Abstract
Information on disease process and pathogenicity mechanisms is important for understanding plant disease. Spring black stem and leaf spot caused by the necrotrophic pathogen Phoma medicaginis var. medicaginis Malbr. & Roum causes large losses to alfalfa. However, till now, little is known about alfalfa-P. medicagnis interactions and the pathogenicity mechanisms of the pathogen. Here, alfalfa inoculated with P. medicaginis was subjected to GC-MS based metabolic profiling. The metabolic response in P. medicaginis-inoculated and mock-inoculated alfalfa leaves was assessed at 2, 4, 6, 8, 12, 16, 20, 24, 26 and 28 days post inoculation. In total, 101 peaks were detected in the control and inoculated groups, from which 70 metabolites were tentatively identified. Using multivariate analysis, 16 significantly regulated compounds, including amino acids, nitrogen-containing compounds and organic acids, polyols, fatty acids, and sugars were tentatively identified (Variable importance values, VIP>1.0 and p <0.05). Among these metabolites, the levels of malate, 5-oxoproline, palmitic acid and stearic acid were increased significantly in P. medicaginis-infected alfalfa leaves compared to the controls. In contrast, the levels ofγ-aminobutyric acid and 2-pyrrolidinone were significantly decreased in infected leaves compared to the controls. Further metabolic pathway analysis of the 16 significantly regulated compounds demonstrated that glycolysis, the tricarboxylic acid cycle, and β-oxidation of fatty acids were significantly induced in the alfalfa leaves at later stages of P. medicaginis infection. The strong induction of tricarboxylic acid cycle pathways at later infection stages caused by the pathogen may induce senescence in the alfalfa leaves, leading to plant death. However, intermediate metabolites of these metabolic pathways, and inositol phosphate, glutathione, the metabolic pathways of some amino acids accumulated rapidly and strongly at early stages of infection, which may enhance the ability of alfalfa to resist necrotrophic P. medicaginis disease. Understanding metabolic pathways is essential for understanding pathogenesis.
Introduction
Alfalfa (Medicago sativa L.) is grown worldwide as a perennial high quality forage [1]. Alfalfa acreage in China has rapidly increased since the Chinese government initiated a program to promote the milk and alfalfa industries in 2012 [2]. The alfalfa acreage in 2015 was 4.7 million ha, which is planned to increase to more than 133 thousand ha in 2020 [3]. However, diseases limit alfalfa production [4]. Spring black stem and leaf spot caused by Phoma medicaginis var. medicaginis Malbr. & Roum commonly occurs in China [5–7], North America, Europe, and Africa [8–10], causing yield losses, decreasing forage quality [11], and affecting the health of livestock [12]. Previous studies showed that one variety, Aohan, bred in Inner Mongolia, China was highly resistant among the 70 alfalfa varieties tested from many counties including China, America, France, Australia, and Canada. [13,14]. In two moderately resistant alfalfa genotypes, the leaves delayed spore germination, penetration, development of mycelium, pycnidia formation, and symptom development compared with leaves from susceptible alfalfa genotypes [11]. In recent studies of mechanisms of pathogenesis and disease-resistance, we showed that P. medicaginis decreased the transfer rate and capture efficiency of photosynthetic electrons, the amount of non-photochemical quenching (qN), and the carboxylation efficiency (CE) in alfalfa leaves [15]. Activity of the antioxidant enzymes, superoxide dismutase (SOD), catalase (CAT), and phenylalanine ammonia 1yase (PAL), increased earlier and to higher levels in resistant alfalfa varieties than in susceptible alfalfa varieties [13]. During infection of alfalfa with P. medicaginis, the fungitoxic phytoalexins, isoflavonoids such as medicarpin, sativan, formononetin 7-O-glucoside, and malonylated formononetin 7-O-glucoside increased in alfalfa leaves infected by P. medicaginis [16–18], and the activity of enzymes, and corresponding transcript level, involved in isoflavonoid biosynthesis including isoflavone reductase (IFR), PAL, and chalcone synthase (CHS) were also upregulated [16]. β-1, 3-glucanase was stimulated in the transcription of leaf tissues and cell suspensions of alfalfa [19]. Currently, little is known about the disease process and mechanism of pathogenesis for P. medicaginis on alfalfa, information which is important for the development of novel disease-control strategies and breeding resistant varieties. In this study, we analyzed the expressions of metabolites in P. medicaginis-inoculated and mock-inoculated alfalfa plant leaves using GC-MS. The up- or down-regulated metabolites were further analysed for their metabolism pathways and a complex metabolic network was developed. This complex metabolic network should extend our understanding of the mechanisms controlling P. medicaginis pathogenicity and alfalfa tolerance to this pathogen.
Materials and methods
Experimental design
The strain of P. medicaginis var. medicaginis, LYZ00164 (Genbank accession KP207577) was isolated from alfalfa leaves collected from the field in Gansu province, China. The isolate was cultured on potato dextrose agar (PDA) media at 25 °C without light in an incubator for 3 weeks. A spore suspension was prepared with sterile distilled water, and the number of spores in the suspension was adjusted to 1–1.5×106 spores⋅mL−1 with a hemocytometer [13]. Seeds of the susceptible alfalfa cultivar, Derby [15] were sown in 70 pots (6 seeds per pot) containing sterilized soil, and grown inside a greenhouse set at 25/15 °C (day/night) for two months. Thirty pots of seedlings were spray inoculated with a spore suspension of P. medicaginis using an atomizer, and the seedlings were covered with black plastic bags for 48 h to promote infection, then, grown without plastic bags in the greenhouse. Thirty additional pots of seedlings were sprayed with sterile water as healthy controls, and 10 pots of seedlings were retained for positive controls to observe symptoms and disease development. Symptoms were observed after inoculation. All plants were grown under the same conditions. Fully expanded leaves at the same development stage were harvested from the P. medicaginis- or water-inoculated plants at 2, 4, 6, 8, 12, 16, 20, 24, 26 and 28 days post inoculation (dpi). Three pots with six plants each were sampled at each sampling date. The harvested leaves were quickly frozen with liquid nitrogen and stored at -80°C until use.
Metabolite extraction, determination and identification
Leaf samples (30 mg each, n = 3) were pulverised individually in mortars, and extracted with MeOH: H2O (3 × 0.5 mL, 4:1, v/v) and 75 μL ribitol (2 μg/μL–1, Shanghai Yuanye Biotechnology Co. LTD), which was used as an internal standard. MeOH soluble fractions were dried individually with nitrogen and each sample was gently mixed with 45 μL methoxyamine hydrochloride (20μg/μL–1, J&K Scientific Ltd.). The mixtures were incubated at 30 °C for 90 min followed by addition of 45 μL N,O-bis-(trimethylsilyl)-trifluoroacetamide (J&K Scientific Ltd.) to each sample and incubation at 70 °C for 30 min. Each sample was diluted with 90 μL hexane prior to injection as described previously [20].
