Abstract
The number of isolated new microorganisms has dramatically increased after the readaption of culture using the culturomics approach. Each of these microorganisms is deposited in an international strain collection institute, with its name being attributed and published by the scientist who isolated it. The attributed name is of Latin or Latinized origin and chosen on the basis of the geographical location of the sample collection, the institute or geographical region where the project was being performed, the name of a concerned scientist, and characteristics of the sample or the microorganism. Our institution has played an important role in the isolation of new microorganisms, with the first effort reporting 468 new bacterial species (3% of the bacterial species isolated at least once worldwide) and 327 species isolated for the first time from human beings, which in turn resulted in an increase of 30% of the total number of microorganisms isolated. Additionally, more than 100 giant viruses, including seven new species, have been isolated at our institute. In the present work, after recalling the rules of nomenclature, we detail the naming of the new microorganisms chosen at our laboratory. The most common species name was massiliensis, attributed 161 times. We consider it imperative for the cultivators, who have frequently made considerable efforts in the field of microbial culture, to be the ones who name the newly isolated microorganisms, taking into consideration the Latinized nomenclature standards.
Keywords: Archaea, Culturomics, Giant virus, Human microbiota, New bacterial species, Taxonogenomics
Introduction
Out of the 10 million predicted bacterial species [1], only about 15<thinsp>000 have been cultured. When comparing the number of isolated bacterial species to the number of known archaeal species, viruses and eukaryotes, the number cultured seems small [1], [2]. However, the number of cultured species has increased dramatically after the reintroduction of culture by Lagier et al. in 2012 [3], [4], and approximately 2776 species are currently predicted to be isolated from human samples [2], [5]. Our laboratory contributed significantly in the past 30 years in enlarging the repertoire of microorganisms by isolating 468 new bacterial species, mainly from human origins (Supplementary Table S1).
We succeeded in isolating 327 bacterial species from human samples that had previously been reported to be isolated from environmental sources [6]. This led to a 30% increase in the bacterial repertoire associated with humans [6]. In addition, we also increased the number of archaeal species isolated from human beings by adding two new species, one of which was the first halophilic archaeal species isolated from humans. Finally, our laboratory contributed significantly to the culture of giant viruses by the isolation of 100 different isolates [6].
Naming these new microorganisms is a challenge because the species' name is proposed by the first publication to report its isolation [7]. The name of the new microorganism is chosen by the author reporting its first culture [7]. In this work, we review the main nomenclature rules for naming microorganisms and discuss the contribution of our laboratory to naming.
Bacteria
Process for identification of a new prokaryote
To confirm the novelty of a bacterial species, 16S rRNA gene sequencing is performed along with a BLAST analysis keyed to the National Center for Biotechnology Information nucleotide database for phylogenetic analyses. A threshold of 98.7% of 16S rRNA gene sequence similarity with the phylogenetically closest species with standing in nomenclature was suggested by Stackebrandt and Ebers [8] to classify a new bacterial species. The 16S rRNA gene sequence of each isolated new bacterial species is submitted to the GenBank database, and its strain type is deposited in a strain collection institute.
However, DNA-DNA hybridization, a technique previously considered to be the reference standard to classify a new bacterial species, should be considered as outdated, as it is not reproducible among laboratories and is not cost-effective [9]. With the advent of both matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and genome sequencing, we recently proposed describing new isolated species by using the taxonogenomic approach [9]. The determination of digital DNA-DNA hybridization as well as the average of genomic identity of orthologous gene sequences are also included [9].
Unfortunately, this process is time-consuming—sometimes several years. Accordingly, we proposed a new format (new species announcement) that reports the 16S GenBank accession number, phylogenetic tree, strain deposit accession numbers and main phenotypic characteristics of the new isolated species [10]. Finally, according to the rules of the International Committee on Systematics of Prokaryotes, the name of the new bacterial species must be officially recognized [6].
