Abstract
The genetic and morphologic similarities between primates and humans means that much information obtained from primates may be applied to humans, and vice versa. However, habitat loss, hunting and the continued presence of humans have a negative effect on the biology and behaviour of almost all nonhuman primates. Noninvasive methods such as stool collection are among the safest alternative ways to study the multiple aspects of the biology of primates. Many epidemiologic issues (e.g. pathogen detection, microbiota studies) may be easily studied using stool samples from primates. Primates are undoubtedly among the first candidates suspected of becoming the source of one of the next emerging epidemic of zoonotic origin, as has already been observed with HIV, malaria and monkeypox. The Institut Hospitalo-Universitaire Méditerranée Infection in Marseille actively participates in the study, mostly epidemiologic, of nonhuman primates, using mostly stool samples.
Keywords: Culturomics, faecal samples, great apes, microbiota, nonhuman primates, zoonoses
Introduction
In 1761, in Lyon, the founder of the first veterinary school in the world, Claude Bourgelat, declared, ‘Knowing the intimacy of the relations that exist between the human machine and the animal machine, the veterinary sciences and the medical sciences can only enlighten and mutually improve each other’ [1]. The concept of One Health is not new; in the last century it has been at the centre of the action led by great innovator and humanist Charles Mérieux. According to him, there are no boundaries between animal medicine and human medicine [2]. For 30 years, after a number of health emergencies (bovine spongiform encephalopathy, avian influenza, coronavirus infection), international organizations such as the World Health Organization, the World Organization for Animal Health and the Food and Agriculture Organization as well as the public have become aware of constant interactions between the environment (including animals and plants), climate and human populations, and their consequences for public health.
Domestic animals are reservoirs of zoonotic agents which are, for the most part, already known and for which the risks of transmission to humans are relatively controlled. This is not the case for wildlife, which, despite declining biodiversity, are important sources of unknown and potentially dangerous infectious agents when they cross the species barrier [3]. Nonhuman primates (NHPs) are the animals most genetically close to humans. We also share with NHPs a similar microbial flora. However, it is thought that they are frequently responsible for the emergence of new human infections. Primates represent an order of about 280 species of mammals, currently distributed in the intertropical zones of the planet. Mostly arboreal and frugivorous, though there are many exceptions, their decline is directly linked to the growing rate of deforestation. In fact, contacts between NHPs and humans have become more frequent than ever. The study of wild NHPs is therefore an important issue to understand and prevent future epidemics. The Institut Hospitalo-Universitaire (IHU) Méditerranée Infection in Marseille, France, specializes in zoonotic and tropical infections, with NHPs being one of the major axes of research.
IHU collection of faecal samples from NHPs
Stool specimens from wild apes have been considered reliable samples for determining the presence of pathogens. This noninvasive detection of pathogens in endangered species opens up new possibilities in the molecular epidemiology and evolutionary analysis of infectious diseases.
In the early 2000s, D. Raoult, director of the IHU Méditerranée Infection, began a collaboration with J.-P. Gonzalez, director of the International Center for Medical Research, in Gabon and then, with E. Delaporte (French Research Institute for Development, IRD, Montpellier-Yaoundé). These collaborations initiated the stool sample collection of IHU (reaching more than 1500 samples) which started out with chimpanzees and gorillas from Cameroon and Gabon, and bonobos from the Democratic Republic of Congo.
During the last 5 years, we have, in partnership with local institutions organized several field missions in Africa and the Amazon to collect faeces. In Senegal, the collection of faeces of green and red monkeys and baboons took place in the Niokolo-Koba National Park and the Sine-Saloum region. At the Dindéfello reserve in southeastern Senegal, with the support of the Jane Goodall Foundation, we collected faeces from wild chimpanzees. In 2015, in Congo Brazzaville, we collaborated with the National Laboratory of Public Health (J. Akiana) and collected gorilla faeces at the Lésio-Louna-Léfini reserve, which were safeguarded by the Aspinall Foundation. In 2017, we began a collaboration with the Association of Friends of Bonobos, founded by C. André. This led to collections of bonobos faeces from the orphan reserve near Kinshasa, Democratic Republic of Congo, as well as the release site of bonobos in the rain forest in the Equator region. Finally, the faeces of wild hamadryas baboons from Saudi Arabia were also collected in collaboration with a Saudi microbiologist (E. Azhar).
