Abstract
We review reviewing our experience of point-of-care and mass spectrometry in Senegal as two disruptive technologies promoting the rapid diagnosis of infection, permitting better medical management of patients.
Keywords: Arthropods, bacteria, borne vectors, MALDI-TOF, point of care
Introduction
In the case of infectious diseases, it is crucial to clearly establish the diagnosis of infections to identify the pathogens as rapidly as possible. Diagnosis is necessary to prescribe the most adequate treatment; it is also a tool for epidemiology survey and prevention. They are four main techniques to identify an infection and eventually the pathogen. The oldest technique is the identification by microcopy. The second is based on phenotypic identification using methods such as the analytical profile index. The third is based on the detection of some microbial antigens or the patient's immune response. The last one is based on the detection of the genetic material from the causative agent using PCR techniques. Sequencing of amplicons can be performed for a more accurate identification. Unfortunately, these techniques are time and money consuming. They also require skilled staff as well as specific equipment and consumables. Indeed, they cannot be performed outside specialized laboratories. Recently a new method of microbe identification has appeared: matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). This is a simple and revolutionary technique that being more widely and routinely used in clinical microbiology [1], [2]. In addition, the IHU-FMI institute has developed a point-of-care (POC) laboratory for diagnosis, integrating systems based on immunology and/or genetic methods. MALDI-TOF MS and POC are complementary and simple; further, they enable nonspecialized staff to perform diagnostic assays at low cost. Consequently, they may be used as first-line tools in clinical microbiology laboratories in tropical countries.
Here we detail MALDI-TOF MS and POC methods and associated developments performed by the IHU-FMI institute in France but also in tropical Africa (Dakar, Senegal).
MALDI-TOF MS analysis
MALDI-TOF MS produces a mass spectrum that represents the masses of each ionized peptide in a sample (more specifically, the mass/charge ratio). When applied to bacteria, mass spectra may be obtained from either colonies in pure culture on agar or in complex samples such as blood. Consequently, MALDI-TOF MS is particularly suitable for the identification of pathogenic microorganisms in the medical field. It allows the identification of a strain by matching its mass spectrum with a database of Main SPectra from numerous well-characterized bacteria. The IHU-FMI institute possesses eight MALDI-TOF mass spectrometers (MicroFlex LT/SH; Bruker Daltonics, Bremen, Germany).
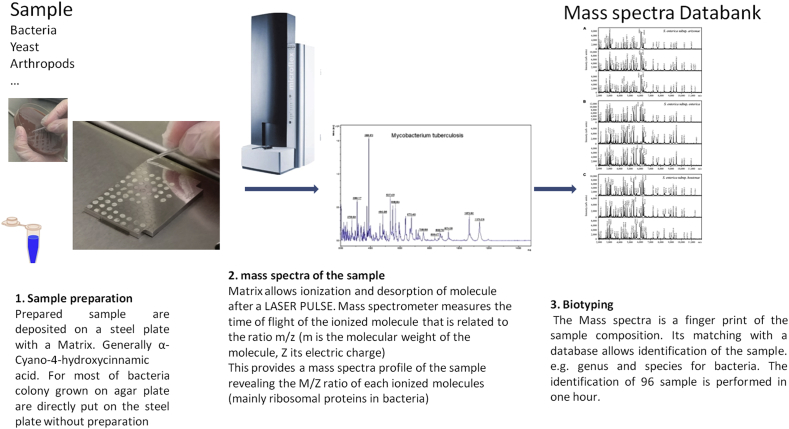
The procedure is simpler and faster than conventional phenotypic and molecular methods (Fig. 1). The cost for analysing a single sample is quite low. Indeed, analysing a 96-sample MALDI plate requires €0.53 of reagents and consumables (alpha-Cyano-4-hydroxycinnamic acid (HCCA), Bacterial Test Standard (BTS), Trifluoroacetic acid (TFA), acetonitrile, ethanol). The plate can be prepared in 10 minutes by a laboratory technician. It is worth noting that a technician can be trained to use MALDI-TOF MS identification (biotyping) in just 2 days. The instrument will take an hour to analyse the plate. The major part of the cost comes from the purchase of the MALDI-TOF MS instrument (€160 k for a complete system) and its maintenance (€25 k per year). One MALDI-TOF mass spectrometer can perform roughly 142<thinsp>000 spot analyses per year at IHU-FMI. From these data, we estimate the consolidated cost of a strain identification at the IHU-FMI at €0.45, including €0.05 for the cost of consumables and labor.
