Abstract
Rationale:
Vascular malformations arise in vessels throughout the entire body. Causative genetic mutations have been identified for many of these diseases; however, little is known about the mutant cell lineage within these malformations.
Objective:
We utilize an inducible mouse model of cerebral cavernous malformations (CCMs) coupled with a multi-color fluorescent reporter to visualize the contribution of mutant endothelial cells (ECs) to the malformation.
Methods and Results:
We combined a Ccm3 mouse model with the confetti fluorescent reporter to simultaneously delete Ccm3 and label the mutant EC with one of four possible colors. We acquired Z-series confocal images from serial brain sections and created 3D reconstructions of entire CCMs to visualize mutant ECs during CCM development. We observed a pronounced pattern of CCMs lined with mutant ECs labeled with a single confetti color (n=42). The close 3D distribution, as determined by the nearest neighbor analysis, of the clonally dominant ECs within the CCM was statistically different than the background confetti labeling of ECs in non-CCM control brain slices as well as a computer simulation (p<0.001). Many of the small (<100μm diameter) CCMs consisted, almost exclusively, of the clonally dominant mutant ECs labeled with the same confetti color whereas the large (>100μm diameter) CCMs contained both the clonally dominant mutant cells and wild type ECs. We propose of model of CCM development in which an EC acquires a second somatic mutation, undergoes clonal expansion to initiate CCM formation, and then incorporates neighboring wild type ECs to increase the size of the malformation.
Conclusions:
This is the first study to visualize, with single-cell resolution, the clonal expansion of mutant ECs within CCMs. The incorporation of wild type ECs into the growing malformation presents another series of cellular events whose elucidation would enhance our understanding of CCMs and may provide novel therapeutic opportunities.
Keywords: Vascular malformation, cerebral cavernous malformations, CCMs, cell lineage, second somatic mutation, clonal expansion, cerebrovascular disease, genetics, transgenic models, mutation, Animal Models of Human Disease, Cerebrovascular Malformations, Genetics, Genetically Altered and Transgenic Models
INTRODUCTION
Vascular malformations occur in arterial, capillary, venous, and lymphatic vessels throughout the entire body. Advances in DNA sequencing capabilities have led to an extensive list of pathogenic loss, or gain, -of-function mutations that cause these diseases, as reviewed previously.1, 2 While specific mutations have been identified for many of these malformations, what is less understood is how a mutant cell, harboring one of these mutations, leads to malformation development and growth. More specifically, it is not known if a single mutation event is sufficient to form these malformations or if multiple different mutation events are necessary to form these complex and structurally abnormal vessels. In this study we utilize an inducible mouse model to investigate this question of mutant endothelial cell (EC) lineage in a specific vascular disease, cerebral cavernous malformations (CCMs).
Cerebral cavernous malformations are ectatic, capillary-venous vessels which are lined with a single layer of endothelial cells and lack surrounding parenchymal cells. Disruption of the endothelial barrier within these slow-flow malformations leads to recurrent hemorrhages, seizures, and focal neurologic deficits. CCMs develop following bi-allelic inactivating mutations in KRIT1/CCM1 (KREV1/RAP1A interaction trapped-1)3, 4, OSM/CCM2 (Osmosensing scaffold for MEKK3)5, 6, or PDCD10/CCM3 (Programmed cell death 10)7. CCMs develop sporadically or through an autosomal dominant pattern of inheritance. Harboring a germline loss-of-function mutation, patients develop the heritable form of disease when a second somatic mutation occurs in the single remaining functional CCM allele. The familial disease often results in 10s to 100s of CCMs developing in adolescent patients while the sporadic form of disease typically presents as a solitary CCM in adult patients. This difference in lesion burden is analogous to Knudson’s observations of tumor burden in sporadic and familial forms of pediatric retinoblastoma8. Knudson’s two-hit model, of the first mutation in germ cells and the second mutation occurring in somatic cells, has been employed to explain CCM pathogenesis. DNA sequencing studies have identified these second somatic mutations, with low allele frequencies, in human CCM samples.9–12 What remains unknown is if a single second somatic mutation is sufficient for CCM formation and precisely where the mutant cells reside within the complex cellular architecture of the vascular malformation. We combined the neonatal Ccm3 mouse model with a multi-color fluorescent reporter to label mutant ECs and directly observe how mutant cells contribute to CCM pathology.
METHODS
The corresponding author will make all data, methods used in the analysis, and materials used to conduct the experiments available upon request. This material will be available at Duke University School of Medicine in Durham, NC.
Please see Online Data Supplement for detailed methods.
RESULTS
Visualization of somatically mutated endothelial cells in cerebral cavernous malformations with an inducible CCM3 mouse model and Confetti reporter.
