Abstract
Surveillance of antibiotic resistance has become a public global concern after the rapid worldwide dissemination of several antibiotic resistance genes. Here we report the role of the Institut Hospitalo-Universitaire Méditerranée Infection created in 2011 in the identification and description of multidrug-resistant bacteria thanks to collaborations and training of students from the Mediterranean basin and from African countries. Since the creation of the institute, 95 students and researchers have come from 19 different countries from these areas to characterize 6359 bacterial isolates from 7280 samples from humans (64%), animals (28%) and the environment (8%). Most bacterial isolates studied were Gram-negative bacteria (n = 5588; 87.9%), mostly from Algeria (n = 4190), Lebanon (n = 946), Greece (n = 610), Saudi Arabia (n = 299) and Senegal (n = 278). Antibiotic resistance was diversified with the detection and characterization of extended-spectrum β-lactamases, carbapenemases and resistance to colistin, vancomycin and methicillin. All those studies led to 97 indexed international scientific papers. Over the last 6 years, our institute has created a huge network of collaborations by training students that plays a major role in the surveillance of resistance to antibiotics in these countries.
Keywords: Africa, Antibiotic resistance, IHU Méditerranée Infection, Mediterranean, Multi-drug resistance
Introduction
During the last decade, antibiotic resistance has become one of the major public health priorities in the world [1] because of the emergence of new mechanisms of resistance. Moreover, the massive media coverage has tended to predict of thousands of human deaths every year [2]. However, recent epidemiologic data from our institution demonstrate that the level of antibiotic resistance for the most common bacterial species of clinical interest did not significantly change over the last 15 years in Marseille, France [3], [4]. Similarly, we found a huge disparity between mortality attributable to antibiotic resistance using simple model estimations and empirical data of true deaths in our institution [5]. Data on the level of antibiotic resistance in Europe show disparities between countries and bacterial species for certain antibiotics; for example, resistance to carbapenems is much more frequent in Romania, Italy and Greece [3]. It appears from those studies that a better understanding and surveillance of antibiotic resistance at the local and national levels is critical to manage antibiotic-resistant bacterial infections in the future [5]. However, data on antibiotic resistance and surveillance of the emergence and spread of new mechanisms of resistance in the Mediterranean basin and in African countries were lacking in most of those countries until now.
Here we report the specific and unique role of the Institut Hospitalo-Universitaire Méditerranée Infection (IHU-MI), created in November 2011, in the identification, description and surveillance of multidrug-resistant bacteria thanks to collaborations among and training of students coming from the Mediterranean basin and from African countries in our institute. The majority of students who come to our institute for surveillance and analysis of antibiotic resistance came with their own bacterial isolates from their countries.
Methods
This study analyses data collected from 2011 (the date of creation of the IHU-MI) through the completion of this article in February 2018. The number of students by level of graduation and by country of origin per year was sorted from our administrative database of students and scientific visitors during the study period from the team dedicated to antibiotic resistance research (JMR team). Students from the Mediterranean basin and Africa were counted from this primary list and were sorted by level of graduation (master's degree, PhD, postdoc and scientific visitors) and by country. Because some students stayed at our institute both for master's and PhD courses, we deduplicated the total count. The number of students present per year in the team was also calculated from this list to show the student kinetics of reception per year.
All students facing antibiotic resistance in their country came with their own isolates to analyse them as a course training. Most of them continued to collaborate with our institute, resulting in real-time surveillance of antibiotic resistance according to their field of research (humans, animals or the environment). Initially there was no rationale for the recruitment and analysis of the samples because no data existed at the beginning of this network. Now, however, the follow-up of antibiotic resistance is mainly focused on the current antibiotic resistance situation. For each epidemiologic study, the number and type of samples and/or bacterial isolates and the country of origin were counted, and data were presented in a single table, with all data provided by country.
Antibiotic resistance for each sample or bacterial isolates was studied using the same procedure. Antibiotic resistance was assessed either directly from samples by PCR or from bacterial isolates by culture and molecular assays. The first step consisted of sample culture and isolation of strains on specific agar media: Columbia agar with 5% sheep's blood, trypticase soy agar or MacConkey (bioMérieux, Marcy l’Etoile, France) with or without addition of antibiotics. All collected strains were subjected to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) for identification [6]. Antibiotic susceptibility testing (AST) was performed using the disc diffusion method on Mueller-Hinton medium agar for phenotypic characterization of the mechanism of resistance. Specific panels of antibiotics were tested according to the bacteria species (e.g. Enterobacteriaceae, nonfermentative Gram-negative bacteria, Gram-positive bacteria). Then AST results were interpreted according to European Committee on Antimicrobial Susceptibility Testing guidelines [7]. Genotypic identification of resistance genes were screened by real-time quantitative PCR and confirmed by standard PCR and sequencing when necessary, and sequences were analysed using ARG-ANNOT software [8] to identify the specific antibiotic resistance gene. Multilocus sequence typing was performed to evaluate genetic relatedness of strains. If necessary, a whole genome sequence study was performed to obtain the complete resistome of a strain [9] or to describe the genetic environment of an antibiotic resistance gene [10], [11].
Each student hosted by our institution received specific training for the study of antibiotic resistance (MALDI-TOF MS, AST, molecular training, genomics, bioinformatics) and presented the progress of their work and their results every week so that we could prepare tables and figures to be used for publication. Finally, each student was trained by the senior member of the team (JMR) to write their scientific papers and to create their own bibliography on the topic. Most of them also wrote a review on their topic while writing their PhD thesis. Weekly seminars or bibliographic sessions were also provided each Friday to improve students' knowledge in the field. The number of indexed international scientific papers per type of sample and per country was also calculated on the basis of published and submitted papers on antibiotic resistance during the study period.
Results
Since the creation of this institute, the JMR team has welcomed a total of 126 students or visiting scientists, including 95 deduplicated students (75.4%) from academic exchanges with 19 countries from the Mediterranean basin, Africa and Middle East. The number of students present in a given year has significantly increased during the study period (ten students in 2011, 15 in 2012, 21 in 2013, 28 in 2014, 27 in 2015, 40 in 2016 and 44 in 2017) to a total of 95 students. Most of the students are from Algeria (52, 55.3%), followed by Lebanon (12, 12.8%), Senegal (7, 7.4%) and Tunisia (5, 5.3%). All these 95 students were from Europe (Spain, 3 students, 3.2%; Italy, 2, 2.1%; Greece, 1, 1.1%), West Africa (Senegal, 7, 7.3%; Benin, Central Africa Republic, Guinea, Ivory Coast, Mali, Nigeria, Togo, 8, 8.4%), North Africa (Algeria, 52, 55.3%; Tunisia, 5, 5.3%; Egypt, 1, 1.1%; Morocco, 1, 1.1%), Middle East (Lebanon, 12, 12.8%; Qatar, Syria, 2, 2.1%) and Madagascar (1, 1.1%).
Each student had a different level of education, including master's degree (n = 15), PhD students (n = 65), scientific visitors and postdocs (n = 22). Overall, the number of PhD students from these countries significantly increased during the study period, from eight in 2011 to 30 in 2017 (2011: 8; 2012: 11; 2013: 16; 2014: 20; 2015: 23; 2016: 28; 2017: 30). The number of postdocs varied from two to four between these different years (2011, 2013, 2015, 2017: 2 students; 2012: 3; 2014, 2016: 4), as did the number of students seeking master's degrees, from one to four (2011, 2012: 1; 2013: 2; 2014, 2015: 3; 2016, 2017: 5). Students trained at the IHU-MI will return to their country of origin and continue to work in this field with our institute, which is now identified as the core laboratory for surveillance of antibiotic resistance and further analysis of new bacterial isolates from those countries.