When stool samples are taken from the wild, environmental contamination before and during sampling may occur. In order to minimize this bias, we collected only very fresh samples (within minutes after being emitted) and used single-use equipment.
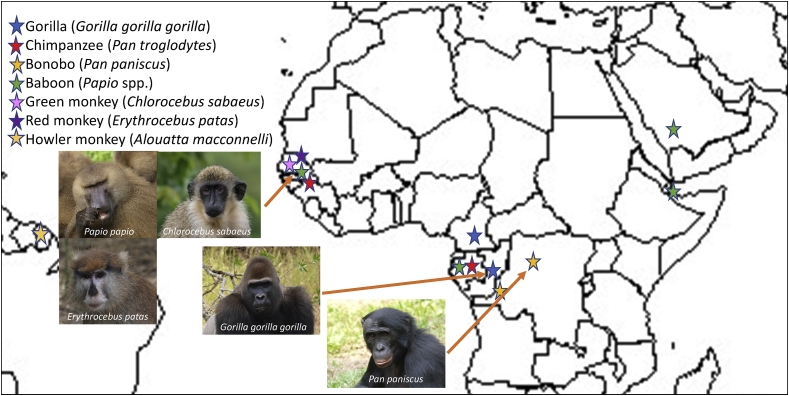
All stool samples were imported from eight countries (Fig. 1) to the IHU, either refrigerated or in alcohol or in a culture media ad hoc, per biosafety rules in application in European countries. For green monkeys and baboons, we could perform other types of sampling after capture of individuals in large traps and teleanesthesia. The collections of samples are therefore various: nasal, rectal, vaginal and oral swabs; collection of ectoparasites (lice, ticks, fleas); skin biopsy samples; and blood, saliva and semen samples. The collection of this type of invasive samples is not possible for great apes from an ethical point of view. Faecal sample collection (as well as necropsy, performed only rarely, and then on animals found dead) is the best approach to study the infectious diseases of great apes. Indeed, the IHU has a veterinary research centre. This centre possesses the material and human resources to intervene in the field in partnership with local institutions.
Fig. 1.
Origin of samples from nonhuman primates studied at Institut Hospitalo-Universitaire Méditerranée Infection, Marseille. Images by O. Mediannikov.
Targeted research of zoonotic agents
The rapid population growth in wooded tropical regions leads to rapid deforestation and the creation of large agricultural areas, mainly in sub-Saharan Africa. This anthropization of the environment stimulates closer contacts between NHPs and humans, which leads to the transmission of pathogens in both directions [4]. The great apes (bonobos, chimpanzees and gorillas) are relevant models of the porosity of the species barrier, enabling or preventing the transmission of microorganisms [5]). The great apes have been identified as a direct source of some emergent pathogens in human populations [6]. The gorilla is the source of human infection with Plasmodium falciparum, the parasite responsible for the most common and virulent form of malaria [7], [8]. Several studies have shown that simian immunodeficiency virus infecting chimpanzees and gorillas can cross the species barrier. These viruses play a role in the origin of the modern HIV infection pandemic [9]. HIV and Plasmodium nucleic acids are often found in gorilla faeces. Studies performed at the IHU allowed us to document enzootic simian immunodeficiency virus infection in West African green monkeys Chlorocebus sabaeus (P. Colson, personal communication).
