the cell lysate. The proteins in the pellet were quantified, indicating 1.5% of proteins existed in the pellet which is supposedly associated with membrane vesicles. Furthermore, we analyzed nucleic acids in the pellet and found that it contained a small amount of DNA (Fig. S1). Together, the combination of nitrogen cavitation and differential centrifugations enables to remove most intracellular contents of their parent cells.
Scheme 1.
Schematic illustrates development of a new vaccine made of DMVs from bacterial membrane
Figure 1. Generation and characterization of double-layered membrane vesicles (DMVs) from P. aeruginosa.
A) Schematic illustrates a procedure to generate DMVs, including cell disruption by nitrogen cavitation and differential centrifugations. B) Cryo-TEM (transmission electron microscopy) images of P. aeruginosa bacteria, DMVs and OMVs using TF20 electron microscope. C) Membrane thickness of P. aeruginosa, DMVs and OMVs measured via cryo-TEM images. D) Hydrodynamic size and E) zeta-potential of P. aeruginosa, DMVs and OMVs using dynamic light scattering. F) Sizes of DMVs and OMVs in HBSS measured by dynamic light scattering in 7 days.
We further analyzed the structure and size of the pellet using cryo-TEM (Fig. 1B). P. aeruginosa-derived OMVs were used as control. Cryo-TEM indicated that the pellet contained many vesicles with a spherical structure and their wall thickness was 13 nm, so-called DMVs. P. aeruginosa was also imaged, showing that the average membrane thickness of P. aeruginosa was observed around 15.1 nm, which was close to that of DMVs. In contrast, the average thickness of OMVs was 4.8 nm (Fig. 1C). Together, the data indicated that the thickness of DMVs was similar to that of their source of P. aeruginosa and was thicker than that of OMVs by almost 2 times. Because a gram-negative bacterium possesses double-layered membrane structure (inner and out membrane), the analysis on the wall size of DMVs suggests that DMVs are directly formed from a whole bacterial membrane (Fig. 1B). Furthermore, the average size and zeta-potential were measured using dynamic light scattering (Fig. 1D and Fig. 1E). The average size of DMVs was about 250 nm in diameter that was much smaller than that of the parent cells. Moreover, OMVs (−10 mV), DMVs (−9 mV) and P. aeruginosa (−5 mV) were negatively charged (Fig. 1E), implying that OMVs and DMVs demonstrated the similar surface properties to their source of P. aeruginosa. The results indicated that DMVs were derived from the membrane of their source cells. We also investigated the stability of DMVs and OMVs over time using dynamic light scattering. The result showed that there was not a significant change in size in 7 days at 4°C, suggesting that OMVs and DMVs were stable (Fig. 1F).
3.2. Proteomics of DMVs:
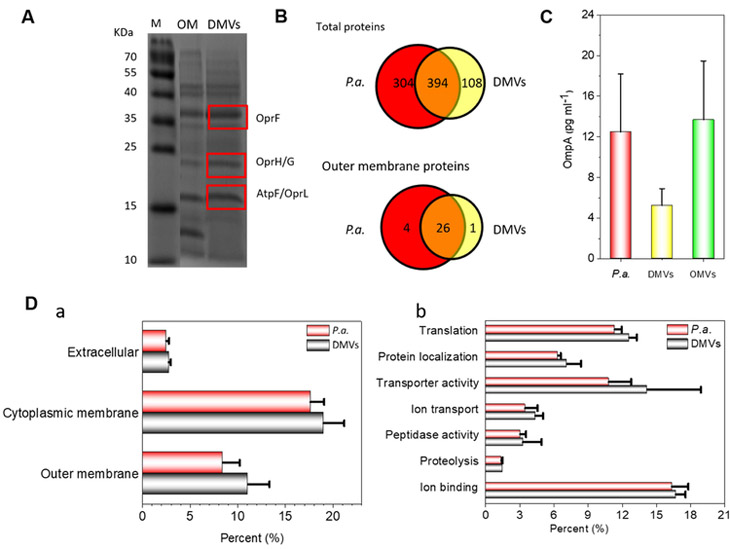
Fig. 1 shows that DMVs possess the whole membrane compartment of P. aeruginosa, so we asked whether DMVs contain antigens located in both inner and outer membranes, and whether these antigens are related to the immunization potential of DMVs. First, we performed SDS-PAGE to analyze proteins in the outer membrane of P. aeruginosa and DMVs (Fig. 2A). Our result showed that DMVs had a similar pattern to that of outer membrane proteins (49,51,66,67). Using LC-MS/MS analyses of in-gel digested tryptic peptides of three major vesicular protein bands, we identified that OprF, OprH/G and AtpF/OprL were the most abundant proteins in DMVs. Interestingly, these proteins were present in both P. aerugniosa and P. aeruginosa-derived OMVs (49,51,66,67). To further determine the protein composition of P. aeruginosa-derived DMVs, we performed an in-solution digestion and LC-MS/MS of tryptic peptides. Using the P. aeruginosa protein database from Uniprot (release 15.12), we identified 698 proteins in P. aeruginosa and 502 proteins in P. aeruginosa-derived DMVs, finding that 394 proteins were detectable and identical in both DMVs and P. aeruginosa. Further analysis of proteins expressed on cell membrane showed that 26 out of 27 outer membrane proteins in DMVs were also shown in P. aeruginosa (Fig. 2B), and nearly 87% of membrane proteins in P. aeruginosa were in DMVs. Collectively, our results suggest that DMVs contain most of membrane proteins of P. aeruginosa, implying that DMVs are made from the whole bacterial membrane, consistent with the double-layer structure of DMVs as shown in Fig. 1B and Fig. 1C.
