Abstract
The receptor-like kinase FERONIA (FER) pleiotropically affects plant reproduction, development, and stress tolerance. We recently showed that the FER ligand RALF1 promotes stomatal closure and inhibits stomatal opening in a G-protein-dependent manner. FER responses have been designated as kinase-dependent or kinase-independent, based largely on fer complementation assays employing a kinase-dead FERK565R. Our quantification of FERK565R-GFP transcript and FERK565R-GFP protein in fer complementation lines reveal that, even within individual complementation lines, different levels of FERK565R expression prevail. FERK565R-GFP expression comparable to that of FER in Col-0 plants fail to elicit complementation of either fer rosette phenotypes or RALF1-elicited stomatal movements, while overexpression levels of FERK565R-GFP result in complementation. These results suggest possible alternative interpretations of previous conclusions regarding kinase-independent FER signaling.
Keywords: FERONIA (FER), guard cell, kinase-dead, RALF, Receptor-Like Kinase (RLK), stomata
One-sentence summary
Lines of a feronia (fer) null mutant transformed with a construct encoding kinase-dead FER show within-line and between-line dose-dependent complementation of rosette morphology and RALF1-mediated stomatal responses.
Introduction
The receptor-like kinase (RLK) superfamily is the largest kinase family in plants, comprising over 600 members in Arabidopsis [1, 2]. FERONIA (FER) is one of the 17 Catharantus roseus RLK1-like (CrRLK1L) RLKs in Arabidopsis [3]. FER was named after the Etruscan goddess of fertility based on its initially identified role in plant reproduction: fer female gametophytes fail to induce extracellular ROS production [4] and appropriate pollen tube rupture, resulting in defective ovule fertilization [4–7]. FER is also implicated in numerous aspects of vegetative plant development. fer mutants exhibit an overall reduction in vegetative growth, accompanied by abnormal root hairs, trichomes, and epidermal cell morphology [8]. FER, in association with other RLKs, also plays central roles in sensing of the biotic environment during immune response [9, 10] and in sensing of the abiotic environment during root mechanical [11] and salinity responses [12–14]. fer cell walls contain reduced cellulose content [15], and interaction of the extracellular domain of FER with pectin [12] has been proposed as a key sensor of cell wall integrity [12, 16].
RALF1 of the Rapid Alkalinization Factor (RALF) family of ~35 peptides is the most extensively studied FER ligand [9, 17]. Exogenous treatment with RALF1 stimulates alkalinization of the apoplast, inhibits cell expansion [9, 17, 18], and elicits changes to the phosphorylation status of FER and other proteins [14, 17].
While FER autophosphorylation [6] and FER transphosphorylation of both a heterologous substrate, myelin basic protein [19] and a receptor-like cytoplasmic kinase (RPM1-induced protein kinase; RIPK) [20] have been documented, one of the most intriguing aspects of FER signaling is the question of whether its kinase activity is required for signal transduction. For example, Kessler et al. [21] demonstrated that the founding fer phenotype, lack of ovule fertilization, was equally restored by complementation of fer null mutants with wild-type FER or with FER encoding a K565R mutation, which has been shown to abolish FER autophosphorylation in an in vitro kinase activity assay [6]. The effectiveness of FERK565R in restoring fertility was replicated in a recent publication from Haruta et al. [19], in which they also confirmed that FERK565R is kinase-dead with respect to transphosphorylation of myelin basic protein. Similarly, some FER-dependent aspects of root mechanosensing, including root agar penetration and skewing angle, and control of strain location, are equally restored by complementation with wild-type FER, with FERK565R, and with a truncated FER lacking the kinase domain [11]. By contrast, root tip angle upon encountering a physical barrier and the second, delayed phase of the biphasic apoplastic alkalinization that follows mechanical bending were partially restored by the kinase-deficient constructs [11]. Apoplastic alkalinization during mechanosensing is paralleled by and a presumed proxy for cytosolic Ca2+ increases [11]. Impairment in cytosolic Ca2+ signaling is consistent with a recent observation from the Sussman group [19] that FERK565R-complemented fer lines show only minor restoration of RALF1-induced cytosolic Ca2+ increase, as measured by aequorin luminescence. They also determined that RALF1 application inhibits primary root growth in wild-type plants, has no effect on the length of fer null mutant roots (as expected), and reduces root length by ~25% when applied to fer plants complemented with FERK565R, suggesting some RALF1-sensitivity of kinase-dead FER; this last effect, however, was not statistically significant due to large variance (see Discussion). Finally, the ability of co-expressed FER to suppress increased water permeability conferred upon oocytes by expression of the Arabidopsis aquaporin PIP2;1 appears to obligately require FER kinase activity: kinase inactive FER (K565R) or kinase domain-deleted FER did not alter the water permeability of PIP2;1-expressing oocytes [22].