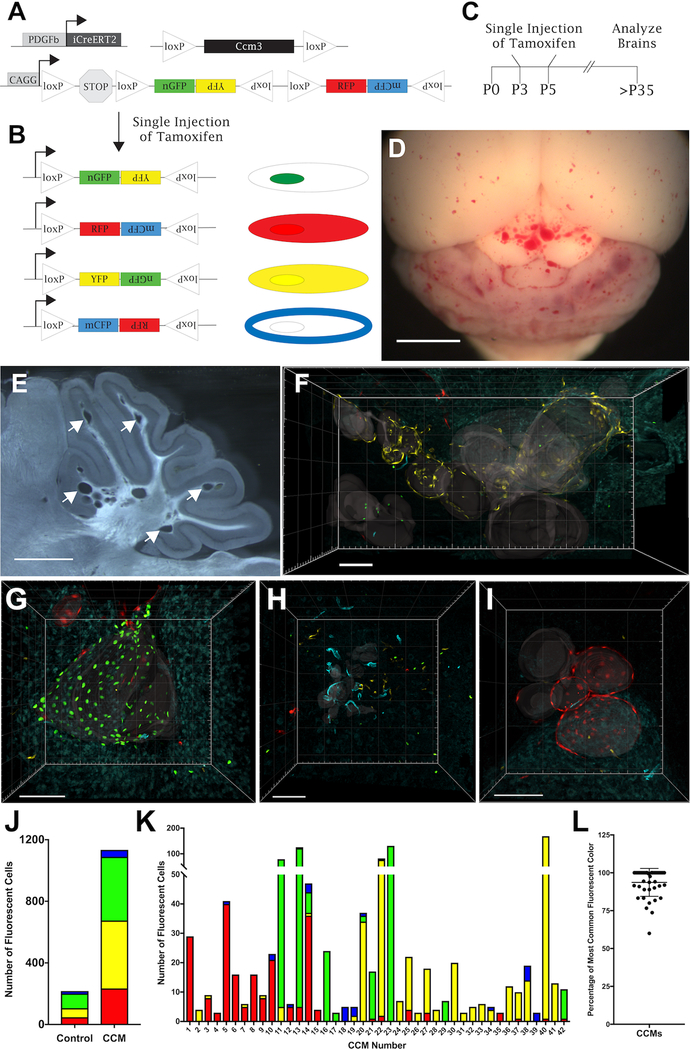
To study the development of cerebral cavernous malformations (CCMs) with respect to the endothelial cells (ECs) that have acquired the second, somatic mutation, we combined an endothelial-specific, tamoxifen-inducible cre recombinase (PDGFb-iCreERT2)13, lox-P flanked and knockout Ccm3 alleles14, and the Brainbow2.1, hereafter referred to as confetti, reporter construct (R26R-Confetti)15, 16 in a transgenic mouse (Figure 1A). The experimental mice (PDGFb-iCreERT2, Ccm3fl/KO, R26R-Confettifl/wt) provide a model in which a single, low dose of tamoxifen (TMX) results in simultaneous cre recombinase-mediated deletion of the single remaining floxed Ccm3 allele and recombination within the R26R-Confetti allele to label mutant ECs and all of their progeny with a fluorescent signal. The confetti reporter is a widely utilized tool to randomly label cells with one of four different fluorescent proteins with varying cellular localizations: nuclear green fluorescent protein (nGFP), cytoplasmic red fluorescent protein (RFP), cytoplasmic yellow fluorescent protein (YFP), and membrane-bound cyan fluorescent protein (mCFP) (Figure 1B). We injected these transgenic mice (n=3) with a single, 2-μg dose of TMX on either postnatal day 3, 4, or 5 to induce transient cre recombinase activity (Figure 1C). Following the single injection of TMX, a subset of endothelial cells within the vascular network acquired the induced second somatic mutation (deletion) and led to CCM development. CCM-null ECs overexpress phospho-myosin light chain (pMLC).17, 18 Due to the lack of a reliable antibody that recognizes Ccm3, and the desire to visualize a gain-of, rather than loss-of signal to mark the deletion of Ccm3, we utilized the increased phosphorylation of MLC as a secondary marker for Ccm3 loss. Endothelial cells lining CCMs exhibit high levels of pMLC and are labeled with one of the confetti colors; thus, recombination at both the Ccm3 and R26R-Confetti loci are observed in the same EC (Online Figure I).
Figure 1. CCMs are composed of clonally dominant ECs labeled with a single confetti color.
(A) Experimental mice contain three transgenes: endothelial-specific, tamoxifen-inducible cre recombinase with platelet derived growth factor b promoter (PDGFb-iCreERT2), loxP-flanked and null Ccm3 alleles (Ccm3fl/KO), and R26R-Confetti reporter allele. (B) Cre recombination of a single R26R-Confetti allele leads to fluorescent labeling with one of four different constructs: nuclear GFP, cytoplasmic YFP, cytoplasmic RFP, and membrane-bound CFP. (C) Experimental design of a single, low dose (2μg) of tamoxifen injected into neonatal pups on postnatal days 3, 4, or 5. (D) Ccm3fl/KO, PDGFb-iCreERT2, R26R-Confettifl/wt experimental mice develop a modest CCM burden in the cerebellum and cerebral hemispheres (n=3, scale bar: 2mm). (E) A representative 80-μm thick brain slice with discrete CCMs dispersed throughout the cerebellum as indicated by arrows (scale bar: 1 mm). (F-I) 3D reconstructions of Z-series confocal images acquired across multiple serial brain slices to visualize the entire volume of CCMs in Ccm3fl/KO, PDGFb-iCreERT2, R26R-Confettifl/wt mice. Gray translucent surfaces have been added to the images to aid in visualizing the vascular lumens of each CCM. Representative CCMs for (F) YFP, (G) nGFP, (H) mCFP, and (I) RFP (scale bars: 100μm). (J) Number of ECs expressing each confetti color in serial brains slices of non-CCM control mice (PDGFb-iCreERT2, R26R-Confettifl/wt, n=1) and CCM mice (Ccm3fl/KO, PDGFb-iCreERT2, R26R-Confettifl/wt, n=3). (K) Number of each confetti color expressed by ECs lining individual CCMs (n=42). CCMs #40, #13, #18, and #s 5–8 are the CCMs shown in panels F, G, H, and I respectively. (L) Percentage of the clonally dominant ECs, labeled with the most common confetti color within a CCM, when considering all confetti labeled ECs within the CCM (94+9 (mean + SD), n = 42).