An Agilent 7890A/5975C gas chromatograph-tandem mass spectrometer equipped with an autosampler 7693 (Agilent Technologies, America) was used for GC-MS detection. The detection conditions were as follows: ion source temperature at 230 °C, electron beam at 70 eV, Agilent 19191S-433 column with 30 m × 0.25 mm internal diameter and 0.25 μm film thickness, helium gas flow rate at 1 ml/min-1, splitless injection, and mass range at m/z 50–650. Constituents were determined under the following conditions: 80 to 240 °C at 6 °C per min, 240 to 280 °C at 10 °C per min, and 280°C for 10 min. The injection temperature was 270 °C. Three replication for GC-MS.
Compounds in leaves were tentatively identified with Automated Mass Spectral Deconvolution and Identification System (AMDIS), our in-house mass spectra library and the standard mass spectral database (National Institute of Standards and Technology, NIST, version 11.0). The signal/noise (S/N) of the tentatively identified compound was higher than 10 in the AMDIS and the compound shared greater than or equal to 80% similarity with the known compounds in the database, indicating that the metabolite identification was reliable [21,22].
The identification of significantly different metabolites and their metabolic pathway construction
The amounts of tentatively identified compounds were normalised with the mean value of the internal standard. The normalised data were imported into SIMCA14.1 (Umetrics, Sweden) for further multi-variate statistical analysis, including principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA). The Q2 value is a prediction error obtained through OPLS-DA analyses, and values greater than or equal to 0.5 are generally considered to have good predictive capability [23]. To validate OPLS-DA models, 200 random permutation tests were performed. The variable importance values (VIP) were obtained through OPLS-DA analyses. When the VIP value of a metabolite exceeded 1.0, this metabolite was considered to be a potentially regulated metabolite and its variables were further analysed using a two-way analysis of variance. When the p value was less than 0.05, this metabolite was considered to be a significantly regulated metabolite compared to the controls [24]. The significantly regulated metabolites were analysed using the MetaboAnalyst platform (www.metaboanalyst.ca/). The final metabolic network was established according to KEGG (http://www.genome.jp/kegg/) [25]. A heat map was drawn with Mev 4.8. Student’s t test was used for comparison between groups at the same time point.
Results
Symptoms and disease development
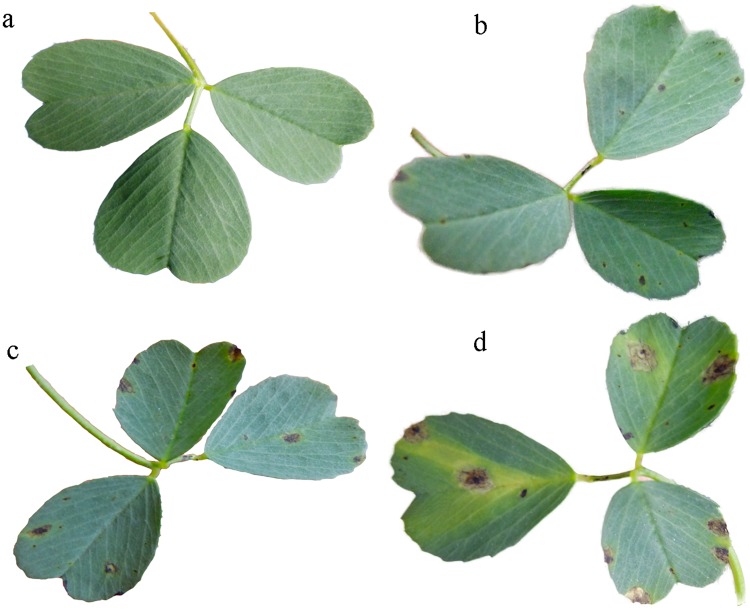
Symptoms were observed as small black spots on inoculated leaf blades on the sixth day post inoculation (Fig 1b). Subsequently, lesions enlarged and coalesced (Fig 1c and 1d), while no such symptoms appeared on the leaf blades of controls (Fig 1a).
Fig 1. Symptoms on alfalfa inoculated with Phoma medicaginis var. medicaginis.
The symptoms observed at 8 d (b), 19 d (c) and 26 d (d) after inoculation, and without inoculation (controls) (a).
Metabolite profiles and metabolite response to pathogen stress
In total, 101 peaks were detected by GC-MS. From the peaks, 70 unique compounds were tentatively identified, including 23 carbohydrates and alcohols, 23 amino acids, 5 nitrogen-containing compounds, 10 organic acids, 5 lipids, 4 other compounds (Table 1).
Table 1. Identification of 70 metabolites from the control and inoculated leaves and the determination of 16 significantly regulated metabolites associated with P. medicaginis infection.