We recently discovered a new role of FER in guard cell signaling [23]. We demonstrated that RALF1 inhibits stomatal opening and promotes stomatal closure in a FER-dependent manner; these responses were not seen with an inactive RALF1 analog [23], RALF1Δ (a deletion of amino acids 2–8 that does not activate FER [17]). In vivo immunoprecipitation, and BiFC analyses revealed that FER interacts with AGB1, the Gβ subunit of the Arabidopsis heterotrimeric G protein. Equivalent lack of RALF1 sensitivity in fer, agb1, and fer agb1 double mutants demonstrated that AGB1 functions downstream of FER in stomatal responses [23]. Our genetic analyses confirmed a loss of RALF1 sensitivity in agg triple Gγ subunit [24–26] and xlg triple Gα subunit mutant stomata [27–29]. Together, these data implicate FER as a plant heterotrimeric G protein-coupled receptor (GPCR) for RALF1 in XLG/AGB1/AGG–based guard cell responses [23].
G proteins have been increasingly linked to RLK signaling [30–34], and a logical hypothesis is that ligand activation of RLKs could be transduced via RLK-mediated phosphorylation of one or more of the downstream G protein subunits. It was therefore of interest to determine the essentiality of FER kinase activity in RALF1 regulation of stomatal apertures. In the process of evaluating this hypothesis, we discovered a dose-dependent effect of fer complementation with kinase-dead FER, a result that has broad implications for understanding the mechanistic underpinnings of the many FER-dependent phenotypes.
Materials and Methods
Plant material and growth
The following Arabidopsis mutants or transgenic lines used in this study are in the Col background: fer-4 [35], fer-4 pFER::FER-GFP and fer-4 pFER::FERK565R-GFP, which is a kinase inactive version of FER (FERK565R) in which Lys at position 565 is substituted for Arg [6]. Lines were generously provided by the Monshausen lab, generated as per Shih et al., in which the FER transgene was coupled to the native promoter [11]. Seeds were sterilized, spread on agar plates with 0.5 × Murashige and Skoog (MS) medium, 1% sucrose and 0.8% agar (Sigma), and stratified at 4 °C in the dark for 48 h. After 10–12 days, uniform seedlings were transferred from agar plates to soil (1:1 mix of Metro Mix 360: Sunshine Mix LC1 (Sun Gro Horticulture). Plants were grown in a growth chamber with an 8 h light/16 h dark cycle with light intensity of 125 μmol m−2 s−1 and day/night temperatures of 21 and 19 °C respectively for four to six weeks before use in whole-plant phenotyping and stomatal aperture assays.
Stomatal aperture experiments
Experiments assaying stomatal movements utilized the protocols described in Yu et al. [23]. In stomatal opening assays, leaf pieces were incubated in 500 μL of 10 mM KCl, 7.5 mM iminodiacetic acid, 10 mM MES-KOH, pH 6.15 in the dark for 2 h to initially close stomata, then treated for 3 h with or without 10 μM synthesized RALF1 (amino acids 72–120 of the precursor) (Biomatik) in the presence of ~150 μmol m−2 s−1 white light, which stimulates stomatal opening. For experiments assaying stomatal closure, excised leaves were incubated in 500 μL of 20 mM KCl, 0.5 mM CaCl2, 5 mM MES-KOH, pH 6.15 for 3 h in the light to initially open stomata, then treated for 2 h with or without addition of 10 μM RALF1 (final concentration). Images of stomatal apertures were taken and measured as previously described [23]. Three independent blinded experiments were performed for each assay.
Real-time qRT-PCR
Total RNA was isolated from rosette leaves using the NucleoSpin RNA Plant kit (Macherey-Nagel) and treated with RQ1 RNase-Free DNase (Promega) to remove DNA contamination following the manufacturers’ instructions. RNA (2 μg) was reverse-transcribed into cDNA using the SuperScript® III Reverse Transcriptase kit (Invitrogen), and the cDNA was diluted 3-fold for use as a template in qRT-PCR. qRT-PCR was performed using SYBR Green (Bio-Rad) to detect synthesized double-stranded DNA in a IQ5 Multicolor Real-Time PCR Detection System (Bio-Rad). The cycling conditions comprised a 5 min denaturation at 95 °C, 40 cycles at 95 °C for 30 s, 62 °C for 30 s and 72 °C for 40 s, and a final extension cycle at 72 °C for 8 min. Actin2/8 was used as the reference to normalize gene expression. Three independent biological experiments with 3 technical replicates each were performed. The gene specific primers used were: Actin2/8 FP: 5’-GGTAACATTGTGCTCAGTGGTGG-3’; Actin2/8 RP: 5’-AACGACCTTAATCTTCATGCTGC-3’; FER/FERK565R FP: 5’-CAAACGGGGTTCCGTTTCAC-3’; FER/FERK565R RP: 5’-AGTAGTCGGGCGTAGGACTT-3’.