Inducible CCM mouse models have been used extensively, with high doses and often multiple injections of TMX, to generate a severe phenotype to investigate the molecular signaling of CCM pathogenesis19–25 and efficacy of proposed therapeutics.26–29 Our approach was to more closely model the more severe familial form of the human disease, in which random second somatic mutations occur in a small population of endothelial cells, by using a very low dose of TMX. We induced a modest phenotype with CCMs dispersed throughout the cerebellum and cerebral hemispheres (Figure 1D). The dispersed CCMs enabled us to visualize individual CCMs to infer mutant EC lineage (Figure 1E). Serial brain sections, confocal microscopy, and 3D reconstruction were utilized to characterize the entire volume of individual CCMs. The Z-series of confocal images from the same brain region of serial slices were aligned and concatenated into 3D reconstructions of CCMs across large brain volumes. It was imperative to visualize an entire CCM, rather than a single section, to more confidently draw conclusions of the mutant EC lineage. Despite the use of a vibratome to section the entire brain into 80-μm thick slices, the sectioning process resulted in slight tissue loss between each slice, which is best demonstrated in a video of a rotating 3D reconstruction (Online Video I). The loss of tissue during brain processing was greatly outweighed by the valuable information gathered by reconstructing images from adjacent slices in order to examine entire CCMs.
Clonal dominance of mutant ECs in CCMs.
Representative CCM images for each confetti color demonstrate that CCMs are lined with a disproportionate number of ECs expressing the same confetti color (Figure 1F-I, Online Videos I-IV). To measure the frequency of recombination within the R26R-Confetti allele with these experimental conditions in the absence of CCMs, we injected control animals (PDGFb-iCreERT2,Ccm3wt/wt, R26R-Confettifl/wt ) with the same dose of TMX on P3 (n=1). Serial brain slices of the P25 control mouse contained a fluorescent signal density of 1.3 confetti labeled ECs per 1×106 μm3 of cerebellar brain volume. This background confetti labeling during normal vascular development established the minimum value that could distinguish between chance recombination at the confetti locus and joint recombination of the Ccm3 and R26R-Confetti alleles. This density of confetti labeling of ECs was the threshold for either including or excluding the CCMs from the clonal analysis. As anticipated, we observed CCMs containing a fluorescent EC density less than the background confetti labeling measured in the control brain slices. The CCMs that contained a confetti labeled EC density less than the control (non-lesion) samples are likely derived from mutant ECs in which the R26R-Confetti allele did not recombine to tag the mutant ECs with a color. We excluded 28 CCMs from the analysis; of the 28 CCMs excluded, 86% (24/28) contained two or fewer ECs labeled with any confetti color. With the great majority of these excluded CCMs consisting of only two or fewer labeled cells, these data, even if included, would have contributed very little towards determining whether or not clonal dominance occurs in lesion genesis.
The CCM and control (non-lesion) samples had approximately equal proportions of ECs expressing each of the confetti colors (Figure 1J). The frequency of the R26R-Confetti allele recombining to tag ECs with each of the confetti colors within CCMs was: Red – 20.6%, Yellow – 38.9% , Green – 36.5%, and Cyan – 4.0%. This low rate of the R26R-Confetti allele tagging ECs with mCFP provided a rare event to more confidently conclude that multiple ECs expressing mCFP within a CCM are derived from a single mutant EC. Furthermore, the ability to detect mCFP in all samples enabled us to confidently conclude that there was no cre recombination event within the confetti locus in the unlabeled ECs. As detailed later, this ability to delineate ECs which have undergone cre recombination, from those ECs which have not, is important in our analysis of CCM development. While the experimental and control samples expressed each of the confetti colors in comparable frequencies, there was an overrepresentation of a single confetti color in labeled ECs within individual CCMs. To gain an accurate view of the molecular events occurring with the developing CCM, the number of ECs labeled with each of the confetti colors within 42 individual CCMs, of varying sizes and stages of maturation, was counted (Figure 1K). The clonal dominance of ECs tagged with the same confetti color was calculated, as the percentage of ECs labeled with the clonally dominant color when considering all ECs tagged with a confetti color within the CCM, to be 94%+9% (n=42) (Figure 1L). The disproportionate number of ECs tagged with the same confetti color in these malformations suggests clonal expansion of a mutant EC during CCM development.
Nearest neighbor analysis validates the clonal dominance of mutant ECs in CCMs.
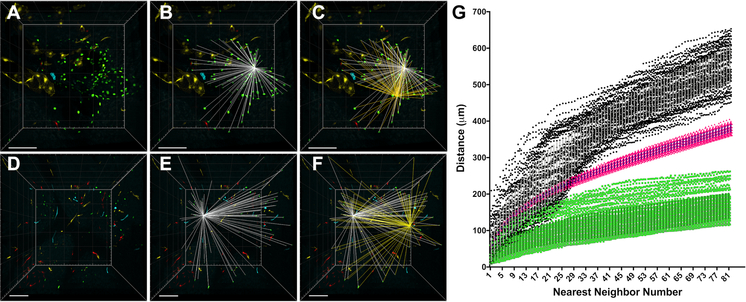
Although even a cursory calculation of the difference in percentage of ECs labeled with each confetti color within individual CCMs showed a strong clonal dominance of one color (somatic mutation), we sought a more rigorous statistical analysis of the data. We performed a nearest neighbor (NN) analysis of labeled ECs using CCM lesion, control (non-lesion) tissue, and computer-generated simulation datasets. The NN algorithm is a method for determining the 3D distance between each member of a dataset. From the perspective of a single data point, the NN algorithm calculates the distance to all of the other data points and then ranks the distances from closest to furthest. The algorithm continues to the next data point within the group until all of the NN distances have been calculated and ranked for each member of the dataset. We applied the NN algorithm to the 3D reconstructions of the entire CCMs gathered from confocal images of serial brain slices. We represented each EC labeled with the clonally dominant confetti color within a CCM as a point with x,y,z coordinates (Figure 2A). The NN distances were calculated and ranked for a single EC in the CCM (visually depicted in Figure 2B, Online Video V). The NN algorithm continued to a second EC in the CCM (visually depicted in Figure 2C, Online Video V), until the NN distances for all of the ECs within the CCM were calculated. The NN distances for all of the ECs within this representative CCM are plotted as distance (μm) vs. NN number (Figure 2G, green data points).