| No. | Compound | RT (min) | Characteristic fragment | Similarity | Inoculated group | Control group | p | VIP |
|---|---|---|---|---|---|---|---|---|
| 1 | Glycineab | 10.452 | 102 | 90 | 0.025±0.001 | 0.035±0.002 | 0.001 | 1.199 |
| 2 | Leucineab | 14.581 | 158 | 95 | 0.270±0.016 | 0.362±0.020 | 0.001 | 1.214 |
| 3 | Isoleucineab | 15.124 | 158 | 92 | 0.475±0.030 | 0.768±0.050 | 0.001 | 1.426 |
| 4 | Tyrosineab | 27.909 | 218 | 96 | 0.119±0.011 | 0.228±0.013 | 0.001 | 1.577 |
| 5 | Methionineab | 20.166 | 176 | 94 | 0.018±0.005 | 0.066±0.006 | 0.001 | 1.532 |
| 6 | 5-oxoprolineab | 20.233 | 156 | 95 | 0.161±0.011 | 0.121±0.006 | 0.001 | 1.146 |
| 7 | Lysineab | 23.843 | 156 | 96 | 0.070±0.004 | 0.093±0.006 | 0.001 | 1.088 |
| 8 | GABAab | 15.201 | 102 | 90 | 0.149±0.016 | 0.398±0.055 | 0.001 | 1.204 |
| 9 | 2-pyrrolidinonea | 11.037 | 142 | 92 | 0.029±0.003 | 0.050±0.002 | 0.001 | 1.592 |
| 10 | Myoinositola | 28.399 | 217 | 85 | 0.155±0.016 | 0.263±0.018 | 0.001 | 1.258 |
| 29.109 | 217 | 80 | ||||||
| 11 | Inositol 1,3,4,5,6-pentakisphosphatea | 30.537 | 554 | 85 | 0.197±0.016 | 0.380±0.047 | 0.001 | 1.142 |
| 12 | Sucrosea | 37.348 | 361 | 94 | 1.919±0.197 | 3.285±0.252 | 0.001 | 1.285 |
| 13 | Malatea | 19.653 | 233 | 91 | 1.585±0.058 | 0.940±0.082 | 0.001 | 1.556 |
| 14 | Malonic acida | 12.828 | 233 | 95 | 0.117±0.014 | 0.381±0.041 | 0.001 | 1.549 |
| 15 | Palmitic acidab | 29.374 | 313 | 95 | 0.530±0.023 | 0.339±0.022 | 0.001 | 1.442 |
| 16 | Stearic acidab | 32.325 | 341 | 99 | 0.537±0.028 | 0.327±0.030 | 0.001 | 1.321 |
| 17 | Acetic acid, hydroxy-a | 9.232 | 205 | 92 | 0.014±0.000 | 0.012±0.000 | 0.001 | 0.901 |
| 18 | Pyruvic acid a | 9.601 | 217 | 93 | 0.014±0.001 | 0.011±0.001 | 0.001 | 0.95 |
| 19 | Alanineab | 10.025 | 116 | 93 | 0.276±0.013 | 0.297±0.017 | 0.163 | 0.762 |
| 20 | Ethanedioic acida | 10.867 | 190 | 94 | 0.016±0.001 | 0.018±0.001 | 0.004 | 0.834 |
| 21 | α-Aminobutyric acida | 11.949 | 130 | 88 | 0.022±0.002 | 0.022±0.002 | 0.907 | 0.776 |
| 22 | Ureaa | 12.250 | 261 | 86 | 0.034±0.002 | 0.028±0.002 | 0.038 | 0.989 |
| 23 | beta-Alaninea | 12.382 | 102 | 86 | 0.127±0.013 | 0.224±0.030 | 0.001 | 0.914 |
| 24 | Valineab | 13.174 | 144 | 89 | 1.094±0.088 | 1.057±0.083 | 0.408 | 0.820 |
| 25 | Benzoic acida | 13.792 | 179 | 80 | 0.081±0.009 | 0.120±0.010 | 0.001 | 0.986 |
| 26 | Phosphatea | 14.658 | 299 | 89 | 0.144±0.018 | 0.111±0.010 | 0.024 | 0.817 |
| 27 | Glycerola | 14.724 | 205 | 93 | 0.327±0.022 | 0.313±0.014 | 0.522 | 0.768 |
| 28 | Succinatea | 15.495 | 247 | 85 | 0.995±0.124 | 0.888±0.060 | 0.108 | 0.772 |
| 29 | Glyceric acida | 16.068 | 189 | 92 | 0.057±0.003 | 0.061±0.003 | 0.069 | 0.756 |
| 30 | Fumaratea | 16.270 | 245 | 83 | 0.020±0.001 | 0.022±0.001 | 0.086 | 0.735 |
| 31 | Pipecolic acida | 16.691 | 156 | 91 | 0.053±0.009 | 0.039±0.002 | 0.076 | 0.884 |
| 32 | Serineab | 16.805 | 204 | 91 | 1.106±0.066 | 1.122±0.065 | 0.747 | 0.723 |
| 33 | Threonineab | 17.420 | 218 | 95 | 0.670±0.049 | 0.670±0.047 | 1 | 0.799 |
| 34 | 2-Piperidone, amino-a | 18.833 | 128 | 80 | 0.045±0.004 | 0.051±0.004 | 0.004 | 0.827 |
| 35 | 2,2-hydroxymalonate a | 19.028 | 308 | 95 | 0.058±0.003 | 0.072±0.003 | 0.001 | 0.801 |
| 36 | Aspartic acidab | 20.256 | 232 | 94 | 0.737±0.130 | 0.487±0.022 | 0.022 | 0.906 |
| 37 | Threonic acida | 21.248 | 292 | 95 | 0.147±0.008 | 0.169±0.006 | 0.001 | 0.946 |
| 38 | Threo-2-pentulosea | 21.703 | 306 | 80 | 1.575±0.098 | 1.369±0.073 | 0.03 | 0.864 |
| 39 | Ornithineab | 22.122 | 142 | 91 | 0.260±0.032 | 0.297±0.027 | 0.112 | 0.801 |
| 40 | Glutamic acidab | 22.235 | 246 | 94 | 0.368±0.017 | 0.360±0.020 | 0.742 | 0.517 |
| 41 | Phenylalanineab | 22.335 | 192 | 94 | 0.792±0.070 | 0.917±0.074 | 0.018 | 0.886 |
| 42 | Arabitola | 22.938 | 147 | 80 | 0.108±0.005 | 0.119±0.004 | 0.05 | 0.812 |
| 43 | Asparagineab | 23.308 | 231 | 95 | 4.276±0.407 | 4.110±0.323 | 0.467 | 0.799 |
| 44 | Arabinosea | 23.586 | 217 | 90 | 0.142±0.007 | 0.126±0.003 | 0.004 | 0.983 |
| 45 | α-Aminoadipic acida | 24.027 | 260 | 89 | 0.052±0.004 | 0.042±0.003 | 0.001 | 0.885 |
| 46 | Phosphoric acid, 2,3- hydroxypropyl estera | 25.049 | 357 | 84 | 0.063±0.005 | 0.071±0.004 | 0.147 | 0.827 |
| 47 | 2-keto-1-guconic acida | 25.193 | 292 | 92 | 0.111±0.005 | 0.114±0.004 | 0.526 | 0.679 |
| 48 | Fructofuranosea | 25.906 | 217 | 85 | 0.294±0.035 | 0.390±0.054 | 0.033 | 0.796 |
| 49 | Citratea | 26.062 | 273 | 94 | 0.436±0.030 | 0.515±0.031 | 0.05 | 0.811 |
| 50 | Pinitola | 26.412 | 217 | 90 | 8.838±0.361 | 8.151±0.154 | 0.017 | 0.839 |
| 51 | Adeninea | 26.701 | 264 | 92 | 0.077±0.005 | 0.078±0.004 | 0.702 | 0.619 |