Protein extractions and immunoblotting
Total protein was extracted from the aerial tissues of the mature plants shown Figure 1A. Ground tissue was homogenized in protein extraction buffer (50 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM DTT, 1 % SDS (w/v)) and debris were cleared by centrifugation (16000 g for 7 minutes). For microsomal protein extraction, ground tissue was homogenized in microsomal buffer (250 mM sucrose, 70 mM Tris pH 8.0, 3 mM EDTA, 15 mM β-mercaptoethanol, 5 mM DTT, 0.5 % PVP 40000 (w/v)) and debris were cleared as above, followed by microsomal pelleting by centrifugation at 100000 g for 30 minutes. Microsomal pellets were solubilized in protein extraction buffer (above). After quantification by BCA assay (Thermo), 30 μg of total protein or 20 μg of microsomal protein for each sample was separated by SDS-PAGE and transferred to a nitrocellulose membrane, which was probed with the JL-8 anti-GFP antibody (Takara) followed by an anti-mouse IgG HRP conjugate (Promega). Bands were detected using SuperSignal West Dura Extended Duration Substrate (Thermo) in a ChemiDoc Imaging System (Bio-Rad). Band intensity was calculated and normalized with identically run total protein gels stained with Gel-Code Blue (Thermo), using Image Lab software (Bio-Rad).
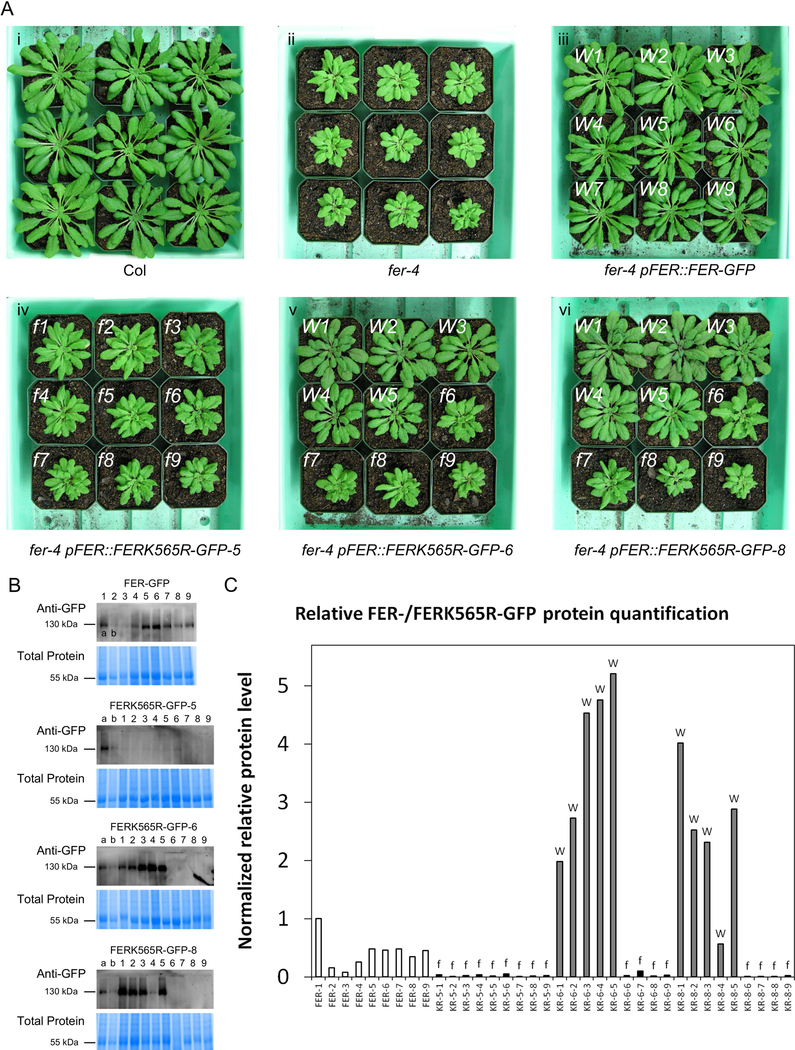
Figure 1. fer-4 lines expressing the kinase-dead FERK565R-GFP protein show different expression levels that correlate with their morphological phenotypes.
(A) Representative plant images of Col (i), fer-4 mutants (ii), fer-4 pFER::FER-GFP plants (iii), and three fer-4 pFER::FERK565R-GFP lines (iv, v and vi) with two types of morphologies. Plants in the fer-4 pFER::FER-GFP line (iii) display wild-type-like morphology (indicated with a white ‘W’ at the top left of each pot) as expected, while plants in the fer-4 pFER::FERK565R-GFP-5 line (iv) display fer morphology (indicated with a white ‘f” at the top left of each pot). Individual plants in the fer-4 pFER::FERK565R-GFP-6 (v) and fer-4 pFER::FERK565R-GFP-8 (vi) lines display WT-like (W) or fer-like (‘f’) morphologies. (B) Anti-GFP immunoblots of total protein samples extracted from plants shown in panel A. Gel Code Blue stained total protein is shown below each blot, and samples labeled ‘a’ and ‘b’ (FER-GFP plants #1 and #2) are included in all blots for comparison. Samples are numbered in accordance with the plant numbering in panel A. (C) Relative quantification of FER-/FERK565R-GFP protein levels from the immunoblots in panel B. Band intensity was normalized to total protein levels from the Gel Code Blue stained total protein in panel B and quantified relative to the FER-GFP plant #1 sample (labeled ‘a’) on each blot. The phenotype of each FERK565R-GFP plant (abbreviated as ‘KR’ on the X-axis) is indicated with an ‘f’ or ‘W’ (based on and corresponding to panel A) above each bar.