Figure 2. Nearest neighbor (NN) analysis as a quantitative comparison of the 3D distribution of confetti labeled ECs in CCMs, control brain slices, and computer simulations.
(A) 3D reconstruction of the entire volume of a CCM containing clonally dominant ECs labeled with nGFP. This CCM corresponds to CCM #23 in Figure 1K. (B) Visual representation of the 3D distances (white) between one EC and all of the other ECs within the CCM. The NN algorithm calculates and ranks these distances from closest to furthest. (C) Visual representation of the 3D distances (yellow) between a second EC and all of the other ECs within the CCM superimposed on the 3D distances of the first EC (white). (D-F) The same NN analysis is applied to non-CCM control brain slices: (D) 3D reconstruction of the control brain slices, (E) NN distances (white) between one nGFP EC and all of the other nGFP ECs, (F) a second set of NN distances (yellow) for a different nGFP EC and all of the nGFP ECs within the sample. (G) All of the NN distances, two of which are visually represented on the 3D reconstructions above, are ranked from closest to furthest and plotted as distance (μm) by NN number. The green data points represent the NN distances for all of the nGFP ECs within the CCM shown in panel A. The black data points represent the NN distances for all of the nGFP ECs within the control brain slices shown in panel D. The pink data points represent the average NN distances from 100 computer simulations in which the x,y,z coordinates of each member of the analysis were assigned by a random number generator. The simulation used the same volume and number of members (ECs) as the experimental CCM sample shown in panel A (error bars represent mean + SD). All scale bars are 100μm.
The same NN algorithm was applied to serial brain slices of the control, PDGFb-iCreERT2, Ccm3wt/wt , R26R-Confettifl/wt, mouse (Figure 2D-F, Online Video VI). These animals develop no CCM lesions and provide data on the background level of confetti recombination in otherwise identical conditions of transient cre recombinase induction. ECs labeled with the same confetti color within the control brain slices were each represented with an x,y,z point and the NN distances were calculated for comparison with the NN distances calculated for the CCM. Given the varying recombination frequencies of the four confetti colors (Figure 1J), each color was analyzed independently in both the CCM and control samples. Thus, the NN distances for nGFP labeled ECs within the control brain slices are plotted, since the representative CCM being analyzed contained clonally dominant ECs expressing nGFP (Figure 2G, black data points). Imaging experiments have the potential for experimenter bias to be introduced when selecting a region of tissue to study. For example, a control region of low confetti labeling would bias in favor of the heavily labeled CCMs. Thus, we deliberately imaged regions of the control brain that contained many ECs expressing all of the confetti colors within the field of view, but we also created a second control data set that was generated randomly by a computer program. Using the x, y, and z dimensions of the 3D reconstruction of the CCM images and the number of ECs labeled with the clonally dominant confetti color of the CCM, the computer code generated a control dataset by randomly assigning the same number of data points into the same 3D volume as the experimental sample. The NN analysis was performed with this randomly generated dataset and then the code repeated the process utilizing the same input parameters for a total of 100 iterations. The average NN distances for 100 simulations are plotted (Figure 2G, pink data points).
Combining the NN analysis for a representative CCM, non-lesional control samples, and a computer-generated random simulation, the stark difference in the NN distances between the control and experimental groups strongly supports the hypothesis that the observed close proximity of the clonally dominant mutant ECs in the CCM is not due to random chance (Figure 2G).
Clonal dominance is observed in CCMs across all confetti colors.
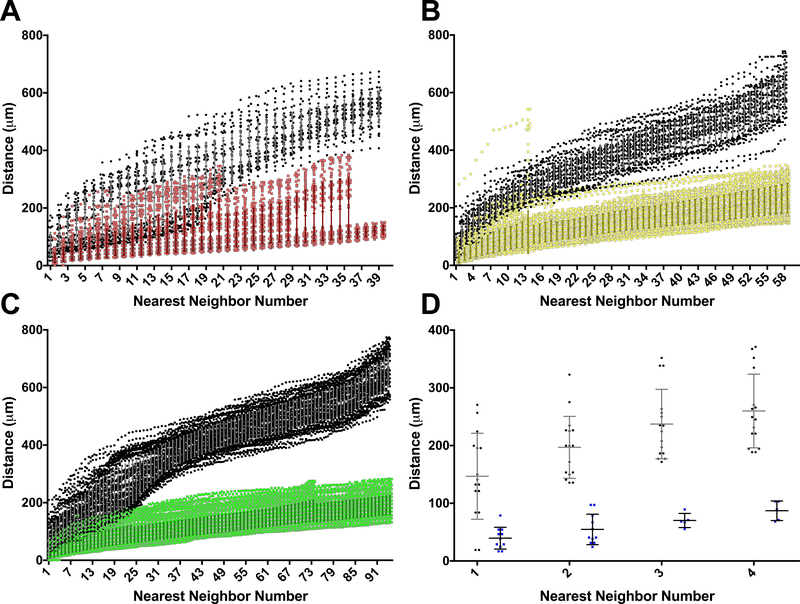
We performed the nearest neighbor (NN) analysis for all CCM and the control (non-lesion) samples; the NN distances are plotted as distance (μm) vs. nearest neighbor number for each of the four confetti colors (Figure 3A-D). All CCMs, regardless of the confetti color expressed by the ECs, contained a clonally dominant population of ECs whose close proximity in 3D was not observed in the control brain slices. The CCM and brain slice control NN distances were statistically different beginning with the 1st NN for the RFP groups (p<0.001), YFP groups (p<0.001), nGFP groups (p<0.001), and mCFP groups (p<0.001) as determined by a Mann-Whitney test. These very low p-values are driven by the large number of NN distances involved in each comparison. As p-values do not comment on the biological process of an observation, the more important result is the clear divergence of the CCM and control NN distances as the NN number increases (Figure 3A-D). The clonally dominant ECs within each of the 42 individual CCMs analyzed were significantly closer in 3D space than their respective confetti color controls. This pattern of clonally dominant ECs was observed across different CCMs as well as different confetti colors and illuminates a fundamental process of clonal expansion of Ccm3-null endothelial cells in CCM development.