Naming rules
The current nomenclature for bacterial species requires a capital letter for the genus name and an epithet beginning by a lowercase letter for the species name [7]. Genera and epithet should be Latin or Latinized; the specific epithet is an adjective that must agree with the gender of the generic name [7]. The name can be derived from a person's name (frequently a microbiologist), a geographical location, a phenotypic characteristic (growth condition, colour, biochemical characteristics) or any other origin. Before being classified as a species (frequently for bacteria not yet cultivated but with genomic criteria), a species may provisionally be classified ‘Candidatus.’ For instance, the genus of Escherichia coli, one of the most commonly found bacteria, is named after Theodor Escherich, who isolated it, and its species epithet name refers to the colon, the source of its isolation [11]. Another example is Staphylococcus aureus. Its genus designation was named after its phenotypic characteristics (the grapelike coccus), and its species was named after the golden colour of its colonies [11]. The four most abundant genera are Bacillus, Paenibacillus, Clostridium and Corynebacterium (Fig. 1).
Fig. 1.
Occurrence of different genera of new species isolated by culturomics (Supplementary Material S1). Word cloud generated by Wordle (http://www.wordle.net/). Name size of each species is relative to its occurrence in list reported in Supplementary Material S1.
In order to highlight the naming preferences of new isolates by their cultivators, Table 1 details the 31 officially recognized Rickettsia spp. Thirteen (42%) were named after a geographic location, 12 (39%) in honour of a zoologist or a microbiologist, four (13%) after a vector name and two (6%) after a clinical characteristic of the disease caused by the microorganism.
Table 1.
Validated species of genus Rickettsia
| Name of bacterium | Named after | Type of naming | Year of description |
|---|---|---|---|
| R. aeschlimannii | Aeschlimann, a Swiss zoologist | Individual | 1997 |
| R. africae | Africa | Geographic location | 1996 |
| R. akari | Greek name of mite vector | Vector | 1946 |
| R. amblyomnatis | Amblyomma, the vector | Vector | 2016 |
| R. asembomensis | Asembo, Western Kenya | Geographic location | 2016 |
| R. asiatica | Asia | Geographic location | 2006 |
| R. australis | Australia | Geographic location | 1950 |
| R. belii | Bell, the person who first isolated the bacteria | Individual | 1983 |
| R. buchneri | Buchner, a German biologist | Individual | 2015 |
| R. canadensis | Canada, the country where the organism was isolated | Geographic location | 1967 |
| R. conorii | Conor, who provided with Bruch the first description of spotted fever | Individual | 1932 |
| R. felis | Ctenocephalides felis, in which the organism was first observed by electron microscopy | Vector | 2001 |
| R. heilongjiangensis | Heilongjiang, the Chinese province where the Dermacentor silvarum tick was collected | Geographic location | 2006 |
| R. helvetica | Helvetia, the New Latin name of Switzerland, where the organism was originally isolated | Geographic location | 1993 |
| R. honei | Frank Sandland Hone, an early pioneer in Australian rickettsiology | Individual | 1998 |
| R. hoogstraalii | Dr Harry Hoogstraal, who contributed significantly to the general knowledge on ticks | Individual | 2010 |
| R. japonica | Japan, the country from which the first isolates were identified | Geographic location | 1992 |
| R. massiliae | Latin name of Marseille, where the organism was first isolated | Geographic location | 1993 |
| R. montanensis | Montana, the American state where the organism was first isolated | Geographic location | 1965/1984 |
| R. parkeri | Parker, named after Ralph R. Parker, a founder of the Rocky Mountain Laboratory | Individual | 1965 |
| R. peacock | M. G. Peacock, a well-respected rickettsiologist | Individual | 1997 |
| R. prowazekii | Stanislav von Prowazek, an early investigator of the aetiology of typhus who died of typhus contracted in the course of his studies | Individual | 1916 |
| R. raoultii | Professor Didier Raoult, founder of the WHO–Collaborative Center for Rickettsioses, Borrelioses and Tick-borne Infections in Marseilles (Marseille), France and a major contributor to the study of rickettsiae | Individual | 2008 |
| R. rhipicephali | Natural tick host Rhipicephalus sanguineus | Vector | 1978 |
