Abstract
Food-borne infections are major causes of public health concern in developing and developed countries. During the past decade, the Institut Hospitalo-Universitaire Méditerranée Infection has conducted or been involved in multiple investigations that aimed at identifying the sources and strains responsible for food-borne diseases and therefore at improving the understanding, diagnosis, prevention and control of these infections. Investigations were conducted in the Mediterranean area and in sub-Saharan Africa on more than 15 food-borne agents, 17 food products and 14 antibiotic resistance-associated genes. Multiple sources, including unexpected ones, and pathogens, including emerging ones, were involved. Travelling in developing countries and zoonoses are major contributors to food-borne infections, while food-borne transmission of resistance-associated genes is increasingly reported. However, risk factors and pathogens associated with food-borne infections likely remain untapped and must be more extensively investigated, monitored and regularly reassessed. Diagnostic tests based on new technologies and real-time surveillance tools based on microbiology laboratory data are promising approaches to detect known food-borne infections and decipher new ones. Studies of the microbiota and its relationships with dietary patterns are also worth being conducted.
Keywords: Africa, bacteria, food, food-borne, fungus, IHU Mediterranée Infection, infections, Mediterranean Basin, virus, zoonosis
Introduction
Food-borne infections are major causes of public health concern in developing and developed countries and can have a considerable social and economic cost [1], [2]. Nevertheless, their global burden remains insufficiently documented. These infections mostly involve bacteria, viruses and parasites that use food as vehicle for their transfer from animals to humans and can generate secondary transmissions between humans [3]. During the past decade the Institut Hospitalo-Universitaire Méditerranée Infection has conducted or been involved in multiple investigations that aimed at identifying the sources and strains responsible for food-borne diseases and therefore at improving the understanding, diagnosis, prevention and control of these infections. We summarize here its contribution to the study of food-borne infections.
Methods
We used the following keywords, cross-matched with names from people of our institution, to search in PubMed, Google Scholar and ISI Web of Science: food, food-borne, alimentary, sausage, shellfish, meat, fish, fruit, vegetable, dairy product or sauce; and infectious, infection, bacteria, virus, parasite, microorganism, microbe, microbiology, virology, parasitology or mycology.
Bacteria
As emphasized by the example of the German outbreak due to Escherichia coli O104:H4 [4], food-borne bacterial infections can be due to the presence of bacterial toxins in food. Alternatively, they can result from the presence of pathogenic bacteria in food products. In our experience, such infections have been suspected several times. In 2015, our laboratory data–based surveillance system, PACASurvE (Provence-Alpes-Côte d’Azur Surveillance Epidemiologic System) [5], detected several consecutive abnormal increases in the number of Enterococcus faecalis infections, especially community-acquired urinary tract infections, in different areas of the Provence-Alpes-Côte d’Azur French region [6]. Investigations led us to strongly suspect a zoonotic origin for the outbreak, with chicken-based food as a vehicle for the pathogen. Another example is that of Enterococcus cecorum, a bacterium rarely involved in human infections but normally present in the intestinal tract of domestic animals, which we identified in two patients receiving immunosuppressive drug regimens hospitalized in Marseille public hospitals [7]. In this case, we finally hypothesized that the two infections originated from food-mediated acquisition of the bacteria facilitated by immunosuppression. Similarly, in 2016, we were the first to identify Vagococcus lutrae, a bacterium initially described in the common otter as a possible human pathogen, in a hospitalized patient [8]. In this case we also hypothesized that bacterial infection was food-borne, especially through consumption of seafood products and possibly promoted by poor hygiene. In another two studies investigating the epidemiology of Coxiella burnetii, the zoonotic agent of Q fever, among children hospitalized in a tertiary-care paediatric hospital in Athens, Greece, and a C. burnetii outbreak in Southern France, we clearly identified the role of cheese and unpasteurized dairy products in human infections [9], [10]. Coxiella burnetii DNA, but not viable bacteria, was also described in dairy products in France [11].
