Abstract
Heparan sulfate (HS) plays an important role in development and disease. It interacts with many growth factors, chemokines and other ligands known to be important for cell growth, motility and differentiation. However, isolating an antibody to HS in mice, rabbits or humans is difficult due to the poor immunogenicity of HS. Phage display is a major antibody engineering technology that allows the selection of antibodies for poorly immunogenic or highly conserved antigens. This protocol contains detailed procedures for HS antigen preparation and isolation of a phage displayed human single chain Fv (HS20) that binds HS on glypican-3 (GPC3), and analysis of the selected phage antibody. It is conceivable that the procedures described in this protocol may be applicable to the isolation of antibodies for a variety of HS molecules.
Keywords: Phage display, Heparan sulfate proteoglycan, Glypican, Single-chain variable fragment or scFv
INTRODUCTION
The purpose of this unit is to isolate and identify single chain variable fragments (scFvs) that recognize heparan sulfate on glypicans by selection and amplification with Tomlinson scFv phage display libraries using recombinant human GPC3 proteins as targets (Fig. 1). We describe the phage antibodies that bind to heparan sulfate on glypicans by the panning and selection in the Basic Protocol. In the Alternate Protocol, we describe another way of isolating binders for heparin sulfate on glypicans. The method of cloning, expression, and purification of GPC3 necessary for panning for phage is described in the Support Protocol 2. It also describes critical steps in troubleshooting and background information about the phage display library used here (see COMMENTARY).
Figure 1.
Flow chart for phage display.
BASIC PROTOCOL 1: PHAGE DISPLAY FOR GPC3
This protocol describes the overall methods required for phage display, including phage library growth, phage concentration by polyethylene glycol (PEG) precipitation, panning and characterization of antibodies using ELISA, FACS, and immunohistochemistry.
Materials
For phage amplification
2YT medium (See Reagents and Solutions)
Ampicillin-100 mg/ml (Teknova)
Kanamycin-50 mg/ml (Teknova)
50% w/v glucose solution (Teknova)
M13KO7 helper phage (NEB)
PEG 6000/NaCl Solution (Teknova)
Elution buffer: 100mM HCl
Trypsin (Sigma-Aldrich)
50 % v/v Glycerol (Teknova)
For ELISA
-
PBST (wash buffer):
1 × PBS containing 0.1% v/v Tween 20 (Sigma).
-
Blocking solution (2 % w/v Skim milk)
Dissolve 0.4 g of Difco skim milk in 20ml of 1 X PBS per one 96 plates, use fresh blocking buffer.
MaxiSoap 96-well plates (Sigma-Aldrich).
HRP-conjugated anti-M13 phage antibodies (GE Healthcare Life Sciences)
Phage display library- Human single-fold scFv libraries Tomlinson I+J (Medical Research Council)
(see Commentary)
1M Tris-HCl (pH 8.0)
E. coli F+ strain: TG1 (for phage production) and HB2151 (for soluble scFV production) were included in Human single-fold scFv libraries Tomlinson I+J. TG1 electrocompetent cells can also be purchased from Lucigen (Middleton, WI)
Isopropyl β-D-thiogalactopyranoside (IPTG)
0.22 μm syringe filter (Millipore)
LB agar plates with 100 μg/ml ampicillin
TMB Chromogen Solution (ThermoFisher Scientific)
For immunohistochemistry
Xylene (Sigma-Aldrich)
Ethanol absolute (Sigma-Aldrich)
For HS20 IgG construction
pFUSE-CHIg-HG1 (Invivogen) for heavy chain cloning
pFUSE2-CLIg-hk (Invivogen) for light chain cloning
Platinum Taq DNA Polymerase High Fidelity (ThermoFisher Scientific)
One Shot TOP10 Chemically Competent E. coli (ThermoFisher Scientific)
293T cells (ThermoFisher Scientific)
DMEM (ThermoFisher Scientific)
FreeStyle293 Expression Medium (ThermoFisher Scientific)
HiTrap Protein A High Performance (GE Healthcare Life Sciences)
Equipment
Bacterial shaker-incubator
50-ml conical tube or flask
96-well U-bottom microtiter plate (Sigma-Aldrich)
Centrifuge with 96-well microtiter plate adapter
NanoDrop (ThermoFisher Scientific)
FACSCalibur (BD Biosciences)
100 mm dish
150 mm dish
Growing the libraries
-
1
Add 100 μl of the library glycerol stock to 50 ml pre-warmed 2YT containing 100 μg/ml ampicillin and 1 % v/v glucose.
-
2
Grow the libraries at 37°C with shaking at 225 rpm until the OD 600 is 0.4 (~1hr).
-
3
Add 50 μl M13KO7helper phage (final concentration 1 × 108 pfu/ml).
-
4
Incubate for 30 minutes at 37 ° C without shaking, then incubate for an additional 30 minutes at 37 ° C with shaking at 250 rpm.
-
5
Centrifuge at 3,000 g for 10 min, decant the supernatant and resuspend the TG1 cells in 100 ml of 2YT containing 100 μg/ml ampicillin, 50 μg/ml kanamycin.
-
6
Incubate cultures overnight at 30 ° C with shaking at 250 rpm.
-
7
Centrifuge the overnight culture at 3,300 g for 30 min, discard pellet and filter the supernatant using 0.22 μm filter.
Phage concentration by PEG precipitation
-
8
Add 12.5 ml PEG/NaCl (20 % w/v Polyethylene glycol 6000, 2.5 M NaCl) to 50 ml of the phage supernatant and mix well and leave for 1 hr on ice to precipitate phage.
-
9
Centrifuge at 3,300 g for 30 min, discard the supernatant and aspirate any remaining phage supernatant carefully.
-
10
Resuspend the pellet in 4 ml PBS to dissolve the phage library thoroughly and aliquot the phage library into 1 ml per vial.
-
11
Store the phage at 4°C for short-term storage (up to a few weeks).
The phage library can also be stored in PBS with 15 % v/v glycerol for longer term at −70°C. -
12
Determine the phage titer (see Support Protocol 1)
Selection on 96-well plates
-
13
Coat maxiSoap 96-well plates overnight with 100 μl of the required antigen (10–100 μg/ml) in each well.
-
14
Wash the plate three times with PBS, then add 300 μl of blocking solution (2 % w/v Skim milk in PBS) in each well.
-
15
Incubate for 1 hr at room temperature for blocking.
-
16
Discard the blocking solution by flipping plate over into the sink.
-
17
Add 1012 phages in 100 μl of 2 % w/v Skim milk. Incubate for 1 hr at room temperature and then continue to incubate for 1 hr at 37°C.
-
18
Discard nonbinding phages by flipping plate over into the sink.
-
19
Wash plate 10–20 times with PBS containing 0.1 % v/v Tween 20.
In the first round of panning, wash wells 10 times and increase it 20 times from the next round of panning. -
20
Elute phage by adding 100 μl of elution buffer (10mM HCl) and incubate for 10 min at room temperature and transfer into new 15 ml conical tube.
-
21
Immediately add 10 μl of 1 M Tris-HCl (pH 8.0) to eluted phages to neutralize.
Alternative elution: add 100 μl of freshly prepared 100mM triethylamine and incubate for 10 min at room temperature. -
22
Take 1.89 ml of TG1 at an OD 600 of 0.4 and add 110 μl of the neutralized phages.
-
23
Incubate for 30 min at 37°C without shaking to enhance transduction and incubate for additional 30 min with shaking at 250 rpm.
In this step, phage containing the gene encoding the scFv of interest attaches to the F-pilus of TG1 and the phage DNA is inserted into E. coli.
Output phages determined by titering
-
24
Spot 100 μl of a 1:102 dilution and 100 μl of a 1:104 dilution on 100 mm 2YT plates containing 100 μg/ml ampicillin and 1% v/v glucose and grow overnight at 37°C to determine the titer of phage.