Figure 3. Divergence of the nearest neighbor (NN) distances of CCMs and non-lesion controls as NN number increases.
Given the difference in recombination frequency among the four confetti colors, the NN comparison is only made between the ECs in CCMs and control brain slices labeled with the same confetti color. (A) NN distances for all CCMs containing clonally dominant ECs labeled with RFP (red) and the NN distances of ECs labeled with RFP in the control brain slices (black) (CCM n=13). (B) NN distances for YFP ECs within CCMs (yellow) and control brain slices (black) (CCM, n=18). (C) NN distances for nGFP ECs within CCMs (green) and control brain slices (black) (CCM, n=8). (D) NN distances for mCFP ECs within CCMs (blue) and control brain slices (black) (CCM, n=3). For each of the four confetti colors, the NN distance was greater for the non-lesion controls than for the CCMs for every NN number pair, beginning with the 1st NN (Mann-Whitney test, p<0.001). Error bars represent mean + SD.
Clonal expansion of mutant ECs and recruitment of neighboring ECs drives CCM development.
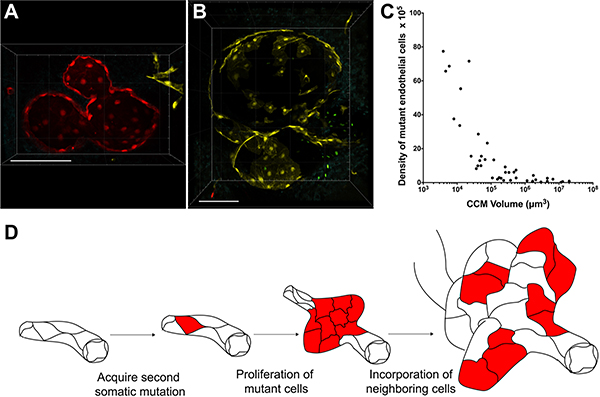
The increased number of confetti labeled ECs lining CCMs, that is not observed in the control samples, supports a clonal expansion of mutant ECs during CCM formation. Upon closer inspection of the CCMs imaged, a difference in the EC composition of small (diameter < 100μm) and large (diameter > 100μm) CCMs emerged. Many of the small CCMs imaged were composed nearly entirely of ECs labeled with the same confetti color (Figure 4A). Small CCMs consisting almost exclusively of clonally dominant ECs suggests that the EC which acquired the second somatic mutation, and was labeled with a confetti color, proliferated to initiate malformation development. We analyzed a small CCM with multiple caverns, some visibly connected within a single brain slice, in which each cavern was lined with ECs tagged with RFP (Figure 4A). Interestingly, these serial brain slice images also suggest that multicavernous CCMs can develop from the same somatic mutation. A rotation of the 3D reconstruction of the confocal images more clearly demonstrates the high proportion or RFP-labeled ECs lining this multicavernous CCM (Online Video VII). In a different mouse, we visualized a larger multicavernous CCM (largest diameter > 200 μm) that contained both confetti-labeled and unlabeled ECs (Figure 4B). The unlabeled ECs appear as blank spaces within the vascular walls for the malformation. These unlabeled ECs within the CCM likely represent wild type ECs that have been incorporated into the growing malformation. The possibility of these unlabeled ECs being Ccm3-null, having undergone cre recombination within the Ccm3 allele and not the R26R-Confetti allele, cannot be excluded. However, as demonstrated in the pMLC staining of CCMs (Online Figure I) CCMs contain a mosaicism of mutant and wild type ECs. The mosaicism of mutant and wild type ECs has been previously shown with staining of human CCMs30 as well as human CCM DNA sequencing studies in which a small percentage of the total DNA reads detected the second somatic mutation of the mutant ECs within the CCM.9–12 In large CCMs we observe the same clonal expansion of mutant ECs as was seen with the small CCMs as well as the recruitment of unlabeled, presumably wild type, neighboring ECs into the growing malformation. These two CCMs represent different stages along the spectrum of CCM development. This spectrum of CCM pathology is well demonstrated in the CCMs shown in Figure 1F. The smaller CCMs in the left half of the 3D reconstruction contain a proportionally higher number of mutant cells than the larger CCMs on the right half of the image. This observation is supported by analysis of all 42 CCM lesions showing that the density of clonally dominant, mutant ECs decreases as CCMs grow larger (Figure 4C). Taken together, we propose a model of CCM pathogenesis by which an EC acquires a second somatic mutation, undergoes clonal expansion through a gain in proliferative capacity, and recruits neighboring ECs as the size and complexity of the malformation increases (Figure 4D).
Figure 4. Clonal expansion of mutant ECs and recruitment of neighboring ECs drives CCM development.
