Abstract
Background
Improving signal-to-noise ratio of chemical-shift-encoded MRI acquisition with complex reconstruction (MRI-C) may improve the accuracy and precision of non-invasive proton density fat fraction (PDFF) quantification in patients with hepatic steatosis.
Purpose
To assess the accuracy of high SNR (Hi-SNR) MRI-C versus standard MRI-C acquisition to estimate hepatic PDFF in adult and pediatric non-alcoholic fatty liver disease (NAFLD) using MR Spectroscopy (MRS) sequence as the reference standard.
Study Type
Prospective
Population/Subjects
231 adult and pediatric patients with known or suspected NAFLD
Field Strength/Sequence
PDFF estimated at 3T by three MR techniques: standard MRI-C; a Hi-SNR MRI-C variant with increased slice thickness, decreased matrix size, and no parallel imaging; and MRS (reference standard).
Assessment
MRI-PDFF was measured by image analysts using a region of interest co-registered with the MRS-PDFF voxel.
Statistical Tests
Linear regression analyses were used to assess accuracy and precision of MRI-estimated PDFF for MRS-PDFF as a function of MRI-PDFF using the standard and Hi-SNR MRI-C for all patients and for patients with MRS-PDFF<10%.
Results
Two hundred seventy-one exams from 231 patients were included (mean MRS-PDFF: 12.6% [SD:10.4]; range: 0.9–41.9). High agreement between MRI-PDFF and MRS-PDFF was demonstrated across the overall range of PDFF with regression slope of 1.035 for the standard MRI-C and 1.008 for Hi-SNR MRI-C. Hi-SNR MRI-C, compared to standard MRI-C, provided small but statistically significant improvements in the slope (respectively, 1.008 vs 1.035, P=0.004) and mean bias (0.412 vs 0.673, P<0.0001) overall. In the low-fat patients only, Hi-SNR MRI-C provided improvements in the slope (1.058 vs 1.190, P=0.002), mean bias (0.168 vs 0.368, P = 0.007), intercept (-0.153 vs -0.796, P<0.0001), and borderline improvement in the R2 (0.888 vs 0.813, P=0.01).
Data Conclusion
Compared to standard MRI-C, Hi-SNR MRI-C provides slightly higher MRI-PDFF estimation accuracy across the overall range of PDFF and improves both accuracy and precision in the low PDFF range.
Keywords: hepatic steatosis, MRI-C, Hi-SNR, MRS, proton density fat fraction, PDFF, optimization, accuracy, quantitative imaging biomarker
Introduction
Proton density fat fraction (PDFF) is a quantitative, confounder-corrected magnetic resonance (MR)-based biomarker for the non-invasive assessment of fat in the liver (1-3). It can be acquired by magnetic resonance imaging (MRI) and magnetic resonance spectroscopy (MRS) techniques, and, although MRS is the current gold standard for PDFF measurement, its use is impractical in clinical settings due to technical requirements and spatial coverage limitation (4-7). Meanwhile, chemical-shift-encoded MRI with complex reconstruction (MRI-C) permits accurate PDFF estimation across the entire liver over a dynamic range of 0 to 100% using a spectrally corrected reconstruction; furthermore, this imaging modality has been demonstrated to be highly accurate with respect to MRS (8-11).
To obtain accurate PDFF estimation, MRI-C corrects or minimizes multiple confounders, including T1 and noise bias (12), spectral complexity (13,14), B0 field inhomogeneity (15,16), phase errors from eddy currents (9), and T2* decay (14,17-19). Nevertheless, MRI-C sequences estimate PDFF with a small bias of about 1% with respect to MRS. This may have clinical implications for the early detection of nonalcoholic fatty liver disease (NAFLD), as PDFF cutoffs ranging from 3.5 to 6.9% have been proposed for the accurate discrimination of any steatosis (defined as grades 1–3 on NASH Clinical Research Network criteria) versus no steatosis (grade 0) diagnosed on biopsy (3,20-22). This relatively wide range for identifying steatosis partly reflects differences in disease severity between study cohorts, but it may also reflect imprecision in the low-fat range of the MRI-PDFF estimation methods. Additionally, many clinical trials in NASH have used PDFF as an eligibility criterion; typically, PDFF values of ≥ 5% are used for this purpose (23,24). Therefore, further improvement in the accuracy and reliability of MRI-based PDFF estimation techniques may be important to narrow and validate the PDFF thresholds for clinical use and for clinical trials.
A recent mixed phantom-and-in-vivo study by Motosugi et al. proposed increasing the signal-to-noise ratio (SNR) of MRI-C for improved PDFF quantification by increasing imaging voxel size, reducing bandwidth, and removing parallel imaging (25). This high-SNR MRI-C sequence (Hi-SNR MRI-C) in healthy adult patients demonstrated improved precision of PDFF quantification across the entire range of fat. However, to date, no study has assessed the accuracy and precision of the Hi-SNR MRI-C sequence specifically in the low-fat fraction range. Moreover, this sequence has not been validated in adults and children with NAFLD.
The purpose of this study was to assess and compare the accuracy and precision of the Hi-SNR MRI-C and standard MRI-C for PDFF estimation with respect to MR spectroscopy as the reference standard. We assessed these acquisitions across the entire range of PDFF and at the low-fat fraction range of MRS-PDFF < 10% in pediatric and adult patients with known or suspected NAFLD.