| 52 | Fructosea | 27.098 | 217 | 93 | 0.524±0.057 | 0.710±0.070 | 0.004 | 0.871 |
| 53 | Sorbosea | 27.279 | 217 | 89 | 0.628±0.070 | 0.899±0.100 | 0.001 | 0.895 |
| 54 | Galactopyranosidea | 27.506 | 204 | 88 | 1.529±0.106 | 2.139±0.054 | 0.002 | 0.896 |
| 55 | Galactosea | 27.542 | 319 | 86 | 0.940±0.120 | 0.946±0.067 | 0.001 | 0.652 |
| 27.765 | 319 | 90 | ||||||
| 56 | Histidineab | 27.627 | 254 | 84 | 0.206±0.027 | 0.262±0.023 | 0.005 | 0.874 |
| 57 | Ethyl-glucopyranosidea | 28.138 | 204 | 84 | 0.404±0.071 | 0.497±0.056 | 0.003 | 0.725 |
| 58 | Glucosea | 28.844 | 204 | 93 | 0.180±0.017 | 0.291±0.045 | 0.005 | 0.881 |
| 31.282 | 319 | 87 | ||||||
| 34.968 | 204 | 91 | ||||||
| 59 | Gluconic acida | 29.110 | 333 | 80 | 0.106±0.005 | 0.132±0.004 | 0.001 | 0.925 |
| 60 | Tryptophanab | 32.253 | 202 | 93 | 0.324±0.029 | 0.343±0.023 | 0.223 | 0.763 |
| 61 | Lactulosea | 33.610 | 204 | 82 | 0.047±0.002 | 0.049±0.002 | 0.472 | 0.812 |
| 62 | Glyceryl-glycosidea | 33.813 | 204 | 90 | 0.175±0.012 | 0.172±0.004 | 0.748 | 0.618 |
| 63 | Glucuronic acida | 34.502 | 204 | 80 | 0.131±0.004 | 0.124±0.004 | 0.118 | 0.834 |
| 64 | Uridinea | 35.220 | 217 | 90 | 0.189±0.008 | 0.164±0.007 | 0.001 | 0.829 |
| 65 | 2-Monopalimitoylglycerola | 36.077 | 129 | 88 | 0.043±0.001 | 0.041±0.002 | 0.254 | 0.719 |
| 66 | Monopalmitina | 36.399 | 371 | 81 | 0.690±0.062 | 0.951±0.053 | 0.001 | 0.968 |
| 67 | Turanosea | 36.585 | 361 | 82 | 0.074±0.003 | 0.082±0.003 | 0.035 | 0.917 |
| 68 | 2-Monostearina | 37.922 | 129 | 80 | 0.077±0.004 | 0.077±0.003 | 0.817 | 0.578 |
| 69 | Monostearina | 38.317 | 399 | 86 | 0.163±0.006 | 0.188±0.007 | 0.003 | 0.915 |
| 70 | Gentiobiosea | 39.690 | 204 | 83 | 0.035±0.002 | 0.039±0.002 | 0.063 | 0.775 |
RT = retention time of compound.
a Identified by NIST data base.
b Identified by our in-house mass spectra library.
Data from inoculated and control groups was the average of the sum of the normalised data of the compound at each time point, which presented as Mean±SE (n = 3). The data were evaluated by a two-way analysis of variance, through which the p value was calculated. The data were determined at 2, 4, 6, 8, 12, 16, 20, 24, 26 and 28 dpi, dpi (day post inoculation). The variable importance values (VIP) were obtained through OPLS-DA analyses.
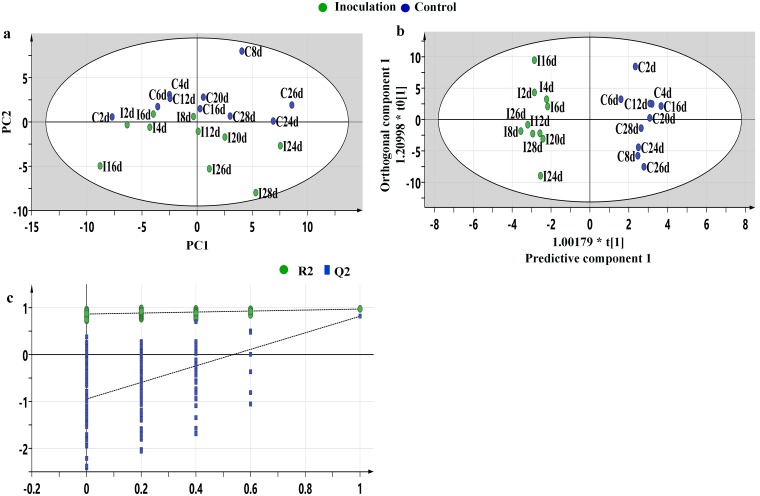
The R2X (cum) value of the PCA model is 0.529, as shown in Fig 2a. The model could not be used to distinguish between control and pathogen-inoculated groups, however, the OPLS-DA results clearly placed the control and pathogen treatments in two distinct groups. The samples collected from control and pathogen-inoculated groups all fell inside the 95% Hotelling T2 ellipse (Fig 2b). The R2Y(cum) and Q2 values of this model were 1.000 and 0.817, respectively. Validation plots showed R2 and Q2 values from the permuted analysis (bottom left) are lower than the corresponding original R2 and Q2 values (top right), indicating a good fit for the model (Fig 2c).
Fig 2. Multivariate statistical analysis of control samples and infected samples.
Principal component analysis (PCA) score plot for the inoculated samples (green) and control samples (blue) collected at 2, 4, 6, 8, 12, 16, 20, 24, 26 and 28 days, PC1 is 35.3% and PC2 is 17.2% (a); Orthogonal partial least-squares discriminant analysis (OPLS-DA) score plot (1+4 components), R2X(cum) = 0.739, R2Y(cum) = 1.000, Q2 = 0.817), R2X(cum) and R2Y(cum) are the cumulative modelled variations in the X and Y matrices, respectively, and Q2 value is a prediction error (b); Validation plots of the orthogonal partial least-squares discriminant analysis (POLS-DA) models acquired through 200 permutation tests for the control vs. inoculated groups. T [1] = scores for predictive component 1, to [1] = scores for orthogonal component 1. The ellipse shows the 95% confidence interval using Hotelling T2 statistics. R and Q were obtained after OPLS-DA permutation tests (n = 200), R2 intercept was 0.861, Q2 intercept was -0.933 (c).
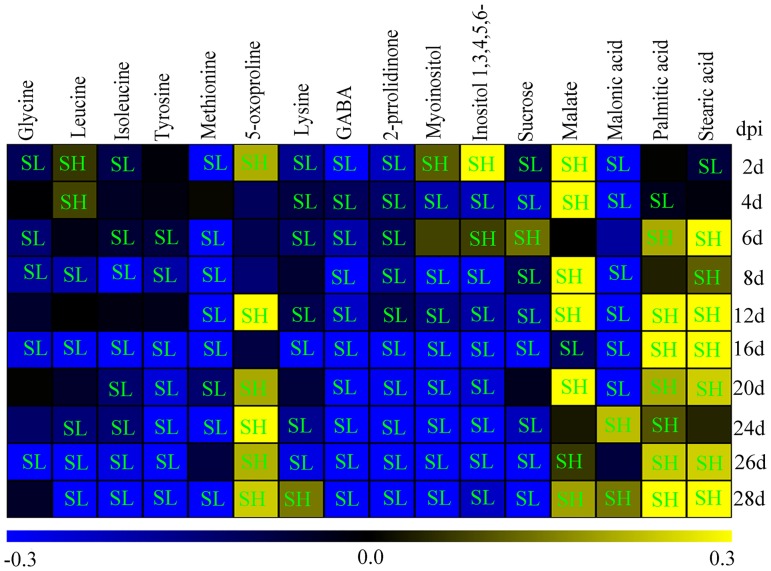
A total of 16 significantly regulated metabolites (VIP > 1 and p < 0.05) associated with the pathogen infection were identified using the OPLS-DA model. These metabolites included eight amino acids (glycine, leucine, isoleucine, lysine, tyrosine GABA, 5-oxoproline, and methionine), one nitrogen-containing compound (2-pyrrolidinone), two organic acids (malonic acid and malate), two polyols (myoinositol and inositol 1,3,4,5,6-pentakisphosphate), one sugar (sucrose) and two fatty acids (palmitic acid and stearic acid) (Fig 3).
