Abstract
Glioblastoma (GBM) is the most common malignant brain tumor in the United States. Incidence of GBM increases with age, and younger age-at-diagnosis is significantly associated with improved prognosis. While the relationship between candidate GBM risk SNPs and age-at-diagnosis has been explored, genome-wide association studies (GWAS) have not previously been stratified by age. Potential age-specific genetic effects were assessed in autosomal SNPs for GBM patients using data from four previous GWAS. Using age distribution tertiles (18–53, 54–64, 65+) datasets were analyzed using age-stratified logistic regression to generate p values, odds ratios (OR), and 95% confidence intervals (95%CI), and then combined using meta-analysis. There were 4,512 total GBM cases, and 10,582 controls used for analysis. Significant associations were detected at two previously identified SNPs in 7p11.2 (rs723527 [p54–63=1.50×10−9, OR54–63=1.28, 95%CI54–63=1.18–1.39; p64+=2.14×10−11, OR64+=1.32, 95%CI64+=1.21–1.43] and rs11979158 [p54–63=6.13×10−8, OR54–63=1.35, 95%CI54–63=1.21–1.50; p64+=2.18×10−10, OR64+=1.42, 95%CI64+=1.27–1.58]) but only in persons >54. There was also a significant association at the previously identified lower grade glioma (LGG) risk locus at 8q24.21 (rs55705857) in persons ages 18–53 (p18–53=9.30×10−11, OR18–53=1.76, 95%CI18–53=1.49–2.10). Within The Cancer Genome Atlas (TCGA) there was higher prevalence of ‘LGG’-like tumor characteristics in GBM samples in those 18–53, with IDH1/2 mutation frequency of 15%, as compared to 2.1% [54–63] and 0.8% [64+] (p=0.0005). Age-specific differences in cancer susceptibility can provide important clues to etiology. The association of a SNP known to confer risk for IDH1/2 mutant glioma and higher prevalence of IDH1/2 mutation within younger individuals 18–53 suggests that more younger individuals may present initially with ‘secondary glioblastoma.’
INTRODUCTION
Glioma is the most common type of malignant primary brain tumor (PBT) in the United States (US), with an average annual age-adjusted incidence of 6.0 per 100,000 from 2010–2014.1 Glioblastoma (GBM) represents the majority of gliomas diagnosed in adults (approximately 62% of all gliomas diagnosed in persons 18+). While these tumors are most common in older adulthood, they also occur in younger adults. Persons diagnosed with GBM at younger ages have significantly better survival than those who are diagnosed at older ages, with 5-year survival of 19.0% in persons 20–44 as compared to 1.8% in persons 65+.
Gliomas are classified using histologic criteria determined by the World Health Organization (WHO) and broadly classified according to apparent cell of origin (astrocyte vs oligodendrocyte), and graded from grade I-IV, where increasing WHO grade is associated with increasing malignant behavior.2 GBMs are WHO grade IV astrocytomas. Recent molecular characterization of gliomas (including both GBM and LGG) has determined that gliomas can be more precisely stratified using two common alterations: mutation in isocitrate dehydrogenase 1/2 (IDH1/2) and loss of the 1p/19q.3 The vast majority of GBMs (~95%) do not have either alteration.4
Many environmental exposures have been investigated as risk factors for glioma, but the only consistently identified risk factors are ionizing radiation (which increases risk), and history of allergies (which decreases risk).5 The contribution of common low-penetrance single nucleotide polymorphisms (SNPs) to the heritability of glioma in persons with no documented family history is estimated to be ~25%.6 A recent glioma genome-wide association study (GWAS) meta-analysis validated 12 previously reported risk loci, and identified 13 new risk loci. These 25 loci in total are estimated to account for ~30% of heritable risk.7 This suggests that there are both undiscovered environmental risk (which accounts for ~75% of risk variance) and genetic risk factors (accounting for ~70% of heritable risk).6, 7
Many previously discovered glioma risk loci have histology-specific associations,7 but the effect of these loci on age-at-diagnosis has not been systematically explored. An age-specific analysis could potentially increase power to detect new variants that may be associated with age-at-diagnosis, and more accurately quantify the age-specific effect sizes of previously-identified variants.8 This analysis aims to explore the associations between genetic risk and age-at-diagnosis in GBM only.