Fig. 1.
Preparation and identification of a sample by MALDI-TOF MS. MALDI-TOF MS, matrix-assisted desorption ionization–time of flight mass spectrometry.
Because the identification by MALDI-TOF MS is simple, fast, reliable and cheap, the IHU-FMI institute has developed protocols and implemented databases for numerous applications (Table 1). In microbiology, MALDI-TOF MS is routinely used to identify bacteria and yeasts [2], [3], [4], [5], [6], [7]. Proofs of concepts have been validated for the Archaea [8] as well as giant viruses [9]. In addition, some bacterial resistances to antibiotics can be surveyed by following the disappearance of the antibiotic peaks degraded by the bacteria [10]. Furthermore, because arthropods are vectors of several tropical diseases, we investigated whether MALDI-TOF MS could be used in entomology [11]. We demonstrated that it is possible to identify at the species level various arthropods [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24] based on the legs at the adult stage for ticks and mosquitoes, or other body parts for fleas and Culicoides, but also at the larval stage [25], [26]. This approach has been further developed for the dual identification of arthropods and vector-borne pathogens such as Plasmodium [27], filariae [28], Borrelia [29] and Rickettsia [30], as well as for the identification of mosquitoes' blood meal origin [31], [32]. This illustrates that MALDI-TOF MS is well suited for vector-borne disease survey. Some developments have been made for cell or tissue identification [33], [34], [35], [36], [37], including tumours [38]. Applications in the field of traceability are also available for fast detection of microorganism contamination in platelet concentrates [39], [40] and animal food origin [41].
Table 1.
MALDI-TOF MS methods developed by IHU-Méditerranée Infection in collaboration with Marseille and Dakar research teams
| MALDI-TOF-MS identification | Reference | |
|---|---|---|
| Bacteria | Mycobacterium | [3] |
| Yersinia | [4] | |
| Legionella | [6] | |
| Bartonella | [5] | |
| Antibiotic resistance | [10] | |
| Archaea | [8] | |
| Yeast | [7] | |
| Giant viruses | [9] | |
| Arthropods | Mosquitoes | [12], [13], [14], [15] |
| Blood meal origin of mosquitoes | [31], [32] | |
| Mosquito larvae | [25], [26] | |
| Vectorized pathogens in mosquitoes | [27], [28] | |
| Fleas | [16], [17] | |
| Triatomines | [18] | |
| Ticks and associated bacteria | [16], [19], [20], [21], [22], [29], [30] | |
| Phlebotomines | [23] | |
| Culicoides | [24] | |
| Tissues or cells ID | Dental pulp | [33] |
| Cancer | [38] | |
| Immune cell | [34], [35], [36], [37] | |
| Traceability | Food origin | [41] |
| Bacterial contaminations of platelets | [39], [40] | |
MALDI-TOF MS, matrix-assisted desorption ionization–time of flight mass spectrometry.
The performance of the identification depends on the sample (protein extraction, molar ratio between sample and matrix), which should be the most reproducible possible, and the quality of the reference spectrum (databases should be exhaustive). Our laboratory notably made huge efforts to cultivate as many bacterial members from the human gut as possible using the culturomics strategy, which enabled the discovery of more than 197 new species [42]. For each of these new species, a reference mass spectrum was added to the in-house bacterial database.