Figure 2. Proteomics of P. aeruginosa DMVs.
A) Coomassie Brilliant Blue-stained SDS-PAGE of P. aeruginosa outer membrane proteins and DMVs. Rectangles indicate three major bands associated with membrane proteins. OprF, OprH/G and atpF/OprL were identified by LC-MS/MS analyses. M: Molecular weight marker; OM: Outer membrane protein. B) Venn diagrams show total proteins and outer membrane proteins identified by LC-MS/MS in P. aeruginosa and P. aeruginosa-derived DMVs. C) OmpA expression in OMVs, DMVs and P. aeruginosa. D) Proteomics and functional categorization of DMVs and P. aeruginosa. Subcellular localizations (a); Specific proteins in DMVs and P. aeruginosa (b).
Furthermore, we performed an ELISA assay to detect the expression of major antigen OmpA (outer membrane protein A). Compared to P. aeruginosa and P. aeruginosa-derived OMVs, OmpA protein was also detected on P. aeruginosa-derived DMVs (Fig. 2C). To determine the origins and locations of proteins identified in Fig. 2B, we categorized proteins in P. aeruginosa and P. aeruginosa-derived DMVs using the Pseudomonas Genome Database (60) and quantified the percentage of each type protein in identified proteins (Fig. 2D). There were the similar composition and content of proteins in both DMVs and their source cells, such as extracellular proteins, cytoplasmic membrane proteins, and outer membrane proteins (Fig. 2D). This result clearly indicates that P. aeruginosa-derived DMVs are double-layered membrane nanovesicles that are directly derived from the membrane of their source. This is consistent with the wall thickness of DMVs imaged by cryo-TEM (Fig. 1B). Furthermore, Gene Ontology (GO) (68,69) was used to analyze several biological functions of proteins such as, transporter activity, ion-binding, protein localization, and peptidase activity and proteolysis in 502 proteins in P. aeruginosa-derived DMVs (Fig. 2D). The percentages of the proteins in DMVs were similar with those in P. aeruginosa, implying that DMVs are derived from the membrane of their source cells. As the immunogenic characteristics of P. aeruginosa mainly depend on the outer membrane proteins and extracellular proteins (70), our proteomics analysis demonstrated that DMVs could be a vaccine candidate.
3.3. Activation of B cells and T cells after immunization of DMVs:
The host defense strongly relies on the activation of both Ag-nonspecific innate immunity and Ag-specific immunity. Dendritic cells (DCs), professional APCs (antigen presentation cells), evoke the Ag-specific adaptive immunity via both the innate and adaptive immune responses (71). DCs encounter a pathogen and activate B cells and T cells to produce antibodies and cytokines, thus leading to a full adaptive immune response. To evaluate DMVs as a potential vaccine candidate, we examined how both immune responses were activated and how DCs recognized and responded to DMVs in eliciting protective effects on acute systemic infections. To determine the innate immunity activated by DMVs in vivo, we examined the functions of DMVs on the acute systemic inflammation. DMVs (0.8 mg/kg) were injected into CD1 mice intraperitoneally and sera were collected at 12 h, 24 h post injections. LPS (10 mg kg−1) injection was conducted as a positive control and mice were injected with a same volume of HBSS as a negative control. Our results (Fig. S2) suggested that serum levels of IL-6, IL-1β and TNF-α were increased after injection of DMVs, indicating that DMVs can evoke the innate immune response in vivo and caused earlier systemic inflammation. Therefore, our findings demonstrate that DMVs were immunogenic, thus providing a basis for utilizing DMVs as a potential vaccine candidate against bacterial infections.