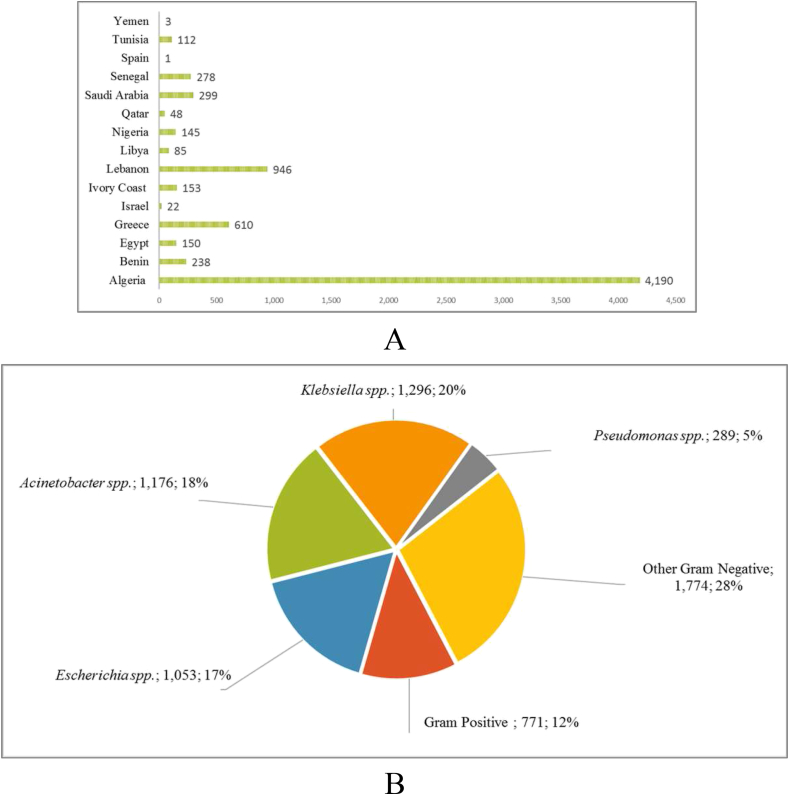
A total of 7280 samples from human (n = 4657; 64%), animal (n = 2058; 28%) or environment (n = 565; 8%) from 15 different countries were analysed during the study period (Fig. 1(A)). More than half of those samples came from Algeria (n = 4190; 57.6%), followed by Lebanon (n = 946; 12.9%), Greece (n = 610; 8.4%), Saudi Arabia (n = 299; 4.1%) and Senegal (n = 278; 3.8%) (Fig. 1(A)). From those 7280 samples, 6359 bacterial isolates were cultured and analysed phenotypically (bacterial identification by MALDI-TOF MS and AST) and genetically (molecular detection of antibiotic resistance genes).
Fig. 1.
(A) Geographic distribution of samples studied in publications of IHU-MI from 2011 to 2017 (n = 7280). (B) Repartition of bacterial species studied from Mediterranean basin or African countries in IHU-MI from 2011 to 2017 (n = 6359). IHU-MI, Institut Hospitalo-Universitaire Méditerranée Infection.
Bacterial genera studied included Enterobacteriaceae (1296 Klebsiella and 1053 Escherichia), followed by Acinetobacter (n = 1176) or other Gram-negative organisms (n = 1774 including bacteria of the genera Serratia, Salmonella, Enterobacter, Salmonella, Raoultella and Shewanella) and Pseudomonas (n = 289) (Fig. 1(B)). Gram-positive strains were represented with 771 strains (12%), including bacteria of the genera Enterococcus, Staphylococcus or Streptococcus (Fig. 1(B)).
All these studies allowed the detection and characterization of specific antibiotic resistance genes in multidrug-resistant bacteria from these genera and led to 97 scientific international indexed publications. Table 1 lists the publications and findings of specific antibiotic resistance determinants by country and bacterial species.
Table 1.
Samples and publications studied according to countries in Institut Hospitalo-Universitaire Méditerranée Infection from 2011 to 2017
| Country | Year | Strain type | Studied strains | No. of samples | No. of positive samples with an AR gene detected | Tested phenotype | Type of antibiotic resistance genes detected (n) | Study |
|---|---|---|---|---|---|---|---|---|
| Algeria | 2008–2011 | Clinical isolates | Klebsiella pneumoniae | 211 | 194 | ESBL Cephalosporinases Penicillinase HLP |
[19] | |
| 2008–2012 | Clinical isolates | K. pneumoniae | 221 | 190 | ESBL Cephalosporinases Penicillinase HLP |
blaTEM (146) blaSHV (154) blaCTX-M (130) |
[20] | |
| 2017 | Animal (45) and human (37) isolates | Salmonella spp. | 92 | 18 | ESBL |
blaCTX-M-1 (12) blaCTX-M-15 (5) blaTEM (8) |
[21] | |
| 2011 | Clinical isolates | Escherichia coli | 1 | 1 | Coli R | mcr-1 | [22] | |
| 2015 | Animal isolates |
E. coli (30) K. pneumoniae (17) |
47 | 47 | Coli R ESBL |
blaCTX-M-15 (47) blaTEM-1 (25) |
[23] | |
| 2014 | Clinical and environmental isolates |
K. pneumoniae Enterobacter cloacae Acinetobacter baumannii Pseudomonas aeruginosa |
89 | 32 | ESBL Carbapenemases |
blaOXA-48 (5) blaNDM-1 (7) blaOXA-43 (2) blaTEM (6) blaSHV + blaCTX-M (7) |
[24] | |
| 2013–2015 | Environmental isolates | A. baumannii | 1 | 1 | Carbapenemases ESBL Fluoroquinolones |
blaNDM-1 (1) | [25] | |
| 2010–2013 | Clinical isolates | Enterococcus spp. | 85 | 85 | Vanco R | vanC | [26] | |
| 2010–2013 | Clinical isolates | A. baumannii | 43 | 43 | Carbapenemases |
blaNDM-1 (7) blaOXA-23 (28) blaOXA-24 (1) blaOXA-58 (6) blaOXA-51 (43) blaOXA-23 + blaNDM-1 (2) blaOXA-58 + blaNDM-1 (1) |
[27] | |
| 2013–2015 | Clinical isolates |
Enterobacteriaceae (161) P. aeruginosa (18) A. baumannii (7) |
186 | 36 | Carbapenemases ESBL |
blaOXA-48 (2) blaVIM-4 (2) blaNDM-1 (2) blaOXA-23 (5) |
[28] | |
| 2011–2013 | Environmental isolates | A. baumannii | 67 | 61 | Carbapenemases |
blaOXA-23 (29) blaNDM-1 (32) |
[29] | |
| 2013–2014 | Clinical isolates | Streptococcus agalactiae | 93 | 74 | MLSB R |
ermB (18) ermA mef(A) mef(E) |
[30] | |
| 2015 | Animal isolates | E. coli | 1 | 1 | Carbapenemase | blaNDM-5 (1) | [31] | |
| 2014–2016 | Animal isolates | Enterobacteriaceae | 380 | 3 | Carbapenemases ESBL Cephalosporinases |
blaOXA-48 (3) | [32] | |
| 2012–2014 | Environmental isolates | K. pneumoniae | 44 | 44 | ESBL Cephalosporinases Fluoroquinolones Aminoglycosides |
blaCTX-M-15 (41) blaCTX-M-3 (3) |
[33] | |
| 2014–2015 | Animal isolates | Samples | 503 | 389 | Carbapenemases ESBL |
blaTEM (128) blaSHV (83) blaCTX-M (46) blaOXA-58 (132) |
[34] | |
| 2015 | Animal isolates | Enterobacteriaceae | 32 | 32 | Carbapenemases ESBL |
blaOXA-48 (32) blaCTX-M-15 (1) blaTEM (2) |
[35] | |
| 2013–2014 | Animal (3) and human (1) isolates | E. coli | 4 | 4 | Coli R | mcr-1 (4) | [36] | |
| 2016 | Animal isolates | E. coli | 8 | 8 | Coli R | In progress | Unpublished results | |
| 2017 | Animal (4) and environmental (5) isolates |
E. coli (8) E. cloacae (1) |
9 | 9 | Coli R | In progress | Unpublished results | |
| 2016 | Clinical isolates | K. pneumoniae | 3 | 3 | Coli R | In progress | Unpublished results | |
| 2015–2016 | Environmental isolates | Staphylococcus aureus | 200 | 153 | Methi R | In progress | Unpublished results | |
| 2011–2013 | Clinical isolates | A. baumannii | 47 | 47 | Carbapenemases Aminoglycosides Fluoroquinolones |
blaOXA-23 (33) blaOXA-24 (10) blaNDM (11) armA (4) aph(3′)VI (24) aadA (6) ant(2′)I (10) aac(3)Ia (33) Mutation in gyrA, parC (45) |
[37] | |
| 2013–2014 | Clinical isolates | A. baumannii | 12 | 12 | Coli R Carbapenemases |
blaOXA-24 (4) blaOXA-23 (6) blaOXA-51 (12) blaNDM-1 (2) |
[38] | |