METHODS
This study was approved locally by the institutional review board (IRB) at University Hospitals Cleveland Medical Center and by each participating study site’s IRB. In this study, data on GBM cases were combined from four prior glioma GWAS.7, 9–11 Due to the molecular heterogeneity of LGGs and the lack of molecular subtype data available within these datasets, this analysis focused on GBM only. After quality control was completed, the combined dataset contained 4,523 GBM cases (See Supplementary Table 1 for study characteristics). Details of data collection, genotyping, and imputation for included GWAS datasets are available in previous publications.7, 9–11 All datasets have previously undergone standard GWAS QC, and duplicate and related individuals have been excluded (as described in Melin, et al.7). Demographic information and somatic features for GBM cases included in The Cancer Genome Atlas (TCGA, see Supplementary Table 1 for characteristics) were obtained from the final dataset used for the TCGA pan-glioma analysis. 4, 12
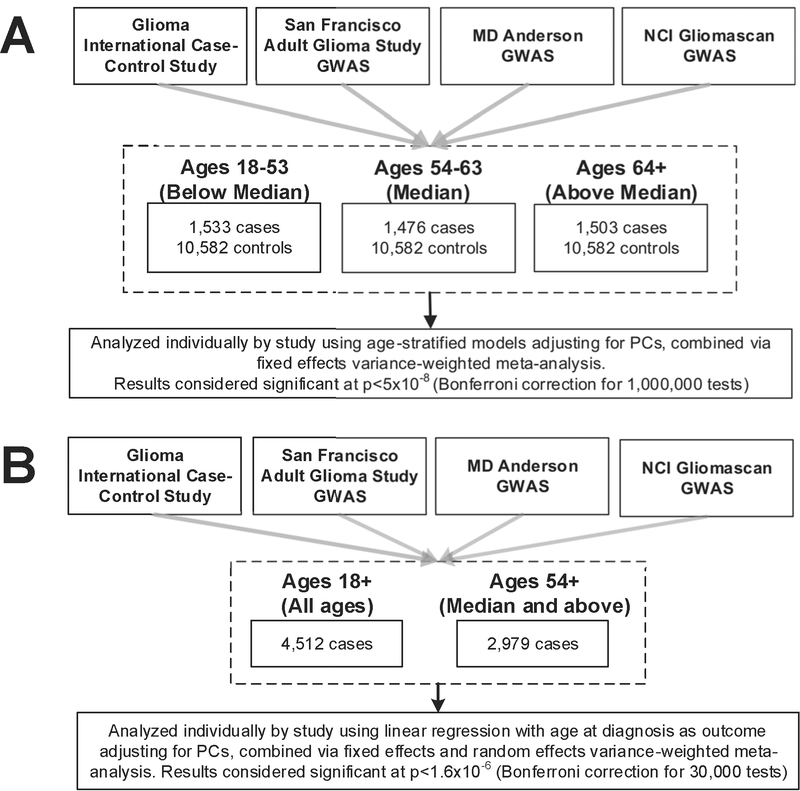
Single-SNP association results were generated for the ~12,000 SNPs (INFO≥0.7, MAF≥0.01) identified as having a nominally significant (p<5×10−4) association with all glioma, GBM, or LGG. SNPs found to have nominally significant association with non-GBM glioma were included to assess potential association of these SNPs with GBM in younger individuals. GBM cases were divided into age strata based on the tertiles of the case age-at-diagnosis distribution (18–53, 54–63, and 64+) for case-control analyses (see Figure 1a for overview of study design) and each age strata was compared against all controls.13 The data were analyzed using age-stratified logistic regression models in SNPTEST adjusted for sex and the number of principal components that significantly differed between cases and controls within each study.7 Single-study results were combined via meta-analysis using an inverse-variance weighted fixed effects method in META. Results from the case-control analysis were considered statistically significant at the p<5×10−8 level (using a Bonferroni correction for 1,000,000 tests).
Figure 1.