Materials and Methods
Study Design
This was a prospective cross-sectional, single-center, secondary analysis of patients with suspected NAFLD who underwent contemporaneous Hi-SNR MRI-C, standard MRI-C, and MRS acquisition; imaging was performed as part of institutional prospective clinical research studies between February 2014 and June 2015. For these clinical research studies, a total of 231 patients with histologically confirmed or suspected NAFLD were recruited at the NAFLD Research Center (adults) or by the Pediatric Fatty Liver Clinic at Rady Children's Hospital-San Diego (children) and referred for MR examination. Suspicion of NAFLD was based on the presence of at least one of the following: (1) elevated liver transaminases in the presence of obesity, (2) presence of diabetes mellitus, (3) family history of NAFLD, or (4) unexplained elevation of liver transaminases. Exclusion criteria were (1) history of liver disease other than NAFLD, (2) history of chronic alcohol consumption or abuse, (3) contraindication to MR examination including claustrophobia, (4) inability to fit inside the MR scanner, and (5) pregnancy or trying to become pregnant.
We included in our study all consecutive pediatric and adult patients with confirmed or suspected NAFLD who underwent standard and Hi-SNR MRI-C, and MRS, as part of clinical research studies at our institution during the study period. If patients underwent more than one exam during the study period, all exams were included to increase sample size for improved assessment and validation. Exams of 14 pediatric and 14 adult patients meeting inclusion criteria were later excluded from our analysis for the following reasons: (1) the presence of imaging artifact(s) at the MRS voxel location on the MR image, (2) the inability to place the required number of regions of interest (ROIs) at the MRS location due to anatomic factors, or (3) MRS technical failure.
This study was approved by our Institutional Review Board and was compliant with the Health Insurance Portability and Accountability Act. All adult patients provided written informed consent, and pediatric patients provided assent through parental informed consent.
MR Examination
Patients were instructed to fast for a minimum of four hours prior to imaging to minimize any confounding effects that might affect liver visualization on imaging. After safety screening, patients underwent a non-contrast MR examination on a 3T scanner (GE Signa EXCITE HDxt, GE Healthcare, Waukesha, WI, USA). Patients were positioned supine, with an 8-channel a/p torso phased-array receive coil centered over the liver. A dielectric pad was positioned between the coil and the abdominal wall. Each MR exam lasted about 45 minutes.
MRI Sequence & Analysis
Hepatic fat quantification was performed using two, 6-echo MRI-C acquisitions; standard MRI-C (14,16) and Hi-SNR MRI-C. The investigational Hi-SNR MRI-C sequence was designed by increasing the slice thickness from 8 mm to 10 mm, increasing the voxel size by decreasing the acquisition matrix from either 192 × 160 to 128 × 128or from 256 × 128 to 128 × 128, reducing the bandwidth from 125 KHz to 100 KHz, and eliminating parallel imaging. Based on these changes, we calculated that the Hi-SNR protocol had an approximately 3.44-fold increase in SNR compared to the standard MRI-C acquisition using the following formula:
Where, X, Y: voxel dimensions, BW: bandwidth, ST: slice thickness, R: parallel imaging acceleration factor/
Our version of the MRI-C sequence software did not permit user control of the repetition time (26) or echo spacing; these were set automatically by the scanner computer, and did not limit the functionality of our validation testing. Hence, the changes in slice thickness and matrix introduced slight differences in TR and echo spacing between the two sequences, as listed in Table 1. The standard MRI-C sequence was imaged over a single breath-hold; however, due to the elimination of parallel imaging, the Hi-SNR sequence was acquired over 2–3 overlapping single breath-holds to image the whole liver.
Table 1. STEAM MRS, Standard MRI-C, and Hi-SNR MRI-C Parameters.
| Parameters | STEAM MRS | Standard MRI-C | Hi-SNR MRI-C |
|---|---|---|---|
| TR (msec) | 3500 | 6-8 | 6-7 |
| Number of Echoes | 5 | 6 | 6 |
| TE (msec) | 10, 15, 20, 25, 30 | 0.94, 1.7, 2.46, 3.21, 3.97, 4.72 | 0.78, 1.4, 2.02, 2.64, 3.26, 3.88 |
| TM (msec) | 5 | n/a | n/a |
| Acquisition Matrix | 192 × 160 or 256 × 128 | 128 × 128 | |
| Slice Thickness (mm) | 8 | 10 | |
| Bandwidth (kHz) | 5 | +/- 125 | +/-100 |
| Flip angle (°) | 90 | 3 | 3 |
| Parallel imaging acceleration factor | 3.18 | 1 | |
| Parallel imaging reconstruction method | Autocalibrating Reconstruction for Cartesian imaging (ARC) | n/a | |
| FOV (cm) | 38-44 × 38-44 | 38-44 × 38-44 | |
| Typical voxel size (mm3) | 8000 | 32 | 78 |
| Plane | Axial | Axial | |
| Dimension | 3D | 3D |
STEAM: Stimulated Echo Acquisition Mode; MRS: Magnetic Resonance Spectroscopy; MRI-C: Complex-based Magnetic Resonance Imaging; Hi-SNR: High Signal-to-Noise Ratio; kHz: TR: Relaxation time; TE: Echo time; TM: Mixing time; kHz: Kilohertz; FOV: Field-of-view
Postprocessing of both standard and Hi-SNR MRI-C sequences was performed automatically by the scanner computer with a specialized reconstruction algorithm (8,14) that generated T2*-corrected multi-fat-peak model parametric PDFF maps online. The resulting parametric PDFF maps were transferred offline for analysis.
All image analyses were performed by three trained image analysts (6–18 months experience) using Osirix imaging software (Pixmeo, Geneva, Switzerland). To prevent reader-related bias, each reader scored both MRI-C sequences in any given patient. For each sequence, a reader manually placed three circular 1-cm radius regions of interest (ROIs) that were co-localized to the MRS voxel location and on the image superior and the image inferior. Readers used the fifth-echo image of the source magnitude images for ROI placement due to the relative ease of hepatic anatomy delineation provided (Figure 1). In addition, placing the ROIs on the source images rather than on PDFF maps prevented feedback bias. All three ROIs were then propagated onto corresponding parametric PDFF maps, and the mean PDFF value was recorded from the average of the three ROIs. Additionally, a T1 bias correction method was applied to the PDFF values obtained from the maps using previously published T1 values at 3 Tesla for liver water (822 ms) and liver fat (312ms) (27), as described by Kuhn el al (28).