The laboratory also helped install a MALDI-TOF MS system (Vitek MS, bioMérieux, Marcy l’Etoile, France) in the Principal Hospital in Dakar in 2012. This was the first MALDI-TOF MS implemented in Africa. This device is currently routinely used in the clinical microbiology laboratory [7], [43], [44]. It has allowed the discovery and description of new bacterial species [45], [46], [47], [48] as well as the creation of a database for the identification of Culicoides biting midges [24]. This illustrates that MALDI-TOF MS is a robust, reliable and appropriate instrument for Africa. We plan to implement MALDI-TOF MS systems in Algeria and Mali.
Point of care
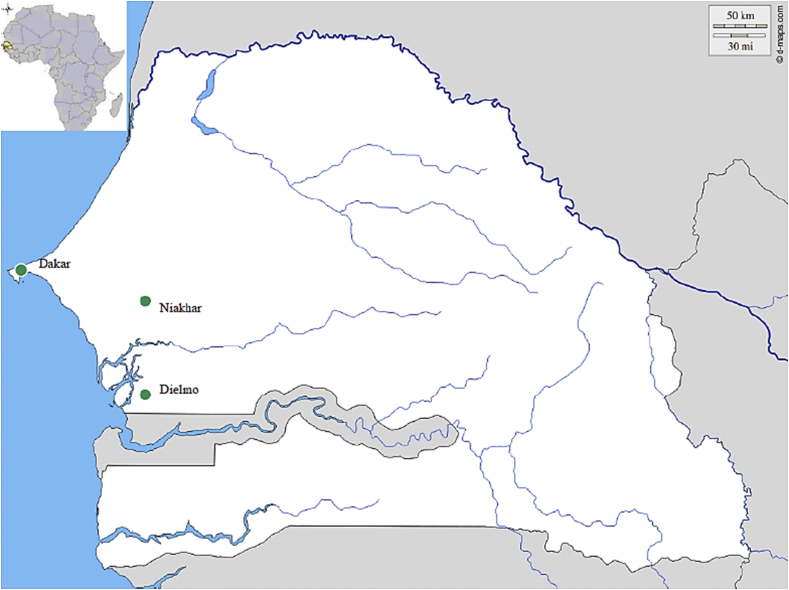
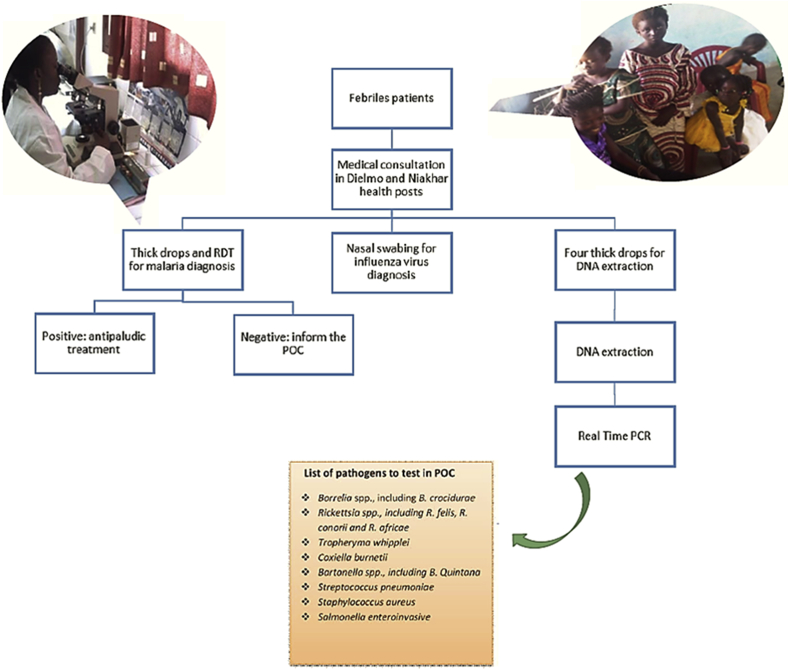
POC laboratories are close-to-patient small laboratories providing rapid (<2 hours) diagnosis of infectious diseases [49]. We have implemented two such POCs in rural Senegal, in Dielmo and Niakhar villages [50] (Fig. 2). Both POCs are operated around the clock by a technician who has been trained in the use of immunochromatographic tests such as the rapid diagnosis test for influenza A and B virus detection and in the use of real-time PCR tests for bacteraemia investigations. Samples are collected in febrile patients who seek care at health posts located in the villages of Dielmo, Ndiop (Dielmo POC) and Toucar, Diohine and Ngayokhem (Niakhar POC). Complete DNA extraction is done on site using the DNA kit for blood extraction. All PCR mixes are prepared monthly in Marseille, then lyophilized and shipped to Senegal. Lyophilization offers the advantage of easy shipment and storage. Before amplification, the mixes are prepared by adding RNAse-free water. Tests are combined in order to provide a syndromic-based diagnosis (Fig. 3) and to guide the medical management of patients. The fact that a total of 5226 tests have been performed within 8 years, including 3352 tests in Dielmo and 1874 in Niakhar (from November 2010 to December 2017), in the two Senegalese POCs testifies to the usefulness of this new approach for rapid and near-patient diagnosis. Moreover, POC data form the basis for the syndrome-based surveillance of infections in rural Senegal [51].
Fig. 2.
Study sites.
Fig. 3.
Chronology of POC diagnosis. POC, point of care.
Conclusion
The implementation and operation of POCs and MALDI-TOF MS in Senegal illustrate the contribution of latest technologies to healthcare in West Africa, as well as the necessity to bypass intermediate technologies and approaches to instead implement cutting-edge methods to promote health.
Acknowledgement
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the program « Investissements d’avenir », reference ANR-10-IAHU-03, the Région Provence Alpes Côte d’Azur and European funding FEDER PRIMI.
Conflict of interest
MD is cofounder and shareholder of Pocramé, a French startup involved in the POC business. The other authors declare that they have no conflict of interest.
References