-
25Calculate total output phages in 2 ml with the following formula.If phage output is less than 1000 phages output after panning, repeat the infection with fresh TG1 culture grown from a single colony (see Troubleshooting).
-
26
Store the plate at 4 °C for monoclonal phage ELISA.
-
27
Add remaining phages on a 150 mm 2YT plate containing 100 μg/ml ampicillin and 1 % v/v glucose and let it dry for 10 min at clean bench and evenly spread with scraper.
-
28
Grow plates at 37°C overnight.
-
29
After overnight growth, add 2 ml of 2YT 25 % v/v glycerol to the plate, loosen the cells with a spreader and collect the cells into a new tube.
-
30
Freeze the cells at −80°C as bacterial library stock.
Further rounds of selection
-
31
Inoculate 10 μl of the bacterial glycerol stock to 10 ml of 2YT containing 100 μg/ml ampicillin and 1 % v/v glucose.
-
32
Grow the TG1 cells by shaking at 37°C until the OD 600 reach 0.4 (about 1 hr) and add 10 μl M13KO7helper phage (final concentration of 1 × 108 pfu/ml).
-
33
Incubate without shaking at 37°C for 30 min and incubate for another 30 min with shaking at 250 rpm.
-
34
Centrifuge at 3,000 g for 10 min, decant the supernatant and resuspend the pellet in 10 ml of 2YT containing 100 μg/ml ampicillin, 50 μg/ml kanamycin.
Do not add glucose in this step -
35
Shake at 250 rpm at 30°C overnight.
-
36
Centrifuge the overnight culture at 3,300 g for 15 min, discard the pellet and filter the supernatant with 0.22 μm filter.
-
37
Add 2.5 ml PEG/NaCl (20% v/v polyethylene glycol 6000, 2.5 M NaCl) to the 10 ml supernatant. Mix well and leave it on ice for 1 hr.
-
38
Centrifuge at 3,300 g for 30 min and discard the supernatant carefully.
-
39
Resuspend the pellet in 0.5 ml PBS.
-
40
Use 0.1 ml of this phage for the next round of selection and store the remaining phages at 4°C for up to 1 month.
-
41
Repeat selection from step 31 for another two rounds.
Screening phage by ELISA
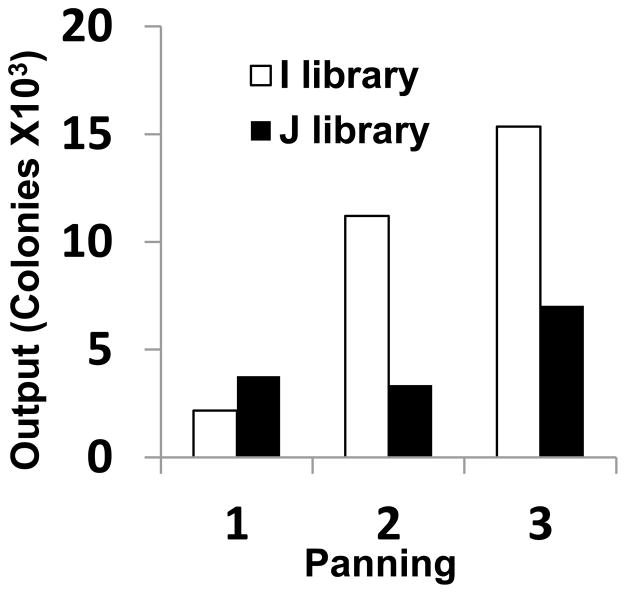
The progress of an affinity selection can be monitored by polyclonal phage ELISA. This affinity panning is usually executed in 3–4 rounds. After 3–4 rounds of panning, the phages obtained from each round of panning can be checked for binding to the target by polyclonal phage ELISA. We screened human scFv phage display library against 100 μg/ml of recombinant GPC3 coated on MaxiSoap 96-well plate for 3 rounds of panning (Fig. 2). This shows enrichment of phage Fvs against GPC3.
Figure 2.
Enrichment of phage antibodies against GPC3. Polyclonal phage ELISA: phage populations selected in each round were analyzed with GPC3. Pooled phages were amplified and 2 X 1012 phage were added to an immunoplate precoated with recombinant GPC3. Bound phages were detected with anti-M13 HRP monoclonal antibody. Output colonies were counted after each round of panning.
Polyclonal phage ELISA
-
42
Coat a maxiSoap 96-well plates overnight with 50 μl per well of antigen in PBS (5–10 μg/ml).
-
43
Wash wells 3 times with PBS. Discard liquid by flipping plate over into the sink.
-
44
Add 200 μl per well of 2% w/v skim milk in PBS to block by incubating for 2 hr at room temperature.
-
45
Mix 25 μl of phage from the end of each round of selection and 25 μl of 4% (w/v) skim milk in a microcentrifuge tube and incubate for 1 hr at room temperature for phage pre-blocking.
-
46
Discard liquid by flipping plate over.
-
47
Add 50 μl pre-blocked phage mixtures to antigen-coated plate and incubate for 1 hr at 37°C.
-
48
Discard phage solution and wash 5 times with PBS-0.1 % v/v Tween 20.
-
49
Add 50 μl of HRP-anti-M13 (1:5000 dilutions) in PBS and incubate for 30 min at 37 °C. Wash 5 times with PBS-0.1 % v/v Tween 20.
-
50
Add 50 μl of TMB substrate solution to each well and incubate for 2–15 min at room temperature until blue color develops. Read the OD at 650 nm.
Alternatively, stop the reaction by adding 25 μl 1 M sulphuric acid. The blue color should turn yellow. Read the OD at 650 nm and at 450 nm. Subtract OD 650 from OD 450.
Monoclonal phage ELISA
Ninety-six randomly picked phage clones at the end of each round of panning are analyzed for GPC3 binding.
-
51
Inoculate individual colonies from each round of selection with sterilized 200 μl pipet tip into 100 μl 2YT containing 100 μg/ml ampicillin and 1% v/v glucose in 96 well culture plate and grow overnight at 37°C.
Keep this plate as a master plate -
52
Transfer 2 μl of culture solution from a master plate to a second 96 deep well plate containing 200 μl of 2YT with 100 μg/ml ampicillin and 1% v/v glucose per well. Grow with shaking at 250 rpm for 2 hr at 37°C.
To make glycerol stocks, add 100 μl of 50% v/v glycerol to master plate, mix well and store the plates at −70°C. -
53
Add 25 μl of 2YT medium containing 100 μg/ml ampicillin, 1% v/v glucose and 1 X 108 helper phage.
-
54
Incubate without shaking for 30 min at 37°C and shake at 250 rpm for additional 30 min at 37°C.
-
55
Centrifuge at 1,800 g for 10 min and aspirate the supernatant.
-
56
Resuspend pellet in 200 μl of 2YT containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. Grow with shaking at 250 rpm overnight at 30°C.
Do not add glucose in this step -
57
Centrifuge at 3,000 g for 10 min and use 50 μl of the supernatant for monoclonal phage ELISA.
Sequencing selected clones
For analysis of Ig variable region, phagemid clones are sequenced using primers VH-R for the heavy chain (5′-CGA CCC GCC ACC GCC GCT G-3′) and Vk-R for light chain (5′-CTA TGC GGC CCC ATT CA-3′). Complementarity-determining regions (CDRs) are determined by aligning with human Ig genes using IMGT/V-QUEST in the International Immunogenetics Database (IMGT, http://imgt.cines.fr/).