Figure 1.
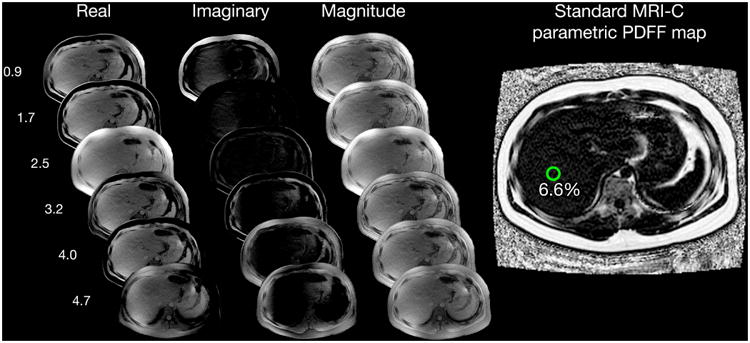
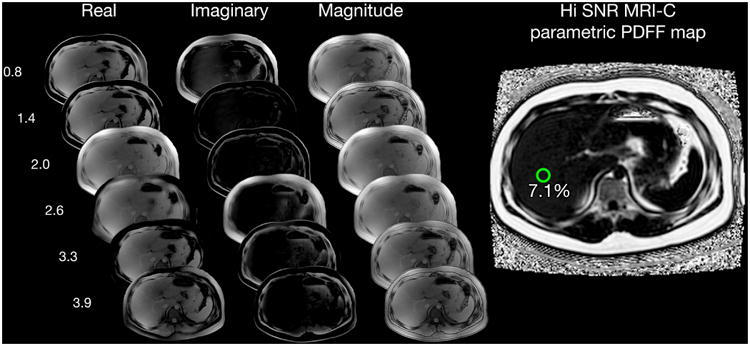
Multi-echo Image Reconstruction of Hi-SNR MRI-C parametric PDFF Map, Six echo source real, imaginary, and magnitude images were reconstructed into a parametric PDFF map using complex-fitting algorithms that utilize fat spectral modeling and T2* correction. A 1-cm circular region of interest was placed on the 5th echo magnitude image at MRS voxel location, then automatically propagated onto the PDFF map to obtain the mean PDFF. Example case of a 17-year-old patient with low-fat content with MRS-PDFF of 7.4%: a) 6.6% measured by standard MRI-C and b) 7.1% measured by Hi-SNR MRI-C. The higher SNR can be visualized in the Hi-SNR MRI-C parametric PDFF map compared to that of the standard MRI-C PDFF map.
MRS Sequence & Analysis
Single-voxel MRS was performed using Stimulated Echo Acquisition Mode (STEAM) (29). A 20 × 20 × 20 mm3 voxel was placed on the right hepatic lobe (Couinaud segment VI or VII) avoiding large blood vessels, biliary ducts, and liver edges. Following automated shimming during free breathing, five TR 3500 ms spectra were collected at echo times of 10, 15, 20, 25, and 30 msec in a single (∼21 sec) breath-hold to permit T2 estimation while minimizing fat-peak-j-coupling (29). The TR and echo spacing for each technique were automatically set by our scanner computer, but were close to the optimal echo spacing for complex reconstruction per Reeder et al (16).MRS acquisition parameters are listed in Table 1.
MR spectral analysis was performed by a single MR spectroscopist with >15 years of experience blinded to MRI-C acquisitions and clinical data. Analysis was performed using custom prior knowledge using the Advanced Method for Accurate, Robust, and Efficient Spectral fitting of MRS data (AMARES) included in Java-based magnetic resonance user interface software package (30,31). T2-corrected areas of the water (4–6 ppm) and the fat (0–3 ppm) were estimated as there is insufficient spectral resolution in vivo to accurately characterize the individual fat peaks, or to distinguish water from two nearby fat peaks (32). The contribution to the water peak from the neighboring fat peaks was corrected using a previously derived fat spectrum post-T2 correction, which reassigns these fat peaks from water to the fat signal (32).
Statistical Analysis
All statistical analyses were performed using the R software package (R Foundation for Statistical Computing, Vienna, Austria, 2016) by a staff biostatistician, supervised by a faculty biostatistician (each with more than 20 years of experience). All summaries and primary analyses were performed on all patients, and on the subgroup of patients with MRS-PDFF < 10%. Bootstrap extension with by-patient resampling was applied to all confidence intervals (CI's) and tests of significance to adjust for patients with multiple scans.
Cohort age and BMI were summarized descriptively
Intra-class correlation coefficient (ICC) and Bland-Altman analysis were used to assess agreement and systematic disagreement between standard MRI-C and Hi-SNR MRI-C sequences, and 95% CI's were computed around ICCs. Bland-Altman plots were generated, and 95% Limits of Agreement (LOA) and Bland-Altman bias were computed, with significance of the bias assessed using a bootstrap extension of a paired t-test.
To assess and compare the accuracy and precision of standard MRI-C and Hi-SNR MRI-C sequences using MRS-PDFF as the reference standard, we performed univariate regression analyses modeling MRS-PDFF as a function of standard MRI-C PDFF and, separately, of Hi-SNR MRI-C PDFF. Regression accuracy metrics (intercept and slope of the regression line, average bias of the regression, and R2) were computed for each model. Bootstrap-based 95% CI's were computed around all regression metrics. Bootstrap-based tests were used to compare the accuracy metrics of the two MRI-C sequences. Bonferroni correction was applied to each set of accuracy metric comparisons (for all patients and for patients with MRS-PDFF < 10%) to ensure a family-wise alpha level of 0.05.