Study schematic for a) age-stratified case-control analyses, and b) case-only analyses
In order to identify individual SNPs with age-at-diagnosis effects, a case-only analysis using age-at-diagnosis as a continuous phenotype was conducted using linear regression in SNPTEST. This analysis assumed an additive model to estimate beta, standard error, and p values (see Figure 1b for overview of study design). All models were adjusted for sex, and both GICC and Gliomascan were adjusted for principal components due to genomic inflation (GICC: λunadjusted=1.02, λadjusted=1.01; SFAGS-GWAS: λunadjusted=1.01, λadjusted=1.01; MDA-GWAS: λunadjusted=0.99, λadjusted=0.99; Gliomascan: λunadjusted=1.04, λadjusted=1.01). Results were combined using both inverse-variance weighted fixed effects and random effects meta-analysis in META. Results were considered statistically significant at p<4.17×10−6 (Bonferroni correction for 12,000 tests). Analyses were also run in two older age-groups (median and above median age, ages 54+) to evaluate whether a dose-response curve exists between genotype and age-at-diagnosis, or if significant associations were identifying differences between the youngest versus oldest age groups. All figures were generated using R 3.3.2, qqman, and ggplot.14–17
TCGA GBM cases were divided into the same three age strata for analysis. Only newly diagnosed, white non-Hispanic cases with no neo-adjuvant treatment or prior cancer were used. Demographic characteristics, molecular classification and somatic alterations data was obtained from Ceccarelli, et al.12 Age-specific differences in frequency of molecular markers by age were tested using chi square tests.
RESULTS
There were 4,512 total GBM cases, and 10,582 controls in the four included GWAS datasets (Supplementary Table 1). Overall, 62.8% of GBM cases were male, with a mean age-at-diagnosis of 57.5.
No new risk loci were identified in an age-stratified genome-wide scan. SNPs in 5/25 previously identified risk loci reached genome wide significance (p<5×10−8, Bonferroni correction for 1,000,000 tests) in at least one age strata, including three previously identified SNPs in 7p11.2 (Table 1, Supplementary Figures 1–2). Results for SNPs in 5p15.33, 9p21.3, 17p.13.1, and 20q13.33 were similar in all three age-groups. Among the three identified SNPs in 7p11.2, results for rs75061358 were similar in all age-groups. Two other SNPs (rs723527 and rs11979158) were nominally significant in the age 18–53 strata, and reached genome-wide significance in the age 54–63, and age 64+ stratum. The risk locus at 8q24.21 reached genome-wide significance in the 18–53 age-group (p=9.30×10−11), while the effect of this SNP was null in the other age-groups. The OR for the 18–53 year age-group was 1.76 (95%CI: 1.49–2.10). There was variability of effect by study, though tests for heterogeneity were not significant (Supplementary Figure 3, Supplementary Table 2). In order to assess whether confounding between age-at-diagnosis and histologic classification may exist, a sensitivity analysis was performed using only individuals diagnosed after 2000 (Supplementary Table 3). Among individuals 18–53 diagnosed after 2000 only, there was still a genome-wide significant signal for the SNP at 8q24.21 (p=5.47×10−10, OR=1.84 [95%CI=1.52–2.23]).
Table 1.
Previously identified glioma risk loci and histology-specific odds ratios (OR) and 95% confidence intervals (95%CI) stratified by age.