| 2016–2017 | Animal isolates | Samples | 200 | 4 | ESBL | In progress | Unpublished results | |
| 2013 | Clinical isolates | Enterococcus hirae | 1 | 1 | [39] | |||
| 2016 | Clinical isolates |
K. pneumoniae (20) A. baumannii (12) P. aeruginosa (9) E. coli (27) |
68 | 68 | Carbapenemases ESBL |
In progress | Unpublished results | |
| 2011–2012 | Clinical isolates | A. baumannii | 30 | 24 | Carbapenemases |
blaOXA-23 (22) blaOXA-58 (1) blaOXA-23 + blaOXA-58 (1) |
[40] | |
| 2011 | Clinical isolates | P. aeruginosa | 17 | 17 | Carbapenemases |
blaVIM-2 (14) Mutation in oprD (3) |
[41] | |
| 2012–2013 | Clinical isolates | E. coli | 105 | 3 | Carbapenemases ESBL Aminoglycosides Fluoroquinolones |
blaNDM-5 (3) blaTEM (3) blaCTX-M (3) aadA (3) |
[42] | |
| 2010–2011 | Clinical isolates |
E. coli (3) K. pneumoniae (24) E. cloacae (11) Serratia marcescens (4) |
42 | 39 | ESBL |
blaCTX-M (10) blaTEM (14) blaSHV (15) |
[43] | |
| 2010–2011 | Clinical isolates | K. pneumoniae | 100 | 100 | ESBL Aminoglycosides Fluoroquinolones |
blaCTX-M (76) blaTEM (74) blaSHV (73) armA (23) aadA (35) aac(6′)Ib (50) qnrB (22) |
[44] | |
| 2013 | Animal isolates | Acinetobacter spp. | 33 | 4 | Carbapenemases |
blaOXA-23 + blaOXA-58 + blaOXA-51 (1) blaOXA-58 + blaOXA-51 (1) blaOXA-58 (2) |
[45] | |
| 2014–2015 | Clinical isolates | K. pneumoniae | 7 | 7 | ESBL Carbapenemases |
blaOXA-48 + blaCTX-M-15 + blaSHV-1 + blaTEM-1D (7) | [46] | |
| 2015 | Environmental isolates | Enterobacteriaceae | 12 | 9 | Carbapenemases ESBL |
blaCTX-M-15 (9) blaOXA-48 (1) blaTEM (1) |
[47] | |
| 2015 | Animal isolates | Pseudomonas putida | 1 | 1 | Carbapenemases ESBL |
blaVIM-2 | [48] | |
| 2017 | Clinical isolates | K. pneumoniae | 1 | 1 | Carbapenemases ESBL |
blaOXA-48 (1) blaSHV-27 (1) |
[49] | |
| 2010–2011 | Clinical isolates | A. baumannii | 71 | 71 | Carbapenemases |
blaOXA-23 (31) blaOXA-24 (5) blaOXA-51 (71) |
[50] | |
| 2010–2011 | Clinical isolates | A. baumannii | 71 | 71 | Carbapenemases ESBL Aminoglycosides Fluoroquinolones |
blaTEM (53) ampC (69) aph(3′)VI (36) aadA (45) ant(2′)I (10) aac(3)Ia (64) aac(6′)Ib (3) Mutation gyrA, parC (67) |
[51] | |
| 2013 | Clinical isolates | K. pneumoniae | 1 | 1 | Carbapenemases ESBL Aminoglycosides Fluoroquinolones |
blaKPC-3 blaTEM blaSHV aac(6′)Ib aadA |
[52] | |
| 2015 | Environmental isolates |
E. coli (12) K. pneumoniae (3) Raoultella ornithinolytica (3) Citrobacter freundii (1) Citrobacter braakii (1) |
20 | 20 | ESBL Carbapenemases |
blaOXA-48 (17) blaOXA-244 (3) blaTEM-1 (9) blaCTX-M-15 + blaTEM-1 (3) |
[13] | |
| 2016 | Environmental isolates | K. pneumoniae | 87 | 3 | ESBL Carbapenemases |
blaOXA-48 (3) blaTEM-1 (1) |
[16] | |
| 2014 | Animal isolates | E. coli | 20 | 20 | Carbapenemases ESBL Aminoglycosides |
blaTEM-1 (20) blaCTX-M-1 (2) blaSHV-12 (14) CMY-2 (4) aadA (20) |
[12] | |
| 2015 | Environmental isolates | Shewanella xiamenensis | 4 | 4 | Carbapenemases |
blaOXA-48 (1) blaOXA-199 (1) blaOXA-181 (2) |
[11] | |
| 2016 | Animal isolates | Enterobacteriaceae | 86 | 1 | Coli R |
mcr-1 blaCTX-M-15 blaTEM-1 qnrB19 |
[17] | |
| 2014 | Clinical isolates | S. aureus | 250 | 171 | Methi R | In progress | Unpublished results | |
| 2005–2007 | Clinical isolates | S. aureus | 64 | 64 | Methi R | mecA (64) | [53] | |
| 2011 | Clinical isolates |
Proteus mirablis Morganella spp. Providencia spp. |
106 | 72 | ESBL Carbapenemases Aminoglycosides Fluoroquinolones |
In progress | Unpublished results | |
| 2014–2015 | Clinical (60) and environmental (39) isolates |
Enterobacteriaceae Acinetobacter spp. |
99 | 10 | Carbapenemases ESBL Cephalosporinases Aminoglycosides |
blaNDM-1 (5) blaOXA-23 (3) blaOXA-48 (2) blaSHV-148 + blaTEM-163 (2) aph(3′)VI–ant(2″)I (2) aac(3)Ia aadA (3) AME-encoding genes (5) |
[54] | |
| 2009–2012 | Clinical isolates | P. aeruginosa | 89 | 39 | Carbapenemases Aminoglycosides |
blaVIM-2 (2) aadA (10) aac(3)Ia (3) |
[55] | |
| 2008–2012 | Clinical isolates | Acinetobacter spp. | 113 | 113 | Carbapenemases ESBL Aminoglycosides Fluoroquinolones |
blaOXA-23 (40) blaOXA-24 (17) blaNDM (5) aph(3′)VI (70) aadA (57) ant(2′)I (60) aac(3)Ia (77) aac(6′)Ib (1) |
[56] | |
| 2012 | Clinical isolates | A. baumannii | 123 | 77 | Carbapenemases ESBL |
blaOXA-23 (40) blaOXA-24 (3) blaOXA-23 + blaOXA-24 (3) |
[57] | |
| Benin | 2015 | Clinical isolates |
Staphylococcus saprophyticus (31) S. aureus (21) Staphylococcus sciuri (17) Staphylococcus conhii (5) Staphylococcus haemolyticus (2) Staphylococcus xylosus (1) Staphylococcus hominis (1) |
78 | 21 | Methi R | mecA (19) | [58] |
| 2015 | Clinical isolates | Enterobacteriaceae | 157 | 103 | ESBL Carbapenemases |
In progress | Unpublished results | |
| 2016 | Clinical isolates | P. aeruginosa | 3 | 3 | Carbapenemases | In progress | Unpublished results | |
| Egypt | 2012–2013 | Clinical isolates | A. baumannii | 150 | 150 | Carbapenemases Aminoglycosides |
blaNDM-1 (59) blaOXA-23 (115) armA (141) blaOXA-51 (150) blaNDM-1 + blaOXA-23 (53) armA + blaNDM-1 + blaOXA-23 (52) |
[59] |
| Greece | 2013–2017 | Clinical isolates |
P. mirablis (4) P. putida (1) C. freundii (1) Enterobacter aerogenes (2) Providencia stuartii (8) P. aeruginosa (79) A. baumannii (158) E. cloacae (10) E. coli (33) K. pneumoniae (314) |
610 | 610 | Carbapenemases ESBL Coli R Fluoroquinolones |
In progress | Unpublished results |
| Israel | 2011 | Clinical isolates | Providencia rettgeri | 1 | 1 | Carbapenemase | blaNDM-1 (1) | [60] |
| 2008–2011 | Clinical isolates | K. pneumoniae | 15 | 15 | Carbapenemases Coli R Aminoglycosides |