| R. rickettsii | Howard Taylor Ricketts, for his classic studies of the aetiology of Rocky Mountain spotted fever | Individual | 1919 |
| R. sennetsu | Japanese word meaning ‘glandular fever’ | Geographic location | 1956 |
| R. sibirica | Siberia, a region in northwestern Asia, the name of which is said to come from Sibir, an ancient Tatar fortress at the confluence of the rivers Tobol and Irtysh | Geographic location | 1948 |
| R. slovaca | Slovakia, the country where the organism was first isolated | Geographic location | 1998 |
| R. tamurae | Japanese rickettsiologist Dr Akira Tamura, who contributed to the knowledge of rickettsiae and rickettsioses in Japan | Individual | 2006 |
| R. tsutsumagushi | Two transliterated Japanese ideographs generally interpreted to mean ‘mite disease’ | Clinical characteristics | 1920 |
| R. typhi | Gr. n. tuphos, the name of four kinds of fever, one of which is accompanied by stupor | Clinical characteristics | 1943 |
Our contribution
Over the last 30 years our laboratory has isolated 468 new bacterial species (more than 3% of the bacterial species isolated at least once), mainly from humans and few from animals and environmental samples (Supplementary Table S1).
Most of the species isolated by our team were named in reference to the place where they were first isolated. For instance, 161 bacterial species were named massiliensis and 22 massiliense for Marseille (Southern France), 38 timonensis and seven timonense for the La Timone hospital in Marseille and eight ihumii or ihuae for our institution (IHU, Institut Hospitalo Universitaire Méditerranée Infection). When comparing species epithet names, massiliensis was the most abundant (Fig. 2). When combining together genus and species names, massiliensis remains the most represented (Fig. 3).
Fig. 2.
Occurrence of different species epithets of new species isolated by culturomics (Supplementary Material S1). Word cloud generated by Wordle (http://www.wordle.net/). Name size of each species is relative to its occurrence in list reported in Supplementary Material S1.
Fig. 3.
Different naming of species isolated by culturomics (Supplementary Material S1). Speci epithet and generic name were taken into consideration. Word cloud generated by Wordle (http://www.wordle.net/). Name size of each species is relative to its occurrence in list reported in Supplementary Material S1.
Additionally, some species were named bouchedurhonensis or bouchedurhnonense (seven species) after the department of Bouches-du-Rhône. Pacaensis or pacaense were also attributed to four species after the name of our region, Provence Alpes Côtes d’Azur. In addition, certain species were named after the geographical region where samples were collected. For example, 15 species were named senegalensis or senegalense for Senegal, Western Africa; five jeddahensis or jeddahense for Jeddah, a city in Saudi Arabia; and three saudii for Saudi Arabia. Geographic locations were also used for naming new genera (e.g. Massilia timonae, Timonella senegalensis, Ihubacter massiliensis, Jeddahella massiliensis) [12], [13]. Also, we named Dielma fastidiosa and Nidopella massiliensis after two rural villages (Dielmo and N'Diop), where our research institution has been involved for the past 10 years (Senegal and Western Africa) [14]. Finally, diverse combinations of the places where the samples were collected and the places where the strains were isolated were used. For example, for Senegalemassilia spp., three species were named massiliosenegalensis, and others were named jeddahmassiliensis, jeddahtimonensis, massiliogabonensis or massilioalgeriensis (Supplementary Table S1).
Eleven new bacterial species were named in honour of some famous microbiologists or technicians particularly involved in culture at our laboratory (e.g. Lactobacillus raoulti, Corynebacterium lascolaense, Afipia birgae) [6]. In addition, 18 new genera were named after famous microbiologists (e.g. Raoultibacter spp., Drancourtella spp., Medianikovella massiliensis), other scientists (e.g. Millonella massiliensis, Khelaifiabacter massiliensis) or students working on culturomics (e.g. Hugonella massiliensis, Ndongobacter massiliensis). Beyond our institute, we honoured other scientists with Legionella rowbothamii, named in honour of Timothy Rowbotham, who isolated the majority of known Legionella-like amoebal pathogen strains, and Gorbachella massiliensis, in honour of the famous microbiologist Sherwood Gorbach at the Tufts University School of Medicine, Boston, MA, USA (Supplementary Table S1).