| SNP (Locus) | Risk Allele | Associated Gene | Diagnosis Ages 18–53 (Below Median Age-at-diagnosis) |
Diagnosis Ages 54–63 (Median Age-at-diagnosis) |
Diagnosis Ages 64+ (Above Median Age-at-diagnosis) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | OR (95%CI) |
Phet | Q | I2 | P | OR (95%CI) |
Phet | Q | I2 | P | OR (95%CI) |
Phet | Q | I2 | |||
| rs10069690 (5p15.33) | C/T | TERT | 3.79×10−18 |
1.51 (1.38–1.66) |
0.7138 | 1.36 | 0.00 | 4.05×10−33 |
1.78 (1.62–1.95) |
0.5182 | 2.27 | 0.00 | 4.59×10−31 |
1.72 (1.57–1.89) |
0.0664 | 7.18 | 58.21 |
| rs75061358 (7p11.2) | T/G | EGFR | 6.78×10−12 |
1.67 (1.44–1.94) |
0.7282 | 1.30 | 0.00 | 2.44×10−12 |
1.70 (1.47–1.97) |
0.4542 | 2.62 | 0.00 | 3.67×10−16 |
1.85 (1.60–2.15) |
0.3607 | 3.21 | 6.48 |
| rs723527 (7p11.2) | A/G | EGFR | 1.37×10−5 | 1.19 (1.10–1.29) |
0.2992 | 3.67 | 18.29 | 1.50×10−9 |
1.28 (1.18–1.39) |
0.6193 | 1.78 | 0.00 | 2.14×10−11 |
1.32 (1.21–1.43) |
0.2263 | 4.35 | 30.99 |
| rs11979158 (7p11.2) | A/G | EGFR | 2.47×10−4 | 1.22 (1.10–1.36) |
0.8084 | 0.97 | 0.00 | 6.13×10−8 | 1.35 (1.21–1.50) |
0.8007 | 1.00 | 0.00 | 2.18×10−10 |
1.42 (1.27–1.58) |
0.8696 | 0.72 | 0.00 |
| rs55705857 (8q24.21) | A/G | CCDC26 | 9.30×10−11 |
1.76 (1.49–2.10) |
0.3040 | 3.63 | 17.41 | 0.4225 | 1.08 (0.90–1.28) |
0.5380 | 2.17 | 0.00 | 0.0280 | 1.21 (1.02–1.44) |
0.0811 | 6.73 | 55.41 |
| rs634537 (9p21.3) | T/G | CDKN2B | 9.50×10−17 |
1.40 (1.29–1.51) |
0.9589 | 0.31 | 0.00 | 3.72×10−19 |
1.44 (1.33–1.56) |
0.2814 | 3.82 | 21.49 | 2.62×10−15 |
1.38 (1.27–1.49) |
0.0503 | 7.80 | 61.54 |
| rs498872 (11q23.3) | A/G | PHLDB1 | 0.0428 | 1.09 (1.00–1.19) |
0.4940 | 2.40 | 0.00 | 0.9587 | 1.00 (0.92–1.09) |
0.9194 | 0.50 | 0.00 | 0.0284 | 0.91 (0.84–0.99) |
0.0537 | 7.66 | 60.82 |
| rs12803321 (11q23.3) | G/C | PHLDB1 | 0.0062 | 1.12 (1.03–1.22) |
0.7465 | 1.23 | 0.00 | 0.1786 | 0.94 (0.87–1.03) |
0.4459 | 2.67 | 0.00 | 0.0795 | 0.93 (0.85–1.01) |
0.2666 | 3.95 | 24.11 |
| rs78378222 (17p13.1) | T/G | TP53 | 8.65×10−16 |
3.80 (2.74–5.25) |
0.7013 | 1.42 | 0.00 | 2.87×10−8 |
2.54 (1.83–3.53) |
0.5256 | 2.23 | 0.00 | 4.13×10−13 |
3.26 (2.37–4.48) |
0.0131 | 10.76 | 72.12 |
| rs2297440 (20q13.33) | T/C | RTEL1 | 8.21×10−13 |
1.41 (1.28–1.55) |
0.5492 | 2.11 | 0.00 | 7.35×10−19 |
1.55 (1.40–1.70) |
0.3556 | 3.24 | 7.50 | 1.49×10−14 |
1.45 (1.32–1.60) |
0.3719 | 3.13 | 4.19 |
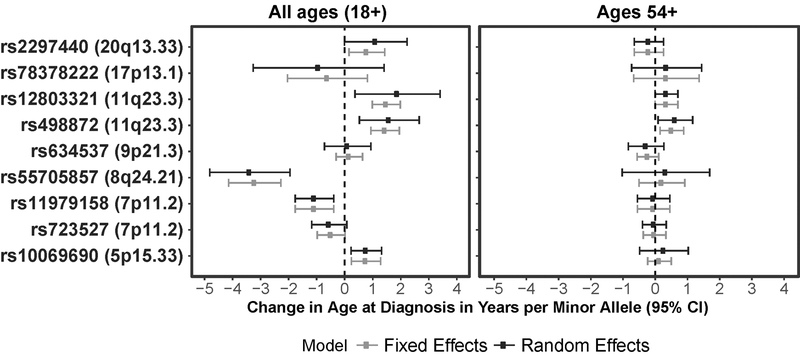
A case-only analysis was conducted in previously-identified nominally significant SNPs with age-at-diagnosis as the outcome variable, and associations were considered significant at p<4.17×10−6 (Bonferroni correction for 12,000 tests). There were two peaks that reached statistical significance in the fixed effects meta-analysis at 8q24.21, and 11q23.3 (Supplementary Figure 4a). When a random effects meta-analysis was carried out, only the peak at 8q24.21 reached the significance threshold (Supplementary Figure 4b). The previously identified SNP at 8p24.21 was significantly associated with younger age-at-diagnosis, with an approximate decrease in age-at-diagnosis of 3 years for each risk allele (prandom=3.70×10−6, pfixed=1.51×10−11) (Figure 2, Supplementary Table 4). When this association was evaluated in cases ages 54+ only, the association was