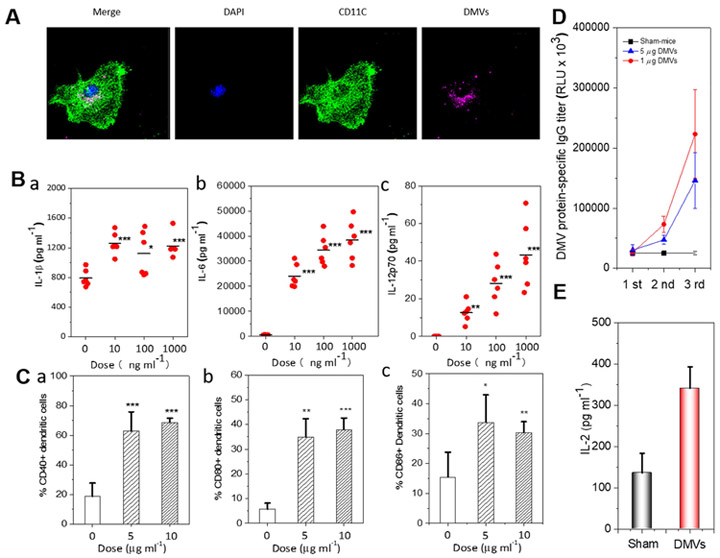
Next, we examined uptake of P. aeruginosa-derived DMVs by DCs. When bone marrow-derived CD11c+ DCs were co-cultured with DiD-labeled DMVs in 37 °Cfor 30 min, DiD-labeled DMVs were detected in the cytoplasm of CD11c+ DCs via a confocal microscope, indicating that DMVs were internalized by CD11c+ DCs (Fig. 3A). To determine the efficiency of P. aeruginosa-derived DMVs in eliciting activation and maturation of CD11c+ DCs, CD11c+ DCs were treated with various doses of DMVs and several cytokines were measured, including IL-12p70, IL-6 (Th-1 and Th-17-polarizing cytokines) and IL-1β (Th-17 cytokines). Our result showed that DMVs can induce the cytokines and their production was dose-dependent (Fig. 3B). When CD11c+ DCs were treated with DMVs (0 μg ml−1, 5 μg ml−1 and 10 μg ml−1 in total protein) for 48 h, we found a shift of CD11c+ DCs from immature to mature phenotype because of upregulation of co-stimulatory molecules, for example, CD40, CD80 and CD86 markers (Fig. S3). Quantification of flow cytometry showed that CD11c+ DCs matured after the stimulation by DMVs in two doses (Fig. 3C). The results are consistent with the previous studies that OMVs can activate DCs in vitro (48). Our studies suggest that immunization with DMVs can effectively promote the maturation and activation of CD11c+ DCs.
Figure 3. P. aeruginosa DMVs evoke the innate and adaptive immune responses.
A) DiD labeled DMVs uptake by DCs studied by confocal fluorescence microscopy. B) Measurement of Th1 (IL-12p70) and Th17 (IL-6, IL-1β) cytokines associated with activation of DCs by ELISA (n=6). C) Maturation of CD11c+DCs analyzed by flow cytometry after DCs were treated with different concentrations of DMVs. D) B cell response to DMVs in vivo. Serum IgG specific to DMVs measured by ELISA (n=4). E) T cell response to DMVs(1μg) in vivo. The production of IL-2 from splenocyte T cells was measured by ELISA after they were challenged with dead P. aeruginosa for 72 h (n=5). *p <0.05, **p<0.01, ***p<0.001
To address whether P. aeruginosa-derived DMVs activate the adaptive immunity, we assessed the effect of P. aeruginosa-derived DMVs immunization on induction of Ag-specific B and T cell responses. We measured the antibody production using ELISA assay (Fig. 3D) after we immunized mice once a week for three times. P. aeruginosa-derived DMVs-protein specific IgG proteins increased with the immunization frequency, however the control of the sham-immunized mice did not produce specific IgG. When increasing the dose of DMVs to 5 μg, we did not observe the increase of IgG production, suggesting that antibody production was not dose-dependent. Therefore, the dose of 1 μg DMVs was used for the survival studies in bacterial infection. Our results show that P. aeruginosa-derived DMVs immunization can evoke B cells to produce DMVs-protein specific IgG antibodies to establish the adaptive immunity.
Next, we evaluated whether T cells respond to DMVs. Splenic cells were isolated from immunized and sham-mice after the last immunization and stimulated with heat-killed P. aerugniosa (1 × 107 CFU) for 72 h. T cell activation was quantified by measuring interleukin 2 (IL-2) in culture medium. The results showed that the production of IL-2 was dramatically increased in the mice immunized with P. aeruginosa-derived DMVs compared to in sham-mice (Fig. 3E). The production of IL-2 is correlated to the activation of T cells, promoting the proliferation and differentiation of naive T cells (72). The result indicated that P. aeruginosa-derived DMVs can effectively activate T cells as compared with the control group. Collectively, the studies (Fig. 3D-E) demonstrate that DMVs can promote the adaptive immunity that produces antibodies from B cells and the cytokine from T cells against P. aeruginosa antigens.