(A) Small (diameter <100μm), multicavernous CCM composed, nearly entirely, of clonally dominant ECs labeled with the same confetti color (RFP) in each cavern (scale bar: 100μm). This CCM corresponds to CCM #1 in Figure 1K. (B) Large (diameter >100μm), multicavernous CCM with a clonally dominant population of ECs labeled with YFP interspersed with unlabeled (appearing as blank spaces) ECs (scale bar: 100μm). This CCM corresponds to CCM #22 in Figure 1K. (C) The mutant EC density (number of clonally dominant ECs per 105 μm3 CCM volume) decreases as CCMs grow larger (D) Working model of CCM development in which a single, second somatic mutation leads to clonal expansion and subsequent recruitment of wild type ECs into growing CCMs.
DISCUSSION
Herein we report a microscopy-based mutant EC lineage study of cerebral cavernous malformations in an inducible mouse model. Utilizing the confetti reporter, serial brain sectioning, confocal microscopy, and 3D reconstructions, we visualized entire CCMs and investigated CCM development at the cellular level. We studied both single-cavern and multicavernous CCMs of varying sizes that contained clonally dominant mutant ECs labeled with the same confetti color. The nearest neighbor analysis comparing CCMs, with non-CCM control brain slices, and a computer simulation determined that the large quantity of ECs labeled with the same confetti color in close 3D space that was seen in the CCMs could not be attributed to background confetti labeling during normal vascular development or random chance. These findings suggest that a single second somatic mutation is sufficient to seed the formation of a CCM. The final important observation from these studies was the incorporation of unlabeled, wild type ECs into the growing CCMs. It appears that CCMs increase in size by incorporating wild type ECs into the vessel wall of the malformation. We propose a model of CCM development by which an EC acquires a second somatic mutation, undergoes clonal expansion, and recruits neighboring ECs to expand the malformation. This study focused on CCMs developing from Ccm3 mutations and additional studies are warranted to determine if the proposed mechanism of CCM development is conserved in malformations arising from Ccm1 and Ccm2 mutations.
These results are consistent with and build upon the findings of several other research groups. Increased proliferation has been observed with astrocytes harvested from this inducible Ccm3 mouse model and maintained in cell culture for several days.31 We published a DNA sequencing study of human CCMs in which one of the cases identified the somatic mutations of a surgically-resected CCM as well as a subsequent CCM that occurred later in the same anatomic location, presumably due to regrowth of a remnant of the original CCM. We found that both the initial and second CCM harbored the identical CCM3 somatic mutation.11, 12 The formation of a second CCM from the same population of mutant ECs strongly supports our model of clonal expansion of a mutant EC in CCM development.
The frequency at which the somatic mutation can be detected in the CCMs is generally less than 10% of all the DNA sequencing reads.12 The low detection rate of the acquired, somatic mutation from large human CCMs highlights the mosaicism of CCM-null and wild type ECs, which was also visualized by immunohistochemistry of human CCMs.30 However, sequencing surgically-resected CCMs includes all of the different cell types present in the tissue, and not solely the endothelial cells lining the caverns. Furthermore, since only one allele will acquire the somatic mutation, the observed mutant allele frequency reflects one half of the mutant cell frequency. In contrast to these sequencing data, our imaging data specifically identifies the mutant endothelial cells in the lesion. Thus, our data more accurately represents the number of mutant cells contributing to the CCM.
Our work has expanded this concept of EC mosaicism by utilizing confocal microscopy to trace mutant ECs, with single-cell resolution, and observe the change in EC composition as CCMs develop. A pattern of CCM development emerged in which many of the small, recently formed CCMs consisted nearly entirely of clonally expanded mutant ECs while the large, multicavernous CCMs consisted of both the clonally expanded mutant ECs as well as a second population of unlabeled ECs. These two stages of CCM development: clonal expansion and incorporation of wild type ECs may offer new opportunities for therapeutic intervention.
While CCMs are not considered vascular tumors, the increase in proliferation and regrowth of a malformation suggests that CCM-null ECs acquire an aggressive phenotype which may be targeted with anti-proliferative therapies. Work by others has targeted cell proliferation in the context of angiogenesis, with anti-angiopoietin-2 antibodies, and observed a reduction in CCM development in this neonatal CCM3 mouse model.24 Additional studies of anti-proliferative therapies, particularly those not directly targeting angiogenesis, may reveal a fundamental roles of CCM3 in both physiologic and pathologic conditions. Although the presence of wild type ECs in CCMs has been inferred by the low mutant allele frequency observed in CCM tissue, the mechanism by which these ECs become incorporated in the growing malformation is completely unknown. As our understanding of this phenomena improves, we can use CCM mouse models like that of the current study to test therapeutics administered to disrupt the cellular signals involved in CCM incorporation of wild type ECs.
In conclusion, this work is the first to provide visual evidence for clonal expansion of mutant ECs in the development of CCMs. By imaging the entire CCM volume with single-cell resolution, we captured the initial clonal burst of mutant ECs in small CCMs followed by the incorporation of wild type ECs into the network of clonally dominant mutant ECs of larger CCMs. This work has illuminated the mutant EC lineage of CCMs and highlighted the need to further investigate the molecular mechanisms by which a single somatic mutation, within a single EC, leads to a large, functional vascular malformation that contains a mixed population of both mutant and wild type cells.
Supplementary Material
NOVELTY AND SIGNIFICANCE
What Is Known?
Cerebral Cavernous Malformations (CCM) are ectatic, capillary-venous vessels within the central nervous system that consist of a single layer of endothelial cells and lack surrounding parenchymal cells.
CCMs can occur sporadically or as an autosomal dominant trait with multiple CCMs in an individual. Three genes have been identified that exhibit germline mutations in autosomal dominant CCM cases.
DNA sequencing analyses of large, surgically-resected CCMs from autosomal dominant CCM cases have identified somatic mutations at very low frequencies of all DNA reads; these findings suggest that the minority of cells in the mature CCM harbor the somatic mutation.