null. There was a nominal association in the case-only analysis between age-at-diagnosis and the previously identified SNP at 5p15.33 (rs10069690) and GBM (pfixed =0.0033, prandom =0.0053) with an increase in age of diagnosis of 0.76 years for each risk allele. When the case-only analysis was limited to the two older age-groups (individuals >53 years old), there was no significant association between this SNP and age-at-diagnosis (pfixed=0.4818, prandom=0.4814). In the fixed effects meta-analysis, two previously identified SNPs in 11q23.3 were significantly associated with older age-at-diagnosis. For both SNPs (rs12803321 and rs498872), each risk allele was associated with an increase of approximately 1.5 years in age-at-diagnosis. There was some heterogeneity by study for both of the evaluated SNPs at 11q23.3 (rs12803321 phet=5.5×10−5; rs498872 phet=0.0138), and in the random effects meta-analysis, the associations at both of these SNPs no longer met the threshold for significance. In the subset of cases ages 54+, rs498872 remained nominally significantly associated with age-at-diagnosis (pfixed=0.0056, prandom=0.0225), with an estimated 0.52-year increase in age of diagnosis for each risk allele. There was no association between rs12803321 and age-at-diagnosis in the older subset. There was a nominal association between rs11979158 and younger age-at-diagnosis (pfixed= 0.0021, prandom= 0.0021), but this association was null in the subset of cases ages 54+.
Figure 2.
Case only estimates for SNPs identified as being significantly associated with age-at-diagnosis in this or prior analyses in a) all ages, b) cases ages 54+
Age-at-diagnosis differed significantly by molecular subgroup within the TCGA GBM dataset. Median age-at-diagnosis was lowest in IDH1/2 mutant GBM samples (38.5) and highest in IDH1/2 wild-type samples (62.0). Within the youngest age-group of individuals 18–53, 15/100 individuals had IDH1/2 mutant tumors (15%), as compared to 2/94 in those 54–63 (2.1%), and 1/121 in those 64+ (0.8%). Frequency of TERT mutation increased with increasing age, with the highest frequency of this feature (94%) occurring in the oldest age-group. Overall, GBM samples in the youngest age-group appeared to be more “LGG-like” as compared to those in the two older groups.3, 12
DISCUSSION
This is the first genome-wide age-specific analysis focused specifically on the relationship between germline risk variants and age-at-diagnosis in GBM, and leverages multiple existing glioma GWAS datasets. This study demonstrated that there are age-related differences in frequency of known heritable genetic risk variants in glioma, largely driven by differences in those less than 54 years of age. While these results replicate some previously identified associations,18 the age-stratified approach reveals potential phenotypic differences between persons diagnosed with these tumors at younger ages. Now that molecular markers are routinely incorporated in glioma diagnosis, incorporation of these more precise phenotype classifications into GWAS is necessary in order to understand the pathways and mechanisms through which these risk variants confer increased susceptibility for GBM, and the extent to which these may vary by age.