Secondary Analyses
We assessed the performance of standard MRI-C and Hi-SNR MRI-C sequences to classify MRS-based PDFF cutoffs of 3%, 4%, 5%, 6%, and 7% using the same thresholds. Performance parameters: sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and total accuracy with 95% bootstrap-based CI's were computed for each threshold.
Results
Cohort Characteristics
Two hundred and seventy-one exams from 231 patients (mean MRS-PDFF: 12.6% [SD: 10.4]; range: 0.9–41.9) were included in this study. A total of 144 exams from 120 patients had MRS-PDFF less than 10%. The cohort demographic is detailed in Table 2. We excluded 28 examinations from our analysis for the following reasons: (1) the presence of parallel imaging artifact at the MRS voxel location on the standard MRI-C image (n= 2), (2) the inability to place all ROIs at the MRS location due to anatomic factors (n = 3), (3) MRI acquisition technical failure (n= 2), or (4) MRS technical failure (n= 21). As the Hi-SNR sequence did not use parallel imaging, no exams were excluded due to parallel imaging artifacts on the Hi-SNR sequence.
Table 2. Cohort Demographics from 271 exams, 144 with MRS-PDFF < 10%.
| Variable | All Patients | Patients with MRS-PDFF < 10% |
|---|---|---|
| No. of Exams | 231 | 120 |
| Age, mean (SD) | 31.1 (21.6) | 28.6 (21.9) |
| Range | 8 – 78 | 8 – 78 |
| Age Group | ||
| Adult | 107 (46.3) | 45 (37.5) |
| Pediatric | 124 (53.7) | 75 (62.5) |
| BMI (kg/m2), mean (SD) | 29.6 (6.5) | 27.9 (6.8) |
| Range | 15.2 – 46.9 | 15.2 – 46.9 |
| Sex, N (%) | ||
| Female | 99 (42.8) | 51 (42.5) |
| Male | 132 (57.1) | 69 (57.5) |
| MRS-PDFF, % (SD) | 12.62 (10.36) | 4.52 (2.64) |
| Range | 0.89 – 41.88 | 0.89 – 9.84 |
| Standard MRI-PDFF, % (SD) | 11.99 (9.91) | 4.45 (1.95) |
| Range | 1.45 – 39.79 | 1.45 – 9.52 |
| Hi-SNR MRI-PDFF, % (SD) | 12.16 (10.22) | 4.34 (2.30) |
| Range | 1.15 – 39.57 | 1.15 – 9.91 |
MRS-PDFF: Magnetic Resonance Spectroscopy Proton Density Fat Fraction; Hi-SNR: High-Signal-to-noise-ratio
Assessment of Agreement Between Standard MRI-C and Hi-SNR MRI-C
Bland-Altman plots are presented in Figure 2. PDFF measurements by the Hi-SNR sequence were minimally but significantly higher than the standard MRI-C values by 0.18% points (P = 0.0018) on average in the overall cohort. Agreement was high with an ICC of 0.995 (95% CI 0.993–0.996); however, in the subset of patients with MRS-PDFF < 10%, no significant bias in the PDFF estimated by the MRI-C sequences was observed (P = 0.505). ICC was 0.895 (95% CI 0.837–0.928).
Figure 2.
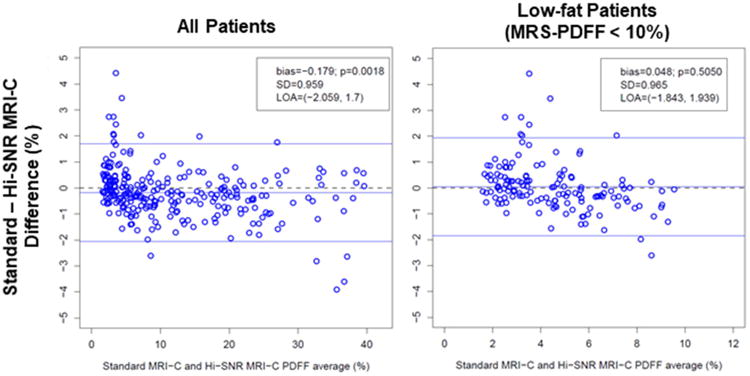
Bland–Altman plots illustrating the difference between Standard MRI-C and Hi-SNR MRI-C PDFF across the entire range of PDFF (left panel) and at the low range of PDFF (right panel). Mean bias between the two MRI-C sequences is small but statistically significant in the all-patient group but was not significant in the low-PDFF patients only.
Accuracy of Standard Versus Hi-SNR MRI-C Sequences
Univariate regression plots demonstrated a high correlation between PDFF estimated by the MRI-C sequences and MRS-PDFF across the overall range of PDFF (Figure 3). In the low-fat range (subset of patients with MRS-PDFF < 10%), standard MRI-C demonstrated relatively greater underestimation with a slope of 1.19, compared to that of Hi-SNR MRI-C (slope of 1.06). All MRI-C PDFF versus MRS-PDFF regression parameters are detailed in Table 3.
Figure 3.