Convert phage to full-length IgG
Because an amber codon was present in the light chain backbone of HS20, the amber codon was changed to glutamine codon by recombinant PCR. The antibody scFv with altered amber codon was subcloned into an appropriate restriction enzyme site in a mammalian expression vector. The heavy chain variable region of the scFv gene was cloned into pFUSE-CHIg-HG1 The forward primer of the VH PCR primer contains the IL-2 signal sequence and the EcoRI restriction site on the 5′ end. The reverse primer contains the NheI restriction site at 5′ end. The PCR product was inserted between the EcoRI and NheI sites of the expression vector pFUSE-CHIg-HG1. The VL region was PCR amplified using a forward primer containing the IL-2 signal sequence and an AgeI restriction enzyme site at the 5′ site and a reverse primer containing the NcoI restriction enzyme site at the 5′ end. The PCR product was inserted into the AgeI and NcoI sites of the expression vector pFUSE2-CLIg-hk. Using Lipofectamine 2000, the plasmids were transiently cotransfected in HEK-293T cells (Invitrogen, Carlsbad, Calif.) in DMEM medium containing 10% BSA and the following day to remove bovine IgG contained in DMEM. The supernatant was replaced with FreeStyle293 expression medium. After 3 days, the medium was collected after centrifugation, further incubated for 3–4 days and collected again. The supernatant was purified using a Protein A Hi-Trap column. The quality and quantity of purified IgG1 was determined by SDS-PAGE and by A280 absorbance using Nanodrop. Purified HS20 IgG was analyzed by ELISA on GPC3-his.
Amplify the HS20 genes
-
58
Use 10 μM of forward and reverse primers described above and 10 ng of HS20 as the template for PCR.
-
59
Add 1 μL of high fidelity Taq DNA polymerase in a 50-μL and cycle using the following profile: 1 cycle at 95°C for 3 min, followed by 30 cycles at 94°C for 15 sec, 55°C for 15 sec, and 72°C for 30 sec, and 1 cycle at 72°C for 10 min.
-
60
Digest the PCR products for heavy and light chain with restriction enzymes, gel purify, and ligate into the pFUSE-CHIg-HG1 and pFUSE2-CLIg-hk, respectively (cut with same restriction enzymes and gel purified).
-
61
Transform the ligation mixture into competent E. coli TOP10 cells and plate on LB agar containing 100 μg/ml ampicillin to select for ligated single clones.
-
62
Incubate the plate at 37°C overnight.
-
63
Pick 5 individual colonies from each plate and culture the individual colonies in 3 ml of LB media and shake at 37°C overnight.
-
64
Purify plasmid from 2 ml of culture using Qiagen miniprep kit.
Save 1 ml of culture for midi-culture after sequence confirmation. -
65
Sequence the inserts of 5 individual clones each and confirm the insert sequence.
-
66
Culture positive clones in 300 ml of LB containing 100 ug/ml of ampicillin by shaking at 150 rpm at 37°C overnight.
-
67
Purify plasmids by using Qiagen plasmid midi kit.
-
68
Co-transfect the plasmids for heavy and light chain into HEK-293T cells (Invitrogen, Carlsbad, CA) using Lipofectamine 2000 in DMEM containing 10 % v/v FBS.
-
69
Next day, change the DMEM media to FreeStyle serum-free medium (Invitrogen, Carlsbad, CA) to eliminate bovine IgG in the purification step. Collect the media after 3 days and add fresh FreeStyle serum-free medium and culture for additional 3–4 days, and collect again.
-
70
Pool supernatants and filter the supernatants using 0.22 μm filter.
-
71
Purify antibody by using a 1-mL recombinant Protein A Hi-Trap column on ÄKTAexplorer as described in the manufacturer’s instruction. Determine the quality and quantity of purified IgG1 by SDS-PAGE and A280 absorbance on a Nanodrop.
Characterization of antibody
ELISA
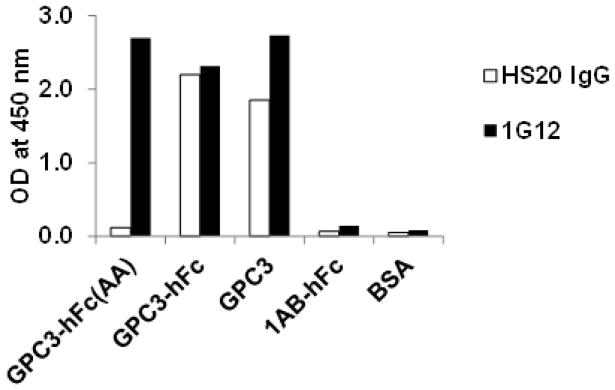
HS-binding ligands include growth factors, cytokines, chemokines, enzymes, enzyme inhibitors, and extracellular matrix proteins (Sarrazin et al. 2011). We describe a phage ELISA protocol that is used to characterize HS20 IgG specific to the target protein, GPC3 (Fig. 3). The GPC3 protein, GPC3-hFc (AA) and GPC3-hFc and GPC3, and negative controls (1AB-hFc or BSA) are immobilized on a 96-well immunoplate. HS20 IgG or anti-GPC3 (1G12) are added to the target protein and control proteins under conditions appropriate for binding. Following a brief incubation period, the immunoplate is washed followed by detection using an appropriate second antibody conjugated to horseradish peroxidase (HRP). Upon addition of the HRP substrate, positive clones are identified using a spectrophotometer.
Figure 3.
Binding properties of HS20. The mAb (HS20) was tested for its binding to GPC3-hFc, GPC3 (AA)-hFc, a mutant GPC3 without HS, and GPC3 alone. Anti-GPC3 (1G12) was used as a positive control. 1AB-hFc (the human Fc control) and BSA were used as negative controls. HS20 bound HS on GPC3, and 1G12 bound on core.
-
72
Coat 96-well ELISA plates with 50 μl of GPC3-hFc (AA), GPC3-hFc and GPC3, 1AB-hFc or BSA at 5–10 μg/ml in PBS at 4°C overnight.
-
73
Block the plate with 100 μl of 1% of BSA in PBS at room temperature for 1 hr.
-
74
Wash the wells with PBS containing 0.1% v/v Tween 20 (PBST).
-
75
Add 50 μl of HS20 IgG or 1G12 (1 μg/ml) and incubate for 1 hr at room temperature.
-
76
Wash the plates four times with PBST.
-
77
Incubate each well with 50 μl of HRP-conjugated second antibody (1:5,000) at room temperature.
-
78
Wash the plates four times with PBST.
-
79
Add 50 μl of TMB/H2O2 and incubate for 15 min at room temperature.
-
80
Read OD at 650 mm or stop the reaction with 50 μL of 1 N sulfuric acid and read the absorbance at 450 nm.
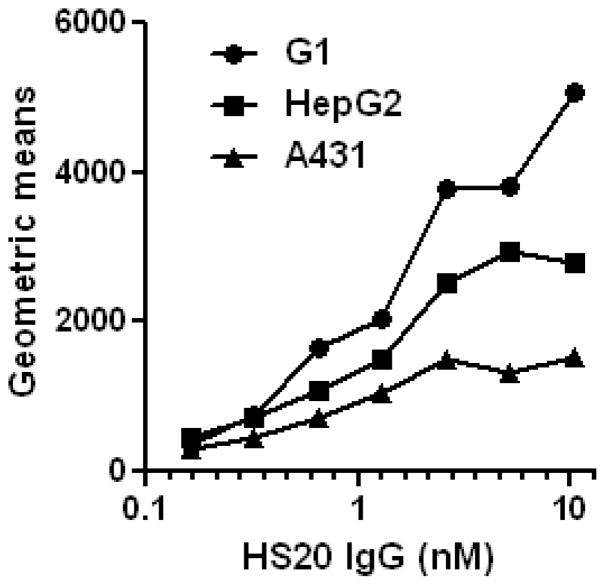
FACS (Fluorescence-activated cell sorting)
To determine the binding of HS20 to cell surface-associated GPC3 proteins, cancer cell lines (A431; human squamous carcinoma cell line, G1; stable cell line expressing recombinant GPC3 in A431, and HepG2; human hepatoblastoma cell line) are incubated with HS20 antibody with various concentrations. The binding was visualized with a goat anti-human IgG PE-conjugated secondary antibody using FACSCalibur (Fig. 4).