Fortunately, all food-borne bacteria do not cause diseases in humans. Most of the time, these bacteria colonize the human gut without any detrimental impact to the host. Thus, yoghurts and probiotic food are major sources of living bacteria [12]. Intriguingly, some of our works on food also allowed us to identify new bacterial species in daily food products. Indeed, we were the first to identify Gracilibacillus massiliensis sp. nov., a moderately halophilic Gram-positive bacterium in commercial table salt originating from the saline of Aigues-Mortes, Southern France [13]. Moreover, we identified in fermented cow's milk products from Algeria a new strain of Lactococcus garvieae, a bacterial species commonly used in the manufacture of fermented milk products and meats [14]. Also, a study conducted among 331 pilgrims departing from France to the 2011 Hajj revealed that 8% of them had previously drunk camel milk, most often in North Africa and Saudi Arabia; such milk has been involved in several zoonotic infections in humans, including brucellosis [15].
Antibiotic resistance genes and antibiotic-resistant bacteria
Antibiotic resistance genes and antibiotic-resistant bacteria in food-producing animals and vegetables
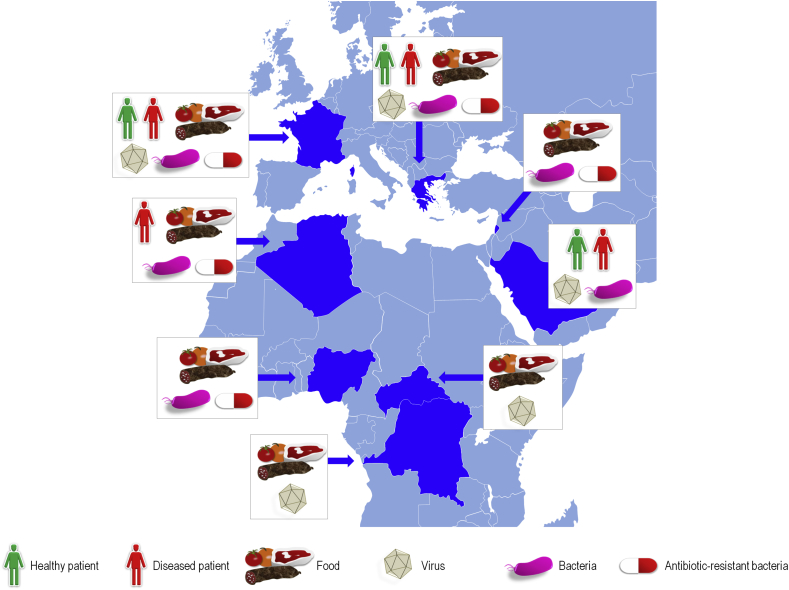
Our institute proved that food products and food-producing animals from the Mediterranean Basin are reservoirs for antibiotic resistance genes (ARGs) and antibiotic-resistant bacteria (ARB) (Fig. 1). Indeed, in Algeria and Lebanon, we identified chicken, cattle and pigs as possible reservoirs for CTX-M, SHV, TEM, CMY, VIM, OXA-23, OXA-58, aadA, qnrA, qnrB and mcr-1 producing bacteria [16], [17], [18], [19], [20], [21], [22]. Moreover, our work performed in Béjaïa, Algeria, allowed us to isolate from tomato, lettuce and parsley freshly purchased in markets three Klebsiella pneumoniae strains harbouring an OXA-48 carbapenemase [23].
Fig. 1.
Countries from Mediterranean Basin, Africa and Middle East where identifications of food-borne pathogens in food and/or humans involved teams from Institut Hospitalo-Universitaire Méditerranée Infection. Virus, bacteria and antibiotic schematics indicate that viruses, bacteria and/or antibiotic-resistant bacteria/genes have been identified in food (food item schematic) and/or in asymptomatic carriers (green-person schematic) and/or in diseased patients (red-person schematic). The food item schematic represents some examples of food items that may be involved in transmission of infectious agents, but they are not necessarily those involved or suspected for a given geographical area/country.