- 1.Seng P., Drancourt M., Gouriet F., La S.B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 2.Seng P., Rolain J.M., Fournier P.E., La S.B., Drancourt M., Raoult D. MALDI-TOF–mass spectrometry applications in clinical microbiology. Future Microbiol. 2010;5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 3.El Kechine A., Couderc C., Flaudrops C., Raoult D., Drancourt M. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayyadurai S., Flaudrops C., Raoult D., Drancourt M. Rapid identification and typing of Yersinia pestis and other Yersinia species by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. BMC Microbiol. 2010;10:285. doi: 10.1186/1471-2180-10-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fournier P.E., Couderc C., Buffet S., Flaudrops C., Raoult D. Rapid and cost-effective identification of Bartonella species using mass spectrometry. J Med Microbiol. 2009;58:1154–1159. doi: 10.1099/jmm.0.009647-0. [DOI] [PubMed] [Google Scholar]
- 6.Moliner C., Ginevra C., Jarraud S., Flaudrops C., Bedotto M., Couderc C. Rapid identification of Legionella species by mass spectrometry. J Med Microbiol. 2010;59:273–284. doi: 10.1099/jmm.0.014100-0. [DOI] [PubMed] [Google Scholar]
- 7.Gouriet F., Ghiab F., Couderc C., Bittar F., Tissot D.H., Flaudrops C. Evaluation of a new extraction protocol for yeast identification by mass spectrometry. J Microbiol Methods. 2016;129:61–65. doi: 10.1016/j.mimet.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Dridi B., Raoult D., Drancourt M. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of Archaea: towards the universal identification of living organisms. APMIS. 2012;120:85–91. doi: 10.1111/j.1600-0463.2011.02833.x. [DOI] [PubMed] [Google Scholar]
- 9.La Scola B., Campocasso A., N’Dong R., Fournous G., Barrassi L., Flaudrops C. Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry. Intervirology. 2010;53:344–353. doi: 10.1159/000312919. [DOI] [PubMed] [Google Scholar]
- 10.Kempf M., Bakour S., Flaudrops C., Berrazeg M., Brunel J.M., Drissi M. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laroche M., Berenger J.M., Delaunay P., Charrel R., Pradines B., Berger F. Medical entomology: a reemerging field of research to better understand vector-borne infectious diseases. Clin Infect Dis. 2017;65:S30–S38. doi: 10.1093/cid/cix463. [DOI] [PubMed] [Google Scholar]
- 12.Yssouf A., Socolovschi C., Flaudrops C., Ndiath M.O., Sougoufara S., Dehecq J.S. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: an emerging tool for the rapid identification of mosquito vectors. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yssouf A., Parola P., Lindstrom A., Lilja T., L’Ambert G., Bondesson U. Identification of European mosquito species by MALDI-TOF MS. Parasitol Res. 2014;113:2375–2378. doi: 10.1007/s00436-014-3876-y. [DOI] [PubMed] [Google Scholar]
- 14.Yssouf A., Almeras L., Raoult D., Parola P. Emerging tools for identification of arthropod vectors. Future Microbiol. 2016;11:549–566. doi: 10.2217/fmb.16.5. [DOI] [PubMed] [Google Scholar]