Figure 4.
Characterization of HS20 on cells by flow cytometric analysis. Cells (G1 or A431) were incubated with an HS20 at a concentration of 0, 10, and 1000 ng/ml. The binding was detected with a goat anti-human IgG PE-conjugated secondary antibody by flow cytometry.
-
81
Incubate 5 × 105 G1, HepG2 and A431 cells in 0.5 mL of FACS blocking buffer (5% v/v BSA in PBS).
-
82
Add various concentrations (0.2 to 10 nM) of HS20 IgG to cells (5 × 105) and incubate on ice for 1 hr.
-
83
Wash cells two times with FACS blocking buffer.
-
84
Add a goat anti-human IgG-PE (1:200), and incubate the cells for 30 min on ice.
-
85
Wash cells two times with FACS blocking buffer, and perform analysis on a FACSCalibur machine in a flow cytometry core facility (NCI CCR Flow Cytometry Core).
-
86
Acquire data using Cell Quest software and analyze the binding of the antibody to cells by measuring the PE signals using FlowJo software.
Measure geometric means to compare the degree of positive. The larger the geometric mean, the more antibodies are bound per cell.
Immunohistochemistry
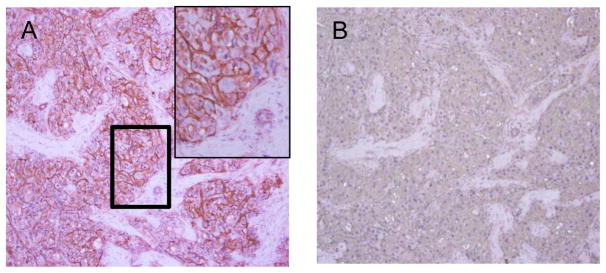
Immunohistochemical staining is a tool for determining the tissue distribution of antigens of interest in health and disease. In order to perform the standard staining procedure, the tissue section should be deparaffinized and then rehydrated before applying the primary antibody. An enzyme-linked secondary antibody can then be applied to visualize specific staining after addition of the enzyme specific substrate. Below we describe immunohistochemical protocols for human HCC tumor specimens using HS20 IgG. Figure 5 shows immunohistochemical analysis of HS20 in HCC tissues.
Figure 5.
Immunohistochemical analysis of HS20 in human hepatocellular carcinoma (HCC) patient’s tissue. Human HCC tissue was immunostained with HS20 (A). The upper left corner in (A) is a magnified view of the bold rectangle. Human IgG was used as isotype control (B). HS20 showed strong immunostaining in HCC cell plasma membrane but not in stromal cells.
Slide Preparation: deparaffinization and rehydration
To perform IHC, paraffin on tissue must be removed from the sample and the sample must be rehydrated.
Place slide-mounted sections in racks, and carry out deparaffinization steps as described below.
-
87
To remove paraffin, place the rack in xylene twice for 3 min each.
-
88
Place the sections in a 1:1 mixture of 100% ethanol and xylene for 3 min
-
89
To start rehydration, place sections in two containers of 100% ethanol for 10 min each.
-
90
Place sections in a container of 95% v/v ethanol for 3 min.
-
91
Place sections in 70 % v/v ethanol for 3 min.
-
92
Place sections in 50 % v/v ethanol for 3 min.
-
93
Rinse the sections two times with deionized (DI) water for 5 min
Pretreatment
If HRP-conjugated antibodies are used for detection, background staining should be blocked by inhibiting endogenous peroxidase activity with H2O2, as described below. Formalin or other aldehyde fixation forms protein cross-links, masking epitope sites recognized by the primary antibody. Such cross-linking can be destroyed by heat-induced and proteolytic inductive epitope retrieval.
Inactivation of endogeneous peroxidase
Since cells have endogenous peroxidase, endogenous peroxidase blockade is a necessary procedure in peroxidase detection systems.
-
94
Soak all slides in hydrogen peroxide block (0.3% v/v hydrogen peroxide) and incubate for 10 minutes.
-
95
Rinse with DI water.
Go to blocking step (step 102) if antigen retrieval is not necessary.
Antigen retrieval
Formaldehyde acts as a fixing agent because it reacts with primary amines on proteins to form crosslinks. Cross-linking of proteins is formed during fixation with formaldehyde, so that antigenic sites that can interfere with antigen recognition by the antibody can be masked. This can be overcome by an antigen retrieval that disrupts protein crosslinking to expose antigenic sites to the antibody.
-
96
Put slide rack in 10 mM Citrate buffer or EDTA.
-
97
Cover the rack and microwave in HIGH (Level 1) power for 2 min 30sec.
-
98
Keep the slides warm by heating for 10 min at LOW (Level 2 or 3) power.
-
99
Cool the slides for 40 min in the microwave.
-
100
Remove slides from microwave and rinse with tap water.
-
101
Submerge rack in 1x PBST.
Blocking non-specific binding
-
102
Apply blocking buffer and incubate for 10 min.
-
103
Rinse 2 times with PBS-0.1 % v/vTween20 (PBST).
Staining
-
104
Dilute the HS20 human antibody with PBS to 1 μg/ml and incubate for 1 hr.
-
105
Rinse two times with PBST.
-
106
Incubate for 10–20 min with anti-human IgG-HRP.
-
107
Rinse three times with PBST.
-
108
Put DAB solution on each slide and incubate for 5–10 min.
-
109
Rinse three times with PBST.
-
110
Soak in hematoxylin for 1 min to counterstain.
Most cells are colorless and transparent, making it difficult to determine the type and location of the cells. Counterstaining after immunostaining helps to find the exact location of the positive cells. A counterstain also provides contrast to make antibody staining of cells more prominent. -
111
Rinse the slide with running tap water immediately.
-
112
Perform dehydration in reverse order of the rehydration steps (step 87–92).
-
113
Cover specimens with mount medium and spread it out evenly.
-
114
Put coverslip on the tissue.
-
115
Allow to dry in fume hood before imaging.
ALTERNATE PROTOCOL: PANNING ON MAGNETIC BEADS
The antigens used for panning can be immobilized to various types of solid supports such as magnetic beads (Walter et al. 2001), column matrix (Noppe et al. 2009), nitrocellulose (Hawlisch et al. 2001) or maxiSoap plates. The advantage of using magnetic beads is that it has more surface area thus allowing screening of antibodies to various conformational epitopes. Phage can interact with biotinylated antigens in solution, and the complexes can then be captured by streptavidin or streptavidin-coated plastic surfaces (Kala et al. 1997)
Materials
10 mM Tris, pH 7.5
0.22 μm filter
μMACS Streptavidin MicroBeads (Miltenyi Biotec Inc.)
PBS (see recipe)
uMACS column (Miltenyi Biotec Inc.)
-
1
Prepare GPC3 (from Support Protocol 2) by biotinylation for phage display using magnetic beads conjugated to streptavidin.
The degree of biotinylation can be determined by measuring the average number of biotin groups per protein molecule after the biotin labeling reaction using the HAVA assay. When HABA is bound to avidin, the complex absorbs light at 500 nm. When a biotin-containing sample is added, biotin strongly binds to avidin and replaces weakly bound HABA; the absorbance decreases relative to the amount of substituted HABA. Moles of biotin per mole of protein can be calculated using HABA calculator (https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-labeling-crosslinking/protein-labeling/biotinylation/biotin-quantitation-kits/haba-calculator.html)
Remove non-specific binders
-
2
Combine 10 μl of 10 mM of Tris (pH 7.5), 100 μl of μMACS streptavidin, and 1011 phages in a [*Author: what size?] tube. Incubate [*Author: at what temperature?] for 5 min.