3.4. Prevention of P.aeruginosa-Induced Sepsis by DMVs:
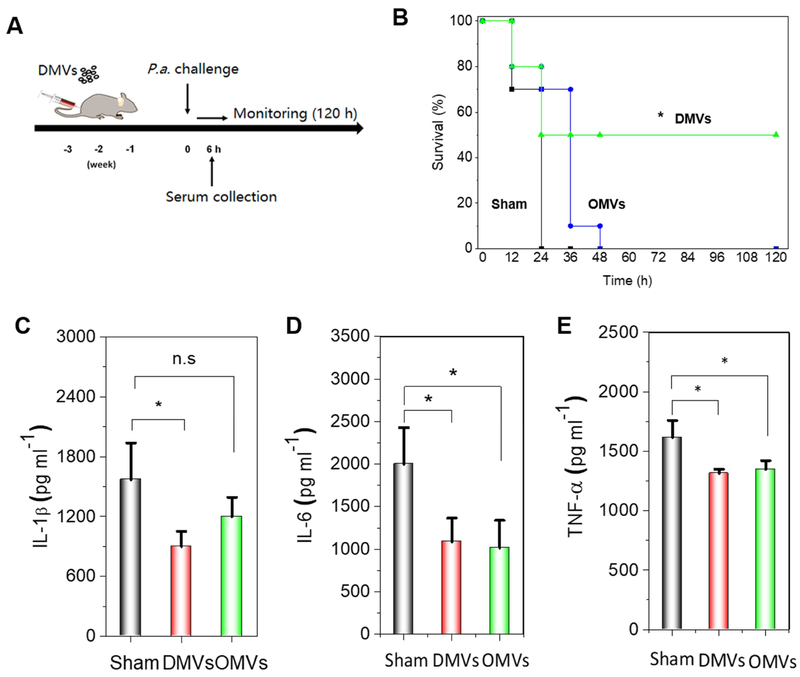
To address the usefulness of P. aeruginosa-derived DMVs as a vaccine, we examined whether they can improve the mouse survival in P. aeruginosa-induced sepsis. CD1 mice were first immunized with P. aeruginosa-derived DMVs and OMVs, respectively (Fig. 4A). Briefly, mice were intraperitoneally administered with P. aeruginosa-derived DMVs or OMVs (1μg in total protein) once a week for three times. 7 days after the final immunization, the mice were challenged with a lethal dose (1×1010 CFU) of live P. aeruginosa. The mice were monitored every 12 h for 5 days (Fig. 4B). It was observed that sham-mice were dead 24 h after bacterial challenge, and P. aeruginosa-derived OMVs immunization extended the mouse life to 48 h. However, P. aeruginosa-derived DMVs allowed 50% mice to live. Collectively, our study suggests that DMVs-based vaccination demonstrates the increased protection of bacterial infections compared to OMVs-based vaccination.
Bacterium-induced sepsis is strongly correlated to the systemic inflammation (73) and upregulation of cytokines would cause the acute cytokine storm, resulting in the tissue damage and organ failure. We measured pro-inflammatory cytokines, such as IL-6, IL-1β and TNF-α in serum 6 h post infection induced by P. aeruginosa. We found that both DMVs or OMVs immunized mice showed the decreased pro-inflammatory cytokines (IL-1 β, TNF- α and IL-6) compared to sham mice (Fig. 4C), suggesting that the reduction of cytokines might contribute the early protection of survival of immunized
However, immunization with DMVs showed the higher protection in sepsis in a long period, but OMVs did not prevent the death from bacterial infection. We asked whether the sepsis protection after immunization might be related to the adaptive immunity. After DMVs or OMVs were administered into CD1 mice at a dose of 1 μg (protein), specific IgG were measured (Fig. 5A). It was observed that both DMVs and OMVs produced specific IgG and the production was dependent on immunization frequency, however, the immune response of DMVs was stronger than that of OMVs. Furthermore, activation of T cells was analyzed after mice were challenged with DMVs or OMVs (Fig. 5B). DMVs increased activation of T cells compared to OMVs (Fig. 5B). Therefore, the long-term protection from sepsis induced by bacteria is related to the strong adaptive response after the immunization of DMVs, thus increasing mouse survival.
Figure 5. Efficacy of DMVs is associated with enhanced adaptive immunity and their trafficking.
A) IgG production in serum after mice were challenged with DMVs or OMVs at 1 μg (protein). B) T cell response after mice were immunized with DMVs or OMVs at1 μ g (protein). IL-2 produced by splenocyte T cells after T cells were challenged with dead P. aeruginosa (n=5) for 72 h. ***p<0.001. In vivo bio-distribution of DiD-labeled DMVs (C) and OMVs (D) in mice after i.p. injections.
The traffic of DMVs might be another factor that contributes the enhanced adaptive immunity. To address this question, we investigated the kinetics of bio-distribution of DMVs and OMVs. DiD-labeled P. aeruginosa-derived DMVs were intraperitoneally administered into CD1 mice and their bio-distribution was analyzed quantitatively in several organs at 3 h and 24 h post injection. To address whether intraperitoneal injection of DMVs disseminated in the blood circulation, we measured their concentrations in peritoneal fluid and serum (Fig. 5C). We observed the high concentrations of DMVs in serum 3h after i.p. injection and we also found most DMVs were detected in spleen and liver. In parallel, we investigated the bio-distribution of OMVs (Fig. 5D), showing a distinct distribution pattern of OMVs compared to DMVs. The results showed OMVs mainly accumulated in liver rather than in spleen in both 3 h and 24 h, consistent with the previous report (74). However, we observed that DMVs in blood were 3-4 folds higher than OMVs after 24h, indicating that DMVs may accumulate in spleen in a long period to increase immunization compared to OMVs. The different bio-distribution of DMVs might be associated with their unique double-layered Membrane structure.
4. Discussion:
Bacterial infections can cause severe diseases, but therapies are limited because of drug-resistance and aging-related deficiency of immunity (75,76). Vaccination is an optional therapy to protect invasion of pathogens, but it is still challenging to develop effective vaccines because bacteria have a wide range of antigens and mutate rapidly. Bacterial membrane antigens play a central role in regulating the innate and adaptive immune systems to respond to invasion of pathogens and to eliminate them. Facile and novel approaches are needed for vaccine developments.