- 15.Tandina F., Almeras L., Kone A.K., Doumbo O.K., Raoult D., Parola P. Use of MALDI-TOF MS and culturomics to identify mosquitoes and their midgut microbiota. Parasit Vectors. 2016;9:495. doi: 10.1186/s13071-016-1776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nebbak A., El H.B., Berenger J.M., Bitam I., Raoult D., Almeras L. Comparative analysis of storage conditions and homogenization methods for tick and flea species for identification by MALDI-TOF MS. Med Vet Entomol. 2017;31:438–448. doi: 10.1111/mve.12250. [DOI] [PubMed] [Google Scholar]
- 17.Yssouf A., Socolovschi C., Leulmi H., Kernif T., Bitam I., Audoly G. Identification of flea species using MALDI-TOF/MS. Comp Immunol Microbiol Infect Dis. 2014;37:153–157. doi: 10.1016/j.cimid.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Laroche M., Berenger J.M., Gazelle G., Blanchet D., Raoult D., Parola P. MALDI-TOF MS protein profiling for the rapid identification of Chagas disease triatomine vectors and application to the triatomine fauna of French Guiana—corrigendum. Parasitology. 2018;145:676. doi: 10.1017/S0031182017001342. [DOI] [PubMed] [Google Scholar]
- 19.Diarra A.Z., Almeras L., Laroche M., Berenger J.M., Kone A.K., Bocoum Z. Molecular and MALDI-TOF identification of ticks and tick-associated bacteria in Mali. PLoS Negl Trop Dis. 2017;11 doi: 10.1371/journal.pntd.0005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumsa B., Laroche M., Almeras L., Mediannikov O., Raoult D., Parola P. Morphological, molecular and MALDI-TOF mass spectrometry identification of Ixodid tick species collected in Oromia, Ethiopia. Parasitol Res. 2016;115:4199–4210. doi: 10.1007/s00436-016-5197-9. [DOI] [PubMed] [Google Scholar]
- 21.Yssouf A., Flaudrops C., Drali R., Kernif T., Socolovschi C., Berenger J.M. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid identification of tick vectors. J Clin Microbiol. 2013;51:522–528. doi: 10.1128/JCM.02665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yssouf A., Almeras L., Berenger J.M., Laroche M., Raoult D., Parola P. Identification of tick species and disseminate pathogen using hemolymph by MALDI-TOF MS. Ticks Tick Borne Dis. 2015;6:579–586. doi: 10.1016/j.ttbdis.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Lafri I., Almeras L., Bitam I., Caputo A., Yssouf A., Forestier C.L. Identification of Algerian field-caught phlebotomine sand fly vectors by MALDI-TOF MS. PLoS Negl Trop Dis. 2016;10 doi: 10.1371/journal.pntd.0004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambou M., Aubadie-Ladrix M., Fenollar F., Fall B., Bassene H., Almeras L. Comparison of matrix-assisted laser desorption ionization–time of flight mass spectrometry and molecular biology techniques for identification of Culicoides (Diptera: Ceratopogonidae) biting midges in Senegal. J Clin Microbiol. 2015;53:410–418. doi: 10.1128/JCM.01855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dieme C., Yssouf A., Vega-Rua A., Berenger J.M., Failloux A.B., Raoult D. Accurate identification of Culicidae at aquatic developmental stages by MALDI-TOF MS profiling. Parasit Vectors. 2014;7:544. doi: 10.1186/s13071-014-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nebbak A., Koumare S., Willcox A.C., Berenger J.M., Raoult D., Almeras L. Field application of MALDI-TOF MS on mosquito larvae identification. Parasitology. 2018;145:677–687. doi: 10.1017/S0031182017001354. [DOI] [PubMed] [Google Scholar]