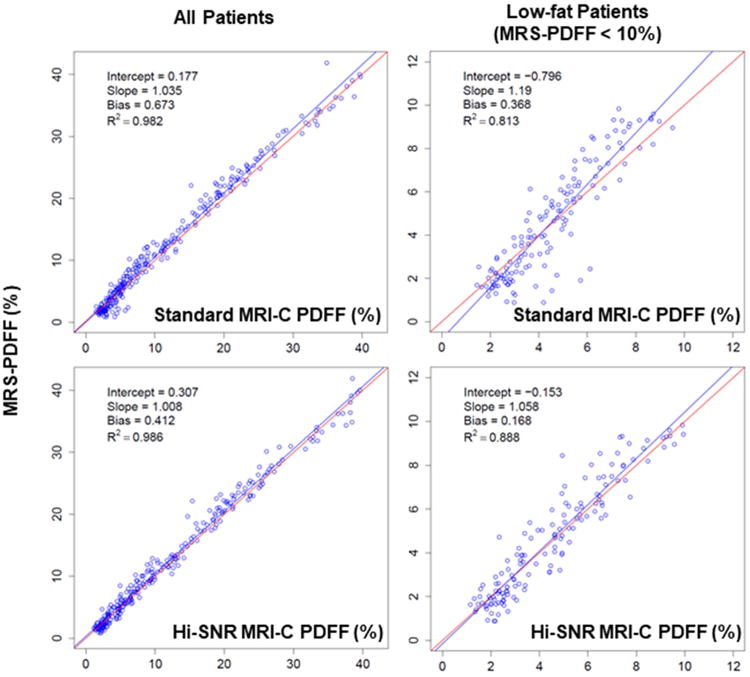
Regression plots of MRS-PDFF as a function of standard MRI-C PDFF (top panels) and Hi-SNR MRI-C PDFF (bottom panels), for all patients (left panels) and low-fat patients (right panels). The line of best fit is shown in blue. The line of perfect MRS to MRI-C PDFF agreement (slope = 1, intercept = 0) is depicted in red.
Table 3. MRI-C PDFF versus MRS-PDFF Regression Parameters in all patients and in low-PDFF patients (MRS-PDFF < 10%).
| Patient group | Sequence | Intercept (95% CI) | Slope (95% CI) | Average Bias (95% CI) | R2 (95% CI) |
|---|---|---|---|---|---|
| All Patients | Standard MRI-C | 0.177 (-0.076, 0.441) | 1.035 (1.015, 1.058) | 0.673 (0.495, 0.865) | 0.982 (0.976, 0.986) |
| Hi-SNR MRI-C | 0.307 (0.111, 0.517) | 1.008 (0.989, 1.027) | 0.412 (0.259, 0.571) | 0.985 (0.979, 0.988) | |
| Low-PDFF | Standard MRI-C | -0.796 (-1.162, -0.455) | 1.190 (1.112, 1.273) | 0.368 (0.202, 0.518) | 0.813 (0.722, 0.886) |
| Hi-SNR MRI-C | -0.153 (-0.403, 0.119) | 1.058 (1.009, 1.118) | 0.168 (0.055, 0.300) | 0.888 (0.840, 0.916) |
MRI-C: Complex-based Magnetic Resonance Imaging; Hi-SNR: High Signal-to-Noise Ratio; PDFF: Proton Density Fat Fraction;
Bonferroni-adjusted bootstrap tests showed that the Hi-SNR MRI-C sequence, compared to the standard MRI-C, was more accurate overall based on regression bias significantly closer to 0 (0.67% for MRI-C vs. 0.41% for Hi-SNR MRI-C, difference of 0.26% [P < 0.0001]), and had and a slope closer to 1 (1.04 for MRI-C vs. 1.01 for Hi-SNR MRI-C, difference of 0.03 [P = 0.004]; these tests similarly showed the improved accuracy of Hi-SNR MRI-C in the low-fat range (bias of 0.37% for MRI-C vs. 0.17% for Hi-SNR MRI-C, difference of 0.20% [P = 0.007]), plus a slope closer to 1 (1.19 for MRI-C vs. 1.06 for Hi-SNR MRI-C, difference of 0.13 [P = 0.002]).
In the low-fat range, all regression-based accuracy metrics were significantly better for the Hi-SNR MRI-C than for the standard MRI-C, with an intercept significantly closer to 0 (-0.80% for MRI-C vs. -1.53% for Hi-SNR MRI-C, difference of 0.64% [P < 0.0001]), and a significantly higher R2 (0.81 for MRI-C vs. 0.89 for Hi-SNR MRI-C, difference of 0.075 [P = 0.010]) (Table 4).
Table 4. (a) Pairwise comparisons of regression parameters between standard MRI-C PDFF or Hi-SNR MRI-C PDFF plotted against MRS-PDFF, for all patients and (b) in patients with MRS-PDFF < 10%.
| Sequence | Comparator | Regression parameter | Difference (98.75% CI) | P | Interpretation |
|---|---|---|---|---|---|
| a) | |||||
| Standard MRI-C | Hi-SNR MRI-C | Abs (Intercept) | -0.130 (-0.369, 0.093) | 0.135 | NS |
| Abs (Slope-1) | 0.027 (0.006, 0.048) | 0.004 | * | ||
| Bias | 0.261 (0.123, 0.444) | <0.0001 | * | ||
| R2 | -0.003 (-0.008, 0.002) | 0.05 | NS | ||
| b) | |||||
| Standard MRI-C | Hi-SNR MRI-C | Abs (Intercept) | 0.643 (0.253, 1.116) | < 0.0001 | * |
| Abs (Slope-1) | 0.132 (0.041, 0.226) | 0.002 | * | ||
| Bias | 0.201 (0.004, 0.386) | 0.007 | * | ||
| R2 | -0.075 (-0.198, 0.000) | 0.010 | NS |
MRI-C: Complex-based Magnetic Resonance Imaging; Hi-SNR: High Signal-to-Noise Ratio; PDFF: Proton Density Fat Fraction; MRS: Magnetic Resonance Spectroscopy
: Significant
NS: Not significant
Because of the Bonferroni's correction, only p values <0.0042 are considered significant at family-wise 0.05 level
Performance of Standard and Hi-SNR MRI-C PDFF in Classifying Equivalent MRS-based PDFF Cutoffs
Table 5 summarizes the performance of the standard and Hi-SNR MRI-C sequences to approximate equivalent MRS-based PDFF cutoffs. Across MRS-PDFF cutoffs of 3–7%, the standard and Hi-SNR MRI-C sequences each demonstrated high accuracy for classifying equivalent MRS-PDFF cutoffs. The two MRI-C sequences demonstrated similar sensitivity, specificity, PPV, NPV, and accuracy at all MRS cutoffs.