ARGs and ARB transmitted by food to humans
Some of our works on food and food-producing animals in the Mediterranean Basin led us to suspect food-mediated transmission of ARB that led to human infections. In a recent work, we identified that seven extended spectrum β-lactamase–producing Salmonella serotype Heidelberg detected in broiler chickens (five strains) and hospitalized humans (two strains) from Northeastern Algeria belonged to the same sequence type (ST15), suggesting a possible food-borne transmission of this strain between chicken and humans through the food chain [20]. However, as mentioned above, not all food-borne bacteria are responsible for diseases in humans. In these cases, ARGs and ARB can be asymptomatically acquired by humans through food consumption [24]. In our institute, asymptomatic food-borne acquisition of ARGs and ARB are particularly studied in pilgrims returning from the Hajj. In these populations, studies of the gut and pharyngeal carriage of ARB and ARGs in pilgrims before, during and after the Hajj enabled us to identify an increase in asymptomatic carriage of some ARGs and ARB during and after this pilgrimage, including blaOXA-51–like Acinetobacter baumannii and plasmid-mediated mcr-1 and New Delhi metallo-β-lactamase 5 ARGs [25]. These observations, coupled with the fact that some pilgrims carried the same ARB clones after returning from the Hajj [25], led us to suspect food as the main common reservoir of the ARG/ARB.
Parasites and fungi
Toxoplasma gondii–specific immunoglobulin G was found in the Var Department, Southeastern France, in 17% of muscle extract samples collected from 841 wild boars [26]. This indicates that consumption of raw or undercooked meat from wild boars is an important risk factor for infection with T. gondii. In addition, a total of 18 fungal species from the Ascomycota, Basidiomycota and Chytridiomycota phyla were detected in faeces collected from a 27-year-old white woman living in Marseille, France (16 by culture, seven by PCR), and many of these fungal species, for which the clinical significance of their presence in the human gut is unknown, may originate in food [27]. Eight fungi (Aspergillus flavipes, Beauveria bassiana, Isaria farinosa, Penicillium brevicompactum, Penicillium dipodomyicola, Penicillium camemberti, Climacocystis sp. and Malassezia restricta) were indeed found for the first time in human gut microbiota.
Viruses
Hepatitis E virus
During the past decade, research has revealed that hepatitis E virus (HEV) has a porcine reservoir and has caused food-borne autochthonous acute hepatitis. In France, we were the first to describe in 2009 the presence of this virus in two thirds of the pigs from a farm located in southeastern France [28]. Then in 2010 we linked HEV infection to consumption of pig liver sausage, which are often consumed uncooked [29]. This was largely confirmed afterward [30], [31]. Consumption of pig liver sausage purchased in southeastern France was linked to hepatitis E in Italy [32]. In southeastern France, consumption of pig liver sausage was documented in approximately half of the HEV infections [33]. We showed the stability of HEV prevalence among farm pigs and the emergence in France of new genotypes in these pigs and concurrently in humans [34]. Although HEV is mostly documented as involved in likely waterborne outbreaks in sub-Saharan Africa, we also detected HEV in farm pigs in this geographical area [35]. Concurrent with being revealed as having an extensive porcine reservoir, HEV was reported to cause acute liver failure and death [36], [37]. Furthermore, HEV was shown to be capable of determining chronic hepatitis in solid organ transplant recipients [38], [39]. We found it to be associated with cirrhosis [39], and it was thereafter confirmed that HEV could cause cirrhosis as soon as 2 years after infection [40]. Recently we described a case of hepatocellular carcinoma in a cirrhotic patient with chronic hepatitis E [41]. Furthermore, HEV infections have been increasingly linked to neurological disorders [42], [43].
Plant viruses
Although animal/human infections and vegetal infections are distinct fields, there are hints that plants and animals can be infected by the same, or at least similar, infectious agents. Plant-associated bacteria and viruses have been found in the gut. Notably, tobamoviruses were the most abundant RNA viruses in human stool in one metagenomic study [44]. We further identified pepper mild mottle virus (PMMoV) in 7% of the stool samples from 304 adult patients from Marseille University hospitals; the presence of this virus was associated with fever, abdominal pain and pruritus [45]. Concomitantly, we found that 57% of pepper samples or pepper-derived food products were positive for PMMoV RNA, and PMMoV load was dramatically high in Tabasco sauce. We then detected tobacco mosaic virus in 100% of tobacco cigarettes and in the saliva of 45% of smokers [46]. These findings, as well as those from other studies, question whether plant viruses from food are only transient passengers in humans, or whether they could interact with them [47].
Giant viruses
Mimiviruses, which are giant viruses of amoebae first described in 2003 in our laboratory, were recently found in oysters from Brazil [48], [49]. These mimiviruses, which are common in water and soil worldwide, have been suspected to cause pneumonia and were isolated from bronchoalveolar fluid and stools from patients with pneumonia [50], [51].