What New Information Does this Article Contribute?
A single somatic mutation in an endothelial cell is sufficient to seed the formation of a CCM.
The early stage CCMs, typically those consisting of a single cavern, consist primarily of somatically-mutated endothelial cells lining the cavern.
Later stage (larger, multi-cavernous) CCMs retain only a single, mutant endothelial cell lineage, but consist of a mixture of normal and somatically-mutant endothelial cells.
We propose a two-stage model of CCM development: a burst of clonal expansion of the somatically-mutated endothelial cells in the early phase of lesion genesis, and subsequent incorporation of normal (non-mutated) endothelial cells as the CCM matures and grows larger.
Here we visualize, with single-cell resolution, the development of CCMs employing a mouse model of an inherited form of the disease and lineage tracing of mutant cells using the Confetti fluorescent reporter. By inducing a somatic mutation in the remaining wild type copy of the CCM gene, we initiate lesion genesis and simultaneously tag the original mutated cell with a fluorescent label. We show that regardless of their size or stage of maturation, CCMs consist of a single mutant lineage, thereby exhibiting clonal dominance. Interestingly, the smaller, early stage lesions consist mostly of mutant endothelial cells, but these grow in size and complexity (multiple caverns) through incorporation of neighboring, normal endothelial cells. This work highlights the need to further investigate the molecular mechanisms by which a somatic mutation within a single endothelial cell leads to a large, complex vascular malformation containing a mixed population of both mutant and wild type cells. Disruption of the cellular signals involved in incorporation of wild type endothelial cells into the growing CCM may be a viable target for future therapies.
ACKNOWLEDGMENTS
We would like to thank Amy Pieper, Heidy Pardo, Erin Griffin, Carol Gallione, and Christian Benavides for animal husbandry, Drs. Lisa Cameron and Benjamin Carlson of the Duke University Light Microscopy Core Facility for microscopy assistance, Dr. Ravi Karra of Duke University for helpful discussions, Dr. Debra Silver of Duke University for use of equipment, Drs Wang Min (Yale University) and Marcus Fruttiger (University College London) for sharing transgenic mice, and Drs. Mark Kahn (University of Pennsylvania), Issam Awad (University of Chicago), and Mark Ginsberg (University of California at San Diego) for helpful discussions.
SOURCES OF FUNDING
This work was supported by the National Institutes of Health (P01 NS092521 to D.A.M., T32 GM007171 to M.R.D), the Fondation Leducq (17 CVD 03 to D.A.M.), and the American Heart Association (18PRE34060061 to M.R.D.).
Nonstandard Abbreviations and Acronyms:
- CCM
cerebral cavernous malformations
- ECs
endothelial cells
- mCFP
membrane-bound cyan fluorescent protein
- nGFP
nuclear green fluorescent protein
- NN
nearest neighbor
- pMLC
phospho-myosin light chain
- RFP
red fluorescent protein
- TMX
tamoxifen
- YFP
yellow fluorescent protein
Footnotes
DISCLOSURES
None.
In August 2018, the average time from submission to first decision for all original research papers submitted to Circulation Research was 12.62 days.
REFERENCES
- 1.Queisser A, Boon LM and Vikkula M. Etiology and Genetics of Congenital Vascular Lesions. Otolaryngologic clinics of North America. 2018;51:41–53. [DOI] [PubMed] [Google Scholar]
- 2.Greene AK and Goss JA. Vascular Anomalies: From a Clinicohistologic to a Genetic Framework. Plastic and reconstructive surgery. 2018;141:709e–717e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahoo T, Johnson EW, Thomas JW, Kuehl PM, Jones TL, Dokken CG, Touchman JW, Gallione CJ, Lee-Lin SQ, Kosofsky B, Kurth JH, Louis DN, Mettler G, Morrison L, Gil-Nagel A, Rich SS, Zabramski JM, Boguski MS, Green ED and Marchuk DA. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1). Human molecular genetics. 1999;8:2325–33. [DOI] [PubMed] [Google Scholar]
- 4.Laberge-le Couteulx S, Jung HH, Labauge P, Houtteville JP, Lescoat C, Cecillon M, Marechal E, Joutel A, Bach JF and Tournier-Lasserve E. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nature genetics. 1999;23:189–93. [DOI] [PubMed] [Google Scholar]
- 5.Liquori CL, Berg MJ, Siegel AM, Huang E, Zawistowski JS, Stoffer T, Verlaan D, Balogun F, Hughes L, Leedom TP, Plummer NW, Cannella M, Maglione V, Squitieri F, Johnson EW, Rouleau GA, Ptacek L and Marchuk DA. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. American journal of human genetics. 2003;73:1459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denier C, Goutagny S, Labauge P, Krivosic V, Arnoult M, Cousin A, Benabid AL, Comoy J, Frerebeau P, Gilbert B, Houtteville JP, Jan M, Lapierre F, Loiseau H, Menei P, Mercier P, Moreau JJ, Nivelon-Chevallier A, Parker F, Redondo AM, Scarabin JM, Tremoulet M, Zerah M, Maciazek J and Tournier-Lasserve E. Mutations within the MGC4607 gene cause cerebral cavernous malformations. American journal of human genetics. 2004;74:326–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, Coubes P, Echenne B, Ibrahim R, Irthum B, Jacquet G, Lonjon M, Moreau JJ, Neau JP, Parker F, Tremoulet M and Tournier-Lasserve E. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. American journal of human genetics. 2005;76:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudson AG Jr., Mutation and cancer: statistical study of retinoblastoma. Proceedings of the National Academy of Sciences of the United States of America. 1971;68:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gault J, Shenkar R, Recksiek P and Awad IA. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke; a journal of cerebral circulation. 2005;36:872–4. [DOI] [PubMed] [Google Scholar]