Here we report a strategy to generate membrane-formed nanovesicles derived from P. aeruginosa (gram-negative) using nitrogen cavitation. Nitrogen cavitation is the mechanic force formed in a chamber to quickly break cells, and afterwards intracellular contents (proteins and genetic molecules) are released and the broken cell membranes form nanovesicles. Cryo-TEM allows us to visualize the intact structure of nanovesicles in suspension. We observed that nanovesicles possess a whole membrane of P. aeruginosa with a thickness of 13 nm comprising the inner and outer membrane (Fig. 1B and Fig. C). In contrast, OMVs showed the wall size with 4.8 nm, consistent with a layer of outer membrane (Fig. 1B and Fig. 1C). The result clearly indicates that double-layered membrane vesicles (DMVs) are formed from the whole bacterial membrane. Furthermore, the analysis on the surface charge of DMVs (Fig. 1D) and their proteomics (Fig. 2) indicate that DMVs possess the integrity of bacterial membrane. The unique structure of DMVs may be a good candidate for vaccination.
Vaccination is strongly associated with activation of adaptive immunity. Dendritic cells (DCs), professional APCs, can promote the Ag-specific adaptive immunity via both innate and adaptive immune responses. We studied the activation and maturation of DCs after DCs were challenged with DMVs (Fig. 3A-C). Activation of B cells and T cells is required to develop the long-term immunity. We observed that B cells can produce specific IgG antibodies to DMVs and T cells released IL-2 required for T cell activation (Fig. 3D-E). Together, the results indicate that DMVs can possibly activate innate and adaptive immune systems to enhance the protection when P. aeruginosa infection occurs. Indeed, we have demonstrated 50% mice survived in P. aeruginosa-induced sepsis after immunization with DMVs compared to OMVs (Fig. 4B).
To address the mechanism in which DMVs increased the survival in mouse sepsis, we investigated the adaptive immune responses (Fig. 5A-B). We observed increased activation of B cells and T cells after mice were immunized with DMVs compared to OMVs. Furthermore, DMVs accumulated in the spleen (a major tissue for the adaptive immunity) (Fig. 5C-D), so it might be a secondary mechanism for more benefit of DMVs. The unique bio-distribution of DMVs might be associated with the double-layered structure of DMVs.
OMVs are comprised of outer membrane of bacteria and contain antigens required for vaccination. OMVs are spontaneously secreted in culture media, and OMVs show heterogeneous sizes and compositions batch by batch. Most importantly, the production of OMVs for translation remains difficult because of lacking scalability.
Here we used nitrogen cavitation to generate bacterial membrane-formed nanovaccines as a vaccine. Compared to generation of OMVs, nitrogen cavitation can quickly generate nanovaccines if pathogens (bacteria) are identified. Although we produced P. aeruginosa-derived nanovaccines with 15-20 ml cell suspension using nitrogen cavitation, the cavitation chamber can be easily scaled up to tens of liters for clinical use without losing reproducibility since the fast and uniform forces are applied to individual cells (77) . Unlike other cell lysis methods, there is no heat or chemical damage to membrane proteins during the nitrogen cavitation procedure. Moreover, nitrogen gas can protect DMVs from oxidation when nitrogen cavitation is performed. Therefore, nitrogen cavitation could be a promising approach to quickly generate bacterial membrane-based vaccines. Further studies should be performed to examine whether our nitrogen cavitation approach is applicable to other bacteria, particularly for fast mutated pathogens.
5. Conclusions:
In summary, we report a simple and novel method to generate membrane nanovesicle-based vaccines made from their source of bacteria via nitrogen cavitation. Quantitative studies using cryo-TEM, biochemistry and proteomics demonstrate the unique double-layered membrane-formed nanovesicles that contain a wide range of antigens expressed on bacterial membrane. The immunization studies in the sepsis model induced by P. aeruginosa show that DMVs can effectively promote the innate and adaptive immunity required for the defense of bacterial infection, thus improving mouse survival. Our top-down approach to design vaccines would shift to a new paradigm: identifying pathogens, generating bacterium-membrane-formed nanovesicles, and utilizing these nanovesicles as a vaccine to prevent infections caused by their source of pathogens. This simple and innovative approach would be a promising technology for vaccine developments.
Supplementary Material
Figure 4. P. aeruginosa-derived DMVs enhance mouse survival in P. aeruginosa-induced sepsis.