Results
Restoration of fer rosette phenotypes by kinase-dead FER is dose-dependent
fer mutants display a characteristic and visually striking aerial morphology, consisting of a compact rosette with smaller leaves and shorter petioles when compared to wild-type plants (Fig. 1Ai vs. Fig. 1Aii) [36]. fer-4 pFER::FER-GFP complemented the morphological phenotypes of fer-4 as expected (Fig. 1Aiii). However, in the fer-4 pFER::FERK565R-GFP lines, we observed morphologically distinct effects of fer-4 complementation with pFER::FERK565R-GFP. Sub-populations were observed within lines, with FERK565R(WT) individuals displaying a restoration of the wild-type aerial morphology (indicated with a white ‘W’), and FERK565R(fer) individuals resembling the fer null mutant (indicated with a white ‘f”) (Fig. 1Aiv, v and vi). Of the lines we examined, the majority displayed these differential phenotypes within each line, while only one line (line #5; Fig. 1Aiv) displayed a mutant-like phenotype in all plants assayed. Because the FER-GFP signal was undetectable under confocal microscopy in mature leaves or guard cells of fer-4 complemented with pFER::FER-GFP, we performed real-time RT-PCR (qRT-PCR) on additional sets of plants that had been categorized as fer-like or WT-like, as in Fig. 1A, to determine the expression levels of FER/FERK565R. We found that in the FERK565R line in which the morphological deficiency of fer-4 was never complemented (fer-4 pFER::FERK565R-GFP-5(fer)) (Fig. 1Aiv), the kinase-dead version of the transgene was expressed at the same level as native FER in Col-0 (Fig. S1A). We found that in the two other fer-4 pFER::FERK565R-GFP lines of Figure 1A (lines 6 and 8, shown in Fig. 1 sub-panels 1Av and 1Avi), plants in the subpopulations in which the wild-type morphology was not restored (fer-4 pFER::FERK565R-GFP-6(fer) and fer-4 pFER::FERK565R-GFP-8(fer)) also expressed the kinase-dead version of the transgene at the same level as native FER in Col-0 (Fig. S1A). By contrast, we found that in the subpopulations of FERK565R lines where the morphological deficiency of fer-4 was complemented (pFER::FERK565R-GFP-6(WT) and pFER::FERK565R-GFP-8(WT)), the transgene was overexpressed (Fig. S1A).
To confirm that transcriptional overexpression resulted in higher levels of protein expression, we conducted immunoblot analyses on total protein extracted from the exact plants shown in Figure 1A. Indeed fer-4 pFER::FERK565R-GFP plants that displayed a wild-type morphology exhibited dramatically higher levels of FERK565R-GFP protein than those that displayed a fer-like morphology (Fig. 1B), which was confirmed by relative quantification (Fig. 1C). Though the Image Lab software used for quantification could detect minor bands in several of the samples from fer-like fer-4 pFER::FERK565R-GFP plants (Fig. 1C), these bands are difficult to visualize (Fig. 1B). Therefore we conducted microsomal enrichment of proteins from the fer-like fer-4 pFER::FERK565R-GFP-6 plants 6, 7, 8 and 9 of Fig. 1Av, and repeated the detection by immunoblot. Upon enrichment, the FERK565R-GFP protein could be detected in these samples (Fig. S1B), confirming that lack of complementation was not due to silencing of the transgene. To further validate the correlation between FERK565R overexpression and morphological complementation, all fer-4 pFER::FERK565R-GFP and fer-4 pFER::FERK565R-GFP plants shown in Figure 1A were genotyped to confirm disruption of the native FER gene, and the FER-/FERK565R-GFP transgenes were sequenced from nucleotides 1427 to 2577 (corresponding to residues 477 to 790) after PCR amplification. All fer-4 pFER::FER-GFP plants contained the wild-type FER sequence, while all fer-4 pFER::FERK565R-GFP plants contained the K565R mutation (examples shown in Fig. S1C). While our data clearly illustrate dose-dependency between FERK565R protein levels and whole plant phenotypes, the exact mathematical and mechanistic relationships between these two parameters will require further investigation.