Fig 3. Heat map of significantly regulated metabolites in the alfalfa leaves between control and inoculated groups.
Metabolite levels in the leaves were shown as log10 ratios of values from infected versus uninfected groups at 2, 4, 6, 8, 12, 16, 20, 24, 26 and 28 days post inoculation (dpi). Yellow color shows upregulated compound levels, and the blue color indicates downregulated levels. Level of compounds in the inoculated leaves was significantly lower (SL) or significantly higher (SH) than in the control leaves. Inositol 1,3,4,5,6- is inositol 1,3,4,5,6-pentakisphosphate. Data were evaluated by student’s t test and p<0.05 has significant difference.
Several amino acids, such as glycine, leucine, isoleucine, methionine and tyrosine, and sucrose, myoinositol and inositol 1, 3, 4, 5, 6-pentakisphosphate were downregulated in the P. medicaginis-inoculated leaves compared to the mock-inoculated leaves after 6 dpi. In contrast, the level of lysine in the P. medicaginis-inoculated leaves was gradually upregulated 39.1% by 28 dpi.
Some compounds showed a dramatic increase in the P. medicaginis-inoculated leaves compared with mock-inoculated leaves; for example, the level of 5-oxoproline in the P. medicaginis-inoculated leaves reached a maximum level of 137.9% by 24 dpi, falling to 74.2% by 28 dpi. Malate dramatically increased by 352.4% at 4 dpi. The levels of palmitic acid and stearic acid reached a maximum level of 266.8% and 291.4%, at 28 dpi, respectively.
Some compounds showed a large decrease in the P. medicaginis-inoculated leaves compared with the mock-inoculated leaves. The levels of GABA (γ-Aminobutyric acid) and 2-pyrrolidinone decreased over time compared with the control groups, and the levels of malonic acid significantly decreased at early infection stages (p<0.05), but increased by 67.2% at 24 dpi and 38.1% at 28 dpi (t test, p<0.05).
Metabolites related to glycolysis and the TCA cycle were significantly induced at later infection stages (t test, p<0.05). For example, at 28 dpi, pyruvic acid increased 1.69 fold, asparagine increased 1.41 fold, succinate increased 1.90 fold, fumarate increased 1.37 fold, and citrate increased by 2.16 fold compared with the control groups (Table 2).
Table 2. Fold change values from pathogen-inoculated versus control groups at each time points.
| Metabolite | Fold change | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2dpi | 4dpi | 6dpi | 8dpi | 12dpi | 16dpi | 20dpi | 24dpi | 26dpi | 28dpi | |
| Pyruvic acid | 1.14 | 1.45* | 1.36* | 1.20** | 1.30 | 0.59* | 1.48* | 1.46* | 0.97 | 1.69** |
| Asparagine | 0.81** | 1.26** | 0.89 | 0.93 | 1.25* | 0.42** | 1.46** | 1.13 | 0.84 | 1.41** |
| Succinate | 0.77 | 0.54** | 0.80 | 0.86 | 1.03 | 0.39** | 1.43** | 1.29** | 1.41** | 1.90** |
| Fumarate | 0.92 | 0.62 | 1.08 | 0.45 | 0.94 | 0.41 | 1.00 | 1.33 | 1.38* | 1.37** |
| Citrate | 0.77** | 0.44* | 0.78 | 1.40** | 0.98 | 0.22** | 1.27** | 0.54** | 1.20** | 2.16** |
Fold change is the ratio of data from inoculated versus control groups at 2, 4, 6, 8, 12, 16, 20, 24, 26 and 28 days post inoculation (dpi). Data were evaluated by student’s t test and indicated by asterisks (*p<0.05 or **p<0.01).
Metabolic pathways
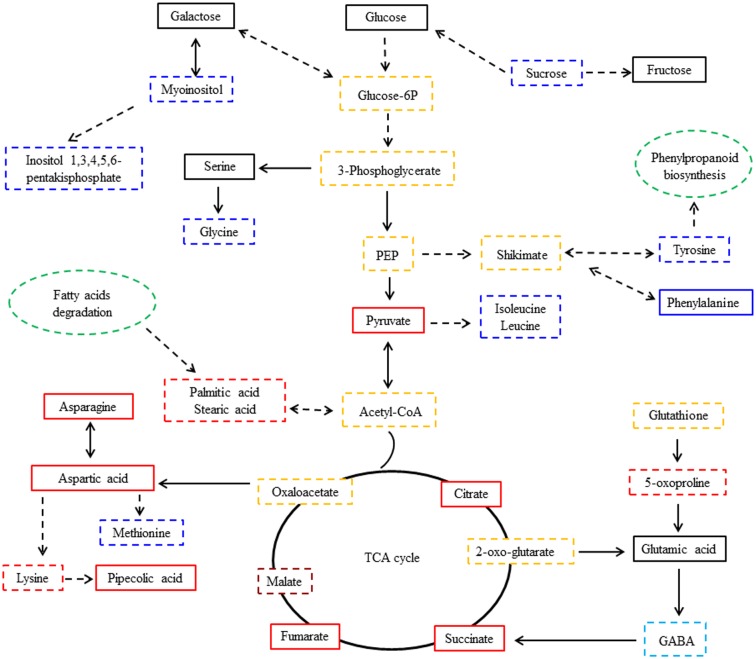
Of the 16 compounds regulated with infection, 14 compounds were associated with 30 primary metabolic pathways (S1 Table). These pathways include metabolism of sugar and alcohol, metabolism of amino acids and organic acids, metabolism of fatty acids and glutathione (S1 Table), of which glycolysis, amino acids, inositol phosphate, TCA cycle, fatty acids, and glutathione are most important (Fig 4). The pathway of 2-pyrrolidinone and malonic acid metabolites were not found to belong to a specific defined metabolic pathway.
Fig 4. Metabolic pathway network of responsive metabolites in alfalfa leaves infected with P. medicaginis.