For Ebola virus, wild great apes can serve as intermediate amplifiers as well as sentinels, because they are susceptible to infection and die quickly [6]. NHPs can rarely transmit serious human diseases such as tuberculosis and rabies [10], [11]. The monkeypox virus is an emergent virus; high risk of dissemination may be linked to the increase in international transport [12]. However, many bacteria and parasites, including Streptococcus pneumoniae, Bacillus anthracis, Schistosoma mansoni and Necator americanus, may affect the great apes just as they affect us [13], [14], [15]. So far, Tropheryma whipplei has not been identified in NHP faeces [16]. To examine whether gorillas harbour Leishmania species, faecal samples from wild western lowland gorillas (Gorilla gorilla gorilla) of Cameroon were screened in the IHU lab [17]. Among the 91 samples analysed, 12 contained Leishmania parasites (molecular identity with L. major). Next, fluorescence in-situ hybridization was performed to visualize L. major parasites in faecal samples from the gorillas. Both promastigote and amastigote forms of the parasite were found. In our lab, for the first time also, Rickettsia felis was detected in wild-living ape faeces [18].
During several expeditions in Senegal, IHU researchers were lucky to discover an enzootic outbreak of venereal treponematosis in Chlorocebus sabaeus and Papio papio baboons. It seems that this evidently venereal emerging outbreak followed the evolution of nonpathogenic or low-pathogenic strain of Treponema pallidum (Fribourg-Blanc) in West African NHPs [19]. This observation may be easily paralleled with the evolution of human syphilis.
Comparative study of microbiota of primates
A diverse range of pathogenic bacteria and/or eukaryotes infects NHPs as well as humans. Studying the microorganisms of the digestive tract of NHPs using stool samples allowed to obtain the most comprehensive diversity possible using both major approaches: high-throughput culture (culturomics) and modern molecular analyses. The renewal of culture, illustrated by culturomics, has proposed for 5 years to study the digestive microbiota using high-throughput culture techniques with identification by matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) or by sequencing the 16S gene for unidentified colonies [20]. By applying culturomics in a study of gorilla faecal samples, 147 bacterial species have been identified, including five new species [21]. Many opportunistic pathogens have been observed. In addition, molecular analyses identified 87 eukaryotic species with 52 fungi, ten protozoa, four nematodes and 21 plant species. We have also reported the presence of pathogenic fungi and parasites [22]. Several new microbial species have been discovered in gorilla faeces, which were cultured and genetically characterized [23], [24], [25]. This demonstrates the complementarity of culture and molecular methods.
The role of diet in primate evolution has not yet been extensively explored. Faeces analysis helps us better understand the diet of NHPs. In our lab, insects have been found in stool samples of gorillas using molecular analysis [26]. We therefore suspect that they ate insects. In Saudi Arabia, the human microbiota and that of monkeys have been compared using 16S rDNA sequencing [27]. The gut microbiome of baboons was more like that of Bedouins than that of urban Saudis, probably due the dietary overlap between baboons and Bedouins.
Metagenomics revolutionized our vision of the microbiome diversity and contributes to elucidate the role of these microorganisms in different diseases [20]. Consequently, using the faeces of NHPs is a reasonable strategy for studying the diversity of the repertoire of viruses and bacteria in wildlife. On the basis of our samples, two successful studies were performed by our laboratory by using metagenomics. Firstly, a large proportion of novel variants of double-stranded RNA viruses (genus Picobirnaviruses) were detected in the stools of wild gorillas [28]. Secondly, our comparative studies of the viromes from the stools of green monkeys (Clorocebus sabeus), baboons (Papio papio) and howler monkeys (Alouatta macconnelli) revealed the heterogeneity of the viromes dominated by bacteriophages for baboons and howler monkey, contrary to the viromes of green monkeys, which contained a majority of eukaryotic viruses of the family Circoviridae (C. Desnues, personal communication). Taken together, our results highlighted the need for virologic surveillance against potential emerging zoonotic viruses.
MALDI-TOF MS is an emerging tool that has dramatically changed the practical use and availability of mass spectrometry methods. It has already been used for identification of bacterial, archaeal and fungal isolates, arthropod species, eukaryotic tissues and many others [29], [30]. In our laboratory, we successfully tested MALDI-TOF MS with stool samples and found that the spectra of such specimens collected from different NHP species were quite specific. Consequently, the origin of the stool sample was identified (D. Raoult and N. Dione, personal communication). This approach is fast, easy and cheap compared to all other methods for the identification of the origin of stool samples collected in the environment.