Restoration of guard cell RALF1 responses by kinase-dead FER is dose-dependent
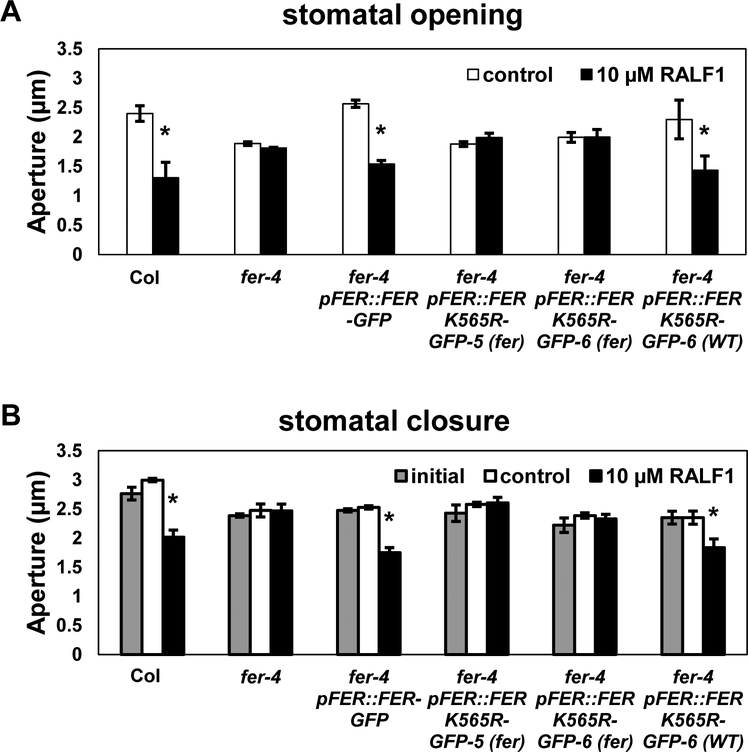
Prior to quantification of transgene expression levels, we performed assays of RALF1 inhibition of stomatal opening on the sets of plants that subsequently yielded the qRT-PCR data of Fig. S1A (the pFER::FERK565R-GFP-8 line was excluded due to constraints on the number of simultaneous assays feasible in stomatal bioassays). Consistent with their FERK565R expression levels (Fig. S1A) and their morphological appearances, the FERK565R-GFP-5(fer) line and the FERK565R-GFP-6(fer) subpopulation were insensitive to RALF1 inhibition of stomatal opening, i.e. these plants showed phenotypes indistinguishable from the fer-4 null mutant (Fig. 2A). By contrast, in the fer-4 pFER::FER-GFP line and in the FERK565R-overexpressing subpopulation (FERK565R-GFP-6(WT)), sensitivity to RALF1 inhibition of stomatal opening was fully restored (Fig. 2A). Similarly, RALF1 promotion of stomatal closure was not observed in either the FERK565R-GFP-5(fer) line or in the FERK565R-GFP-6(fer) subpopulation, but was fully restored in the fer-4 pFER::FER-GFP line and in the FERK565R-overexpressing subpopulation (FERK565R-GFP-6(WT)) (Fig. 2B). We then performed these same assays on the specific pFER::FERK565R-GFP line originally used by Shih et al. [11] in analyses of root mechanosensing, and fully replicated (Fig. S2) the phenotypic and qRT-PCR results of Figures 1, 2 and S1A. Therefore three of the four pFER::FERK565R-GFP lines we analyzed showed divergent populations in morphological and RALF1 responses, while one line (pFER::FERK565R-GFP-5) showed no complementation. In summary, fer-4 plants that express the kinase-dead transgene at wild-type FER levels fail to show complementation of rosette or guard cell phenotypes. However, overexpression of FERK565R can overcome the deficiency of the kinase-inactive FER, at least for the phenotypes assayed here.
Figure 2. RALF1-mediated regulation of stomatal movement is not restored in fer-4 pFER::FERK565R-GFP plants with fer-like morphology, but is restored in pFER::FERK565R-GFP plants with wild-type morphology, corresponding to FERK565R-GFP transgene overexpression.
(A) RALF1 inhibition of stomatal opening. Leaf pieces were incubated in the dark to close the stomata for 2 h and then treated with or without 10 μM RALF1 in the light for another 3 h. Plants used in this assay are the same plants as were used for transcript quantification in Fig. S1A. (B) RALF1 promotion of stomatal closure. Leaf pieces were incubated in the light for 3 h to open the stomata and then treated with or without 10 μM RALF1 in the light for another 2 h. The results are mean ± S.E. for 3 independent experiments with over 80 apertures measured per genotype per treatment. Significant differences between control and treatment (Student’s t-test) are indicated (*P < 0.05).
Discussion
Our results indicate that in the native condition, i.e. in wild-type plants, the kinase activity of FER is required for FER control of aerial development and for RALF1-mediated effects on stomatal movement. RALF1 is known to promote phosphorylation of FER and other downstream proteins [17]. Evidence is accumulating that RLK-based phosphorylation of G proteins is a new paradigm in G protein signaling regulation [34, 37, 38].
There are several possible explanations for why overexpression of the kinase-dead transgene sufficed to restore wild-type phenotypes. Formally, it is possible that a low level of kinase activity is retained by the FERK565R protein, and that overexpression provides sufficient kinase activity and thus substrate phosphorylation to elicit wild-type phenotypes. While recombinant FERK565R proteins are documented to lack auto- [6] and transphosphorylation activity [19], those assays were conducted for relatively short periods of time, which may not have allowed detection of weak residual activity. However, this explanation seems less likely for two reasons; first, because the lysine (K) at position 565 has been shown to be involved in ATP co-ordination [39] and second, because the aspects of root mechanosensing that were restored upon complementation with the FERK565R protein were also equally restored by a truncated version of FER lacking the kinase domain [11].