The characteristics of the individuals from whom the samples were collected have been used to name some new species. For example, three bacteria were isolated from children with marasmus (Bacillus marasmi, Blautia marasmi, Paenibacillus marasmiensis), and one was isolated from a child with kwashiorkor (B. kwashiorkori). Other clinical characteristics, such as obesity, were used for three new species (obesi or obesiensis). In addition, a new genus was named Enorma massiliensis because it was isolated from the stool sample of an obese woman. Finally, we named Kallypiga massiliensis after the Greek epithet kallipygos, referring to a statue of Aphrodite having beautifully proportioned buttocks [15]. Other names pertained to the type of sample tested, such as Merdibacter massiliensis or Clostridium merdae isolated from stool samples, Actinomyces urinae from urine samples and Colinsella vaginalis from vaginal samples (Supplementary Table S1).
Finally, phenotypic characteristics such as the type of atmosphere required for growth (Senegalemassilia anaerobia) or the form of the bacteria observed during Gram staining or its shape (Soleaferrea massiliensis is horseshoe shaped) were taken into account for naming. Sometimes a combination of a phenotypic characteristic and the type of sample (Duodenibacillus massiliensis) or the geographic place (Libanicoccus massiliensis, Gabonibacter massiliensis) was used (Supplementary Table S1).
Occurrence of new bacterial species among different microbiota
Among the large panel of new species described by culturomics [16], 80 have been isolated in at least one other type of sample. For example, Ezakiella massiliensis [17], which was isolated for the first time from a vaginal sample, has been then cultivated from stool, urine and respiratory samples. Actinomyces ihumii [18] and Butyricimonas phoceensis [19] were both first isolated from stool samples and then later isolated from vaginal and respiratory samples. Olegusella massiliensis [20] has been isolated in vaginal, stool and urine samples. Enterococcus massiliensis [21] was originally isolated from a stool sample and afterwards from urine and vaginal samples.
From commensal to potentially pathogenic bacteria and vice versa
The microorganisms' repertoire should include the totality of species isolated from human body at different sites or environment because any commensal can become pathogenic in certain conditions. For example, Rickettsia parkerii was detected for the first time in a clinical case in 2004—in other words, 39 years after its first isolation [22]. Being able to detect new species isolated by culturomics in clinical samples or vice versa emphasizes the fact that any commensal can acquire pathogenicity at a certain stage. For instance, among the new bacterial species first isolated from the human gut microbiota by culturomics, 12 were isolated 57 times from clinical samples at our clinical microbiology laboratory, with Peptoniphilus grossensis isolated 18 times from diverse samples (including abscesses) [23]. On the other hand, Paenibacillus provencensis, first isolated from the urine sample of a patient with a urinary infection [24], has been isolated from a stool sample by culturomics. Similarly, Paenibacillus massiliensis, initially isolated from a blood culture bottle in a clinical microbiology laboratory [25], has since been isolated in a stool sample analysed by culturomics. Indeed, the boundary between commensal and pathogenic bacteria remains indistinct, so efforts must be sustained by both clinical microbiology laboratories and culturomics studies in order to increase the identification of new microorganisms. This also highlights the need to update and share MALDI-TOF MS databases for the optimal identification of bacterial species during clinical or research studies [26].
Archaea
Our laboratory isolated seven methanogenic Archaea and two halophilic Archaea, including three new archaeal species that were named after a geographical location (Haloferax massiliensis, Methanobrevibacter massiliense (for Marseille, the city where the strain was first isolated) and Methanomassilicoccus luminyensis (for Luminy, the place where the species was isolated in Marseille)) [27], [28], [29] (Supplementary Table S1). In addition, we first isolated in humans two other Archaea (Methanobrevibacter arboriphilicus and Haloferax alexandrines), and we isolated Methanobrevibacter oralis and Methanobrevibacter millerae for the first time from the human gut [6].