In progress | Unpublished results | |
| 2010–2011 | Clinical isolates |
K. pneumoniae (1) E. coli (1) P. mirabilis (1) P. rettgeri (1) Morganella morganii (1) |
5 | 5 | Carbapenemases | blaNDM-1 (5) | [61] | |
| 2014 | Clinical isolates | M. morganii | 1 | 1 | Carbapenemases | blaNDM-1 (1) | [62] | |
| Ivory Coast | 2012–2015 | Clinical isolates | Enterobacteriaceae | 153 | 153 | ESBL Aminoglycosides Fluoroquinolones |
In progress | Unpublished results |
| Lebanon | 2013 | Clinical isolates | P. aeruginosa | 35 | 35 | Carbapenemases Cephalosporinases |
blaVIM-2 (16) blaIMP-15 (2) ampC (8) |
[14] |
| 2013 | Animal isolates | E. coli | 1 | 1 | ESBL Carbapenemases |
blaOXA-48 (1) blaCTX-M-14 (1) blaTEM-1 (1) |
[63] | |
| 2013 | Animal isolates |
P. aeruginosa (4) A. baumannii (5) |
9 | 9 | Carbapenemases |
blaOXA-23 (4) blaOXA-58 (1) blaVIM-2 (4) |
[64] | |
| 2015 | Clinical isolates | R. ornithinolytica | 1 | 1 | Carbapenemases Cephalosporinase BLSE MLSB R Chloramphenicol Fluoroquinolones |
blaOXA-48 ampC ampH blaTEM-166 macA macB cmr cat gyrA mutated |
[10] | |
| 2015 | Animal isolates | E. coli | 1 | 1 | ESBL Coli R |
blaTEM-135-like (1) mcr-1 (1) |
[18] | |
| 2015 | Animal isolates | E. cloacae | 1 | 1 | Cephalosporinase Coli R |
In progress | Unpublished results | |
| 2015 | Animal isolates |
E. coli (217) K. pneumoniae (8) Escherichia fergusonii (1) A. baumannii (1) P. mirabilis (3) E. cloacae (2) |
235 | 235 | ESBL Cephalosporinases |
In progress | Unpublished results | |
| 2017 | Animal isolates |
E. coli (105) E. fergusonii (2) K. pneumoniae (4) |
111 | 111 | ESBL Cephalosporinase Coli R |
In progress | Unpublished results | |
| 2017 | Animal (346) Environmental (53) Human (11) isolates |
E. coli (341) K. pneumoniae (31) Enterobacter asburiae (1) Stenotrophomonas maltophilia (4) Serratia rubidae (1) A. baumannii (4) Acinetobacter genomospecies (4) Pseudomonas spp. (8) Ochrobactrum spp. (1) E. cloacae (4) |
399 | 399 | ESBL Cephalosporinase Coli R |
In progress | Unpublished results | |
| 2016–2017 | Clinical isolates | Enterobacter faecium | 4 | 4 | Glycopeptides | In progress | Unpublished results | |
| 2016 | Clinical isolates | A. baumannii | 31 | 31 | ESBL Carbapenemases |
In progress | Unpublished results | |
| 2016 | Clinical isolates | Campylobacter jejuni | 1 | 1 | ESBL Carbapenemases |
In progress | Unpublished results | |
| 2010–2016 | Clinical isolates | E. coli | 43 | 43 | ESBL | In progress | Unpublished results | |
| 2015 | Clinical isolates | K. pneumoniae | 3 | 3 | ESBL Coli R |
blaCTX-M-15 + blaTEM-12 + blaSHV-5 (2) blaSHV-5 (1) Mutation mgrB (2) phoQ (1) pmrA (1) |
[65] | |
| 2016–2017 | Clinical isolates |
Enterobacteriaceae (8) P. aeruginosa (1) |
9 | 9 | ESBL Carbapenemases Coli R |
In progress | Unpublished results | |
| 2012 | Clinical isolates | A. baumannii | 4 | 4 | Carbapenemases |
blaNDM-1 (4) blaOXA-94 (4) |
[15] | |
| 2016 | Clinical isolates | Neisseria meningitidis | 58 | 1 | In progress | Unpublished results | ||
| Libya | 2013–2014 | Clinical isolates |
P. aeruginosa (24) A. baumannii (25) |
49 | 43 | Carbapenemases |
blaOXA-24 (3) blaOXA-23 (19) blaVIM-2 (19) |
[66] |
| 2015 | Clinical isolates | A. baumannii | 36 | 36 | Carbapenemases |
blaOXA-23 (29) blaNDM-1 (8) blaNDM (36) |
[67] | |
| Nigeria | 2012 | Clinical isolates | A. baumannii | 3 | 3 | Carbapenemases | blaOXA-23 (3) | [68] |
| 2012–2013 | Clinical isolates | Klebsiella spp. | 139 | 1 | Coli R | mgrB (1) | [69] | |
| 2012 | Animal (2) and human (1) isolates | E. coli | 3 | 1 | Coli R | mcr-1 (1) | [70] | |
| Qatar | 2011–2012 | Clinical isolates | A. baumannii | 48 | 48 | Carbapenemases | blaOXA-23 (48) | [68] |
| Saudi Arabia | 2013–2014 | Clinical isolates |
E. coli (10) K. pneumoniae (1) |
11 | 11 | Coli R ESBL |
mcr-1 (11) blaTEM-1 (10) blaSHV-1 (1) blaCTX-M-15 (1) |
[71] |
| 2014 | Clinical isolates | A. baumannii | 42 | 28 | Carbapenemases ESBL Aminoglycosides |
oxa-72 (1) blaNDM-5 (1) blaNDM-1 (1) blaOXA-48 (1) blaOXA-58 (22) blaOXA-51 + blaOXA-72 (1) blaNDM-5 + blaCTX-M-15 + blaTEM-1 + aadA2 (1) blaNDM-5 + blaTEM-1 + aadA2 (1) |
[72] | |
| 2013–2014 | Clinical isolates |
E. coli (23) K. pneumoniae (5) |
28 | 28 | ESBL |
blaCTX-M (27) blaTEM (19) blaSHV (4) |
[73] | |
| 2013–2014 | Clinical isolates | [74] | ||||||
| 2013–2014 | Clinical isolates | Samples | 218 | 73 | ESBL | blaCTX-M (73) | [75] | |
| Senegal | 2011 | Clinical isolates | A. baumannii | 5 | 3 | Carbapenemases | blaOXA-23 (3) | [76] |
| 2014 | Clinical isolates | M. morganii | 112 | 112 | ESBL |
blaCTX (112) blaTEM (86) blaSHV (63) |
[77] | |
| 2015–2017 | Clinical isolates | Enterobacteriaceae | 161 | 120 | Carbapenemases ESBL Aminoglycosides Fluoroquinolones Coli R |
In progress | Unpublished results | |
| Spain | 2015 | Clinical isolates | Acinetobacter nosocomialis | 1 | 1 | Coli R | [78] | |
| Tunisia | 2013–2016 | Clinical isolates | A. baumannii | 25 | 25 | Carbapenemases |
blaOXA-51 + blaOXA-23 (25) blaOXA-58 (1) |
[79] |
| 2015 | Clinical isolates |
E. coli (51) K. pneumoniae (36) |
87 | 68 | Carbapenemases ESBL |
blaCTX (47) blaTEM-1 (31) blaSHV (18) blaOXA (10) |
[80] | |
| Yemen | 2013 | Clinical isolates | A. baumannii | 3 | 3 | Carbapenemases Aminoglycosides Fluoroquinolones |
blaOXA-23 (3) armA (3) aac(6′)Ib (1) Mutated gyrA (3) |
[81] |
| Total studied samples | 7280 | Total no. of publications | 97 | |||||
| Total studied strains | 6359 |
‘Samples’ indicates that no strains were isolated but samples were directly tested by PCR.