Conclusion
Food-borne infections appear to involve multiple sources and agents, and can be unpredictable in some cases. They require researchers to conduct investigations to decipher or confirm their sources and modes of transmission. Travel abroad is highly involved in the transfer of pathogens from one geographical area to another [52]. This is well exemplified by investigations conducted on the carriage and transmission of bacteria and viruses promoted by mass gatherings such as the Hajj [25], and it warrants collaborations between countries on both sides of the Mediterranean Sea, such as those (REMEDIER, GIRAFE) involving our institute.
Zoonoses are other major contributors to food-borne infections. Animal-derived products potentially harbouring various bacteria and viruses can also travel from one continent to another, as recently exemplified in our laboratory using metagenomics for African simian bushmeat seized at a French airport [53]. Thus, in this study, metagenomic sequencing of the DNA and RNA viromes detected sequences related to bacteriophages from families Siphoviridae and Myoviridae that can infect bacteria potentially pathogenic for humans such as Bacillus spp., Enterococcus spp. or Staphylococcus spp. Many sequences related to DNA from parasites such as Spirometra erinaceieuropaei, a tapeworm pathogenic for humans, were also identified. In addition, food-borne transmission of resistance-associated genes is increasingly reported.
Taken together, previous findings suggest that risk factors and pathogens associated with food-borne infections remain largely untapped and must be more extensively investigated, monitored and regularly reassessed. Furthermore, they warrant the implementation of new technology-based diagnostic tests, approaches and strategies [12], [54], [55], including culturomics, proteomics and genomics, to detect known infectious agents and discover new ones. Moreover, real-time surveillance tools, including some based on microbiology laboratory data such as those set up by our institution in southeastern France and in sub-Saharan Africa, deserve implementation [5], [56]. Finally, the exploration of the microbiota and its relationships with dietary patterns is another important research field being explored at the Institut Hospitalo-Universitaire Méditerranée Infection [57], [58].
Acknowledgements
Supported in part by the French government under the ‘Investissements d'avenir’ (Investments for the Future) programme managed by the Agence Nationale de la Recherche (Méditerranée Infection 10-IAHU-03).
Conflict of interest
None declared.
References
- 1.Newell D.G., Koopmans M., Verhoef L., Duizer E., Aidara-Kane A., Sprong H. Food-borne diseases—the challenges of 20 years ago still persist while new ones continue to emerge. Int J Food Microbiol. 2010;139(Suppl. 1):S3–S15. doi: 10.1016/j.ijfoodmicro.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study, 2016. Lancet. 2017;390:1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wheeler C., Vogt T.M., Armstrong G.L., Vaughan G., Weltman A., Nainan O.V. An outbreak of hepatitis A associated with green onions. N Engl J Med. 2005;353:890–897. doi: 10.1056/NEJMoa050855. [DOI] [PubMed] [Google Scholar]
- 4.Frank C., Werber D., Cramer J.P., Askar M., Faber M., an der H M. Epidemic profile of Shiga-toxin–producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 5.Huart M., Bedubourg G., Abat C., Colson P., Rolain J.M., Chaudet H. Implementation and initial analysis of a laboratory-based weekly biosurveillance system, Provence-Alpes-Cote d’Azur, France. Emerg Infect Dis. 2017;23:582–589. doi: 10.3201/eid2304.161399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abat C., Huart M., Garcia V., Dubourg G., Raoult D. Enterococcus faecalis urinary-tract infections: do they have a zoonotic origin? J Infect. 2016;73:305–313. doi: 10.1016/j.jinf.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Delaunay E., Abat C., Rolain J.M. Enterococcus cecorum human infection. France New Microbe New Infect. 2015;7:50–51. doi: 10.1016/j.nmni.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia V., Abat C., Rolain J.M. Report of the first Vagococcus lutrae human infection, Marseille, France. New Microbe New Infect. 2016;9:56–57. doi: 10.1016/j.nmni.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fishbein D.B., Raoult D. A cluster of Coxiella burnetii infections associated with exposure to vaccinated goats and their unpasteurized dairy products. Am J Trop Med Hyg. 1992;47:35–40. doi: 10.4269/ajtmh.1992.47.35. [DOI] [PubMed] [Google Scholar]