Giant viruses
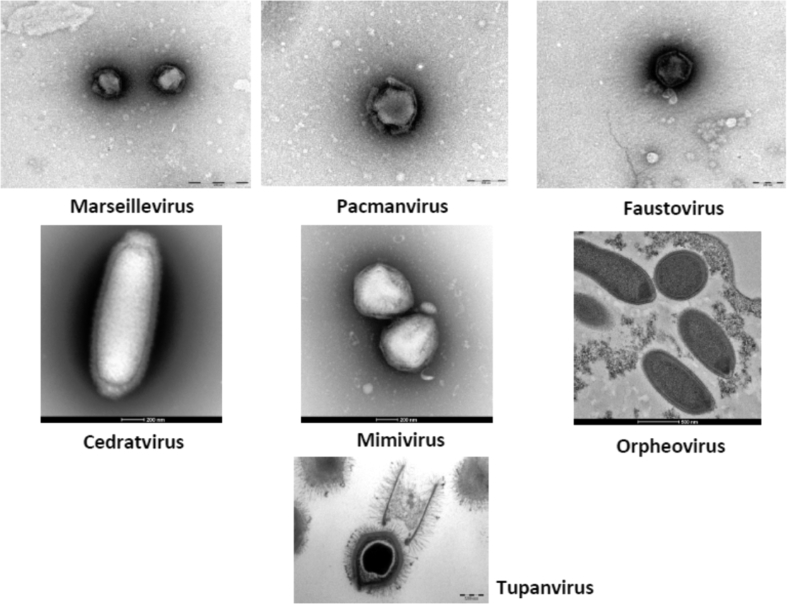
Our laboratory has developed the culture of giant viruses and has succeeded in isolating more than 100 different isolates [6]. Among these isolates, Mimivirus was named after its ability to ‘mimic’ microbes, which previously led to its misidentification as a bacterium [30], and after ‘Mimi the amoeba,’ a tale about evolution invented by the father of one of our scientists (DR) and told to him when he was a child (Fig. 4). Marseillevirus was named after the geographical location of its first culture and description, Marseille [31] (Fig. 4). In addition, we cultured Faustovirus, which was isolated from sewage by a sewage worker named Fausto [32], as well as Kaumoebavirus, which was isolated in a sample from Saudi Arabia by a PhD student using amoeba co-culture and which was supported by a grant from King Abdulaziz University [33]. Cedratvirus was named after its shape, similar to a lemon known as a cedrat (Citrus medica) [34] and Pacmanvirus, which, given its broken-looking capsid, resembles Pac-Man, from the arcade game [35] (Fig. 4). Recently a student isolated a virus from a sewage sample, and because the student compared himself to Orpheus, the hero of the Greek legend who travelled to the underworld to bring back his dead wife, Eurydice [36], the virus was named Orpheovirus. Finally, Tupanvirus, an extraordinary giant virus isolated in Brazil, was named in honour of the Indian Amazonian god Tupa (Abrahão et al., personal communication).
Fig. 4.
Electron microscopy of seven new species of giant viruses isolated by our laboratory.
Conclusion
It is impossible to predict infectious diseases and consequently, as demonstrated here, to anticipate the evolution of a new microorganism. It is essential to allow the cultivators to choose the name of new isolates because their considerable efforts made their cultivation possible. Contrary to the beliefs of some scientists [37], the use of geographical locations in the naming of bacteria has been known for more than a century. The genera Rickettsia is a perfect example, with 13 bacteria named after geographical locations, including five species isolated more than 50 years ago and three others isolated more than 25 years ago (Table 1). Naming species on the basis of a geographical location does not represent a contradiction with the literature or previous efforts within the field.