- 27.Laroche M., Almeras L., Pecchi E., Bechah Y., Raoult D., Viola A. MALDI-TOF MS as an innovative tool for detection of Plasmodium parasites in Anopheles mosquitoes. Malar J. 2017;16:5. doi: 10.1186/s12936-016-1657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahir D., Almeras L., Varloud M., Raoult D., Davoust B., Parola P. Assessment of MALDI-TOF mass spectrometry for filariae detection in Aedes aegypti mosquitoes. PLoS Negl Trop Dis. 2017;11 doi: 10.1371/journal.pntd.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fotso Fotso A., Mediannikov O., Diatta G., Almeras L., Flaudrops C., Parola P. MALDI-TOF mass spectrometry detection of pathogens in vectors: the Borrelia crocidurae/Ornithodoros sonrai paradigm. PLoS Negl Trop Dis. 2014;8 doi: 10.1371/journal.pntd.0002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yssouf A., Almeras L., Terras J., Socolovschi C., Raoult D., Parola P. Detection of Rickettsia spp. in ticks by MALDI-TOF MS. PLoS Negl Trop Dis. 2015;9 doi: 10.1371/journal.pntd.0003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niare S., Almeras L., Tandina F., Yssouf A., Bacar A., Toilibou A. MALDI-TOF MS identification of Anopheles gambiae Giles blood meal crushed on Whatman filter papers. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niare S., Berenger J.M., Dieme C., Doumbo O., Raoult D., Parola P. Identification of blood meal sources in the main African malaria mosquito vector by MALDI-TOF MS. Malar J. 2016;15:87. doi: 10.1186/s12936-016-1152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran T.N., Aboudharam G., Gardeisen A., Davoust B., Bocquet-Appel J.P., Flaudrops C. Classification of ancient mammal individuals using dental pulp MALDI-TOF MS peptide profiling. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouedraogo R., Daumas A., Capo C., Mege J.L., Textoris J. Whole-cell MALDI-TOF mass spectrometry is an accurate and rapid method to analyze different modes of macrophage activation. J Vis Exp. 2013;(82):50926. doi: 10.3791/50926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouedraogo R., Textoris J., Daumas A., Capo C., Mege J.L. Whole-cell MALDI-TOF mass spectrometry: a tool for immune cell analysis and characterization. Methods Mol Biol. 2013;1061:197–209. doi: 10.1007/978-1-62703-589-7_12. [DOI] [PubMed] [Google Scholar]
- 36.Ouedraogo R., Daumas A., Ghigo E., Capo C., Mege J.L., Textoris J. Whole-cell MALDI-TOF MS: a new tool to assess the multifaceted activation of macrophages. J Proteomics. 2012;75:5523–5532. doi: 10.1016/j.jprot.2012.07.046. [DOI] [PubMed] [Google Scholar]
- 37.Ouedraogo R., Flaudrops C., Ben A.A., Capo C., Raoult D., Mege J.L. Global analysis of circulating immune cells by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brioude G., Bregeon F., Trousse D., Flaudrops C., Secq V., De D.F. Correction: rapid diagnosis of lung tumors, a feasibility study using MALDI-TOF mass spectrometry. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chetouane Y., Dubourg G., Gallian P., Flaudrops C., Chiaroni J., Chabriere E. Rapid identification of microorganisms from platelet concentrates by matrix-assisted laser desorption ionization time-of-flight mass spectrometry after short-term incubation on liquid medium. Transfusion. 2018;58:766–773. doi: 10.1111/trf.14430. [DOI] [PubMed] [Google Scholar]