Previous studies have consistently reported that alterations in telomerase-related genes (e.g. TERT and RTEL1) are associated with increased age-at-diagnosis for gliomas.18 This analysis found that variants in TERT and RTEL1 reached genome-wide significance among all three age cohorts, with no substantial difference in estimated effect by age. In the case-only analysis, there was a nominally significant association between rs10069690 (TERT, pRandom=0.0053) and increased age-at-diagnosis, with an increase of 0.76 years of age per risk allele. There was no significant association between rs2297440 (RTEL1, pRandom=0.0512) and increased age-at-diagnosis. The analysis also reported association between risk alleles in PHLDB1 and decreased age-at-diagnosis.18 There was a nominally significant association between these SNPs in PHLDB1 and age-at-diagnosis in a case-only analysis, but when analysis was limited to those 54 years of age and older, the effect of these SNPs on age-at-diagnosis was null, suggesting that phenotypic differences between the younger and older cohort may be driving the observed allele frequency differences rather than a true effect on age-at-diagnosis.
Two SNPs at the 7p11.2 locus showed variation in association by age, with similar effects in all included studies (Supplementary Figures 4–5). These variants were within one of two previously identified independent glioma risk loci located near EGFR, which is most strongly associated with risk for GBM.7, 19 Variation in an observed association by age with a SNP at 8q24.21 was present. This SNP has been previously associated with non-GBMs—in particular low-grade tumors that have IDH1/2 mutation and 1p/19q co-deletion—and a significant association between this SNP and GBM has not been previously reported. While IDH1/2 mutation is most common in LGG (where ~80% of tumors have this alteration), only a minority of GBM (~5%) have this alteration.12 Among individuals with tumors that appear histologically to be GBM, IDH1/2 mutation is known to occur more commonly in younger individuals.20 Among the TCGA GBM set, IDH1/2 mutation occurred in 15% of cases in individuals 18–53, as compared to 2.1% and 0.8% in individuals 54–63 and 64+, respectively (p=0.0005). These mutations occur very early in gliomagenesis, and as a result represent an entity that is distinct from IDH1/2 wild-type GBM. IDH1/2 mutation is generally considered to be a marker of ‘secondary GBM’, or GBM that has progressed from a previously undiagnosed LGG.21 Individuals with these ‘secondary GBMs’ have been consistently reported to be younger than those with primary GBM.21
All GBM cases from the included four GWAS datasets were recruited at time of first diagnosis, and the assigned diagnoses represent the primary tumor type. Most glioma GWAS have categorized phenotype based on histologic criteria only. The significant inter-rater variability in diagnosis of glioma has been well documented,22 though the extent of misclassification is more significant in LGG.23 Diagnostic technologies and histologic classification criteria of GBM has changed over the recruitment periods for the four included studies (1974–2013, varying). While classification using IDH1/2 mutation and 1p19q co-deletion was not codified until the release of the 2016 WHO classification scheme, these markers were gradually adopted for use in glioma diagnosis prior to the release of this scheme. Changing classification criteria over time as well as gradual adoption of molecular markers could have resulted in a small number of some earlier cases that may not have been classified as GBM if diagnosed during a later period. The incorporation of molecular markers into classification criteria significantly improves diagnostic accuracy and consistency.24 These markers should be incorporated into glioma GWAS in order to better understand the ways that germline markers confer risk, and for which tumor types they increase susceptibility.
An alternate approach to this analysis could be to include age-at-diagnosis as an interaction term in logistic regression. Use of interaction models often requires that the effect on the association reach a stringent p-value to correct for multiple testing, and as a result these methods may miss effects that are not very large.25 Stratified analyses decrease sample size and therefore power, but these analyses may more successfully identify associations of small effect size that may not be identified using interaction models.
CONCLUSIONS
Age-specific differences in cancer susceptibility can provide important clues to etiology, and these differences can be leveraged for discovery in genetic association studies. This analysis identified potential age-specific effects in two previous identified glioma risk loci (7p11.2, and 8q24.21). The association of a SNP known to confer risk for IDH1/2 mutant glioma with GBM within individuals 18–53 suggests that a substantial portion of younger individuals included in GBM research may present initially with ‘secondary GBM.’ The higher prevalence of IDH1/2 mutant GBM within this younger age-group is also evident within TCGA GBMs. While age is known to be a strong factor associated with differences in incidence and prognosis for GBM, the results of this analysis suggest that younger age is associated with phenotype and risk of GBM.