Conclusion
The IHU continues to study NHPs, and specifically their role in harbouring potential emerging pathogens, which could affect both humans and animals. As soon as there was undeniable evidence that habitat loss, hunting and the continued presence of humans had a negative effect on the biology and behaviour of almost all NHPs, noninvasive methods such as the study of stool samples (e.g. pathogens detection, microbiota studies) became of the utmost importance.
By taking into consideration the great genetic and morphologic similarities between primates and humans, much information obtained from primates may be applied to humans, and vice versa. Moreover, primates might potentially become a source of a future emerging epidemic of zoonotic origin, as has already been seen with HIV, malaria and monkeypox.
Acknowledgements
This work has received financial support from the French Government through the Agence Nationale pour la Recherche, including the “Programme d'Investissement d'Avenir” under the reference Méditerranée Infection 10-IAHU-03. This work was supported also by the Région Provence-Alpes-Côte d'Azur and European funding FEDER PRIMI.
Conflict of interest
None declared.
References
- 1.Bourgelat C. Imprimerie royale; 1777. Réglements pour les écoles royales vétérinaires de France. [Google Scholar]
- 2.Francioli M. Editions du Rocher; Paris: 2017. Charles Mérieux: l'homme qui voulait vacciner tous les enfants du monde. [Google Scholar]
- 3.Morse S.S., Mazet J.A., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380(9857):1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mossoun A., Pauly M., Akoua-Koffi C., Couacy-Hymann E., Leendertz S.A., Anoh A.E. Contact to non-human primates and risk factors for zoonotic disease emergence in the Tai region, Cote d'Ivoire. Ecohealth. 2015;12:580–591. doi: 10.1007/s10393-015-1056-x. [DOI] [PubMed] [Google Scholar]
- 5.Keita M.B., Hamad I., Bittar F. Looking in apes as a source of human pathogens. Microb Pathog. 2014;77:149–154. doi: 10.1016/j.micpath.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Calvignac-Spencer S., Leendertz S.A., Gillespie T.R., Leendertz F.H. Wild great apes as sentinels and sources of infectious disease. Clin Microbiol Infect. 2012;18:521–527. doi: 10.1111/j.1469-0691.2012.03816.x. [DOI] [PubMed] [Google Scholar]
- 7.Duval L., Fourment M., Nerrienet E., Rousset D., Sadeuh S.A., Goodman S.M. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc Natl Acad Sci U S A. 2010;107:10561–10566. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W., Li Y., Learn G.H., Rudicell R.S., Robertson J.D., Keele B.F. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467(7314):420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keele B.F., Van H.F., Li Y., Bailes E., Takehisa J., Santiago M.L. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313(5786):523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghodbane R., Drancourt M. Non-human sources of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2013;93:589–595. doi: 10.1016/j.tube.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Gautret P., Blanton J., Dacheux L., Ribadeau-Dumas F., Brouqui P., Parola P. Rabies in nonhuman primates and potential for transmission to humans: a literature review and examination of selected French national data. PLoS Negl Trop Dis. 2014;8 doi: 10.1371/journal.pntd.0002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radonic A., Metzger S., Dabrowski P.W., Couacy-Hymann E., Schuenadel L., Kurth A. Fatal monkeypox in wild-living sooty mangabey, Cote d'Ivoire, 2012. Emerg Infect Dis. 2014;20:1009–1011. doi: 10.3201/eid2006.131329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raoult D. The apes as reservoir of human pathogens. Clin Microbiol Infect. 2012;18:513. doi: 10.1111/j.1469-0691.2012.03810.x. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann C., Zimmermann F., Biek R., Kuehl H., Nowak K., Mundry R. Persistent anthrax as a major driver of wildlife mortality in a tropical rainforest. Nature. 2017;548(7665):82–86. doi: 10.1038/nature23309. [DOI] [PubMed] [Google Scholar]