Alternatively, over-expressed FERK565R might genuinely lack kinase activity but still complement a kinase-dependent phenotype. One hypothesis is that FER-based phosphorylation of downstream targets (e.g. G protein subunits) normally promotes their association with additional proteins, but that assembly of this multi-protein complex can alternatively be mediated, albeit more weakly, by FERK565R in place of target phosphorylation. A second hypothesis is that phosphorylation of downstream target proteins is mediated by FER in the wild-type situation, but that overexpression of FERK565R in the fer background suffices to bring other kinases into the complex, and these other kinases then perform the requisite role in target phosphorylation. This hypothesis is consistent with observations that FER has been found in association with several other RLKs [9], as well as with the cytoplasmic receptor-like kinase, RIPK [20]. In our own studies, we identified four other RLKs in AGB1 immunoprecipitates: MIK2, AT5G39000, AT1G48480 and AT5G63410 [23], although such identification does not suffice to conclude direct physical interaction with FER. A third hypothesis is that the kinase activity of FER is largely superfluous, i.e. that FER does not phosphorylate downstream targets but plays a scaffolding role. According to this hypothesis, FERK565R simply scaffolds more weakly than native FER, thus requiring FERK565R overexpression for restoration of the wild-type phenotype. With regard to this hypothesis, it is interesting to note that a significant number of other RLK family members have amino acid substitutions in their active sites, suggesting the possibility of kinase-independent functions [40].
Regardless which of these, or alternative, hypotheses, proves to be correct, the dose-dependency that we observe suggests that appropriate interpretation of phenotypes arising from fer complementation with FERK565R ideally requires assessment of FERK565R mRNA and protein levels both across and within lines. While it is customary, and usually quite appropriate, to average results from all plants within a single transgenic line, our observation of different phenotypic subpopulations within single FERK565R-GFP lines suggests the possibility that results showing “partial” complementation with FER565R, e.g. as observed by Haruta et al. [19] in root growth and Ca2+ signaling, as well as by Shih et al. [11] in aspects of root mechanosensing, may actually arise from averaging “true” lack of complementation in a subpopulation of plants expressing wild-type levels of the FERK565R transgene and “false” complementation in another subpopulation of plants with FERK565R transgene overexpression. In such cases, larger variation in the results (larger error bars) are expected for the FERK565R complemented lines than for, e.g. FER complemented lines. As noted above, greater variance in RALF1-inhibition of root growth was indeed reported by Haruta et al. [19] in their fer lines complemented with FERK565R. It would therefore be of interest to determine whether subpopulations of plants, of the type reported here, were also present in their lines. In summary, our analyses suggest that rosette development and RALF1-mediated stomatal responses require FER kinase activity. In the future, it will be of interest to determine whether FER kinase-in/dependence of the many FER phenotypes correlates with FER association with specific partner RLKs, or with FER association with particular classes of downstream effectors.
Supplementary Material
(A) Transcript levels of FER/FERK565R in Col, fer-4, fer-4 pFER::FER-GFP and three independent fer-4 pFER::FERK565R-GFP lines. qRT-PCR of FER/FERK565R was performed in Col, fer-4, fer-4 pFER::FER-GFP and three independent lines of fer-4 pFER::FERK565R-GFP. Both fer-4 pFER::FERK565R-GFP-6 and pFER::FERK565R-GFP-8 lines display fer or WT morphology in different plants (as designated in parentheses in the x-axis labels, and exemplified in Fig. 1A). The expression level of FERK565R correlates with the whole plant morphologies seen in Fig. 1A: plants that express the kinase-dead construct at wild-type levels fail to restore the wild-type rosette phenotypes, while plants that over-express the kinase-dead construct restore the wild-type phenotypes. qRT-PCR measurements were repeated on three biological replicates with three technical replicates each, and statistical groupings were determined by one way ANOVA with post-hoc Tukey’s HSD test (P < 0.05). (B) Immunoblotting of proteins from enriched microsomal fractions shows detection of FERK565R-GFP in fer-4 pFER::FERK565R-GFP-6 plants #6–9 (Fig. 1Av). Presumably due to low abundance of FERK565R-GFP, these bands were not clear when aliquots of total protein were probed (Fig. 1B). Gel Code Blue stained microsomal protein is shown below the blot. (C) Examples of sequencing chromatographs of FER PCR products from fer-4 pFER::FER-GFP and fer-4 pFER::FERK565R-GFP plants, demonstrating that the intended K565R mutation (arrows indicate position) was incorporated in the fer-4 pFER::FERK565R-GFP plants.
(A) Representative plant images of Col, fer-4, fer-4 pFER::FER-GFP, and a fer-4 pFER::FERK565R-GFP line with subpopulations of plants showing either fer-like or wild-type-like morphologies, as indicated in parentheses. (B) Transcript levels of FER/FERK565R of the genotypes shown in (A). Transcript levels correlate with morphologies, with wild-type morphologies conferred by FERK565R-GFP overexpression. (C) RALF1 inhibition of stomatal opening. Leaf pieces were incubated in the dark to close the stomata for 2 h and treated with or without 10 μM RALF1 in the light for another 3 h. The results are mean ± S.E. for 3 independent experiments with over 80 apertures measured per genotype per treatment. Significant differences between control and treatment (Student’s t-test) are indicated (*P < 0.05). (D) RALF1 promotion of stomatal closure. Leaf pieces were incubated in the light for 3 h to open the stomata and then treated with or without 10 μM RALF1 in the light for another 2 h. The results are mean ± S.E. for 3 independent experiments with over 80 apertures measured per genotype per treatment. Significant differences between control and treatment (Student’s t-test) are indicated (*P < 0.05).