Table 5. Diagnostic test characteristics of Hi-SNR MRI-C in diagnosing steatosis at different MRS-based PDFF cutoffs.
| MRS-PDFF cutoff using equivalent MRI-C thresholds | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | PPV (%) (95% CI) | NPV (%) (95% CI) | Accuracy (%) (95% CI) | |
|---|---|---|---|---|---|---|
| 3% | Standard MRI-C | 99.1 (96.7 – 99.9) | 72.2 (58.4 – 83.5) | 93.5 (89.5 – 96.3) | 95.1 (83.5 – 99.4) | 93.7 (90.1 – 96.3) |
| Hi-SNR MRI-C | 98.2 (95.3 – 99.5) | 90.7 (79.7 – 96.9) | 97.7 (94.7 – 99.3) | 92.5 (81.8 – 97.9) | 96.7 (93.8 – 98.5) | |
| 4% | Standard MRI-C | 98.0 (94.9 – 99.4) | 83.3 (72.7 – 91.1) | 94.2 (90.1 – 97.0) | 93.8 (84.8 – 98.3) | 94.1 (90.6 – 96.6) |
| Hi-SNR MRI-C | 97.0 (93.6 – 98.9) | 90.3 (81.0 – 96.0) | 96.5 (92.9 – 98.6) | 91.5 (82.5 – 96.8) | 95.2 (91.9 – 97.4) | |
| 5% | Standard MRI-C | 93.6 (89.1 – 96.6) | 94.0 (86.7 – 98.0) | 97.2 (93.6 – 99.1) | 86.8 (78.1 – 93.0) | 93.7 (90.1 – 96.3) |
| Hi-SNR MRI-C | 95.7 (91.7 – 98.1) | 98.8 (93.5 – 100) | 99.4 (96.9 – 100) | 91.2 (83.4 – 96.1) | 96.7 (93.8 – 98.5) | |
| 6% | Standard MRI-C | 92.9 (87.9 – 96.3) | 98.0 (93.1 – 99.8) | 98.7 (95.5 – 99.8) | 89.3 (82.0 – 94.3) | 94.8 (91.5 – 97.1) |
| Hi-SNR MRI-C | 95.3 (90.9 – 97.9) | 97.1 (91.6 – 99.4) | 98.2 (94.7 – 99.6) | 92.5 (85.8 – 96.7) | 95.9 (92.9 – 98.0) | |
| 7% | Standard MRI-C | 91.7 (86.3 – 95.5) | 99.1 (95.2 – 100) | 99.3 (96.2 – 100) | 89.7 (83.0 – 94.4) | 94.8 (91.5 – 97.1) |
| Hi-SNR MRI-C | 93.6 (88.6 – 96.9) | 99.1 (95.2 – 100) | 99.3 (96.3 – 100) | 91.9 (85.6 – 96.0) | 95.9 (92.9 – 98.0) |
PPV: Positive predictive value; NPV: Negative predictive value
Discussion
In this study, a Hi-SNR MRI-C acquisition protocol provided slightly higher PDFF estimation accuracy in adult and pediatric NAFLD patients overall. In patients with low range of PDFF values (0–10%), we found that the Hi-SNR MRI-C sequence provided slightly higher PDFF estimation accuracy and precision. However, the observed increases in accuracy and precision were small and possibly not clinically meaningful, suggesting that the current standard MRI-C sequence is already well optimized for PDFF estimation. Although the HI-SNR MRI-C sequence represents a slight improvement in PDFF estimation using MRS as the reference standard, this sequence sacrifices spatial resolution to increase the SNR and may reduce imaging's ability to delineate fine details within the liver. This potential limitation has not yet been assessed, and may prove to be a minor concern if the primary purpose of the imaging sequence is to quantify liver fat.
In addition, the Hi-SNR MRI-C sequence that we tested may be disadvantageous in clinical settings due to the multi-breath-hold acquisition (versus single-breath-hold acquisition of the standard MRI-C). However, the need for whole-liver coverage for PDFF estimation is unknown, and a single acquisition through the widest portion of the liver—typically requiring a single breath-hold—is likely to provide sufficient anatomic coverage to sample the liver adequately.
Finally, an unanticipated benefit of our Hi-SNR MRI-C technique was the elimination of parallel imaging artifacts, which occasionally corrupt PDFF parametric maps using the standard MRI-C sequence with the implemented parallel imaging acceleration factors. More robust parallel imaging methods would be required to address this limitation in standard MRI-C. Alternatively, compressed imaging and other methods to reduce acquisition time might prove helpful in the clinical setting.
Our techniques and findings are similar to those described in a recently published study by Motosugi et al., in which the High-SNR MRI-C sequence performed on 1.5 T incrementally improved the precision of PDFF estimation across the entire range of PDFF in a cohort of 20 adult volunteers and 28 adult patients (25). Our study demonstrated similar findings on 3.0 T and with a larger cohort of adult and pediatric NAFLD patients. In contrast with Motosugi et al., our study also demonstrated improvements in the regression bias (i.e., accuracy) using Hi-SNR MRI-C. We expect that this difference may be due to a larger sample size used in our study and is unlikely to be clinically significant.
While the effects of varying acquisition parameters such as voxel size and slice thickness may be of interest for further investigation, our result suggests that the effect of varying these acquisition parameters is small.
Prior studies have proposed various PDFF cutoffs ranging from 3.5 to 6.9 % to discriminate mild steatosis from no steatosis with respect to histology (3,20-22,33). As standard MRI-C and Hi-SNR MRI-C provide the same performance at these ranges of PDFF cutoffs, it is unlikely that the various cutoffs in the literature are caused by differences in the PDFF sequence parameters, but rather can be linked to differences in subject groups (e.g., different PDFF cutoff for adults and children). More technically, in a chemical-shift-encoded MRI sequence with complex reconstruction that utilizes fat spectral modeling, T2* and T1 correction that is acquired using parameters that are within the boundaries of those reasonable to estimate PDFF, we found that the results are the same regardless of changing the parameters, and that accuracy is not limited by SNR.