- 15.Standley C.J., Mugisha L., Dobson A.P., Stothard J.R. Zoonotic schistosomiasis in non-human primates: past, present and future activities at the human–wildlife interface in Africa. J Helminthol. 2012;86:131–140. doi: 10.1017/S0022149X12000028. [DOI] [PubMed] [Google Scholar]
- 16.Fenollar F., Trani M., Davoust B., Salle B., Birg M.L., Rolain J.M. Prevalence of asymptomatic Tropheryma whipplei carriage among humans and nonhuman primates. J Infect Dis. 2008;197:880–887. doi: 10.1086/528693. [DOI] [PubMed] [Google Scholar]
- 17.Hamad I., Forestier C.L., Peeters M., Delaporte E., Raoult D., Bittar F. Wild gorillas as a potential reservoir of Leishmania major. J Infect Dis. 2015;211:267–273. doi: 10.1093/infdis/jiu380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keita A.K., Socolovschi C., Ahuka-Mundeke S., Ratmanov P., Butel C., Ayouba A. Molecular evidence for the presence of Rickettsia felis in the feces of wild-living African apes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054679. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Knauf S., Gogarten J.F., Schuenemann V.J., de Nys H.M., Düx A., Strouhal M. Nonhuman primates across sub-Saharan Africa are infected with the yaws bacterium Treponema Pallidum Subsp. pertenue. Emerg Microbes Infect. 2018;7:157. doi: 10.1038/s41426-018-0156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La S.B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bittar F., Keita M.B., Lagier J.C., Peeters M., Delaporte E., Raoult D. Gorilla gorilla gorilla gut: a potential reservoir of pathogenic bacteria as revealed using culturomics and molecular tools. Sci Rep. 2014;4:7174. doi: 10.1038/srep07174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamad I., Keita M.B., Peeters M., Delaporte E., Raoult D., Bittar F. Pathogenic eukaryotes in gut microbiota of western lowland gorillas as revealed by molecular survey. Sci Rep. 2014;4:6417. doi: 10.1038/srep06417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadjadj L., Rathored J., Keita M.B., Michelle C., Levasseur A., Raoult D. Non contiguous-finished genome sequence and description of Microbacterium gorillae sp. nov. Stand Genomic Sci. 2016;11:32. doi: 10.1186/s40793-016-0152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keita M.B., Padhmanabhan R., Caputo A., Robert C., Delaporte E., Raoult D. Non-contiguous finished genome sequence and description of Gorillibacterium massiliense gen. nov, sp. nov., a new member of the family Paenibacillaceae. Stand Genomic Sci. 2014;9:807–820. doi: 10.4056/sigs.5199182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keita M.B., Padhmananabhan R., Caputo A., Robert C., Delaporte E., Raoult D. Non-contiguous finished genome sequence and description of Paenibacillus gorillae sp. nov. Stand Genomic Sci. 2014;9:1031–1045. doi: 10.4056/sigs.5189179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamad I., Delaporte E., Raoult D., Bittar F. Detection of termites and other insects consumed by African great apes using molecular fecal analysis. Sci Rep. 2014;4:4478. doi: 10.1038/srep04478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angelakis E., Yasir M., Bachar D., Azhar E.I., Lagier J.C., Bibi F. Gut microbiome and dietary patterns in different Saudi populations and monkeys. Sci Rep. 2016;6:32191. doi: 10.1038/srep32191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duraisamy R., Akiana J., Davoust B., Mediannikov O., Michelle C., Robert C. Detection of novel RNA viruses from free-living gorillas, Republic of the Congo: genetic diversity of picobirnaviruses. Virus Genes. 2018;54:256–271. doi: 10.1007/s11262-018-1543-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fall B., Lo C.I., Samb-Ba B., Perrot N., Diawara S., Gueye M.W. The ongoing revolution of MALDI-TOF mass spectrometry for microbiology reaches tropical Africa. Am J Trop Med Hyg. 2015;92:641–647. doi: 10.4269/ajtmh.14-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yssouf A., Almeras L., Raoult D., Parola P. Emerging tools for identification of arthropod vectors. Future Microbiol. 2016;11:549–566. doi: 10.2217/fmb.16.5. [DOI] [PubMed] [Google Scholar]