Acknowledgements
This research was supported by NSF-MCB grant 1715826 to S.M.A. and by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM126079. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Prof. Gabriele Monshausen (Pennsylvania State University) for providing seeds of the fer mutants and the FER and FERK565R transgenic lines. We thank Prof. Alice Cheung for fertile discussions on FER signaling mechanisms.
References
- 1.Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran L-SP: Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. Journal of Experimental Botany 2013, 64(2):445–458. [DOI] [PubMed] [Google Scholar]
- 2.Zulawski M, Schulze G, Braginets R, Hartmann S, Schulze WX: The Arabidopsis Kinome: phylogeny and evolutionary insights into functional diversification. BMC Genomics 2014, 15(1):548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindner H, Müller LM, Boisson-Dernier A, Grossniklaus U: CrRLK1L receptor-like kinases: not just another brick in the wall. Current Opinion in Plant Biology 2012, 15(6):659–669. [DOI] [PubMed] [Google Scholar]
- 4.Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu H-M, Cheung AY: Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nature Communications 2014, 5:3129. [DOI] [PubMed] [Google Scholar]
- 5.Huck N, Moore JM, Federer M, Grossniklaus U: The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 2003, 130(10):2149–2159. [DOI] [PubMed] [Google Scholar]
- 6.Escobar-Restrepo J-M, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang W-C, Grossniklaus U: The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 2007, 317(5838):656–660. [DOI] [PubMed] [Google Scholar]
- 7.Rotman N, Rozier F, Boavida L, Dumas C, Berger F, Faure J-E: Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Current Biology 2003, 13(5):432–436. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Wu H-M, Cheung AY: FERONIA and her pals: functions and mechanisms. Plant Physiology 2016, 171(4):2379–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C: The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355(6322):287–289. [DOI] [PubMed] [Google Scholar]
- 10.Masachis S, Segorbe D, Turrà D, Leon-Ruiz M, Fürst U, El Ghalid M, Leonard G, López-Berges MS, Richards TA, Felix G: A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nature Microbiology 2016, 1(6):16043. [DOI] [PubMed] [Google Scholar]
- 11.Shih H-W, Miller ND, Dai C, Spalding EP, Monshausen GB: The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Current Biology 2014, 24(16):1887–1892. [DOI] [PubMed] [Google Scholar]
- 12.Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu M-C, Maman J, Steinhorst L, Schmitz-Thom I: The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Current Biology 2018, 28(5):666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y, Assmann SM: Inter‐relationships between the heterotrimeric Gβ subunit AGB1, the RLK FERONIA and RALF1 in salinity response. Plant, Cell & Environment 2018: doi: 10.1111/pce.13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Yu F, Liu Y, Du C, Li X, Zhu S, Wang X, Lan W, Rodriguez PL, Liu X: FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proceedings of the National Academy of Sciences 2016, 113(37):E5519–E5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeats TH, Sorek H, Wemmer DE, Somerville CR: Cellulose Deficiency Is Enhanced on Hyper Accumulation of Sucrose by a H+-Coupled Sucrose Symporter. Plant Physiology 2016, 171(1):110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf S: Plant cell wall signalling and receptor-like kinases. Biochemical Journal 2017, 474(4):471–492. [DOI] [PubMed] [Google Scholar]
- 17.Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR: A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343(6169):408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy E, De Smet I: Understanding the RALF family: a tale of many species. Trends in Plant Science 2014, 19(10):664–671. [DOI] [PubMed] [Google Scholar]
- 19.Haruta M, Gaddameedi V, Burch H, Fernandez D, Sussman MR: Comparison of the effects of a kinase dead mutation of FERONIA on ovule fertilization and root growth of Arabidopsis. FEBS Letters 2018, 592(14):2395–2402. [DOI] [PubMed] [Google Scholar]
- 20.Du C, Li X, Chen J, Chen W, Li B, Li C, Wang L, Li J, Zhao X, Lin J: Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proceedings of the National Academy of Sciences 2016, 113(51):E8326–E8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler SA, Lindner H, Jones DS, Grossniklaus U: Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Reports 2015, 16(1):107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellati J, Champeyroux C, Hem S, Rofidal V, Krouk G, Maurel C, Santoni V: Novel aquaporin regulatory mechanisms revealed by interactomics. Molecular & Cellular Proteomics 2016, 15(11):3473–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y, Chakravorty D, Assmann SM: The G protein β subunit, AGB1, interacts with FERONIA in RALF1-regulated stomatal movement. Plant Physiology 2018, 176(3):2426–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mason MG, Botella JR: Completing the heterotrimer: isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proceedings of the National Academy of Sciences 2000, 97(26):14784–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason MG, Botella JR: Isolation of a novel G-protein γ-subunit from Arabidopsis thaliana and its interaction with Gβ. Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression 2001, 1520(2):147–153. [DOI] [PubMed] [Google Scholar]
- 26.Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR: An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. The Plant Journal 2011, 67(5):840–851. [DOI] [PubMed] [Google Scholar]