Coli R, colistin resistance; ESBL, extended-spectrum β-lactamase; HLP, high-level penicillinase; Methi
R, methicillin resistance; MLSB, macrolide–lincosamide–streptogramin B phenotype; Vanco R, vancomycin resistance.
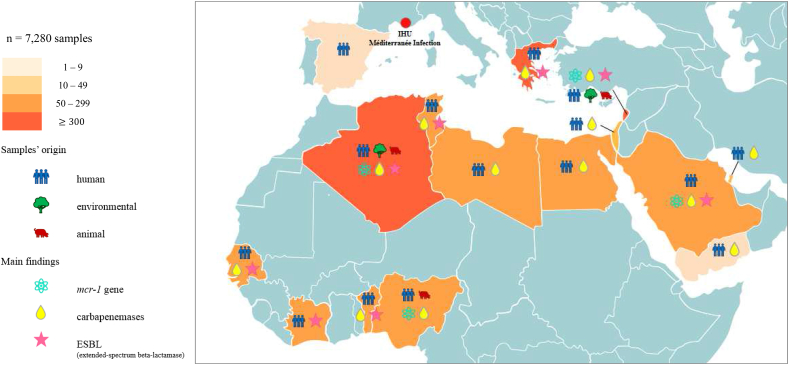
The main antibiotic resistance determinants detected and characterized were extended-spectrum β-lactamases (ESBLs) and carbapenemases including blaCTX-M, blaSHV, blaTEM [12], blaOXA-48 [13], blaNDM [82] and blaVIM [14] genes. Genes encoding for resistance to aminoglycosides were also reported, including, for example, armA or aac(6′)-Ib in Acinetobacter baumannii [81]. Resistance to colistin mediated by the newly plasmid-mediated mcr-1 gene in human and animal isolates has been tested to date in 21 studies from eight countries (Algeria, Greece, Israel, Lebanon, Nigeria, Saudi Arabia, Senegal and Spain), leading to ten scientific publications (Table 1). An overview of the global distribution of the main findings of antibiotic resistance determinants in the 7280 samples studied per country and type of samples is provided in the map in Fig. 2.
Fig. 2.
Global distribution of samples in Mediterranean basin and African countries.
Discussion
Here we show the unique role of IHU-MI in training about 100 students working in the field of antibiotic resistance from the Mediterranean basin and Africa over the last 6 years. This has led to the description and surveillance of new mechanisms of resistance to antibiotics in 15 various countries reported in 97 scientific publications, including 24 different peer-reviewed journals. The majority of the publications have reported the first detection of antibiotic resistance genes, mainly ESBLs [47], carbapenemases [15], [16], [31] and the mcr-1 plasmid-mediated colistin resistance gene [17], [18], [22] in these countries in both humans and animals. One of the main contributions in the field is the description of a strong link between antibiotic consumption in animals and emergence and spread of antibiotic resistance genes in animals as well as the transfer to humans [83].
Antibiotics are widely used in agricultural settings in these countries, without clear control policies; this situation has affected human health and is implicated in the evolution of new mechanisms of resistance [84]. Epidemiologic descriptions are essential, and our results confirmed that surveillance should continue in Africa and in the Mediterranean basin to monitor and control the emergence and spread of antibiotic resistance genes. Thanks to these students and their training at the IHU-MI, the institute has created a unique collaborative network for surveillance and study of antibiotic resistance in Africa and in the Mediterranean basin because most of these students returned to their country of origin and created microbiology laboratories to study and survey antibiotic resistance in collaboration with the IHU-MI institute. Further engagements with key individuals are ongoing to create new partnerships to study antibiotic resistance in humans and animals in Italy, Morocco, Turkey and in the Balkans, and should be reinforced in Egypt and Libya as well as the Middle East, although the political situation is currently complex. The current migrant crisis in Europe should prompt us to survey antibiotic resistance in humans and animals from these countries, including Syria and Iraq, to avoid the possible spreading of specific clones, as previously reported in Greece [85], [86] and Israel [87] for Klebsiella pneumoniae carbapenemase producers.
Because of its special location as a seaport in the Mediterranean basin, Marseille has historically always been a critical place for the entrance of infectious diseases such as plague or cholera [88]. Because antibiotic-resistant bacteria and antibiotic resistance genes that could spread in the Mediterranean basin do not have borders, the IHU-MI in Marseille plays a critical role in the surveillance of resistance in these areas as well as in African countries that historically have links to France. Thus, over the last 6 years, the institute has become a reference centre for the surveillance of antibiotic resistance and the training of students from countries in the Mediterranean basin and Africa. Such a collaborative network will expand in the future, permitting real-time surveillance of antibiotic resistance determinants that may emerge and spread in these areas [89].
Acknowledgement
We are grateful to IHU-MI.
Conflict of interest
None declared.
References
- 1.World Health Organization; Organisation mondiale de la Santé. Résistance aux antibiotiques. Updated 5 February 2018. Available at: http://www.who.int/fr/news-room/fact-sheets/detail/résistance-aux-antibiotiques.
- 2.Neill J.O. December 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Review on antimicrobial resistance.https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf Available at: [Google Scholar]
- 3.Rolain J.M., Abat C., Jimeno M.T., Fournier P.E., Raoult D. Do we need new antibiotics? Clin Microbiol Infect. 2016;22:408–415. doi: 10.1016/j.cmi.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Abat C., Raoult D., Rolain J.M. Antibiotic resistance: are we living a nightmare? Clin Microbiol Infect. 2018 doi: 10.1016/j.cmi.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abat C., Rolain J.M., Dubourg G., Fournier P.E., Chaudet H., Raoult D. Evaluating the clinical burden and mortality attributable to antibiotic resistance: the disparity of empirical data and simple model estimations. Clin Infect Dis. 2017;65:S58–S63. doi: 10.1093/cid/cix346. [DOI] [PubMed] [Google Scholar]
- 6.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 7.European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint tables for interpretation of MICs and zone diameters, version 8. http://www.eucast.org/clinical_breakpoints/ Available at:
- 8.Gupta S.K., Padmanabhan B.R., Diene S.M., Lopez-Rojas R., Kempf M., Landraud L. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rolain J.M., Diene S.M., Kempf M., Gimenez G., Robert C., Raoult D. Real-time sequencing to decipher the molecular mechanism of resistance of a clinical pan–drug-resistant Acinetobacter baumannii isolate from Marseille, France. Antimicrob Agents Chemother. 2013;57:592–596. doi: 10.1128/AAC.01314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Bayssari C., Olaitan A.O., Leangapichart T., Okdah L., Dabboussi F., Hamze M. Whole-genome sequence of a blaOXA-48–harboring Raoultella ornithinolytica clinical isolate from Lebanon. Antimicrob Agents Chemother. 2016;60:2548–2550. doi: 10.1128/AAC.02773-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tafoukt R., Leangapichart T., Hadjadj L., Bakour S., Diene S.M., Rolain J.M. Characterization of blaOXA-538, a new variant of blaOXA-48 in Shewanella xiamenensis isolated from river water in Algeria. J Glob Antimicrob Resist. 2017;13:70–73. doi: 10.1016/j.jgar.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Belmahdi M., Bakour S., Al Bayssari C., Touati A., Rolain J.M. Molecular characterisation of extended-spectrum β-lactamase– and plasmid AmpC–producing Escherichia coli strains isolated from broilers in Béjaïa, Algeria. J Glob Antimicrob Resist. 2016;6:108–112. doi: 10.1016/j.jgar.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Tafoukt R., Touati A., Leangapichart T., Bakour S., Rolain J.M. Characterization of OXA-48–like–producing Enterobacteriaceae isolated from river water in Algeria. Water Res. 2017;120:185–189. doi: 10.1016/j.watres.2017.04.073. [DOI] [PubMed] [Google Scholar]