A) Experimental protocol for DMVs immunization in P. aeruginosa infection. B) Survival rates of DMVs-, OMVs-and sham-immunized mice after mice were challenged with a lethal dose of P. aeruginosa (1×1010 CFU) (n=10). The procedure was shown in Figure 4A, and the immunization of DMVs or OMVs was performed at 1 μg injection each time. * p<0.05 shows the comparison of DMVs to either OMVs-immunized mice or sham mice. No significance between OMVs and sham mice. C-E) Serum IL-1β, IL-6 and TNF-α 6 h after P. aeruginosa challenge (n=4). The procedure was shown in Figure 4A. *p <0.05, **p<0.01, ***p<0.001
Acknowledgments:
The work was supported by NIH grants RO116823 to Z. W., and Robbers Research Awards (17A-2950-9842) to S.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Dreyfuss D, and Ricard JD (2005) Acute lung injury and bacterial infection. Clin Chest Med 26, 105–112 [DOI] [PubMed] [Google Scholar]
- 2.Ware LB, and Matthay MA (2005) Clinical practice. Acute pulmonary edema. N Engl J Med 353, 2788–2796 [DOI] [PubMed] [Google Scholar]
- 3.Matthay MA, Ware LB, and Zimmerman GA (2012) The acute respiratory distress syndrome. J Clin Invest 122, 2731–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert PA (2002) Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med 95 Suppl 41, 22–26 [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher RC (2004) New concepts of the pathogenesis of cystic fibrosis lung disease. Eur Respir J 23, 146–158 [DOI] [PubMed] [Google Scholar]
- 6.Rodewohl A, Scholbach J, Leichsenring A, Koberle M, and Lange F (2017) Age-dependent cellular reactions of the human immune system of humanized NOD scid gamma mice on LPS stimulus. Innate Immun 23, 258–275 [DOI] [PubMed] [Google Scholar]
- 7.Arias CA, and Murray BE (2009) Antibiotic-resistant bugs in the 21st century--a clinical super-challenge. N Engl J Med 360, 439–443 [DOI] [PubMed] [Google Scholar]
- 8.Wright GD, and Sutherland AD (2007) New strategies for combating multidrug-resistant bacteria. Trends Mol Med 13, 260–267 [DOI] [PubMed] [Google Scholar]
- 9.Ahmed A, Li J, Shiloach Y, Robbins JB, and Szu SC (2006) Safety and immunogenicity of Escherichia coli O157 O-specific polysaccharide conjugate vaccine in 2-5-year-old children. J Infect Dis 193, 515–521 [DOI] [PubMed] [Google Scholar]
- 10.Simon R, Tennant SM, Wang JY, Schmidlein PJ, Lees A, Ernst RK, Pasetti MF, Galen JE, and Levine MM (2011) Salmonella enterica serovar enteritidis core O polysaccharide conjugated to H:g,m flagellin as a candidate vaccine for protection against invasive infection with S. enteritidis. Infect Immun 79, 4240–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowers Y, Kirschner J, Keller N, Barshack I, Bar-Meir S, Ashkenazi S, Schneerson R, Robbins J, and Passwell JH (2007) O-specific [corrected] polysaccharide conjugate vaccine-induced [corrected] antibodies prevent invasion of Shigella into Caco-2 cells and may be curative. Proc Natl Acad Sci U S A 104, 2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zariri A, and van der Ley P (2015) Biosynthetically engineered lipopolysaccharide as vaccine adjuvant. Expert Rev Vaccines 14, 861–876 [DOI] [PubMed] [Google Scholar]
- 13.Santos MF, New RR, Andrade GR, Ozaki CY, Sant'Anna OA, Mendonca-Previato L, Trabulsi LR, and Domingos MO (2010) Lipopolysaccharide as an antigen target for the formulation of a universal vaccine against Escherichia coli O111 strains. Clin Vaccine Immunol 17, 1772–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross AS, Opal S, Cook P, Drabick J, and Bhattacharjee A (2004) Development of an anti-core lipopolysaccharide vaccine for the prevention and treatment of sepsis. Vaccine 22, 812–817 [DOI] [PubMed] [Google Scholar]
- 15.Nagy G, and Pal T (2008) Lipopolysaccharide: a tool and target in enterobacterial vaccine development. Biol Chem 389, 513–520 [DOI] [PubMed] [Google Scholar]
- 16.Zhang W, and Sack DA (2015) Current Progress in Developing Subunit Vaccines against Enterotoxigenic Escherichia coli-Associated Diarrhea. Clin Vaccine Immunol 22, 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair M (2012) Protein conjugate polysaccharide vaccines: challenges in development and global implementation. Indian J Community Med 37, 79–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon RJ, and Lowy FD (2008) Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46 Suppl 5, S350–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cremer N, and Watson DW (1960) Host-parasite factors in group A streptococcal infections. A comparative study of streptococcal pyrogenic toxins and gram-negative bacterial endotoxin. J Exp Med 112, 1037–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin LC, Chattopadhyay S, Lin JC, and Hu CJ (2018) Advances and Opportunities in Nanoparticle- and Nanomaterial-Based Vaccines against Bacterial Infections. Adv Healthc Mater 7,e1701395. [DOI] [PubMed] [Google Scholar]
- 21.Farokhzad OC, and Langer R (2009) Impact of nanotechnology on drug delivery. ACS Nano 3, 16–20 [DOI] [PubMed] [Google Scholar]