Supplementary Material
Novelty and Impact:
This is the first genome-wide association analysis examining age-at-diagnosis--which is strongly associated with incidence and prognosis--and germline risk variants in glioblastoma. We detected a higher frequency of germline variants associated with lower grade gliomas (LGG) in the younger cohort, as well as high frequency of LGG-like somatic variants in The Cancer Genome Atlas GBM cohort. Age-specific differences in phenotype should be taken into consideration when molecular classification data is not available
ACKNOWLEDGEMENTS
The GICC was supported by grants from the National Institutes of Health, Bethesda, Maryland (R01CA139020, R01CA52689, P50097257, P30CA125123). Additional support was provided by the McNair Medical Institute and the Population Sciences Biorepository at Baylor College of Medicine.
In Sweden work was additionally supported by Acta Oncologica through the Royal Swedish Academy of Science (BM salary) and The Swedish Research council and Swedish Cancer foundation. We are grateful to the National clinical brain tumor group, all clinicians and research nurses throughout Sweden who identified all cases.
The UCSF Adult Glioma Study was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257, R01CA126831, and R01CA139020), the Loglio Collective, the National Brain Tumor Foundation, the Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, the Robert Magnin Newman Endowed Chair in Neuro-oncology, and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. This project also was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1 RR024131. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement # U58DP003862–01 awarded to the California Department of Public Health. The ideas and opinions expressed herein are those of the author(s) and endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors is not intended nor should be inferred. Other significant contributors for the UCSF Adult Glioma Study include: M Berger, P Bracci, S Chang, J Clarke, A Molinaro, A Perry, M Pezmecki, M Prados, I Smirnov, T Tihan, K Walsh, J Wiemels, S Zheng.
Glioma scan group comprised: Laura E. Beane Freeman, Stella Koutros, Demetrius Albanes, Kala Visvanathan, Victoria L. Stevens, Roger Henriksson, Dominique S. Michaud, Maria Feychting, Anders Ahlbom, Graham G. Giles Roger Milne, Roberta McKean-Cowdin, Loic Le Marchand, Meir Stampfer, Avima M. Ruder, Tania Carreon, Goran Hallmans, Anne Zeleniuch-Jacquotte, J. Michael Gaziano, Howard D. Sesso, Mark P. Purdue, Emily White, Ulrike Peters, Howard D. Sesso, Julie Buring.
UK10K data generation and access was organized by the UK10K consortium and funded by the Wellcome Trust. The results here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
We are grateful to all the patients and individuals for their participation and we would also like to thank the clinicians and other hospital staff, cancer registries and study staff in respective centers who contributed to the blood sample and data collection.
Financial Support: QTO is supported by a Research Training Grant from the Cancer Prevention and Research Institute of Texas (CPRIT; RP160097T). The GICC was supported by grants from the National Institutes of Health, Bethesda, Maryland (R01CA139020, R01CA52689, P50097257, P30CA125123, P30CA008748, C. Thompson PI). Additional support was provided by the McNair Medical Institute and the Population Sciences Biorepository at Baylor College of Medicine. In Sweden work was additionally supported by Acta Oncologica through the Royal Swedish Academy of Science (BM salary) and The Swedish Research council and Swedish Cancer foundation.
The UCSF Adult Glioma Study was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257, R01CA126831, and R01CA139020), the Loglio Collective, the National Brain Tumor Foundation, the Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, the Robert Magnin Newman Endowed Chair in Neuro-oncology, and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. This project also was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1 RR024131. The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute's Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention's National Program of Cancer Registries, under agreement # U58DP003862-01 awarded to the California Department of Public Health. UK10K data generation and access was organized by the UK10K consortium and funded by the Wellcome Trust
Footnotes
Conflict of Interest: There are no conflicts of interest to report.