Filter phage to prevent clogging the columns
-
3
Place the μMACS column in the MACS magnetic stand and rinse the column with 200 μl of PBS buffer.
-
4
Load [*Author: is the column still in the magnetic stand at this point?] the phage-microbeads mix onto the column and let it pass through the column.
-
5
Collect the flow-through in a tube.
-
6
Add another 50 μl of PBS to the column and collect the flow-through in the same tube.
Isolate binders
-
7
Add biotinylated antigen (from step 1) to collected phages and incubate for 15 min at room temperature with rotation.
-
8
Add 100 μl of μMACS streptavidin microbeads to the phage solution and incubate for 10 min at room temperature.
-
9
Place the μMACS column in the magnet of the MACS stand and rinse the column two times with 200 μl PBS buffer.
-
10
Apply the mixture (from step 2) on the column. Pass the phage mixtures through the column and wash the column 10 times with 200 μl PBS.
-
11
Add 20 μl of elution buffer on the column and incubate 5 min at room temperature. Add an additional 200 μl of elution buffer and collect the flow through in the new tube. This fraction contains antigen-bound phages.
Phage can also be eluted by adding excess soluble antigen to compete with phage of immobilized antigen.Add 20 μl of 100 μg/ml of the free target on the column and incubate 5 min at room temperature.
Add an additional 20 μl of the free target (100 μg/ml) and collect the flow through in the new tube.
SUPPORT PROTOCOL 1: Determine phage titer
Phage titers can be determined by serial diluting the bacteriophage culture. Spread the diluted solution of bacteriophage onto the agar plate containing ampicillin. Bacteria grow into small colonies on the agar plates. By counting the number of colonies and multiplying by a serial dilution factor, the number of phage particles in the original phage culture can be determined. Here we also describe a simple alternative to determine the phage titer by measuring OD at 260 nm.
Materials
PBS (see REAGENTS AND SOLUTIONS)
E.Coli. TG1 (see REAGENTS AND SOLUTIONS)
2YT (see REAGENTS AND SOLUTIONS)
PEG 6000/NaCl Solution (Teknova)
Ampicillin-100 mg/ml (Teknova)
50% w/v glucose solution (Teknova)
-
Prepare dilutions using 100 fold serial dilutions of phage in PBS range 1 to 10−10.
For example, dilute 2 μl phage in 200 μl PBS, 2 μl of this in 200 μl PBS and so on until there are 6 dilutions in total including original stock. Suggested dilution ranges: for amplified phage stocks and infected culture supernatants, 108–1010; for unamplified panning elutes, 10–104. Add 10 μl to 90 μl of TG1 at an OD 600 of 0.4 to each tube and incubate at 37°C for 30 min. Plate 100 μl of each dilution on a 2YT agar plate including 100 μg/ml ampicillin and 1% v/v glucose.
Incubate the plate overnight at 37°C. Phage stock should be 1011–1013 cfu/ml.
-
Count colonies on plates that have approximately several hundred colonies.
Calculate the colony forming units (cfu) using the following formula:e.g., if colony number is 50 in a plate with 100 μl of 108 dilution,
50 X 108 X 10= 5 × 1010 pfu/ml.
Alternatively, phages precipitated with PEG are measured for OD at 260 nm and the number of phage particles are quantified by UV spectrophotometry according to the following formula (Lee et al., 2007):
SUPPORT PROTOCOL 2: ANTIGEN PREPARATION
To make GPC3 antigen for screening for GPC3 binder, pReceiver vector containing a full-length GPC3 cDNA followed by a His6 tag at C-terminal was used for transfection. Histidine-tagged GPC3 can be purified using HisTrap HP column as described in GPC3 protein preparation and its characterization was performed by western blot and ELISA in this unit.
Materials
Plasmid DNA (pPESVER-M77, GeneCopia Inc.)
Lipofectamine (Thermo Fisher Scientific)
PLUS Reagent (Thermo Fisher Scientific)
Serum-free DMEM
FBS (fetal bovine serum)
HisTrap HP column (GE Healthcare Life Sciences)
Binding buffer (see REAGENTS AND SOLUTIONS)
Wash buffer (see REAGENTS AND SOLUTIONS)
Elution buffer (see REAGENTS AND SOLUTIONS)
Immidazol solution (see REAGENTS AND SOLUTIONS)
Amicon Ultra-15 Centrifugal Filter Unit
PBS-0.1 % v/v Tween 20
HRP/Anti-M13 Monoclonal Conjugate (GE Lealthcare Life Sciences)
TMB (3,3′, 5,5;-tetramethylbenzidine) chromogen
PVDF (Polyvinylidene difluoride) blotting membrane (GE Lealthcare Life Sciences)
Ponceau S (Sigma-Aldrich)
Peroxidase AffiniPure Goat Anti-Human IgG (Jackson ImmunoResearch Laboratories, Inc.)
TBST (Tris-buffered saline, 0.1% v/v Tween 20)
Preparation of immunogen
Transfection
Transfect pReceiver vector containing a full-length GPC3 cDNA followed by a His6 tag at the C-terminus (pRESEVER-M77) into 500 ml of 293F cells (1X106/ml).
-
1
Plate five 100-mm tissue culture dishes at 2 × 106 cells per plate for 24 hr before transfection.
-
2
Make a dilution of DNA with serum-free DMEM (4 μg GPC3 DNA in 750 μL of DMEM per dish), mix well.
-
3
Add 20 μL of PLUS Reagent to the 750 μL of DNA solution, mix well, and incubate at room temperature for 15 min.
-
4
Dilute Lipofectamine reagent with serum-free DMEM in the second tube (30 μL in 750 μL for 1 dish).
-
5
Combine the DNA solution and the diluted Lipofectamine (1.5 mL total volume per dish). Mix well, and incubate at room temperature for more than 15 min.
-
6
While complexes are forming, replace the medium on cells with 5 mL of serum-free DMEM per 100 mm dish.
-
7
Gently add the DNA-Lipofectamine mixture (from step 5) to the dishes (1.5 mL mixture per dish). Gently shake the dish several times.
-
8
Incubate the dishes in CO2-incubator for 5 hr.
-
9
Gently add 6 mL of 20% v/v FBS DMEM to each dish and return the dish to CO2 incubator.
-
10
Add 10 mL of 10% v/v FBS DMEM growth medium into each dish 24 hr after transfection.
-
11
Incubate the cells for 7 days in a shaking CO2 incubator at 37°C.
-
12
Harvest from the culture supernatant by centrifugation and filter the supernatant through 0.22 μm filter.
Purification
Histidine-tagged GPC3 can be purified using HisTrap HP column, which is prepacked with Ni Sepharose by immobilized metal ion affinity chromatography (IMAC).
-
13
Fill the pump tubing with distilled water. Remove the stopper and connect the column to the pump tubing.
-
14
Remove the twist-off end of HisTrap HP column and wash the column with 5 ml of distilled water.
-
15
Equilibrate the column with at least 5 ml of binding buffer with 1 ml/min flow rate.
The binding capacity of Ni-NTA Agarose is 5–10 mg of protein per mL of resin. -
16
Apply the pretreated sample using a pump.
-
17
Wash with wash buffer until the absorbance reaches a steady baseline.
Alternatively, a low concentration of imidazole (up to 20 mM) can be used to increase the purity of recombinant His-tagged proteins. -
18
Elute with 5 ml of elution buffer.