That said, other methods (e.g., acquiring images at a high flip angle in the hepatobiliary phase following gadolinium administration) have been previously described for improving the SNR of acquisition while avoiding the loss of resolution (34,35). The method described in this paper represents one approach to increase the SNR for higher fat quantification accuracy without the need for contrast agent administration in the low-PDFF range.
A limitation of this study is that we did not assess the diagnostic performance of Hi-SNR MRI-C with respect to liver biopsy. However, we demonstrated that the standard MRI-C and Hi-SNR MRI-C modalities have similar performance for diagnosing steatosis using equivalent MRS-based PDFF cutoffs (from 3 to 7%) as proxy, suggesting that the two MRI-C PDFF sequences are also likely to demonstrate similar accuracy for diagnosing steatosis with respect to histology. Our study did not investigate optimal PDFF thresholds for diagnosing steatosis in adults or children with NAFLD. Another limitation was that the TR and echo spacing were altered automatically by our scanner computer. While all echo spacings were close to the optimal echo spacing for complex reconstruction as defined by Reeder et al. (16), we could not completely exclude effects due to changes in TR and echo spacing. As TRs were different between the standard and Hi-SNR MRI-C techniques, we applied T1 correction. Other limitations include: use of a single scanner from a single manufacturer, use of single field strength, and the highly-specialized, single-center setting of this study; all factors that might limit the generalizability of our findings in other clinical settings.
In conclusion, the Hi-SNR MRI-C acquisition provides an incremental improvement in the accuracy and precision of PDFF estimation in the low PDFF range. Further protocol optimization for Hi-SNR MRI-C may be warranted in order to optimize the applicability of this technique in a clinical setting.
Acknowledgments
Grant Support: The project described was partially supported by the National Institutes of Health, Grants R01DK075128, R01DK088925, K24 DK102595, R01 DK100651, R01 DK083380 and TL1 TR00098. Further, the authors wish to acknowledge support from the GE Healthcare who provides research support to UCSD and UW-Madison. Dr. Reeder is a Romnes Faculty Fellow, and has received an award provided by the University of Wisconsin-Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH
Footnotes
Conflict of Interest: Dr. Sirlin consults, advises, and is on the speakers' bureau for Bayer. He has received grants from GE Healthcare. All other authors report no other relevant conflict of interests.
References
- 1.Permutt Z, Le TA, Peterson MR, et al. Correlation between liver histology and novel magnetic resonance imaging in adult patients with non-alcoholic fatty liver disease - MRI accurately quantifies hepatic steatosis in NAFLD. Aliment Pharmacol Ther. 2012;36(1):22–29. doi: 10.1111/j.1365-2036.2012.05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeder SB, Hu HH, Sirlin CB. Proton density fat-fraction: a standardized MR-based biomarker of tissue fat concentration. J Magn Reson Imaging. 2012;36(5):1011–1014. doi: 10.1002/jmri.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang A, Desai A, Hamilton G, et al. Accuracy of MR imaging-estimated proton density fat fraction for classification of dichotomized histologic steatosis grades in nonalcoholic fatty liver disease. Radiology. 2015;274(2):416–425. doi: 10.1148/radiol.14140754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson NA, Walton DW, Sachinwalla T, et al. Noninvasive assessment of hepatic lipid composition: Advancing understanding and management of fatty liver disorders. Hepatology. 2008;47(5):1513–1523. doi: 10.1002/hep.22220. [DOI] [PubMed] [Google Scholar]
- 5.Mehta SR, Thomas EL, Bell JD, Johnston DG, Taylor-Robinson SD. Non-invasive means of measuring hepatic fat content. World J Gastroenterol. 2008;14(22):3476–3483. doi: 10.3748/wjg.14.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szczepaniak LS, Babcock EE, Schick F, et al. Measurement of intracellular triglyceride stores by H spectroscopy: validation in vivo. Am J Physiol. 1999;276(5 Pt 1):E977–989. doi: 10.1152/ajpendo.1999.276.5.E977. [DOI] [PubMed] [Google Scholar]
- 7.Thomsen C, Becker U, Winkler K, Christoffersen P, Jensen M, Henriksen O. Quantification of liver fat using magnetic resonance spectroscopy. Magn Reson Imaging. 1994;12(3):487–495. doi: 10.1016/0730-725x(94)92543-7. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, Shimakawa A, Hines CD, et al. Combination of complex-based and magnitude-based multiecho water-fat separation for accurate quantification of fat-fraction. Magn Reson Med. 2011;66(1):199–206. doi: 10.1002/mrm.22840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernando D, Hines CD, Yu H, Reeder SB. Addressing phase errors in fat-water imaging using a mixed magnitude/complex fitting method. Magn Reson Med. 2012;67(3):638–644. doi: 10.1002/mrm.23044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meisamy S, Hines CD, Hamilton G, et al. Quantification of hepatic steatosis with T1-independent, T2-corrected MR imaging with spectral modeling of fat: blinded comparison with MR spectroscopy. Radiology. 2011;258(3):767–775. doi: 10.1148/radiol.10100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokoo T, Serai SD, Pirasteh A, et al. Linearity, Bias, and Precision of Hepatic Proton Density Fat Fraction Measurements by Using MR Imaging: A Meta-Analysis. Radiology. 2017:170550. doi: 10.1148/radiol.2017170550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu CY, McKenzie CA, Yu H, Brittain JH, Reeder SB. Fat quantification with IDEAL gradient echo imaging: correction of bias from T(1) and noise. Magn Reson Med. 2007;58(2):354–364. doi: 10.1002/mrm.21301. [DOI] [PubMed] [Google Scholar]