REFERENCES
- 1.Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2010–2014. Neuro-oncology 2017;19: v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica 2016;131: 803–20. [DOI] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, Robertson AG, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England journal of medicine 2015;372: 2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155: 462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostrom QT, Bauchet L, Davis F, Deltour I, Eastman C, Fisher JL, Pekmezci M, Turner M, Schwartzbaum J, Walsh KM, Wrensch MR, Barnholtz-Sloan JS. The epidemiology of glioma in adults: a “state of the science” review. Neuro-oncology 2014;16: 896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinnersley B, Mitchell JS, Gousias K, Schramm J, Idbaih A, Labussiere M, Marie Y, Rahimian A, Wichmann HE, Schreiber S, Hoang-Xuan K, Delattre JY, et al. Quantifying the heritability of glioma using genome-wide complex trait analysis. Scientific reports 2015;5: 17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melin BS, Barnholtz-Sloan JS, Wrensch MR, Johansen C, Il’yasova D, Kinnersley B, Ostrom QT, Labreche K, Chen Y, Armstrong G, Liu Y, Eckel-Passow JE, et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nature genetics 2017;49: 789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raynor LA, Pankratz N, Spector LG. An analysis of measures of effect size by age of onset in cancer genomewide association studies. Genes, chromosomes & cancer 2013;52: 855–9. [DOI] [PubMed] [Google Scholar]
- 9.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nature genetics 2009;41: 905–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nature genetics 2009;41: 899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajaraman P, Melin BS, Wang Z, McKean-Cowdin R, Michaud DS, Wang SS, Bondy M, Houlston R, Jenkins RB, Wrensch M, Yeager M, Ahlbom A, et al. Genome-wide association study of glioma and meta-analysis. Human Genetics 2012;131: 1877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016;164: 550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, Benlloch S, Hazelett DJ, Wang Z, Saunders E, Leongamornlert D, Lindstrom S, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nature genetics 2014;46: 1103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team. R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing, 2017. [Google Scholar]
- 15.Wickham H ggplot2: elegant graphics for data analysis.: Springer; New York, 2009. [Google Scholar]
- 16.Karssen LC, van Duijn CM, Aulchenko YS. The GenABEL Project for statistical genomics. F1000Research 2016;5: 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv DOI: 10.1101/005165, 2014. [DOI] [Google Scholar]
- 18.Walsh KM, Rice T, Decker PA, Kosel ML, Kollmeyer T, Hansen HM, Zheng S, McCoy LS, Bracci PM, Anderson E, Hsuang G, Wiemels JL, et al. Genetic variants in telomerase-related genes are associated with an older age at diagnosis in glioma patients: evidence for distinct pathways of gliomagenesis. Neuro Oncol 2013;15: 1041–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanson M, Hosking FJ, Shete S, Zelenika D, Dobbins SE, Ma Y, Enciso-Mora V, Idbaih A, Delattre JY, Hoang-Xuan K, Marie Y, Boisselier B, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Human molecular genetics 2011;20: 2897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, et al. IDH1 and IDH2 mutations in gliomas. New England journal of medicine 2009;360: 765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clinical cancer research : an official journal of the American Association for Cancer Research 2013;19: 764–72. [DOI] [PubMed] [Google Scholar]
- 22.van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta neuropathologica 2010;120: 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott CB, Nelson JS, Farnan NC, Curran Jr., Murray KJ, Fischbach AJ, Gaspar LE, Nelson DF. Central pathology review in clinical trials for patients with malignant glioma. A Report of Radiation Therapy Oncology Group 83–02. Cancer 1995;76: 307–13. [DOI] [PubMed] [Google Scholar]
- 24.Kim BY, Jiang W, Beiko J, Prabhu SS, DeMonte F, Gilbert MR, Sawaya R, Aldape KD, Cahill DP, McCutcheon IE. Diagnostic discrepancies in malignant astrocytoma due to limited small pathological tumor sample can be overcome by IDH1 testing. Journal of neuro-oncology 2014;118: 405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powers MS, Smith PH, McKee SA, Ehringer MA. From sexless to sexy: Why it is time for human genetics to consider and report analyses of sex. Biology of sex differences 2017;8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.