Acknowledgements
We thank all the contributors involved in culture and those who work in our laboratory. This work has benefited from the support of the French state, managed by the ‘Agence Nationale de la Recherche,’ including the ‘Programme d’Investissement d'avenir’ under the reference Méditerranée Infection 10-IAHU-03. This work was supported by Région Provence Alpes Côte d'Azur and European funding FEDER PRIMI.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.nmni.2018.08.006.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Youle M., Haynes M., Rohwer F. Scratching the surface of biology’s dark matter. In: Witzany G., editor. Viruses: essential agents of life. Springer; New York: 2002. [Google Scholar]
- 2.Hugon P., Dufour J.C., Colson P., Fournier P.E., Sallah K., Raoult D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis. 2015;15:1211–1219. doi: 10.1016/S1473-3099(15)00293-5. [DOI] [PubMed] [Google Scholar]
- 3.Hugon P., Lagier J.C., Colson P., Bittar F., Raoult D. Repertoire of human gut microbes. Microb Pathog. 2017;106:103–112. doi: 10.1016/j.micpath.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La S.B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilen M., Dufour J.C., Lagier J.C., Cadoret F., Daoud Z., Dubourg G. The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. Microbiome. 2018;6:94. doi: 10.1186/s40168-018-0485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagier J.C., Drancourt M., Charrel R., Bittar F., La Scola B., Ranque S. Many more microbes in humans: enlarging the microbiome repertoire. Clin Infect Dis. 2017;65(Suppl. 1):S20–S29. doi: 10.1093/cid/cix404. [DOI] [PubMed] [Google Scholar]
- 7.Truper H.G. How to name a prokaryote: etymological considerations, proposals and practical advice in prokaryotes nomenclature. FEMS Microbiol Rev. 1999;23:231–249. [Google Scholar]
- 8.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 9.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabanh R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64(Pt 2):384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 10.Fournier P.E., Raoult D., Drancourt M. New species announcement: a new format to prompt the description of new human microbial species. New Microbe. New Infect. 2017;15:136–137. doi: 10.1016/j.nmni.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LSPN: List of prokaryotic names with standing in nomenclature. Available at: http://www.bacterio.net/. [DOI] [PMC free article] [PubMed]
- 12.La S.B., Birtles R.J., Mallet M.N., Raoult D. Massilia timonae gen. nov., sp. nov., isolated from blood of an immunocompromised patient with cerebellar lesions. J Clin Microbiol. 1998;36:2847–2852. doi: 10.1128/jcm.36.10.2847-2852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ndongo S., Lagier J.C., Fournier P.E., Raoult D., Khelaifia S. ‘Ihubacter massiliensis’: a new bacterium isolated from the human gut. New Microbes New Infect. 2016;13:104–105. doi: 10.1016/j.nmni.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramasamy D., Lagier J.C., Nguyen T.T., Raoult D., Fournier P.E. Non contiguous–finished genome sequence and description of Dielma fastidiosa gen. nov., sp. nov., a new member of the Family Erysipelotrichaceae. Stand Genom Sci. 2013;8:336–351. doi: 10.4056/sigs.3567059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugon P., Ramasamy D., Robert C., Couderc C., Raoult D., Fournier P.E. Non-contiguous finished genome sequence and description of Kallipyga massiliensis gen. nov., sp. nov., a new member of the family Clostridiales incertae sedis XI. Stand Genomic Sci. 2013;8:500–515. doi: 10.4056/sigs.4047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagier J.C., Dubourg G., Million M., Cadoret F., Bilen M., Fenollar F. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018 doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 17.Diop K., Andrieu C., Michelle C., Armstrong N., Bittar F., Bretelle F. Characterization of a new Ezakiella isolated from the human vagina: genome sequence and description of Ezakiella massiliensis sp. nov. Curr Microbiol. 2018;75:456–463. doi: 10.1007/s00284-017-1402-z. [DOI] [PubMed] [Google Scholar]