The black, red, blue, light blue and crimson full and dotted box indicates that metabolites were detected by GC-MS from the alfalfa leaves, and the dotted box presents significant regulated metabolites in the inoculated leaves compared with the controls (VIP>1.0 and p<0.05). Yellow dotted box indicates that the metabolites were not detected by GC-MS from the alfalfa leaves, but were involved in the corresponding pathway. Green dotted ovals indicates that the metabolic pathway was associated with the regulated metabolites. (VIP, the variable importance for the projection; PEP, phosphoenolpyruvate; TCA cycle, tricarboxylic acid cycle). Red color indicates that the compound was significantly upregulated at later infection stages. Blue color indicates that the compound was significantly downregulated at later infection stages. Light blue color indicates that the compound was significantly downregulated over time. Crimson color indicates that the compound was almost significantly upregulated over time.
Discussion
During antagonistic interactions between plants and pathogenic fungi, the pathogens try to modulate plant carbon and nitrogen metabolism to ensure their proliferation and survival, while plants need nutrients and energy to synthesize phytoalexins, defense related protein, antioxidants and many other metabolites to resist pathogen infection [26].
As disease progressed, lesions enlarged and coalesced, leaves became yellow, and the levels of 16 metabolites in the infected plants were significantly changed compared to the non-infected plants during P. medicaginis colonization. These metabolites included amino acids (glycine, leucine, isoleucine, tyrosine, lysine, 5-oxoproline, methionine, and GABA), 2-pyrrolidinone and organic acids (malonic acid and malate), sucrose, alcohols (myoinositol and inositol 1,3,4,5,6-pentakisphosphate), and fatty acids (palmitic acid and stearic acid).
The levels of glycine, isoleucine, leucine and tyrosine in the P. medicaginis-infected alfalfa leaves decreased compared to the uninfected plants. These amino acids are not only the basic units of protein synthesis, but also enhance defense responses to biotic stresses including strengthening of plant cell walls, regulation of jasmonic acid (JA), and biosynthesis of phytoalexins. [27–30]. This suggests that the decrease in these amino acids levels in the infected leaves may promote the susceptibility of alfalfa to the pathogen infection.
As shown in Fig 4, 5-oxoproline and glutathione (GSH) are involved in the glutathione metabolism pathway; 5-oxoproline is a degradation product of glutathione. Upon P. medicaginis infection, 5-oxoproline levels significantly increased in the inoculated alfalfa leaves in different infection stages, but especially in the later infection (p<0.05). This suggests that the antioxidant system based on the glutathione-mediated detoxification pathway may be induced mostly later in infection. High accumulation of 5-oxoproline may increase plant resistance, and lower levels of the metabolite at early infection stages may help the pathogen colonize the plant. Previous studies showed that antioxidant enzyme activity was induced earlier and to higher levels in the resistant varieties of alfalfa than in the susceptible varieties [13]. These results reinforce the expectation that antioxidant compounds and enzymes play a synergistic role in plant disease resistance.
With P. medicaginis infection, lysine and its degradation product pipecolic acid were induced later and strongly in infected alfalfa leaves, compared with control leaves, which could be related to alfalfa susceptibility. Pipecolic acid was previously shown to induce systemic acquired resistance (SAR) [31]. Pipecolic acid was produced faster and higher in Arabidopsis leaves inoculated with the incompatible, HR-inducing Psm avrRpm1 strain than in Arabidopsis leaves infected with the compatible Psm strain, and pipecolic acid accumulated to higher levels during later stages of the compatible interaction [32]. Therefore, later strong induction of lysine degradation in the infected alfalfa leaves may be a way for the pathogen to escape host resistance.
For some pathogens, they need to assimilate methionine or its derivatives from host tissue to support their growth, which is related to their pathogenicity. For instance, methionine auxotrophic mutants of the pathogens Ustilago maydis and Fusairum graminearum, showed only reduced pathogenicity on plant leaves and were still able to penetrate and colonize host tissues [33,34]. Under P. medicaginis infection, methionine levels decreased, which may be related to the assimilation of P. medicaginis. The pathogen may need to acquire sufficient amounts of methionine from alfalfa leaves to support its growth. Inhibiting the use of methionine by the pathogen can increase alfalfa resistance.
As shown in S1 Table and Fig 4, myoinositol (inositol) and inositol-1, 2, 3, 4, 5, 6-hexakisphosphate are involved in the inositol phosphate metabolism pathway. With P. medicaginis infection, myoinositol and inositol derivatives (Inositol 1, 3, 4, 5, 6-pentakisphosphate) were induced early but decreased significantly at later infection stages in the infected leaves (p<0.01), which suggested that ability of alfalfa to resist disease decreased. Conversely, early strong induction of myoinositol and inositol derivatives (Inositol 1, 3, 4, 5, 6-pentakisphosphate) may enhance alfalfa resistance. The amounts of myoinositol and inositol derivatives in resistant wheat cultivars (Sumai3) were higher than that in susceptible wheat cultivars (Roblin) [35].
In this research, malonic acid decreased in the infected leaves significantly at early infection stages (p<0.05), and accumulated at later infection stages. Malonic acid metabolism likely provides a carbon source for the survival of microorganisms [36,37]. Therefore, the changes of malonic acid in the alfalfa detected may have provided carbon for P. medicaginis to invade, increasing host susceptibility.
As shown in Fig 4, metabolism of sucrose relates to the glycolysis pathway; its end product is pyruvic acid (pyruvate), which was induced significantly at later infection stages (p<0.05) (Table 2). The decrease in sucrose levels may also be involved in promoting the glycolysis pathway in the later infection stages, except when it serves as a carbon source for fungal mycelium growth [5,11].
In this study, the TCA cycle intermediates malate, fumarate, succinate, and citrate all accumulated significantly in infected leaves at later infection stages (p<0.05) (Fig 3 and Table 2). Moreover, as shown in Fig 4, GABA and asparagine are synthesized by the TCA cycle, asparagine were induced strongly at late infection stages when compared with controls (Table 2). Asparagine is one of the major metabolic products in senescing leaves [38], and it could increase the P. medicaginis spore germination rate [39]. GABA also likely provided an extra source of translocated nitrogen for the growth of the fungus [40], 2-Pyrrolidinone (pyrrolidinone) is one important metabolites of GABA [41]. These results suggest that impaired photosynthesis of alfalfa leaves caused by the infection of P. mecaginis may lead to a very strong induction in respiration, TCA cycle and glycolysis in the infected leaves, which could compensate for reduced ATP to the host cells as a result of the impaired photosystem [15]. In addition, the decrease in GABA and 2-Pyrrolidinone in the infected leaves may relate to depletion of P. medicaginis, since these compounds may also provide nitrogen for the pathogen growth and survival. The induction of asparagine through the TCA cycle following P. medicaginis infection provides a rich nitrogen reservoir to support fungal growth in alfalfa and also facilitate pathogen-initiated host senescence. However, the strong induction of plant respiration by the pathogen may induce senescence leading to host plant death.