The elution buffer contains a high concentration (250 mM) of imidazole. -
19
Perform buffer exchange to remove the imidazole and concentrate to 500 μl using an Amicon Ultra-15 Centrifugal Filter Unit.
Antigen characterization
ELISA
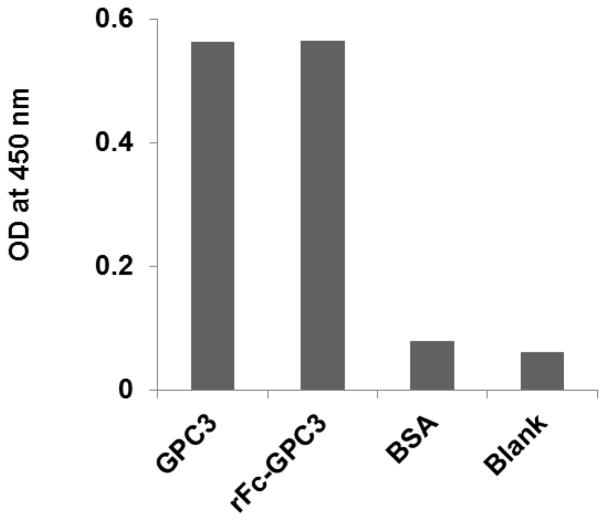
The characterization of GPC3 protein preparation was performed by ELISA (Fig. 6). One microgram of the purified recombinant proteins (GPC3 and rFc GPC3, rabbit Fc-GPC3 fusion) were coated on 96-well plates and probed with the 1G12 anti-GPC3 mAb.
Figure 6.
Characterization of recombinant GPC3 protein by ELISA. One microgram of purified recombinant proteins (GPC3 and rFc GPC3) were coated on 96-well plates and probed with the 1G12 mouse anti-GPC3 IgG.
-
20
Add 50 μl of 1 μg/ml of purified GPC3 protein (from SUPPORT PROTOCOL 2) to [*Author: each?] well of immuno plate.
-
21
Incubate the plate for 2 hr at room temperature, or overnight at 4°C.
-
22
Wash the plate with PBS three times.
-
23
Block the well with 2% w/v skim milk in PBS for 1 hr at room temperature. Meanwhile, prepare samples by mixing the samples with 4% w/v skim milk in PBS with 1:1 dilution and incubate 1 hr at room temperature.
-
24
Add 50 μL of pre-incubated phages to wells and incubate at room temperature for 2 hr.
-
25
Wash wells four times with PBS-0.1 % v/v Tween 20 using a squirt wash bottle or an automated 96-well plate washer.
-
26
Add 50 μL of diluted detection antibody to the wells and incubate at room temperature for 1 hr.
-
27
Wash wells 5 times with PBS-0.1 % v/v Tween 20.
-
28
Add 50 μL of diluted HRP conjugated goat anti-M13 IgG (1:5000 dilution) to each well and incubate at room temperature for 30 min.
-
29
Wash wells five times with PBS-0.1 % v/v Tween 20.
-
30
Add 50 μL of a chromogenic substrate (TMB) to each well.
-
31
Incubate the plate at room temperature for 10 to 30 min until the color changes to blue.
Option: add 50 μL of stop solution to each well. The stop solution is to quench the reaction produced by HRP and TMB and reaction will continue to proceed until all substrate is consumed. For an end-point measurement, the reaction can be stopped by 1N sulfuric acid equal to the volume of the substrate reaction and read OD at 450 mm. -
32
Read the absorbance of each well at 650 nm.
If the OD is three times that of the negative control, the sample is considered positive. The absorbance must be measured within 30 minutes after stopping the reaction.
Western blot
Figure 7 shows a western blot of purified recombinant GPC3 proteins. GPC3-hFc, GPC3-human Fc fusion; GPC3 (AA)-hFc, GPC3-human Fc fusion without the HS chains. We recommend reducing and denaturing the samples for western blot using the following method:
Figure 7.
Characterization of recombinant GPC3 protein by western blot analysis. GPC3 is a human GPC3 protein expressed in a mouse myeloma cell line (NS0 cell) having a 6-His tag at its C-terminus. GPC3-hFc was generated by expressing in HEK-293F cells with fused human Fc and GPC3 core protein. GPC3-hFc (AA) is expressed in HEK-293F cells with mutants without HS chain by replacing serine with alanine at positions 495 and 509, which are binding sites for HS on GPC3. Due to the heterogeneity of glycosylation, purified GPC3 and GPC-hFc appear as smears on SDS-PAGE.
-
33
Add an equal volume 2X Laemmli sample containing 10% v/v mercaptoethanol (or 100 mM DTT) buffer to 1 μg of protein.
Reducing reagents (mercaptoethanol and DTT) rapidly oxidize in air so must be prepared fresh. -
34
Boil each cell lysate in sample buffer at 100°C for 5 min.
Lysates can be stored at −20°C for future use. -
35
Load boiled protein into the wells of the SDS-PAGE gel, along with molecular weight marker.
-
36
Run the gel for 1–2 hr at 100 V using an electrophoresis power supply (Bio-Rad).
-
37
Transfer the protein from the gel to the membrane (Bio-Rad) for 1 hr at 100V at 4°C using an electrophoresis power supply (Bio-Rad).
The membrane can be either nitrocellulose or PVDF. For PVDF membrane, activate the PVDF membrane in methanol for 1 min and rinse with transfer buffer before preparing the stack. For nitrocellulose membrane, soak the membrane in transfer buffer for 10 min. Transfer of proteins to the membrane can be checked using Ponceau S staining before the blocking step. -
38
Block the membrane for 1 hr at room temperature or overnight at 4°C using 5% w/v skim milk in TBST.
-
39
Incubate the membrane with HS20 IgG (1 μg/ml) in TBST at room temperature for 1 hr or overnight at 4°C.
-
40
Wash the membrane three washes with TBST for 5 min each time.
-
41
Incubate the membrane with the HRP-conjugated secondary antibody (goat anti-human IgG-HRP) in TBST at room temperature for 1 hr.
-
42
Wash the membrane three washes with TBST for 5 min each time.
-
43
For signal development, follow the kit manufacturer’s recommendations. Remove excess reagent and cover the membrane with transparent plastic wrap.
-
44
Acquire image using with films in a darkroom for chemiluminescence, or use image scanning methods for colorimetric detection.
Production of recombinant human GPC3 proteins
The human GPC3 fused with rabbit Fc (named rFc-GPC3) and two human Fc fusion clones [GPC3-hFc and GPC3 (AA)-hFc] were constructed in pSecTag2 and pVRC vector containing the IL-2 signal sequence at the N-terminus. GPC3-hFc (AA) was a mutant without the HS chains created by replacement of Ser495 and Ser509 with Alanine. The plasmid for rFc-GPC3 was produced in CHO cells. GPC3-hFc and GPC3-hFc (AA) were made in HEK-293F cells. The protein was harvested from the cell culture supernatant and purified with a HisTrap Nickel Sepharose HP column. GPC3-hFc and GPC3 (AA)-hFc were purified with a Protein A Sepharose HP column. The purified recombinant GPC3 proteins were analyzed by western blot with 1G12 and by ELISA using 1G12 mouse anti-GPC3 antibodies.
SUPPORT PROTOCOL 3: Production of soluble antibody fragments
Individual colonies can be expressed in HB215 (non-amber suppressor strain) into soluble antibody fragments. The amber suppressor strain, TG-1, does not undergo transcription termination because it converts this codon into a glutamate residue in the TAG (amber) stop codon located between the scFv and gIII genes. Thus, the scFv-pIII fusion protein is made, in which scFv expression tends to be lowered because gIII is toxic. However, when the selected phage is made into soluble antibody fragments in HB2151 strain, transcription occurs at the amber codon between the scFv and gIII genes, producing a higher level of soluble scFv in TG-1 cells. The expressed scFvs can check binding by ELISA.