- 18.Ndongo S., Khelaifia S., Bittar F., Fournier P.E., Raoult D. ‘Actinomyces ihumii,’ a new bacterial species isolated from the digestive microbiota of a HIV-infected patient. New Microbes New Infect. 2016;12:71–72. doi: 10.1016/j.nmni.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Togo A.H., Diop A., Dubourg G., Nguyen T.T., Andrieu C., Caputo A. Butyricimonas phoceensis sp. nov., a new anaerobic species isolated from the human gut microbiota of a French morbidly obese patient. New Microbes New Infect. 2016;14:38–48. doi: 10.1016/j.nmni.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diop K., Diop A., Bretelle F., Cadoret F., Michelle C., Richez M. Olegusella massiliensis gen. nov., sp. nov., strain KHD7(T), a new bacterial genus isolated from the female genital tract of a patient with bacterial vaginosis. Anaerobe. 2017;44:87–95. doi: 10.1016/j.anaerobe.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Le P.S., Cimmino T., Togo A., Million M., Michelle C., Khelaifia S. Noncontiguous finished genome sequence and description of Enterococcus massiliensis sp. nov. New Microbes New Infect. 2016;12:90–95. doi: 10.1016/j.nmni.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parola P., Paddock C.D., Socolovschi C., Labruna M.B., Mediannikov O., Kernif T. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26:657–702. doi: 10.1128/CMR.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Roux V., Fenner L., Raoult D. Paenibacillus provencensis sp. nov., isolated from human cerebrospinal fluid, and Paenibacillus urinalis sp. nov., isolated from human urine. Int J Syst Evol Microbiol. 2008;58(Pt 3):682–687. doi: 10.1099/ijs.0.65228-0. [DOI] [PubMed] [Google Scholar]
- 25.Roux V., Raoult D. Paenibacillus massiliensis sp. nov., Paenibacillus sanguinis sp. nov. and Paenibacillus timonensis sp. nov., isolated from blood cultures. Int J Syst Evol Microbiol. 2004;54(Pt 4):1049–1054. doi: 10.1099/ijs.0.02954-0. [DOI] [PubMed] [Google Scholar]
- 26.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khelaifia S., Raoult D. Haloferax massiliensis sp. nov., the first human-associated halophilic archaea. New Microbes New Infect. 2016;12:96–98. doi: 10.1016/j.nmni.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dridi B., Fardeau M.L., Ollivier B., Raoult D., Drancourt M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol. 2012;62(Pt 8):1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- 29.Huynh H.T.T., Pignoly M., Drancourt M., Aboudharam G. A new methanogen ‘Methanobrevibacter massiliense’ isolated in a case of severe periodontitis. BMC Res Notes. 2017;10:657. doi: 10.1186/s13104-017-2980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raoult D., Audic S., Robert C., Abergel C., Renesto P., Ogata H. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306(5700):1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 31.Boyer M., Yutin N., Pagnier I., Barrassi L., Fournous G., Espinosa L. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reteno D.G., Benamar S., Khalil J.B., Andreani J., Armstrong N., Klose T. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol. 2015;89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bajrai L.H., Benamar S., Azhar E.I., Robert C., Levasseur A., Raoult D. Kaumoebavirus, a new virus that clusters with faustoviruses and Asfarviridae. Viruses. 2016;8:E278. doi: 10.3390/v8110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreani J., Aherfi S., Bou Khalil J.Y., Di Pinto F., Bitam I., Raoult D. Cedratvirus, a double-cork structured giant virus, is a distant relative of pithoviruses. Viruses. 2016;8:E300. doi: 10.3390/v8110300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andreani J., Khalil J.Y.B., Sevvana M., Benamar S., Di Pinto F., Bitam I. Pacmanvirus, a new giant icosahedral virus at the crossroads between Asfarviridae and faustoviruses. J Virol. 2017;91:e00212–e00217. doi: 10.1128/JVI.00212-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreani J., Khalil J.Y.B., Baptiste E., Hasni I., Michelle C., Raoult D. Orpheovirus IHUMI-LCC2: a new virus among the giant viruses. Front Microbiol. 2017;8:2643. doi: 10.3389/fmicb.2017.02643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truper H.G. Is ‘localimania’ becoming a fashion for prokaryote taxonomists? Int J Syst Evol Microbiol. 2005;55(Pt 5):1753. doi: 10.1099/ijs.0.63953-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.