Palmitic acid involves fatty acid degradation pathways (S1 Table and Fig 4), the primary manner of fatty acid degradation is β-oxidation, which not only provides a large amount of energy and carbon sources needed for the life activities of plant, but also provides abundant fatty acids and carbon source for pathogen invasion and growth. Under P. medicaginis infection, genes in the octadecanoid pathway were induced significantly and accumulated more rapidly in a resistant M. truncatula accession than in a susceptible M. truncatula accession [30]. In our studies, large amounts of palmitic acid (C16) and stearic acid (C18) accumulated in later P. medicaginis infection stages, suggesting that the induction of β-oxidation in the later infection stages may have provided nutrition for P. medicaginis for invasion and growth, while the rapid accumulation of these two fatty acids at early infection stages may enhance plant resistance to the pathogen infection.
This research was carried out on a susceptible cultivar. We anticipate that a resistant cultivar would induce intermediate metabolites of glycolysis, TCA cycle and β-oxidation metabolism pathways, and inositol phosphate, glutathione, some amino acids metabolic pathways, such as glycine, leucine, isoleucine, tyrosine and lysine, rapidly and strongly at an early infection stage. When Kamphuis and coworkers compared susceptible and resistant cultivars of M. truncatula infected by P. medicaginis, they found expression of genes associated with disease resistance was induced to lower levels in a resistant cultivar later in infection than for a susceptible cultivar. Their experiment showed the greatest upregulation in PR10 gene expression, lipoxygenase transcripts and isoflavone 2’-hydroxylase in the octadecanoid pathway from resistant plants earlier and to a higher level [30]. Our work was done with a different system at the metabolic level, however, we would anticipate that glycine, leucine, isoleucine, tyrosine, lysine, 5-oxoproline, myoinositol and inositol 1,3,4,5,6-pentakisphosphate, palmitic acid and stearic acid would likely be induced early and higher in resistant cultivars. These compounds and their related metabolic pathways could provide information for the identification and characterization of resistant cultivars.
Conclusion
A detailed primary metabolic profile of the response of alfalfa leaves to P. medicaginis infection was presented. In the process of P. medicaginis colonization of the alfalfa leaves, glycolysis, TCA cycle and β-oxidation metabolism pathways in the infected leaves accumulated strongly at later infection stages, which may provide ATP, a N source and nutrition for P. medicaginis to colonize alfalfa leaves. Moreover very strong induction of the TCA cycle pathways by the pathogen at later infection stage may induce senescence in alfalfa leaves, leading to plant death. Intermediates metabolites of these metabolic pathways, and inositol phosphate, glutathione, some amino acids metabolic pathways, such as glycine, leucine, isoleucine, tyrosine and lysine, accumulated rapidly and strongly at an early infection stage may enhance the ability of alfalfa to resist necrotrophic P. medicaginis. The tentatively identified differential metabolites and related pathways may serve as potential markers for developing novel disease-control strategies and breeding resistant varieties.
Supporting information
Relevant pathways based on MetaboAnalyst 4.0 analysis.
(DOCX)
Amount of individual metabolite normalised to internal standard is shown.
(XLSX)
Acknowledgments
We thank Dr. Huan Yang Qi from Xiamen Health and Medical Big Data Management Center/Xiamen Institute of Medicine for data analysis and assistance of gas chromatography-mass spectrometry determination and thank Dr. Xin Shun Ding for his comments and edit during manuscript preparation.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was financially supported by Special Fund for Agro-scientific Research in the Public Interest (No. 201303057) to Yanzhong Li. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yang QC, Sun Y. The history, current situation and development of alfalfa breeding in China. Chinese Journal of Grrassland 2011; 33: 95–101. [Google Scholar]
- 2.Lu XS. The development status and thinking of the commercial grass industry of alfalfa in China. China Dairy Cattle. 2013: 3–6 [Google Scholar]
- 3.Wang YT. The Ministry of Agriculture issued the national alfalfa industry development plan (2016–2020). China Dairy. 2017: 74–74. [Google Scholar]
- 4.Nan ZB. Alfalfa disease and its comprehensive control system in China. Animal science and veterinary medicine. 2001; 18: 81–84. [Google Scholar]
- 5.Zhang L, Pan LQ, Wang SR, Yuan QH, Wang Y, Miao LH. Study on the biological characteristics of the alfalfa Phoma leaf spot pathogen. Journal of China Agricultural University. 2015; 20: 158–166. [Google Scholar]
- 6.Wang Y, Yuan QH, Miao LH, Zhang L, Pan LQ. The major types and epidemic trends of alfalfa diseases in Northeast and North China. Acta Prataculturae Sinica. 2016; 25: 52–59. [Google Scholar]
- 7.Li YZ, Yu BH, Xu LB. Alfalfa Disease Atlas: Peking: China agricultural science and technology press; 2016.