Materials
E.Coli. HB215 (see REAGENTS AND SOLUTIONS)
2YT (see REAGENTS AND SOLUTIONS)
Ampicillin-100 mg/ml (Teknova)50% w/v glucose solution (Teknova)
Isopropyl β-D-1-thiogalactopyranoside (IPTG)
50% v/v glycerol (50 ml): 25 mL of glycerol in a 500 mL distilled water Horse Radish Peroxidase conjugated Protein A (Thermo Fisher Scientific)
Horseradish Peroxidase-conjugated Protein L (Thermo Fisher Scientific)
-
1
Add 10 μl of eluted phage from each selection to 200 μl exponentially growing HB2151 bacteria (OD 600 of 0.4) and incubate for 30 min at 37°C. Plate 50 μl, 50 μl of a 1:102 dilution, 50 μl of a 1:104 dilution and 50 μl of a 1:106 dilution on 100 mm 2YT plates containing 100 μg/ml ampicillin and 1 % glucose and incubate at 37°C overnight.
-
2
Pick individual colonies to inoculate in 100 μl of 2YT medium containing 100 μg/ml of ampicillin and 1 % w/v glucose in 96 cell-wells plates and grow at 37 °C with shaking at 250 rpm overnight.
-
3
Transfer 2 μl of culture solution from this plate to a second 96 cell well plate containing 200 μl 2YT containing 100 μg/ml ampicillin per well. Grow with shaking (250 rpm) at 37°C until the OD 600 is approximately 0.8 (about 3 hr).
A master plate stock can be made with the first plate, by adding glycerol to a final concentration of 25 % v/v and storing the plate at −70°C. -
4
Add 25 μl 2YT containing 100 μg/ml ampicillin and IPTG (final concentration 1 mM IPTG). Continue shaking (250 rpm) at 30°C overnight.
-
5
Centrifuge at 3,000 g for 10 min and use 50 μl of the supernatant for ELISA.
ELISA
-
6
Coat a maxiSoap 96-well plate overnight with 50 μl per well of antigen (5–10 μg/ml) in PBS.
-
7
Wash the plate with PBS three times and incubate with 100 μl of blocking buffer for 1 hr at room temperature.
-
8
Mix 25 μl of samples and 25 μl of 6 % w/v BSA in PBS, and incubate at 37°C for 1 hr
This method can prevent nonspecific binding of soluble scFv to the plate. -
9
Discard the solution and wash the plate 5 times with PBS-0.1 % v/v Tween 20.
-
10
Add 50 μl of 1:5000 dilution of protein A or L-HRP 2nd antibody and incubate for 30 min at 37 °C.
-
11
Wash the plate five times with PBS-0.1 % Tween 20.
-
7
Add 50 μl of TMB substrate solution to each well and incubate for 2–15 min at room temperature. A blue color should develop and read the OD at 650 nm.
Alternatively, stop the reaction by adding 25 μl 1 M sulphuric acid. The blue color should turn yellow. Read the OD at 650 nm and at 450 nm. Subtract OD 650 from OD 450.
REAGENTS AND SOLUTIONS
-
E. Coli. TG1 genotype:
K12 Δ(lac-proAB) supE thi hsdD5/F′ traD36 proA+B lacIq lacZΔM15
-
E. Coli. HB2151 genotype:
K12 ara Δ(lac-proAB) thi/F′ proA+B lacIq lacZΔM15
2YT, 1 liter
-
16g Tryptone,
10g Yeast Extract
5g NaCl in 1 liter distilled water and autoclave
Phosphate-buffered saline (PBS) 10X, 1 liter
-
8 g NaCl
2.4 g Na2HPO4
14.4 g NaH2PO4.2H2O and
2 g KCl and in 800 ml of distilled water
Adjust the pH to 7.4 with HCl.
Add distilled water to a total volume of 1 liter.
For purification of recombinant GPC3 fused with histidine taq
Sodium phosphate stock solutions for His-conjugated protein purification
-
Stock Solution A (10X): 200 mM sodium phosphate, monobasic (NaH2PO4) and 5 M NaCl.
Dissolve 27.6 g sodium phosphate, monobasic (NaH2PO4) and 292.9 g NaCl in 900 mL of deionized water.
Mix well and adjust the volume to 1 L with deionized water.
-
Stock Solution B (10X): 200 mM sodium phosphate, dibasic (Na2HPO4) 5 M NaCl.
Dissolve 28.4 g sodium phosphate, dibasic (Na2HPO4) and 292.9 g of NaCl in 900 mL of deionized water.
Mix well and adjust the volume to 1 L with deionized water.
-
Purification Buffer (5X), pH 8.0 (250 mM NaH2PO4 and 2.5 M NaCl) −200 ml
Dissolve 7 g of Sodium phosphate, monobasic and 29.2 g of NaCl in 180 ml of distilled water
Mix well and adjust the pH with NaOH to pH 8.0.
Make the final volume to 200 mL with water.
-
3 M Imidazole pH 6.0 (3 M Imidazole 500 mM NaCl 20 mM Sodium Phosphate Buffer)-100 ml
Add 20.6 g of imidazole
8.77 ml of Stock Solution A (10X)
1.23 mL of Stock Solution B (10X)
Disolve well and adjust the pH to 6.0 with concentrated HCl or NaOH.
Make the final volume to 100 mL with water. If the precipitate is formed, dissolve the solution by heating.
-
Wash Buffer with 20 mM imidazole-50ml
50 mL of 1X Native Purification Buffer
335 μL of 3 M imidazole, pH 6.0
Mix well and adjust pH to 8.0 with NaOH or HCl.
-
Elution Buffer with 250 mM imidazole-15 ml
13.75 mL of 1X Native Purification Buffer
1.25 mL of 3 M imidazole, pH 6.0
Mix well and adjust pH to 8.0 with NaOH or HCl.
COMMENTARY
Background Information
M13 filamentous bacteriophages were developed by George P. Smith in the mid-1980s (Smith 1985). The M13 filamentous bacteriophages incorporate a virus that does not kill E. coli. With this technique it became possible to insert the gene encoding the foreign protein into the phage coat protein gene. Foreign peptides are expressed and displayed on phage, and screening is possible. In 1990, McCafferty et al. used this technique for the expression of antibody fragments on the surface of the phage (McCafferty et al., 1990). Hybridoma technology is a useful method that has been used to develop antibodies in a variety of applications (Milstein, 1999). However, hybridoma technology suffers from the limitation in that it is difficult to produce antibodies to poorly immunogenic antigens (such as HS described here) or highly conserved/self-antigens. Phage display is used to select targets in immune libraries, naive, and synthetic libraries with a large repertoire. Phage display can be used to isolate antibodies against strongly toxic antigens, poorly immunogenic antigens regardless of the immunogenicity of the antigen in all species.
Two Tomlinson libraries (I and J) were produced by the MRC Center for Protein Engineering (de Wildt et al. 2000). These are two semi-synthetic scFv libraries (the I library and the J library) that randomize complementarity-determining regions (CDRs) that are important for binding to antigen to diversify single VH and V kappa framework sequences. Structural studies have identified important positions for interaction with the antigen. H50, H52, H52a, H53, H55, H56, H58, H95, H96, H97 and H98 were diversified in the heavy chain. In the light chain, amino acids were randomized at positions L50, L53, L91, L92, L93, L94 and L96. Library I was changed to DVT (A, G or T; A, C or G; T) and library J was changed to NNK (any base; any base; G or T). Clone integrity and diversity can be analyzed by PCR or sequencing clone samples. The size of each library is approximately 1 × 108. All scFvs were cloned to the phagemid vector called pIT2 which contains the replication origin of M13 origin and sequences of ampicillin resistance gene, His-tag, c-myc and gIII. CDR 3 was determined using IMGT/V-QUEST at International Immunogenetics Database (IMGT, http://www.imgt.org/IMGT_vquest/vquest).