- 40.Chetouane Y., Dubourg G., Gallian P., Delerce J., Levasseur A., Flaudrops C. In vitro detection of bacterial contamination in platelet concentrates by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: a preliminary study. J Med Microbiol. 2017;66:1523–1530. doi: 10.1099/jmm.0.000533. [DOI] [PubMed] [Google Scholar]
- 41.Flaudrops C., Armstrong N., Raoult D., Chabriere E. Determination of the animal origin of meat and gelatin by MALDI-TOF-MS. J Food Comp Anal. 2015;41:104–112. [Google Scholar]
- 42.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Lo C.I., Fall B., Sambe-Ba B., Diawara S., Gueye M.W., Mediannikov O. MALDI-TOF mass spectrometry: a powerful tool for clinical microbiology at Hopital Principal de Dakar, Senegal (West Africa) PLoS One. 2015;10 doi: 10.1371/journal.pone.0145889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fall B., Lo C.I., Samb-Ba B., Perrot N., Diawara S., Gueye M.W. The ongoing revolution of MALDI-TOF mass spectrometry for microbiology reaches tropical Africa. Am J Trop Med Hyg. 2015;92:641–647. doi: 10.4269/ajtmh.14-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo C.I., Sankar S.A., Fall B., Sambe-Ba B., Diawara S., Gueye M.W. High-quality draft genome sequence and description of Haemophilus massiliensis sp. nov. Stand Genomic Sci. 2016;11:31. doi: 10.1186/s40793-016-0150-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Lo C.I., Sankar S.A., Fall B., Sambe-Ba B., Mediannikov O., Robert C. High-quality genome sequence and description of Paenibacillus dakarensis sp. nov. New Microbe. New Infect. 2016;10:132–141. doi: 10.1016/j.nmni.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sankar S.A., Lo C.I., Fall B., Sambe-Ba B., Mediannikov O., Diallo I. Noncontiguous finished genome sequence and description of Weeksella massiliensis sp. nov. New Microbe. New Infect. 2015;8:89–98. doi: 10.1016/j.nmni.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo C.I., Padhamanabhan R., Fall B., Sambe-Ba B., Mediannikov O., Nguyen T.T. Noncontiguous finished genome sequence and description of Necropsobacter massiliensis sp. nov. New Microbe. New Infect. 2015;8:41–50. doi: 10.1016/j.nmni.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drancourt M., Michel-Lepage A., Boyer S., Raoult D. The point-of-care laboratory in clinical microbiology. Clin Microbiol Rev. 2016;29:429–447. doi: 10.1128/CMR.00090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokhna C., Mediannikov O., Fenollar F., Bassene H., Diatta G., Tall A. Point-of-care laboratory of pathogen diagnosis in rural Senegal. PLoS Negl Trop Dis. 2013;7 doi: 10.1371/journal.pntd.0001999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abat C., Colson P., Chaudet H., Rolain J.M., Bassene H., Diallo A. Implementation of syndromic surveillance systems in two rural villages in Senegal. PLoS Negl Trop Dis. 2016;10 doi: 10.1371/journal.pntd.0005212. [DOI] [PMC free article] [PubMed] [Google Scholar]