- 14.Al Bayssari C., Diene S.M., Loucif L., Gupta S.K., Dabboussi F., Mallat H. Emergence of VIM-2 and IMP-15 carbapenemases and inactivation of oprD gene in carbapenem-resistant Pseudomonas aeruginosa clinical isolates from Lebanon. Antimicrob Agents Chemother. 2014;58:4966–4970. doi: 10.1128/AAC.02523-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rafei R., Dabboussi F., Hamze M., Eveillard M., Lemarié C., Mallat H. First report of blaNDM-1–producing Acinetobacter baumannii isolated in Lebanon from civilians wounded during the Syrian war. Int J Infect Dis. 2014;21:21–23. doi: 10.1016/j.ijid.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Touati A., Mairi A., Baloul Y., Lalaoui R., Bakour S., Thighilt L. First detection of Klebsiella pneumoniae producing OXA-48 in fresh vegetables from Béjaïa city, Algeria. J Glob Antimicrob Resist. 2017;9:17–18. doi: 10.1016/j.jgar.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Bachiri T, Lalaoui R, Bakour S, Allouache M, Belkebla N, Rolain JM, et al. First report of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli ST405 isolated from wildlife in Bejaia, Algeria. Microb Drug Resist 2017:mdr.2017.0026. 10.1089/mdr.2017.0026. [DOI] [PubMed]
- 18.Dandachi I., Leangapichart T., Daoud Z., Rolain J.M. First detection of mcr-1 plasmid-mediated colistin-resistant Escherichia coli in Lebanese poultry. J Glob Antimicrob Resist. 2018;12:137–138. doi: 10.1016/j.jgar.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Berrazeg M., Diene S.M., Drissi M., Kempf M., Richet H., Landraud L. Biotyping of multidrug-resistant Klebsiella pneumoniae clinical isolates from France and Algeria using MALDI-TOF MS. PLoS One. 2013;8:e61428. doi: 10.1371/journal.pone.0061428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berrazeg M., Drissi M., Medjahed L., Rolain J.M. Hierarchical clustering as a rapid tool for surveillance of emerging antibiotic-resistance phenotypes in Klebsiella pneumoniae strains. J Med Microbiol. 2013;62:864–874. doi: 10.1099/jmm.0.049437-0. [DOI] [PubMed] [Google Scholar]
- 21.Djeffal S., Bakour S., Mamache B., Elgroud R., Agabou A., Chabou S. Prevalence and clonal relationship of ESBL-producing Salmonella strains from humans and poultry in northeastern Algeria. BMC Vet Res. 2017;13:132. doi: 10.1186/s12917-017-1050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berrazeg M., Hadjadj L., Ayad A., Drissi M., Rolain J.M. First detected human case in Algeria of mcr-1 plasmid-mediated colistin resistance in a 2011 Escherichia coli isolate. Antimicrob Agents Chemother. 2016;60:6996–6997. doi: 10.1128/AAC.01117-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachiri T., Bakour S., Ladjouzi R., Thongpan L., Rolain J.M., Touati A. High rates of CTX-M-15–producing Escherichia coli and Klebsiella pneumoniae in wild boars and Barbary macaques in Algeria. J Glob Antimicrob Resist. 2017;8:35–40. doi: 10.1016/j.jgar.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Bouguenoun W., Bakour S., Bentorki A.A., Al Bayssari C., Merad T., Rolain J.M. Molecular epidemiology of environmental and clinical carbapenemase-producing Gram-negative bacilli from hospitals in Guelma, Algeria: multiple genetic lineages and first report of OXA-48 in Enterobacter cloacae. J Glob Antimicrob Resist. 2016;7:135–140. doi: 10.1016/j.jgar.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Chaalal W., Chaalal N., Bakour S., Kihal M., Rolain J.M. First occurrence of NDM-1 in Acinetobacter baumannii ST85 isolated from Algerian dairy farms. J Glob Antimicrob Resist. 2016;7:150–151. doi: 10.1016/j.jgar.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Bourafa N., Abat C., Loucif L., Olaitan A.O., Bentorki A.A., Boutefnouchet N. Identification of vancomycin-susceptible major clones of clinical Enterococcus from Algeria. J Glob Antimicrob Resist. 2016;6:78–83. doi: 10.1016/j.jgar.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Ramoul A., Loucif L., Bakour S., Amiri S., Dekhil M., Rolain J.M. Co-occurrence of blaNDM-1 with blaOXA-23 or blaOXA-58 in clinical multidrug-resistant Acinetobacter baumannii isolates in Algeria. J Glob Antimicrob Resist. 2016;6:136–141. doi: 10.1016/j.jgar.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Mellouk F.Z., Bakour S., Meradji S., Al-Bayssari C., Bentakouk M.C., Zouyed F. First detection of VIM-4–producing Pseudomonas aeruginosa and OXA-48–producing Klebsiella pneumoniae in northeastern (Annaba, Skikda) Algeria. Microb Drug Resist. 2017;23:335–344. doi: 10.1089/mdr.2016.0032. [DOI] [PubMed] [Google Scholar]
- 29.Zenati K., Touati A., Bakour S., Sahli F., Rolain J.M. Characterization of NDM-1– and OXA-23–producing Acinetobacter baumannii isolates from inanimate surfaces in a hospital environment in Algeria. J Hosp Infect. 2016;92:19–26. doi: 10.1016/j.jhin.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 30.Bergal A., Loucif L., Benouareth D.E., Bentorki A.A., Abat C., Rolain J.M. Molecular epidemiology and distribution of serotypes, genotypes, and antibiotic resistance genes of Streptococcus agalactiae clinical isolates from Guelma, Algeria and Marseille, France. Eur J Clin Microbiol Infect Dis. 2015;34:2339–2348. doi: 10.1007/s10096-015-2487-6. [DOI] [PubMed] [Google Scholar]
- 31.Yousfi M., Mairi A., Bakour S., Touati A., Hassissen L., Hadjadj L. First report of NDM-5–producing Escherichia coli ST1284 isolated from dog in Bejaia, Algeria. New Microbe. New Infect. 2015;8:17–18. doi: 10.1016/j.nmni.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachiri T., Bakour S., Lalaoui R., Belkebla N., Allouache M., Rolain J.M. Occurrence of carbapenemase-producing Enterobacteriaceae isolates in the wildlife: first report of OXA-48 in wild boars in Algeria. Microb Drug Resist. 2018;24:337–345. doi: 10.1089/mdr.2016.0323. [DOI] [PubMed] [Google Scholar]
- 33.Zenati K., Sahli F., Garcia V., Bakour S., Belhadi D., Rolain J.M. Occurrence and clonal diversity of multidrug-resistant Klebsiella pneumoniae recovered from inanimate surfaces in Algerian hospital environment: first report of armA, qnrB and aac(6′)-Ib-cr genes. J Glob Antimicrob Resist. 2017;10:148–153. doi: 10.1016/j.jgar.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Selma C., Hamza L., Bernard D., Atef A., Rolain J.M. Prevalence of extended-spectrum beta lactamase and carbapenemase-encoding genes in poultry feces from Algeria and Marseille, France. J Glob Antimicrob Resist. 2017;13:28–32. doi: 10.1016/j.jgar.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Bouaziz A., Loucif L., Ayachi A., Guehaz K., Bendjama E., Rolain J.M. Migratory white stork (Ciconia ciconia): a potential vector of the OXA-48–producing Escherichia coli ST38 clone in Algeria. Microb Drug Resist. 2018;24:461–468. doi: 10.1089/mdr.2017.0174. [DOI] [PubMed] [Google Scholar]
- 36.Hadjadj L., Riziki T., Zhu Y., Li J., Diene S., Rolain J.M. Study of mcr-1 gene–mediated colistin resistance in Enterobacteriaceae isolated from humans and animals in different countries. Genes (Basel) 2017;8:394. doi: 10.3390/genes8120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakour S., Touati A., Bachiri T., Sahli F., Tiouit D., Naim M. First report of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and rapid spread of metallo-β-lactamase NDM-1 in Algerian hospitals. J Infect Chemother. 2014;20:696–701. doi: 10.1016/j.jiac.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 38.Bakour S., Olaitan A.O., Ammari H., Touati A., Saoudi S., Saoudi K. Emergence of colistin- and carbapenem-resistant Acinetobacter baumannii ST2 clinical isolate in Algeria: first case report. Microb Drug Resist. 2015;21:279–285. doi: 10.1089/mdr.2014.0214. [DOI] [PubMed] [Google Scholar]