- 10.Maltezou H.C., Constantopoulou I., Kallergi C., Vlahou V., Georgakopoulos D., Kafetzis D.A. Q fever in children in Greece. Am J Trop Med Hyg. 2004;70:540–544. [PubMed] [Google Scholar]
- 11.Eldin C., Angelakis E., Renvoise A., Raoult D. Coxiella burnetii DNA, but not viable bacteria, in dairy products in France. Am J Trop Med Hyg. 2013;88:765–769. doi: 10.4269/ajtmh.12-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angelakis E., Million M., Henry M., Raoult D. Rapid and accurate bacterial identification in probiotics and yoghurts by MALDI-TOF mass spectrometry. J Food Sci. 2011;76:M568–M572. doi: 10.1111/j.1750-3841.2011.02369.x. [DOI] [PubMed] [Google Scholar]
- 13.Diop A., Khelaifia S., Armstrong N., Labas N., Fournier P.E., Raoult D. Microbial culturomics unravels the halophilic microbiota repertoire of table salt: description of Gracilibacillus massiliensis sp. nov. Microb Ecol Health Dis. 2016;27:32049. doi: 10.3402/mehd.v27.32049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moumene M., Drissi F., Croce O., Djebbari B., Robert C., Angelakis E. Complete genome sequence and description of Lactococcus garvieae M14 isolated from Algerian fermented milk. New Microbe New Infect. 2016;10:122–131. doi: 10.1016/j.nmni.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautret P., Benkouiten S., Gaillard C., Parola P., Brouqui P. Camel milk–associated infection risk perception and knowledge in French Hajj pilgrims. Vector Borne Zoonotic Dis. 2013;13:425–427. doi: 10.1089/vbz.2012.1010. [DOI] [PubMed] [Google Scholar]
- 16.Al B.C., Dabboussi F., Hamze M., Rolain J.M. Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J Antimicrob Chemother. 2015;70:950–951. doi: 10.1093/jac/dku469. [DOI] [PubMed] [Google Scholar]
- 17.Al B.C., Olaitan A.O., Dabboussi F., Hamze M., Rolain J.M. Emergence of OXA-48–producing Escherichia coli clone ST38 in fowl. Antimicrob Agents Chemother. 2015;59:745–746. doi: 10.1128/AAC.03552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olaitan A.O., Chabou S., Okdah L., Morand S., Rolain J.M. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16:292–293. doi: 10.1016/S1473-3099(15)00540-X. [DOI] [PubMed] [Google Scholar]
- 19.Belmahdi M., Bakour S., Al B.C., Touati A., Rolain J.M. Molecular characterisation of extended-spectrum beta-lactamase– and plasmid AmpC-producing Escherichia coli strains isolated from broilers in Bejaia, Algeria. J Glob Antimicrob Resist. 2016;6:108–112. doi: 10.1016/j.jgar.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Djeffal S., Bakour S., Mamache B., Elgroud R., Agabou A., Chabou S. Prevalence and clonal relationship of ESBL-producing Salmonella strains from humans and poultry in northeastern Algeria. BMC Vet Res. 2017;13:132. doi: 10.1186/s12917-017-1050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selma C., Hamza L., Bernard D., Atef A., Rolain J.M. Prevalence of extended-spectrum beta lactamase and carbapenemase-encoding genes in poultry feces from Algeria and Marseille, France. J Glob Antimicrob Resist. 2018;13:28–32. doi: 10.1016/j.jgar.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Dandachi I., Leangapichart T., Daoud Z., Rolain J.M. First detection of mcr-1 plasmid-mediated colistin-resistant Escherichia coli in Lebanese poultry. J Glob Antimicrob Resist. 2018;12:137–138. doi: 10.1016/j.jgar.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Touati A., Mairi A., Baloul Y., Lalaoui R., Bakour S., Thighilt L. First detection of Klebsiella pneumoniae producing OXA-48 in fresh vegetables from Bejaia city, Algeria. J Glob Antimicrob Resist. 