- 22.Mitragotri S, Burke PA, and Langer R (2014) Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat Rev Drug Discov 13, 655–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Li J, Cho J, and Malik AB (2014) Prevention of vascular inflammation by nanoparticle targeting of adherent neutrophils. Nat Nanotechnol 9, 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, and Malik AB (2013) Nanoparticles squeezing across the blood-endothelial barrier via caveolae. Ther Deliv 4, 131–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Tiruppathi C, Cho J, Minshall RD, and Malik AB (2011) Delivery of nanoparticle: complexed drugs across the vascular endothelial barrier via caveolae. IUBMB Life 63, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu D, Gao J, and Wang Z (2015) Neutrophil-Mediated Delivery of Therapeutic Nanoparticles across Blood Vessel Barrier for Treatment of Inflammation and Infection. ACS Nano 9, 11800–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irvine DJ, Swartz MA, and Szeto GL (2013) Engineering synthetic vaccines using cues from natural immunity. Nat Mater 12, 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li WA, and Mooney DJ (2013) Materials based tumor immunotherapy vaccines. Curr Opin Immunol 25, 238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart JM, and Keselowsky BG (2017) Combinatorial drug delivery approaches for immunomodulation. Adv Drug Deliv Rev 114, 161–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo JW, Irvine DJ, Discher DE, and Mitragotri S (2011) Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat Rev Drug Discov 10, 521–535 [DOI] [PubMed] [Google Scholar]
- 31.Hu CM, Zhang L, Aryal S, Cheung C, Fang RH, and Zhang L (2011) Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc Natl Acad Sci U S A 108, 10980–10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu CM, Fang RH, Copp J, Luk BT, and Zhang L (2013) A biomimetic nanosponge that absorbs pore-forming toxins. Nat Nanotechnol 8, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu CM, Fang RH, Luk BT, and Zhang L (2013) Nanoparticle-detained toxins for safe and effective vaccination. Nat Nanotechnol 8, 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nochi T, Yuki Y, Takahashi H, Sawada S, Mejima M, Kohda T, Harada N, Kong IG, Sato A, Kataoka N, Tokuhara D, Kurokawa S, Takahashi Y, Tsukada H, Kozaki S, Akiyoshi K, and Kiyono H (2010) Nanogel antigenic protein-delivery system for adjuvant-free intranasal vaccines. Nat Mater 9, 572–578 [DOI] [PubMed] [Google Scholar]
- 35.Gahan ME, Webster DE, Wesselingh SL, Strugnell RA, and Yang J (2009) Bacterial Antigen Expression Is an Important Component in Inducing an Immune Response to Orally Administered Salmonella-Delivered DNA Vaccines. Plos One 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strugnell R, Dougan G, Chatfield S, Charles I, Fairweather N, Tite J, Li JL, Beesley J, and Roberts M (1992) Characterization of a Salmonella-Typhimurium-Aro Vaccine Strain Expressing the P.69 Antigen of Bordetella-Pertussis. Infection and Immunity 60, 3994–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W, Wang S, Yao Y, Xia Y, Yang X, Li K, Sun P, Liu C, Sun W, Bai H, Chu X, Li Y, and Ma Y (2016) Employing Escherichia coli-derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against Acinetobacter baumannii infection. Sci Rep 6, 37242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Priebe GP, Walsh RL, Cederroth TA, Kamei A, Coutinho-Sledge YS, Goldberg JB, and Pier GB (2008) IL-17 is a critical component of vaccine-induced protection against lung infection by lipopolysaccharide-heterologous strains of Pseudomonas aeruginosa. J Immunol 181, 4965–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doring G, and Pier GB (2008) Vaccines and immunotherapy against Pseudomonas aeruginosa. Vaccine 26, 1011–1024 [DOI] [PubMed] [Google Scholar]
- 40.Roberts R, Moreno G, Bottero D, Gaillard ME, Fingermann M, Graieb A, Rumbo M, and Hozbor D (2008) Outer membrane vesicles as acellular vaccine against pertussis. Vaccine 26, 4639–4646 [DOI] [PubMed] [Google Scholar]
- 41.Sierra GV, Campa HC, Varcacel NM, Garcia IL, Izquierdo PL, Sotolongo PF, Casanueva GV, Rico CO, Rodriguez CR, and Terry MH (1991) Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. NIPH Ann 14, 195–207; discussion 208-110 [PubMed] [Google Scholar]
- 42.Holst J, Oster P, Arnold R, Tatley MV, Naess LM, Aaberge IS, Galloway Y, McNicholas A, O'Hallahan J, Rosenqvist E, and Black S (2013) Vaccines against meningococcal serogroup B disease containing outer membrane vesicles (OMV): lessons from past programs and implications for the future. Hum Vaccin Immunother 9, 1241–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsay L, Dold C, Green CA, Rollier CS, Norheim G, Sadarangani M, Shanyinde M, Brehony C, Thompson AJ, Sanders H, Chan H, Haworth K, Derrick JP, Feavers IM, Maiden MC, and Pollard AJ (2015) A novel meningococcal outer membrane vesicle vaccine with constitutive expression of FetA: A phase I clinical trial. J Infect 71, 326–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schild S, Nelson EJ, and Camilli A (2008) Immunization with Vibrio cholerae outer membrane vesicles induces protective immunity in mice. Infect Immun 76, 4554–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S, Gao J, and Wang Z (2018) Outer membrane vesicles for vaccination and targeted drug delivery. Wiley Interdiscip Rev Nanomed Nanobiotechnol, e1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bos MP, Robert V, and Tommassen J (2007) Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 47.Choi DS, Kim DK, Kim YK, and Gho YS (2015) Proteomics of extracellular vesicles: Exosomes and ectosomes. Mass spectrometry reviews 34, 474–490 [DOI] [PubMed] [Google Scholar]