- 13.Reeder SB, Robson PM, Yu H, et al. Quantification of hepatic steatosis with MRI: the effects of accurate fat spectral modeling. J Magn Reson Imaging. 2009;29(6):1332–1339. doi: 10.1002/jmri.21751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H, Shimakawa A, McKenzie CA, Brodsky E, Brittain JH, Reeder SB. Multiecho water-fat separation and simultaneous R2* estimation with multifrequency fat spectrum modeling. Magn Reson Med. 2008;60(5):1122–1134. doi: 10.1002/mrm.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernando D, Haldar JP, Sutton BP, Ma J, Kellman P, Liang ZP. Joint estimation of water/fat images and field inhomogeneity map. Magn Reson Med. 2008;59(3):571–580. doi: 10.1002/mrm.21522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeder SB, Pineda AR, Wen Z, et al. Iterative decomposition of water and fat with echo asymmetry and least-squares estimation (IDEAL): application with fast spin-echo imaging. Magn Reson Med. 2005;54(3):636–644. doi: 10.1002/mrm.20624. [DOI] [PubMed] [Google Scholar]
- 17.Bydder M, Yokoo T, Hamilton G, et al. Relaxation effects in the quantification of fat using gradient echo imaging. Magn Reson Imaging. 2008;26(3):347–359. doi: 10.1016/j.mri.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chebrolu VV, Hines CD, Yu H, et al. Independent estimation of T*2 for water and fat for improved accuracy of fat quantification. Magn Reson Med. 2010;63(4):849–857. doi: 10.1002/mrm.22300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, McKenzie CA, Shimakawa A, et al. Multiecho reconstruction for simultaneous water-fat decomposition and T2* estimation. J Magn Reson Imaging. 2007;26(4):1153–1161. doi: 10.1002/jmri.21090. [DOI] [PubMed] [Google Scholar]
- 20.Tang A, Tan J, Sun M, et al. Nonalcoholic fatty liver disease: MR imaging of liver proton density fat fraction to assess hepatic steatosis. Radiology. 2013;267(2):422–431. doi: 10.1148/radiol.12120896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Martino M, Pacifico L, Bezzi M, et al. Comparison of magnetic resonance spectroscopy, proton density fat fraction and histological analysis in the quantification of liver steatosis in children and adolescents. World J Gastroenterol. 2016;22(39):8812–8819. doi: 10.3748/wjg.v22.i39.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paparo F, Cenderello G, Revelli M, et al. Diagnostic value of MRI proton density fat fraction for assessing liver steatosis in chronic viral C hepatitis. Biomed Res Int. 2015;2015:758164. doi: 10.1155/2015/758164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui J, Philo L, Nguyen P, et al. Sitagliptin vs. placebo for non-alcoholic fatty liver disease: A randomized controlled trial. J Hepatol. 2016;65(2):369–376. doi: 10.1016/j.jhep.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10(11):686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 25.Motosugi U, Hernando D, Wiens C, Bannas P, Reeder SB. High SNR Acquisitions Improve the Repeatability of Liver Fat Quantification Using Confounder-corrected Chemical Shift-encoded MR Imaging. Magn Reson Med Sci. 2017 doi: 10.2463/mrms.mp.2016-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.European Association For The Study Of The L, European Organisation For R, Treatment Of C. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton G, Middleton MS, Hooker JC, et al. In vivo breath-hold (1) H MRS simultaneous estimation of liver proton density fat fraction, and T1 and T2 of water and fat, with a multi-TR, multi-TE sequence. J Magn Reson Imaging. 2015;42(6):1538–1543. doi: 10.1002/jmri.24946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn JP, Jahn C, Hernando D, et al. T1 bias in chemical shift-encoded liver fat-fraction: role of the flip angle. J Magn Reson Imaging. 2014;40(4):875–883. doi: 10.1002/jmri.24457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton G, Middleton MS, Bydder M, et al. Effect of PRESS and STEAM sequences on magnetic resonance spectroscopic liver fat quantificfation. J Magn Reson Imaging. 2009;30(1):145–152. doi: 10.1002/jmri.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naressi A, Couturier C, Devos JM, et al. Java-based graphical user interface for the MRUI quantitation package. MAGMA. 2001;12(2-3):141–152. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]
- 31.Vanhamme L, van den Boogaart A, Van Huffel S. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. J Magn Reson. 1997;129(1):35–43. doi: 10.1006/jmre.1997.1244. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton G, Yokoo T, Bydder M, et al. In vivo characterization of the liver fat (1)H MR spectrum. NMR Biomed. 2011;24(7):784–790. doi: 10.1002/nbm.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szczepaniak LS, Nurenberg P, Leonard D, et al. Magnetic resonance spectroscopy to measure hepatic triglyceride content: prevalence of hepatic steatosis in the general population. Am J Physiol Endocrinol Metab. 2005;288(2):E462–468. doi: 10.1152/ajpendo.00064.2004. [DOI] [PubMed] [Google Scholar]
- 34.Park CC, Hamilton G, Desai A, et al. Effect of intravenous gadoxetate disodium and flip angle on hepatic proton density fat fraction estimation with six-echo, gradient-recalled-echo, magnitude-based MR imaging at 3T. Abdom Radiol (NY) 2016 doi: 10.1007/s00261-016-0992-4. [DOI] [PubMed] [Google Scholar]
- 35.Frydrychowicz A, Nagle SK, D'Souza SL, Vigen KK, Reeder SB. Optimized high-resolution contrast-enhanced hepatobiliary imaging at 3 tesla: a cross-over comparison of gadobenate dimeglumine and gadoxetic acid. J Magn Reson Imaging. 2011;34(3):585–594. doi: 10.1002/jmri.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]