2017;9:17–18. doi: 10.1016/j.jgar.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Rolain J.M. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol. 2013;4:173. doi: 10.3389/fmicb.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leangapichart T., Rolain J.M., Memish Z.A., Al-Tawfiq J.A., Gautret P. Emergence of drug resistant bacteria at the Hajj: a systematic review. Travel Med Infect Dis. 2017;18:3–17. doi: 10.1016/j.tmaid.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Roqueplo C., Blaga R., Jean-Lou M., Vallee I., Davoust B. Seroprevalence of Toxoplasma gondii in hunted wild boars (Sus scrofa) from southeastern France. Folia Parasitol (Praha) 2017;64 doi: 10.14411/fp.2017.003. [DOI] [PubMed] [Google Scholar]
- 27.Gouba N., Raoult D., Drancourt M. Plant and fungal diversity in gut microbiota as revealed by molecular and culture investigations. PLoS One. 2013;8:e59474. doi: 10.1371/journal.pone.0059474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaba M., Davoust B., Marie J.L., Barthet M., Henry M., Tamalet C. Frequent transmission of hepatitis E virus among piglets in farms in Southern France. J Med Virol. 2009;81:1750–1759. doi: 10.1002/jmv.21553. [DOI] [PubMed] [Google Scholar]
- 29.Colson P., Borentain P., Queyriaux B., Kaba M., Moal V., Gallian P. Pig liver sausage as a source of hepatitis E virus transmission to humans. J Infect Dis. 2010;202:825–834. doi: 10.1086/655898. [DOI] [PubMed] [Google Scholar]
- 30.Mansuy J.M., Bendall R., Legrand-Abravanel F., Saune K., Miedouge M., Ellis V. Hepatitis E virus antibodies in blood donors, France. Emerg Infect Dis. 2011;17:2309–2312. doi: 10.3201/eid1712.110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavio N., Merbah T., Thebault A. Frequent hepatitis E virus contamination in food containing raw pork liver, France. Emerg Infect Dis. 2014;20:1925–1927. doi: 10.3201/eid2011.140891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garbuglia A.R., Alessandrini A.I., Pavio N., Tesse S., Grignolo S., Viscoli C. Male patient with acute hepatitis E in Genoa, Italy: figatelli (pork liver sausage) as probable source of the infection. Clin Microbiol Infect. 2015;21:e4–e6. doi: 10.1016/j.cmi.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Colson P., Romanet P., Moal V., Borentain P., Purgus R., Benezech A. Autochthonous infections with hepatitis E virus genotype 4, France. Emerg Infect Dis. 2012;18:1361–1364. doi: 10.3201/eid1808.111827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colson P., Saint-Jacques P., Ferretti A., Davoust B. Hepatitis E virus of subtype 3a in a pig farm, south-eastern France. Zoonoses Public Health. 2015;62:593–598. doi: 10.1111/zph.12211. [DOI] [PubMed] [Google Scholar]
- 35.Kaba M., Colson P., Musongela J.P., Tshilolo L., Davoust B. Detection of hepatitis E virus of genotype 3 in a farm pig in Kinshasa (Democratic Republic of the Congo) Infect Genet Evol. 2010;10:154–157. doi: 10.1016/j.meegid.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Borentain P., Colson P., Gregoire E., Allard G., Gerolami R. Letter: liver transplantation for acute HEV infection in cirrhotic patients in France. Aliment Pharmacol Ther. 2017;46:477–478. doi: 10.1111/apt.14188. [DOI] [PubMed] [Google Scholar]
- 37.Colson P., Raoult D. Autochthonous hepatitis E: a common and fatal but neglected emerging disease in France. Clin Microbiol Infect. 2017;23:898–899. doi: 10.1016/j.cmi.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamar N., Selves J., Mansuy J.M., Ouezzani L., Peron J.M., Guitard J. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med. 2008;358:811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 39.Gerolami R., Moal V., Colson P. Chronic hepatitis E with cirrhosis in a kidney-transplant recipient. N Engl J Med. 2008;358:859–860. doi: 10.1056/NEJMc0708687. [DOI] [PubMed] [Google Scholar]