- 8.Akamatsu HO, Chilvers MI, Peever TL. First report of spring black stem and leaf spot of alfalfa in Washington State caused by Phoma medicaginis. Plant Disease. 2008; 92: 833–833. [DOI] [PubMed] [Google Scholar]
- 9.Barbetti MJ. Resistance in annual Medicago species to Phoma medicaginis under controlled environment and field conditions. Australian Journal of Experimental Agriculture. 1990; 30: 209–214. [Google Scholar]
- 10.Djebali N. Aggressiveness and host range of Phoma medicaginis isolated from Medicago species growing in Tunisia. Phytopathologia Mediterranea. 2013; 52: 3–15. [Google Scholar]
- 11.Castell-Miller CV, Zeyen RJ, Samac DA. Infection and development of Phoma medicaginis on moderately rersistant and susceptible alfalfa genotypes. Canadian Journal of Plant Pathology. 2007; 29: 290–298. [Google Scholar]
- 12.Barbetti MJ. Resistance in annual Medicago spp. to Phoma medicaginis and Leptosphaerulina trifolii and its relationship to induced production of a phytoestrogen. Plant Disease. 2007; 91: 239–244. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Pan LQ, Yuan QH, Wang Y, Miao LH. Resistance evaluation of different alfalfa materials on Phoma leaf spot. Acta Agrestia Sinica. 2016; 24: 652–657. [Google Scholar]
- 14.Wang Y, Liu Y, Zhou BB, Yuan QH, Zhang L, Pan LQ. An evaluation of the resistance of alfalfa cultivars to Stemphylium and Phoma leaf spot diseases. Acta Prataculturae Sinica. 2015; 24: 155–162. [Google Scholar]
- 15.Fan Q, Li YZ. The effect of Phoma medicaginis on the photosynthetic physiology of Medicago sativa. Acta Prataculturae Sinica. 2017; 26: 112–121 [Google Scholar]
- 16.Paiva NL, Oommen A, Harrison MJ, Dixon RA. Regulation of isoflavonoid metabolism in alfalfa. Plant Cell Tissue & Organ Culture. 1994; 38: 213–220. [Google Scholar]
- 17.Blount JW, Dixon RA, Paiva NL. Stress responses in alfalfa (Medicago sativa L.) XVI. Antifungal activity of medicarpin and its biosynthetic precursors; implications for the genetic manipulation of stress metabolites. Physiological & Molecular Plant Pathology. 1992; 41: 333–349. [Google Scholar]
- 18.Jasiński M, Kachlicki P, Rodziewicz P, Figlerowicz M, Stobiecki M. Changes in the profile of flavonoid accumulation in Medicago truncatula leaves during infection with fungal pathogen Phoma medicaginis. Plant Physiology and Biochemistry. 2009: 847–853 10.1016/j.plaphy.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 19.Maher EA, Lamb CJ, Dixon RA. Stress responses in alfalfa (Medicago sativa L). XVII. Identification of multiple hydrolases and molecular characterization of an acidic glucanase. Physiological & Molecular Plant Pathology. 1993; 43: 329–342. [Google Scholar]
- 20.Schliemann W, Ammer C, Strack D. Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry. 2008; 69: 112–146. 10.1016/j.phytochem.2007.06.032 [DOI] [PubMed] [Google Scholar]
- 21.Kai PL, Han TL, Yang Y, Zhang H. Analytical challenges of untargeted GC-MS-based metabolomics and the critical issues in selecting the data processing strategy. F1000research. 2017; 6: 967 doi: 10.12688/f1000research.11823.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahdavi V, Farimani MM, Fathi F, Ghassempour A. A targeted metabolomics approach toward understanding metabolic variations in rice under pesticide stress. Analytical Biochemistry. 2015; 478: 65–72. 10.1016/j.ab.2015.02.021 [DOI] [PubMed] [Google Scholar]
- 23.Westerhuis JA, Hoefsloot HCJ, Smit S, Vis DJ, Smilde AK, Velzen EJJV, et al. Assessment of PLSDA cross validation. Metabolomics. 2008; 4: 81–89. [Google Scholar]
- 24.Sun HZ, Wang DM, Wang B, Wang JK, Liu HY, Guan LL, et al. Metabolomics of four biofluids from dairy cows: potential biomarkers for milk production and quality. Journal of Proteome Research. 2015; 14: 1287–1298. 10.1021/pr501305g [DOI] [PubMed] [Google Scholar]
- 25.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0-a comprehensive server for metabolomic data analysis. Nucleic Acids Research. 2012; 40: 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger S, Roitsch T. Plant physiology meets phytopathology: plant primary metabolism and plant-pathogen interactions. Journal of Experimental Botany. 2007; 58: 4019–4026. 10.1093/jxb/erm298 [DOI] [PubMed] [Google Scholar]
- 27.Molina A, Mena M, Carbonero P, Garcíaolmedo F. Differential expression of pathogen-responsive genes encoding two types of glycine-rich proteins in barley. Plant Molecular Biology. 1997; 33: 803–810. [DOI] [PubMed] [Google Scholar]
- 28.Matteo AD, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, et al. The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci U S A. 2003; 100: 10124–10128. 10.1073/pnas.1733690100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenzo O, Chico JM, Sánchezserrano JJ, Solano R. JASMONATE-INSENSITIVE1 Encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004; 16: 1938–1950. 10.1105/tpc.022319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamphuis LG, Williams AH, Küster H, Trengove RD, Singh KB, Oliver RP, et al. Phoma medicaginis stimulates the induction of the octadecanoid and phenylpropanoid pathways in Medicago truncatula. Molecular Plant Pathology. 2012; 13: 593–603. 10.1111/j.1364-3703.2011.00767.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song JT, Lu H, McDowell JM, Greenberg JT. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant Journal. 2004; 40: 200–212. 10.1111/j.1365-313X.2004.02200.x [DOI] [PubMed] [Google Scholar]
- 32.Návarová H, Bernsdorff F, Döring AC, Zeier J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell. 2012; 24: 5123–5141. 10.1105/tpc.112.103564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer JA, McCann MP, Snetselaar KM. Methylation is involved in the Ustilago maydis mating response. Fungal Genetics & Biology. 2001; 34: 21–35. [DOI] [PubMed] [Google Scholar]
- 34.Seong K, Hou Z, Tracy M, Kistler HC, Xu JR. Random insertional mutagenesis identifies genes associated with virulence in the wheat scab fungus Fusarium graminearum. Phytopathology. 2005; 95: 744–750. 10.1094/PHYTO-95-0744 [DOI] [PubMed] [Google Scholar]
- 35.Hamzehzarghani H, Kushalappa AC, Dion Y, Rioux S, Comeau A, Yaylayan V, et al. Metabolic profiling and factor analysis to discriminate quantitative resistance in wheat cultivars against Fusarium head blight. Physiological & Molecular Plant Pathology. 2005; 66: 119–133. [Google Scholar]
- 36.Karunakaran R, East AK, Poole PS. Malonate catabolism does not drive N2 fixation in legume nodules. Applied and Environmental Microbiology. 2013; 79: 4496–4498. 10.1128/AEM.00919-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatnaparat T, Prathuangwong S, Lindow S. Global pattern of gene expression of Xanthomonas axonopodis pv. glycines within soybean leaves. Mol Plant Microbe Interact. 2016; 29: 508–522. 10.1094/MPMI-01-16-0007-R [DOI] [PubMed] [Google Scholar]
- 38.Herrerarodríguez MB, Maldonado JM, Pérezvicente R. Role of asparagine and asparagine synthetase genes in sunflower (Helianthus annuus) germination and natural senescence. Journal of Plant Physiology. 2006; 163: 1061–1070. 10.1016/j.jplph.2005.10.012 [DOI] [PubMed] [Google Scholar]
- 39.Renfro BL, Wilcoxson RD. Spring black stem of alfalfa in relation to temperature, moisture, wounding, and nutrients and some observations on pathogen dissemination. Phytopathology. 1963; 53: 1340–1345 [Google Scholar]
- 40.Bouché N, Fromm H. GABA in plants: just a metabolite? Trends in Plant Science. 2004; 9: 110–115. 10.1016/j.tplants.2004.01.006 [DOI] [PubMed] [Google Scholar]
- 41.Petroff O, Hyder FR, Rothman D. Topiramate increases brain GABA, homocarnosine, and pyrrolidinone in patients with epilepsy. Neurology. 1999; 52: 473–478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relevant pathways based on MetaboAnalyst 4.0 analysis.
(DOCX)
Amount of individual metabolite normalised to internal standard is shown.
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.