- 10.Gault J, Awad IA, Recksiek P, Shenkar R, Breeze R, Handler M and Kleinschmidt-DeMasters BK. Cerebral cavernous malformations: somatic mutations in vascular endothelial cells. Neurosurgery. 2009;65:138–44; discussion 144–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akers AL, Johnson E, Steinberg GK, Zabramski JM and Marchuk DA. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Human molecular genetics. 2009;18:919–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald DA, Shi C, Shenkar R, Gallione CJ, Akers AL, Li S, De Castro N, Berg MJ, Corcoran DL, Awad IA and Marchuk DA. Lesions from patients with sporadic cerebral cavernous malformations harbor somatic mutations in the CCM genes: evidence for a common biochemical pathway for CCM pathogenesis. Human molecular genetics. 2014;23:4357–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claxton S, Kostourou V, Jadeja S, Chambon P, Hodivala-Dilke K and Fruttiger M. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis (New York, NY : 2000). 2008;46:74–80. [DOI] [PubMed] [Google Scholar]
- 14.He Y, Zhang H, Yu L, Gunel M, Boggon TJ, Chen H and Min W. Stabilization of VEGFR2 signaling by cerebral cavernous malformation 3 is critical for vascular development. Sci Signal. 2010;3:ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR and Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. [DOI] [PubMed] [Google Scholar]
- 16.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD and Clevers H. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. Cell. 2010;143:134–144. [DOI] [PubMed] [Google Scholar]
- 17.Stockton RA, Shenkar R, Awad IA and Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. The Journal of experimental medicine. 2010;207:881–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borikova AL, Dibble CF, Sciaky N, Welch CM, Abell AN, Bencharit S and Johnson GL. Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. The Journal of biological chemistry. 2010;285:11760–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitehead KJ, Chan AC, Navankasattusas S, Koh W, London NR, Ling J, Mayo AH, Drakos SG, Jones CA, Zhu W, Marchuk DA, Davis GE and Li DY. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nature medicine. 2009;15:177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan AC, Drakos SG, Ruiz OE, Smith AC, Gibson CC, Ling J, Passi SF, Stratman AN, Sacharidou A, Revelo MP, Grossmann AH, Diakos NA, Davis GE, Metzstein MM, Whitehead KJ and Li DY. Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J Clin Invest. 2011;121:1871–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maddaluno L, Rudini N, Cuttano R, Bravi L, Giampietro C, Corada M, Ferrarini L, Orsenigo F, Papa E, Boulday G, Tournier-Lasserve E, Chapon F, Richichi C, Retta SF, Lampugnani MG and Dejana E. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498:492–6. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Tang AT, Wong WY, Bamezai S, Goddard LM, Shenkar R, Zhou S, Yang J, Wright AC, Foley M, Arthur JS, Whitehead KJ, Awad IA, Li DY, Zheng X and Kahn ML. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 2016;532:122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuttano R, Rudini N, Bravi L, Corada M, Giampietro C, Papa E, Morini MF, Maddaluno L, Baeyens N, Adams RH, Jain MK, Owens GK, Schwartz M, Lampugnani MG and Dejana E. KLF4 is a key determinant in the development and progression of cerebral cavernous malformations. EMBO molecular medicine. 2015;8:6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou HJ, Qin L, Zhang H, Tang W, Ji W, He Y, Liang X, Wang Z, Yuan Q, Vortmeyer A, Toomre D, Fuh G, Yan M, Kluger MS, Wu D and Min W. Endothelial exocytosis of angiopoietin-2 resulting from CCM3 deficiency contributes to cerebral cavernous malformation. Nature medicine. 2016;22:1033–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang AT, Choi JP, Kotzin JJ, Yang Y, Hong CC, Hobson N, Girard R, Zeineddine HA, Lightle R, Moore T, Cao Y, Shenkar R, Chen M, Mericko P, Yang J, Li L, Tanes C, Kobuley D, Vosa U, Whitehead KJ, Li DY, Franke L, Hart B, Schwaninger M, Henao-Mejia J, Morrison L, Kim H, Awad IA, Zheng X and Kahn ML. Endothelial TLR4 and the microbiome drive cerebral cavernous malformations. Nature. 2017;545:305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bravi L, Rudini N, Cuttano R, Giampietro C, Maddaluno L, Ferrarini L, Adams RH, Corada M, Boulday G, Tournier-Lasserve E, Dejana E and Lampugnani MG. Sulindac metabolites decrease cerebrovascular malformations in CCM3-knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:8421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson CC, Zhu W, Davis CT, Bowman-Kirigin JA, Chan AC, Ling J, Walker AE, Goitre L, Delle Monache S, Retta SF, Shiu YT, Grossmann AH, Thomas KR, Donato AJ, Lesniewski LA, Whitehead KJ and Li DY. Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation. 2015;131:289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura S, Mishra-Gorur K, Park J, Surovtseva YV, Sebti SM, Levchenko A, Louvi A and Gunel M. Combined HMG-COA reductase and prenylation inhibition in treatment of CCM. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:5503–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Ramirez MA, Fonseca G, Zeineddine HA, Girard R, Moore T, Pham A, Cao Y, Shenkar R, de Kreuk BJ, Lagarrigue F, Lawler J, Glass CK, Awad IA and Ginsberg MH. Thrombospondin1 (TSP1) replacement prevents cerebral cavernous malformations. The Journal of experimental medicine. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pagenstecher A, Stahl S, Sure U and Felbor U. A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Human molecular genetics. 2009;18:911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Louvi A, Chen L, Two AM, Zhang H, Min W and Gunel M. Loss of cerebral cavernous malformation 3 (Ccm3) in neuroglia leads to CCM and vascular pathology. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3737–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.