- 40.Dalton H.R., Bendall R.P., Keane F.E., Tedder R.S., Ijaz S. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med. 2009;361:1025–1027. doi: 10.1056/NEJMc0903778. [DOI] [PubMed] [Google Scholar]
- 41.Borentain P., Colson P., Bolon E., Gauchez P., Coso D., Gerolami R. Hepatocellular carcinoma complicating hepatitis E virus–related cirrhosis. Hepatology. 2018;67:446–448. doi: 10.1002/hep.29508. [DOI] [PubMed] [Google Scholar]
- 42.van Eijk J.J.J., Dalton H.R., Ripellino P., Madden R.G., Jones C., Fritz M. Clinical phenotype and outcome of hepatitis E virus–associated neuralgic amyotrophy. Neurology. 2017;89:909–917. doi: 10.1212/WNL.0000000000004297. [DOI] [PubMed] [Google Scholar]
- 43.Despierres L.A., Kaphan E., Attarian S., Cohen-Bacrie S., Pelletier J., Pouget J. Neurologic disorders and hepatitis E, France, 2010. Emerg Infect Dis. 2011;17:1510–1512. doi: 10.3201/eid1708.102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang T., Breitbart M., Lee W.H., Run J.Q., Wei C.L., Soh S.W. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colson P., Richet H., Desnues C., Balique F., Moal V., Grob J.J. Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PLoS One. 2010;5:e10041. doi: 10.1371/journal.pone.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balique F., Colson P., Raoult D. Tobacco mosaic virus in cigarettes and saliva of smokers. J Clin Virol. 2012;55:374–376. doi: 10.1016/j.jcv.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 47.Balique F., Lecoq H., Raoult D., Colson P. Can plant viruses cross the kingdom border and be pathogenic to humans? Viruses. 2015;20:2074–2098. doi: 10.3390/v7042074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Assis F.L., Bajrai L., Abrahao J.S., Kroon E.G., Dornas F.P., Andrade K.R. Pan-genome analysis of Brazilian lineage A amoebal mimiviruses. Viruses. 2015;7:3483–3499. doi: 10.3390/v7072782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrade K.R., Boratto P.P., Rodrigues F.P., Silva L.C., Dornas F.P., Pilotto M.R. Oysters as hot spots for mimivirus isolation. Arch Virol. 2015;160:477–482. doi: 10.1007/s00705-014-2257-2. [DOI] [PubMed] [Google Scholar]
- 50.Saadi H., Pagnier I., Colson P., Cherif J.K., Beji M., Boughalmi M. First isolation of Mimivirus in a patient with pneumonia. Clin Infect Dis. 2013;57:e127–e134. doi: 10.1093/cid/cit354. [DOI] [PubMed] [Google Scholar]
- 51.Colson P., La Scola B., Levasseur A., Caetano-Anolles G., Raoult D. Mimivirus: leading the way in the discovery of giant viruses of amoebae. Nat Rev Microbiol. 2017;15:243–254. doi: 10.1038/nrmicro.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parola P., Soula G., Gazin P., Foucault C., Delmont J., Brouqui P. Fever in travelers returning from tropical areas: prospective observational study of 613 cases hospitalised in Marseilles, France, 1999–2003. Travel Med Infect Dis. 2006;4:61–70. doi: 10.1016/j.tmaid.2005.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Temmam S., Davoust B., Chaber A.L., Lignereux Y., Michelle C., Monteil-Bouchard S. Screening for viral pathogens in African simian bushmeat seized at a French airport. Transbound Emerg Dis. 2017;64:1159–1167. doi: 10.1111/tbed.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersen S.C., Hoorfar J. Surveillance of foodborne pathogens: towards diagnostic metagenomics of fecal samples. Genes (Basel) 2018;9:E14. doi: 10.3390/genes9010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abat C., Colson P., Chaudet H., Rolain J.M., Bassene H., Diallo A. Implementation of syndromic surveillance systems in two rural villages in Senegal. PLoS Negl Trop Dis. 2016;10:e0005212. doi: 10.1371/journal.pntd.0005212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Angelakis E., Yasir M., Bachar D., Azhar E.I., Lagier J.C., Bibi F. Gut microbiome and dietary patterns in different Saudi populations and monkeys. Sci Rep. 2016;6:32191. doi: 10.1038/srep32191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Million M., Diallo A., Raoult D. Gut microbiota and malnutrition. Microb Pathog. 2017;106:127–138. doi: 10.1016/j.micpath.2016.02.003. [DOI] [PubMed] [Google Scholar]