- 27.Lee Y-RJ, Assmann SM: Arabidopsis thaliana “extra-large GTP-binding protein” (AtXLG1): a new class of G-protein. Plant Molecular Biology 1999, 40(1):55–64. [DOI] [PubMed] [Google Scholar]
- 28.Ding L, Pandey S, Assmann SM: Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. The Plant Journal 2008, 53(2):248–263. [DOI] [PubMed] [Google Scholar]
- 29.Chakravorty D, Gookin TE, Milner M, Yu Y, Assmann SM: Extra-large G proteins (XLGs) expand the repertoire of subunits in Arabidopsis heterotrimeric G protein signaling. Plant Physiology 2015, 169(1):512–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aranda-Sicilia MN, Trusov Y, Maruta N, Chakravorty D, Zhang Y, Botella JR: Heterotrimeric G proteins interact with defense-related receptor-like kinases in Arabidopsis. Journal of Plant Physiology 2016, 188:44–48. [DOI] [PubMed] [Google Scholar]
- 31.Je BI, Xu F, Wu Q, Liu L, Meeley R, Gallagher JP, Corcilius L, Payne RJ, Bartlett ME, Jackson D: The CLAVATA receptor FASCIATED EAR2 responds to distinct CLE peptides by signaling through two downstream effectors. eLife 2018, 7:e35673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR, Zhang Y: Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiology 2013, 161(4):2146–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tunc-Ozdemir M, Urano D, Jaiswal DK, Clouse SD, Jones AM: Direct modulation of heterotrimeric G protein-coupled signaling by a receptor kinase complex. Journal of Biological Chemistry 2016, 291(27):13918–13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakravorty D, Assmann SM: Phosphorylation as a regulatory mechanism in heterotrimeric G protein signaling in mammals, yeast, and plants. Biochemical Journal submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan Q, Kita D, Li C, Cheung AY, Wu H-M: FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proceedings of the National Academy of Sciences 2010, 107(41):17821–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y: Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences 2009, 106(18):7648–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trusov Y, Botella JR: Plant G-proteins come of age: breaking the bond with animal models. Frontiers in Chemistry 2016, 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S: Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 2016, 5:e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minkoff BB, Makino S-i, Haruta M, Beebe ET, Wrobel RL, Fox BG, Sussman MR: A cell-free method for expressing and reconstituting membrane proteins enables functional characterization of the plant receptor-like protein kinase FERONIA. Journal of Biological Chemistry 2017, 292(14):5932–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castells E, Casacuberta JM: Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. Journal of Experimental Botany 2007, 58(13):3503–3511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Transcript levels of FER/FERK565R in Col, fer-4, fer-4 pFER::FER-GFP and three independent fer-4 pFER::FERK565R-GFP lines. qRT-PCR of FER/FERK565R was performed in Col, fer-4, fer-4 pFER::FER-GFP and three independent lines of fer-4 pFER::FERK565R-GFP. Both fer-4 pFER::FERK565R-GFP-6 and pFER::FERK565R-GFP-8 lines display fer or WT morphology in different plants (as designated in parentheses in the x-axis labels, and exemplified in Fig. 1A). The expression level of FERK565R correlates with the whole plant morphologies seen in Fig. 1A: plants that express the kinase-dead construct at wild-type levels fail to restore the wild-type rosette phenotypes, while plants that over-express the kinase-dead construct restore the wild-type phenotypes. qRT-PCR measurements were repeated on three biological replicates with three technical replicates each, and statistical groupings were determined by one way ANOVA with post-hoc Tukey’s HSD test (P < 0.05). (B) Immunoblotting of proteins from enriched microsomal fractions shows detection of FERK565R-GFP in fer-4 pFER::FERK565R-GFP-6 plants #6–9 (Fig. 1Av). Presumably due to low abundance of FERK565R-GFP, these bands were not clear when aliquots of total protein were probed (Fig. 1B). Gel Code Blue stained microsomal protein is shown below the blot. (C) Examples of sequencing chromatographs of FER PCR products from fer-4 pFER::FER-GFP and fer-4 pFER::FERK565R-GFP plants, demonstrating that the intended K565R mutation (arrows indicate position) was incorporated in the fer-4 pFER::FERK565R-GFP plants.
(A) Representative plant images of Col, fer-4, fer-4 pFER::FER-GFP, and a fer-4 pFER::FERK565R-GFP line with subpopulations of plants showing either fer-like or wild-type-like morphologies, as indicated in parentheses. (B) Transcript levels of FER/FERK565R of the genotypes shown in (A). Transcript levels correlate with morphologies, with wild-type morphologies conferred by FERK565R-GFP overexpression. (C) RALF1 inhibition of stomatal opening. Leaf pieces were incubated in the dark to close the stomata for 2 h and treated with or without 10 μM RALF1 in the light for another 3 h. The results are mean ± S.E. for 3 independent experiments with over 80 apertures measured per genotype per treatment. Significant differences between control and treatment (Student’s t-test) are indicated (*P < 0.05). (D) RALF1 promotion of stomatal closure. Leaf pieces were incubated in the light for 3 h to open the stomata and then treated with or without 10 μM RALF1 in the light for another 2 h. The results are mean ± S.E. for 3 independent experiments with over 80 apertures measured per genotype per treatment. Significant differences between control and treatment (Student’s t-test) are indicated (*P < 0.05).