- 39.Bourafa N., Loucif L., Boutefnouchet N., Rolain J.M. Enterococcus hirae, an unusual pathogen in humans causing urinary tract infection in a patient with benign prostatic hyperplasia: first case report in Algeria. New Microbe. New Infect. 2015;8:7–9. doi: 10.1016/j.nmni.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Touati M., Diene S.M., Racherache A., Dekhil M., Djahoudi A., Rolain J.M. Emergence of blaOXA-23 and blaOXA-58 carbapenemase-encoding genes in multidrug-resistant Acinetobacter baumannii isolates from University Hospital of Annaba, Algeria. Int J Antimicrob Agents. 2012;40:89–91. doi: 10.1016/j.ijantimicag.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 41.Touati M., Diene S.M., Dekhil M., Djahoudi A., Racherache A., Rolain J.M. Dissemination of a class I integron carrying VIM-2 carbapenemase in Pseudomonas aeruginosa clinical isolates from a hospital intensive care unit in Annaba, Algeria. Antimicrob Agents Chemother. 2013;57:2426–2427. doi: 10.1128/AAC.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sassi A., Loucif L., Gupta S.K., Dekhil M., Chettibi H., Rolain J.M. NDM-5 carbapenemase-encoding gene in multidrug-resistant clinical isolates of Escherichia coli from Algeria. Antimicrob Agents Chemother. 2014;58:5606–5608. doi: 10.1128/AAC.02818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labid A., Gacemi-Kirane D., Timinouni M., Amoura K., Rolain J.M. High prevalence of extended spectrum beta-lactamase (ESBL) producers in fatal cases of pediatric septicemia among the Enterobacteriaceae in the pediatric hospital of Annaba, Algeria. Afr J Microbiol Res. 2014;8:947–954. [Google Scholar]
- 44.Belbel Z., Chettibi H., Dekhil M., Ladjama A., Nedjai S., Rolain J.M. Outbreak of an armA methyltransferase-producing ST39 Klebsiella pneumoniae clone in a pediatric Algerian hospital. Microb Drug Resist. 2014;20:310–315. doi: 10.1089/mdr.2013.0193. [DOI] [PubMed] [Google Scholar]
- 45.Morakchi H., Loucif L., Gacemi-Kirane D., Rolain J.M. Molecular characterisation of carbapenemases in urban pigeon droppings in France and Algeria. J Glob Antimicrob Resist. 2017;9:103–110. doi: 10.1016/j.jgar.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Loucif L., Kassah Laouar A., Saidi M., Messala A., Chelaghma W., Rolain J.M. Outbreak of OXA-48–producing Klebsiella pneumoniae involving an ST 101 clone in Batna University Hospital, Algeria. Antimicrob Agents Chemother. 2016;60:7494–7497. doi: 10.1128/AAC.00525-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loucif L., Gacemi-Kirane D., Cherak Z., Chamlal N., Grainat N., Rolain J.M. First report of German cockroaches (Blattella germanica) as reservoirs of CTX-M-15 extended-spectrum-β-lactamase- and OXA-48 carbapenemase-producing Enterobacteriaceae in Batna University Hospital, Algeria. Antimicrob Agents Chemother. 2016;60:6377–6380. doi: 10.1128/AAC.00871-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loucif L., Cherak Z., Chamlal N., Bendjama E., Gacemi-Kirane D., Grainat N. First detection of VIM-2 metallo-β-lactamase–producing Pseudomonas putida in Blattella germanica cockroaches in an Algerian hospital. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.00357-17. e00357–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loucif L., Chelaghma W., Helis Y., Sebaa F., Baoune R.D., Zaatout W. First detection of OXA-48–producing Klebsiella pneumoniae in community-acquired urinary tract infections in Algeria. J Glob Antimicrob Resist. 2018;12:115–116. doi: 10.1016/j.jgar.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 50.Bakour S., Kempf M., Touati A., Ameur A.A., Haouchine D., Sahli F. Carbapenemase-producing Acinetobacter baumannii in two university hospitals in Algeria. J Med Microbiol. 2012;61:1341–1343. doi: 10.1099/jmm.0.045807-0. [DOI] [PubMed] [Google Scholar]
- 51.Bakour S., Touati A., Sahli F., Ameur A.A., Haouchine D., Rolain J.M. Antibiotic resistance determinants of multidrug-resistant Acinetobacter baumannii clinical isolates in Algeria. Diagn Microbiol Infect Dis. 2013;76:529–531. doi: 10.1016/j.diagmicrobio.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 52.Bakour S., Sahli F., Touati A., Rolain J.M. Emergence of KPC-producing Klebsiella pneumoniae ST512 isolated from cerebrospinal fluid of a child in Algeria. New Microbe. New Infect. 2015;3:34–36. doi: 10.1016/j.nmni.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ouchenane Z., Agabou A., Smati F., Rolain J.M., Raoult D. Staphylococcal cassette chromosome mec characterization of methicillin-resistant Staphylococcus aureus strains isolated at the military hospital of Constantine/Algeria. Pathol Biol. 2013;61:280–281. doi: 10.1016/j.patbio.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Yagoubat M., Ould El-Hadj-Khelil A., Malki A., Bakour S., Touati A., Rolain J.M. Genetic characterisation of carbapenem-resistant Gram-negative bacteria isolated from the University Hospital Mohamed Boudiaf in Ouargla, southern Algeria. J Glob Antimicrob Resist. 2017;8:55–59. doi: 10.1016/j.jgar.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 55.Sefraoui I., Berrazeg M., Drissi M., Rolain J.M. Molecular epidemiology of carbapenem-resistant Pseudomonas aeruginosa clinical strains isolated from western Algeria between 2009 and 2012. Microb Drug Resist. 2014;20:156–161. doi: 10.1089/mdr.2013.0161. [DOI] [PubMed] [Google Scholar]
- 56.Mesli E., Berrazeg M., Drissi M., Bekkhoucha S.N., Rolain J.M. Prevalence of carbapenemase-encoding genes including New Delhi metallo-β-lactamase in Acinetobacter species, Algeria. Int J Infect Dis. 2013;17:739–743. doi: 10.1016/j.ijid.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 57.Kempf M., Bakour S., Flaudrops C., Berrazeg M., Brunel J.M., Drissi M. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization–time of flight mass spectrometry. PLoS One. 2012;7:e31676. doi: 10.1371/journal.pone.0031676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koudokpon H., Dougnon T.V., Setondji Islamiath K., Alidah A., Brice Armand F.V., Frédéric L. Methicillin-resistance of Staphylococcus species in southern Benin : resistance gene, virulence factor associated and staphylococcal chromosomal cassette distribution. Int J Microbiol Res. 2017;9:976–980. [Google Scholar]
- 59.El-Sayed-Ahmed M.A.E.G., Amin M.A., Tawakol W.M., Loucif L., Bakour S., Rolain J.M. High prevalence of blaNDM-1 carbapenemase-encoding gene and 16S rRNA armA methyltransferase gene among Acinetobacter baumannii clinical isolates in Egypt. Antimicrob Agents Chemother. 2015;59:3602–3605. doi: 10.1128/AAC.04412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olaitan A.O., Diene S.M., Assous M.V., Rolain J.M. Genomic plasticity of multidrug-resistant NDM-1 positive clinical isolate of Providencia rettgeri. Genome Biol Evol. 2016;8:723–728. doi: 10.1093/gbe/evv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lachish T., Elimelech M., Arieli N., Adler A., Rolain J.M., Assous M.V. Emergence of New Delhi metallo-β-lactamase in Jerusalem, Israel. Int J Antimicrob Agents. 2012;40:566–567. doi: 10.1016/j.ijantimicag.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 62.Olaitan A.O., Diene S.M., Gupta S.K., Adler A., Assous M.V., Rolain J.M. Genome analysis of NDM-1 producing Morganella morganii clinical isolate. Expert Rev Anti Infect Ther. 2014;12:1297–1305. doi: 10.1586/14787210.2014.944504. [DOI] [PubMed] [Google Scholar]