Critical Parameters
Phage can be sticky and bind to immunoplate wells non-specifically. Before panning, it is necessary to pre-block the plate wells.
The phage is expressed under the control of the lac promoter. Glucose suppresses lacZ promoter activity, and decreases gIII expression which is toxic to bacteria. Inhibition of gIII expression facilitates the expression of bacterial pilus, allowing infection of helper phage. Therefore it is important to add glucose before infection with helper phage. After infection with helper phage, glucose must be removed to increase the expression of gIII in order to display scFv in the phage.
Troubleshooting (Table 1)
Table 1.
Troubleshooting
| Problem | Possible cause | Solution |
|---|---|---|
| No phagemid after the growth of individual clones or library | Helper phage not functional | Try fresh helper phage |
| TG1 no longer has pilus | TG1 need to be grown on minimal plates to maintain the pilus and use fresh TG1 | |
| Use wrong antibiotic for selection | Check and use fresh antibiotics | |
|
| ||
| No ELISA signals | Problems with substrate or secondary antibodies | Use appropriate positive controls. Add 1 μl of HRP-conjugated secondary antibody in 100 μl of the substrate, if the color does not change use new substrates or antibodies. |
| Antigen did not coat well | If you have positive control, use these to test coating conditions | |
| Too much background | Phage bound on plate Blocking is not sufficient |
Increase pre-block time with skim-milk Increase blocking time with skim-milk; increase the concentration of skim-milk (up to 10%) |
| TG1 contamination | Check for contamination by culturing with tet/amp media. It should not grow on the culture media. | |
| Washing is not enough | Wash more | |
|
| ||
| Small number of output after selection | Wrong selection used | Check the antibiotic resistance of the vector used |
| The selected phage are weak binders or phages are not bound | Optimize coating and blocking condition for panning Use fresh cultured TG1 cells |
|
| Poor clone integrity and diversity | Analyze by sequencing. In late rounds (round 3 or later) of panning, enrichment of phage after elusion should be expected. |
|
|
| ||
| No selective binders | Low stringency | Refine stringency by using low antigen concentrations, short incubation time in the room temperate or cold room with phage, and/or more washes. We normally use 5–10 μg/ml of antigen for phage panning. The antigen concentrations are suitable for most high affinity phage binders. To increase the stringency, lower antigen concentrations such as 1 μg/ml or 0.1 μg/ml can be used. May also put the MaxiSoap 96-well plates on a shaker when incubating with phage to increase stringency |
| No selective binders in the library | Use another phage library to repeat phage panning. In this project, if no binders can be found in the I library, try the J library. | |
The first round of panning is the most important. At this point the errors will be amplified in the next round of selection. After each round of panning, you should get at least 1000 phages. If fewer than 1000 phages are obtained, check all reagents and repeat the infection with freshly grown TG1 at OD 600 of 0.4 and optimize coating and blocking conditions for panning. After each round of panning, if there are few or no colonies on the plate, it is possible that TG1 cells lost F pilus and were not infected with phage. If there are too many output phages (more than 107) after the first panning, optimize the blocking conditions and pre-block the phages by diluting the phages to blocking solution for longer time.
Anticipated Results (Table 2)
Table 2.
Time Considerations and Antipicated Results
| Days | Time | Procedures |
|---|---|---|
| 1 | 5 h | Grow libraries I and J and make phage |
| 2 | 6 h | Grow libraries I and J and make phage (cont.) |
| Coat immunoplate for 1st round of selection | ||
| 3 | 7 h | 1st round of selection |
| 4 | 3 h | Make phage from 1st round of selection |
| Coat immunoplate for 2nd round of selection | ||
| 5 | 3 h | Make phage from the 2nd round of selection (cont.) |
| 7 h | 2nd round of selection | |
| 6 | 3 h | Make phage from 2nd round of selection |
| Coat immunoplate for 3rd round of selection | ||
| 7 | Make phage from the 3rd round of selection (cont.) | |
| 7 h | 3rd round of selection | |
| 8 | 3 h | Make phage from 3rd round of selection |
| Coat 96 well plate for polyclonal phage ELISA | ||
| 9 | 3 h | Make phage from the 3rd round of selection (cont.) |
| 3 h | Polyclonal phage ELISA | |
| Coat 96 well plate for monoclonal phage ELISA | ||
| 2 h | Culture randomly selected individual clones for monoclonal ELISA | |
| 10 | 3 h | Monoclonal phage ELISA |
| 11 | Bacteria growth from positive clones | |
| 12 | 2 h | DNA prep. From positive clones for sequencing |
| 2 days | sequencing | |
| 14 | 1 day | IMGT analysis |
| 15–25 | 10 days | Antibody expression in mammalian cells |
| 26–33 | 7 days | Antibody characterization |
If panning is successful, you can find the enrichment of phage after each round of panning (in particular after the third round). Enrichment can be confirmed by an increase in the polyclonal phages ELISA signal or an increased outputs per each round of panning. After any round of panning, at least 1000 infectious phage can be obtained. If fewer are obtained, repeat the infection with freshly grown TG1 culture.
After the second panning, the number of phages may not increase as compared to the first panning, because it was subjected to more stringent washing than in the first panning.
Acknowledgments
This research was supported by the Intramural Research Program of NIH, NCI, Center for Cancer Research (Z01 BC 010891 and ZIA BC 010891 to MH). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. The authors disclose no conflicts of interest.
Footnotes
INTERNET RESOURCES
http://www.imgt.org/IMGT_vquest/vquest
Complementarity-determining region (CDR3) was determined using IMGT/V-QUEST in the International Immunogenetics Database (IMGT).
LITERATURE CITED
- de Wildt RM, Mundy CR, Gorick BD, Tomlinson IM. Antibody arrays for high-throughput screening of antibody-antigen interactions. Nat Biotechnol. 2000;18:989–994. doi: 10.1038/79494. [DOI] [PubMed] [Google Scholar]
- Hawlisch H, Muller M, Frank R, Bautsch W, Klos A, Kohl J. Site-specific anti-C3a receptor single-chain antibodies selected by differential panning on cellulose sheets. Anal Biochem. 2001;293:142–145. doi: 10.1006/abio.2001.5120. [DOI] [PubMed] [Google Scholar]
- Kala M, Bajaj K, Sinha S. Magnetic bead enzyme-linked immunosorbent assay (ELISA) detects antigen-specific binding by phage-displayed scFv antibodies that are not detected with conventional ELISA. Anal Biochem. 1997;254:263–266. doi: 10.1006/abio.1997.2378. [DOI] [PubMed] [Google Scholar]
- Lee CM, Iorno N, Sierro F, Christ D. Selection of human antibody fragments by phage display. Nat Protoc. 2007;2:3001–3008. doi: 10.1038/nprot.2007.448. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- Milstein C. The hybridoma revolution: an offshoot of basic research. Bioessays. 1999;21:966–973. doi: 10.1002/(SICI)1521-1878(199911)21:11<966::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Noppe W, Plieva F, Galaev IY, Pottel H, Deckmyn H, Mattiasson B. Chromato-panning: an efficient new mode of identifying suitable ligands from phage display libraries. BMC Biotechnol. 2009;9:21. doi: 10.1186/1472-6750-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- Walter G, Konthur Z, Lehrach H. High-throughput screening of surface displayed gene products. Comb Chem High Throughput Screen. 2001;4:193–205. doi: 10.2174/1386207013331228. [DOI] [PubMed] [Google Scholar]