- 48.Kim OY, Hong BS, Park KS, Yoon YJ, Choi SJ, Lee WH, Roh TY, Lotvall J, Kim YK, and Gho YS (2013) Immunization with Escherichia coli outer membrane vesicles protects bacteria-induced lethality via Th1 and Th17 cell responses. J Immunol 190, 4092–4102 [DOI] [PubMed] [Google Scholar]
- 49.Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, and Gho YS (2011) Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 11, 3424–3429 [DOI] [PubMed] [Google Scholar]
- 50.Gao W, Fang RH, Thamphiwatana S, Luk BT, Li J, Angsantikul P, Zhang Q, Hu CM, and Zhang L (2015) Modulating antibacterial immunity via bacterial membrane-coated nanoparticles. Nano Lett 15, 1403–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bauman SJ, and Kuehn MJ (2006) Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect 8, 2400–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyczak JB, Cannon CL, and Pier GB (2000) Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 53.Driscoll JA, Brody SL, and Kollef MH (2007) The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368 [DOI] [PubMed] [Google Scholar]
- 54.Priebe GP, and Goldberg JB (2014) Vaccines for Pseudomonas aeruginosa: a long and winding road. Expert Rev Vaccines 13, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pier GB (2003) Promises and pitfalls of Pseudomonas aeruginosa lipopolysaccharide as a vaccine antigen. Carbohydr Res 338, 2549–2556 [DOI] [PubMed] [Google Scholar]
- 56.Haghbin M, Armstrong D, and Murphy ML (1973) Controlled prospective trial of Pseudomonas aeruginosa vaccine in children with acute leukemia. Cancer 32, 761–766 [DOI] [PubMed] [Google Scholar]
- 57.Pennington JE, Reynolds HY, Wood RE, Robinson RA, and Levine AS (1975) Use of a Pseudomonas Aeruginosa vaccine in pateints with acute leukemia and cystic fibrosis. Am J Med 58, 629–636 [DOI] [PubMed] [Google Scholar]
- 58.Horstman AL, and Kuehn MJ (2000) Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem 275, 12489–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Couto N, Schooling SR, Dutcher JR, and Barber J (2015) Proteome Profiles of Outer Membrane Vesicles and Extracellular Matrix of Pseudomonas aeruginosa Biofilms. J Proteome Res 14, 4207–4222 [DOI] [PubMed] [Google Scholar]
- 60.Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock RE, and Brinkman FS (2009) Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res 37, D483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, and Schuler G (1999) An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods 223, 77–92 [DOI] [PubMed] [Google Scholar]
- 62.Dalseg R, Wedege E, Holst J, Haugen IL, Hoiby EA, and Haneberg B (1999) Outer membrane vesicles from group B meningococci are strongly immunogenic when given intranasally to mice. Vaccine 17, 2336–2345 [DOI] [PubMed] [Google Scholar]
- 63.Oliver KJ, Reddin KM, Bracegirdle P, Hudson MJ, Borrow R, Feavers IM, Robinson A, Cartwright K, and Gorringe AR (2002) Neisseria lactamica protects against experimental meningococcal infection. Infection and Immunity 70, 3621–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao J, Chu D, and Wang Z (2016) Cell membrane-formed nanovesicles for disease-targeted delivery. J Control Release 224, 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao J, Wang S, and Wang Z (2017) High yield, scalable and remotely drug-loaded neutrophil-derived extracellular vesicles (EVs) for anti-inflammation therapy. Biomaterials 135, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Renelli M, Matias V, Lo RY, and Beveridge TJ (2004) DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150, 2161–2169 [DOI] [PubMed] [Google Scholar]
- 67.Tashiro Y, Ichikawa S, Shimizu M, Toyofuku M, Takaya N, Nakajima-Kambe T, Uchiyama H, and Nomura N (2010) Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl Environ Microbiol 76, 3732–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang da W, Sherman BT, and Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 69.Huang da W, Sherman BT, and Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee EY, Choi DS, Kim KP, and Gho YS (2008) Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom Rev 27, 535–555 [DOI] [PubMed] [Google Scholar]
- 71.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, and Palucka K (2000) Immunobiology of dendritic cells. Annu Rev Immunol 18, 767–811 [DOI] [PubMed] [Google Scholar]
- 72.Murphy K (2012) Immuno Biology. [Google Scholar]
- 73.Nau R, and Eiffert H (2002) Modulation of release of proinflammatory bacterial compounds by antibacterials: potential impact on course of inflammation and outcome in sepsis and meningitis. Clin Microbiol Rev 15, 95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jang SC, Kim SR, Yoon YJ, Park KS, Kim JH, Lee J, Kim OY, Choi EJ, Kim DK, Choi DS, Kim YK, Park J, Di Vizio D, and Gho YS (2015) In vivo kinetic biodistribution of nano-sized outer membrane vesicles derived from bacteria. Small 11, 456–461 [DOI] [PubMed] [Google Scholar]
- 75.Busse PJ, and Mathur SK (2010) Age-related changes in immune function: effect on airway inflammation. J Allergy Clin Immunol 126, 690–699; quiz 700-691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luepke KH, Suda KJ, Boucher H, Russo RL, Bonney MW, Hunt TD, and Mohr JF 3rd. (2017) Past, Present, and Future of Antibacterial Economics: Increasing Bacterial Resistance, Limited Antibiotic Pipeline, and Societal Implications. Pharmacotherapy 37, 71–84 [DOI] [PubMed] [Google Scholar]
- 77.Simpson RJ (2010) Disruption of cultured cells by nitrogen cavitation. Cold Spring Harb Protoc 2010, pdb prot5513 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.