- 63.Al Bayssari C., Olaitan A.O., Dabboussi F., Hamze M., Rolain J.M. Emergence of OXA-48–producing Escherichia coli clone ST38 in fowl. Antimicrob Agents Chemother. 2015;59:745–746. doi: 10.1128/AAC.03552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al Bayssari C., Dabboussi F., Hamze M., Rolain J.M. Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J Antimicrob Chemother. 2015;70:950–951. doi: 10.1093/jac/dku469. [DOI] [PubMed] [Google Scholar]
- 65.Okdah L., Leangapichart T., Hadjadj L., Olaitan A.O., Al-Bayssari C., Rizk R. First report of colistin-resistant Klebsiella pneumoniae clinical isolates in Lebanon. J Glob Antimicrob Resist. 2017;9:15–16. doi: 10.1016/j.jgar.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 66.Mathlouthi N., Areig Z., Al Bayssari C., Bakour S., Ali El Salabi A., Ben Gwierif S. Emergence of carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates collected from some Libyan hospitals. Microb Drug Resist. 2015;21:335–341. doi: 10.1089/mdr.2014.0235. [DOI] [PubMed] [Google Scholar]
- 67.Mathlouthi N., El Salabi A.A., Ben Jomàa-Jemili M., Bakour S., Al-Bayssari C., Zorgani A.A. Early detection of metallo-β-lactamase NDM-1– and OXA-23 carbapenemase–producing Acinetobacter baumannii in Libyan hospitals. Int J Antimicrob Agents. 2016;48:46–50. doi: 10.1016/j.ijantimicag.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 68.Olaitan A.O., Berrazeg M., Fagade O.E., Adelowo O.O., Alli J.A., Rolain J.M. Emergence of multidrug-resistant Acinetobacter baumannii producing OXA-23 carbapenemase, Nigeria. Int J Infect Dis. 2013;17:e469–e470. doi: 10.1016/j.ijid.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 69.Olaitan A.O., Diene S.M., Kempf M., Berrazeg M., Bakour S., Gupta S.K. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents. 2014;44:500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 70.Olaitan A.O., Chabou S., Okdah L., Morand S., Rolain J.M. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16:147. doi: 10.1016/S1473-3099(15)00540-X. [DOI] [PubMed] [Google Scholar]
- 71.Leangapichart T., Gautret P., Brouqui P., Mimish Z., Raoult D., Rolain J.M. Acquisition of mcr-1 plasmid-mediated colistin resistance in Escherichia coli and Klebsiella pneumoniae during Hajj 2013 and 2014. Antimicrob Agents Chemother. 2016;60:6998–6999. doi: 10.1128/AAC.01486-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leangapichart T., Gautret P., Griffiths K., Belhouchat K., Memish Z., Raoult D. Acquisition of a high diversity of bacteria during the Hajj pilgrimage, including Acinetobacter baumannii with blaOXA-72 and Escherichia coli with blaNDM-5 carbapenemase genes. Antimicrob Agents Chemother. 2016;60:5942–5948. doi: 10.1128/AAC.00669-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leangapichart T., Dia N.M., Olaitan A.O., Gautret P., Brouqui P., Rolain J.M. Acquisition of extended-spectrum β-lactamases by Escherichia coli and Klebsiella pneumoniae in gut microbiota of pilgrims during the Hajj pilgrimage of 2013. Antimicrob Agents Chemother. 2016;60:3222–3226. doi: 10.1128/AAC.02396-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leangapichart T., Rolain J.M., Memish Z.A., Al-Tawfiq J.A., Gautret P. Emergence of drug resistant bacteria at the Hajj: a systematic review. Travel Med Infect Dis. 2017;18:3–17. doi: 10.1016/j.tmaid.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 75.Leangapichart T., Tissot-Dupont H., Raoult D., Memish Z.A., Rolain J.M., Gautret P. Risk factors for acquisition of CTX-M genes in pilgrims during Hajj 2013 and 2014. J Antimicrob Chemother. 2017;72:2627–2635. doi: 10.1093/jac/dkx155. [DOI] [PubMed] [Google Scholar]
- 76.Diene S.M., Fall B., Kempf M., Fenollar F., Sow K., Niang B. Emergence of the OXA-23 carbapenemase-encoding gene in multidrug-resistant Acinetobacter baumannii clinical isolates from the principal hospital of Dakar, Senegal. Int J Infect Dis. 2013;17:e209–e210. doi: 10.1016/j.ijid.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 77.Diene S.M., Fenollar F., Fall B., Sow K., Niang B., Samba Ba P. CTX-M-15–producing Morganella morganii from Hôpital Principal de Dakar, Senegal. New Microbe. New Infect. 2014;2:46–49. doi: 10.1002/nmi2.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vila-Farrés X., Ferrer-Navarro M., Callarisa A.E., Martí S., Espinal P., Gupta S. Loss of LPS is involved in the virulence and resistance to colistin of colistin-resistant Acinetobacter nosocomialis mutants selected in vitro. J Antimicrob Chemother. 2015;70:2981–2986. doi: 10.1093/jac/dkv244. [DOI] [PubMed] [Google Scholar]
- 79.Mathlouthi N., Ben Lamine Y., Somai R., Bouhalila-Besbes S., Bakour S., Rolain J.-M. Incidence of OXA-23 and OXA-58 carbapenemases coexpressed in clinical isolates of Acinetobacter baumannii in Tunisia. Microb Drug Resist. 2018;24:136–141. doi: 10.1089/mdr.2016.0306. [DOI] [PubMed] [Google Scholar]
- 80.Mathlouthi N., Al-Bayssari C., El Salabi A., Bakour S., Ben Gwierif S., Zorgani A.A. Carbapenemases and extended-spectrum β-lactamases producing Enterobacteriaceae isolated from Tunisian and Libyan hospitals. J Infect Dev Ctries. 2016;10:718–727. doi: 10.3855/jidc.7426. [DOI] [PubMed] [Google Scholar]
- 81.Bakour S., Alsharapy S.A., Touati A., Rolain J.M. Characterization of Acinetobacter baumannii clinical isolates carrying blaOXA-23 carbapenemase and 16S rRNA methylase armA genes in Yemen. Microb Drug Resist. 2014;20:604–609. doi: 10.1089/mdr.2014.0018. [DOI] [PubMed] [Google Scholar]
- 82.Yagoubat M., Ould El-Hadj-Khelil A., Malki A., Bakour S., Touati A., Rolain J.M. Genetic characterisation of carbapenem-resistant Gram-negative bacteria isolated from the University Hospital Mohamed Boudiaf in Ouargla, southern Algeria. J Glob Antimicrob Resist. 2017;8:55–59. doi: 10.1016/j.jgar.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 83.Olaitan A.O., Thongmalayvong B., Akkhavong K., Somphavong S., Paboriboune P., Khounsy S. Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos. J Antimicrob Chemother. 2015;70:3402–3404. doi: 10.1093/jac/dkv252. [DOI] [PubMed] [Google Scholar]
- 84.Zhu Y.G., Johnson T.A., Su J.Q., Qiao M., Guo G.X., Stedtfeld R.D. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci USA. 2013;110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Papagiannitsis C.C., Di Pilato V., Giani T., Giakkoupi P., Riccobono E., Landini G. Characterization of KPC-encoding plasmids from two endemic settings, Greece and Italy. J Antimicrob Chemother. 2016;71:2824–2830. doi: 10.1093/jac/dkw227. [DOI] [PubMed] [Google Scholar]
- 86.Karampatakis T., Antachopoulos C., Iosifidis E., Tsakris A., Roilides E. Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in Greece. Future Microbiol. 2016;11:809–823. doi: 10.2217/fmb-2016-0042. [DOI] [PubMed] [Google Scholar]
- 87.Geffen Y., Adler A., Paikin S., Khabra E., Gorenshtein S., Aronov R. Detection of the plasmid-mediated KPC-2 carbapenem-hydrolysing enzyme in three unusual species of the Enterobacteriaceae family in Israel. J Antimicrob Chemother. 2013;68:719–720. doi: 10.1093/jac/dks443. [DOI] [PubMed] [Google Scholar]
- 88.Bataille J., Brouqui P. Building an intelligent hospital to fight contagion. Clin Infect Dis. 2017;65:S4–S11. doi: 10.1093/cid/cix402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Raoult D. Alice’s living croquet theory. Int J Antimicrob Agents. 2016;47:249. doi: 10.1016/j.ijantimicag.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]