Abstract
Proximity ligation assay (PLA), also referred to as Duolink® PLA technology, permits detection of protein-protein interactions in situ (at distances < 40 nm) at endogenous protein levels. It exploits specific antibodies identifying (either directly or indirectly) the two proteins of interest and takes advantage of specific DNA primers covalently linked to the antibodies. A hybridization step followed by a PCR amplification with fluorescent probes then permit visualization of spots of proximity by fluorescence microscopy. Since the development of PLA in 2002, it has been increasingly popular to detect the interaction between two proteins with high sensitivity and specificity. It is a very simple and sensitive technique to study protein-protein interaction in cells.
Keywords: Protein-protein interaction, Antibody, In situ assays, Duolink®
INTRODUCTION
Protein-protein interactions are fundamental to cellular responses (Braun and Gingras, 2012). There are many methods one can use to investigate such interactions in cells, ranging from commonly-used co-immunoprecipitation (Co-IP) (Ni et al., 2016; Lee, 2007) to more advanced fluorescence-based methods such as FRET (Fluorescence Resonance Energy Transfer) (Edidin, 2003; Cheng, 2006; Zheng, 2006). Each method has limitations, and most depend on genetic modification of the cells or cloning of genes, which are not feasible with patient samples. Therefore, a suitable and sensitive assay is needed to detect direct protein-protein interactions at endogenous levels of expression and without genetic manipulation.
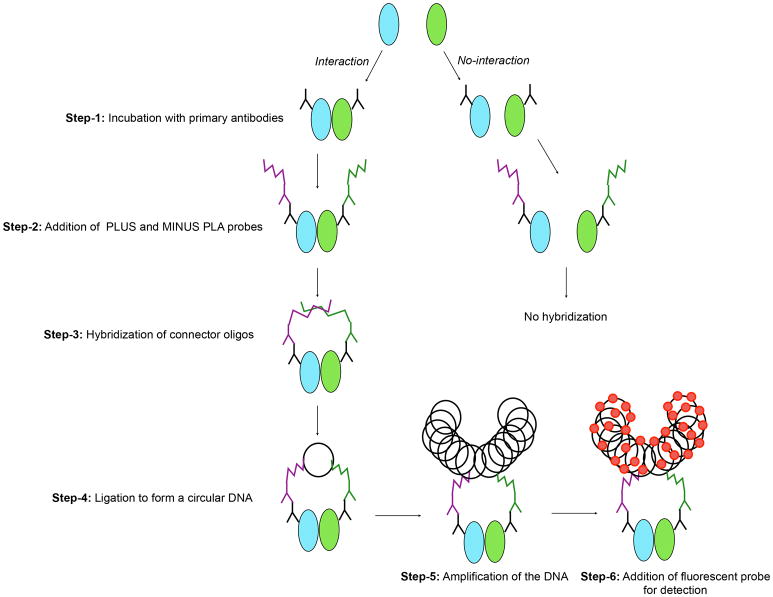
Duolink® PLA technology, allows one to detect protein-protein interactions at endogenous protein levels with high sensitivity and specificity (Fredriksson et al., 2002; Gullberg et al., 2004; Söderberg et al., 2006). Cells need to be stained with two immunohistochemistry- or immunofluorescence-compatible primary antibodies to the target proteins. The two primary antibodies must be generated from different species (i. e., mouse/rabbit, rabbit/goat, or mouse/goat). Cells are then stained with secondary antibodies (2°-Ab) known as PLA probes (one PLUS and one MINUS). The PLA probes that bind to the constant regions of the primary antibodies contains a unique DNA strand. If the proteins of interest interact with each other, the DNA probes hybridize to make circular DNA. This DNA can be amplified and visualized by fluorescently-labeled complementary oligonucleotide probes. The schematic diagram of Duolink® PLA is shown in . interactions Figure-1 are visualized as dots, and the number and intensity of the dots can be quantified by fluorescence microscopy.
Figure-1. Representative diagram of the Duolink® proximity ligation assay (PLA).
Step by step representation of the PLA.
BASIC PROTOCOL
Materials
Primary antibodies for detecting the desired protein-protein interaction raised in two different species, e.g., rabbit anti-X and mouse anti-Y; since these will be detected with species specific primer-tagged secondary antibodies.
-
Duolink® in Situ Orange Starter Kit, Sigma (for Mouse/Rabbit antibody combination, Cat. No. DUO92102; for Mouse/Goat antibody combination, Cat. No. DUODUO92104; for Goat/Rabbit antibody combination, Cat. No. DUO92106).
The kits contain the indicated reagents. Different species and colors and can be ordered separately. The catalog number is given as an example for the combination of mouse and rabbit in orange. The ordering details for different combinations can be found in Table-1.
Duolink® in situ reagents: a) Duolink® in situ complementary oligonucleotide PLA probe PLUS (Sigma-Aldrich, anti-rabbit PLUS: Cat. No. DUO92002) and MINUS (Sigma-Aldrich, anti-mouse MINUS: Cat. No. DUO92004); b) Blocking solution (Sigma-Aldrich, Cat. No. DUO82007); c) Antibody diluent (Sigma-Aldrich, Cat. No. DUO82008). The reagents need to be stored at 4 °C.
Duolink®in situ detection reagents (Sigma-Aldrich, Cat. No. DUO92007): a) Ligation reagent (5x) (Sigma-Aldrich, Cat. No. 82009), b) Ligase (1 unit/μl) (Sigma-Aldrich, Cat. No. DUO82029), c) Amplification reagents (5x) (Sigma-Aldrich, Cat. No. DUO82010), d) Polymerase (10 unit/μl) (Sigma-Aldrich, Cat. No. DUO82030). The reagents need to be stored at -20 °C.
-
Duolink® Wash Buffers:
Although commercially available the buffers can be prepared in the laboratory.
-
Duolink® in situ wash buffer A (Sigma-Aldrich, Cat. No. DUO82046)
10 mM Tris, pH-7.4, 150 mM NaCl and 0.05% Tween.
Filter the buffer with 0.45 μm. The buffer can be stored at 4 °C. Bring the solution at room temperature before use.
-
Duolink® in situ wash buffer B (Sigma-Aldrich, Cat. No. DUO82048)
200 mM Tris, pH-7.5, 100 mM NaCl.
Filter the buffer with 0.45 μm. The buffer can be stored at 4 °C. Bring the solution at room temperature before use.
-
Duolink® in situ mounting medium with DAPI (Sigma-Aldrich, Cat. No. DUO82040). The solution needs to be stored at 4 °C.
μ-Slide Angiogenesis (ibiTreat, Cat. No. 81506)
Alexa Fluor 488 conjugated wheat germ agglutinin (WGA) (ThermoFisher Scientific, Cat. No. W11261), optional
4% paraformaldehyde solution in PBS (Affymatrix, Cat. No. 19943)
Methanol
Phosphate-buffered saline (PBS)
Bovine serum albumin (BSA)
Distilled water
Tris-HCl, pH 7.4, optional
Tris-HCl, pH 7.5, optional
Tween 20, optional
Table-1.
A typical Duolink® starter kit for antibodies of different host available in Sigma
| Product# | Kit | Website |
|---|---|---|
| DUO92101 | Duolink® In Situ Red starter kit Mouse/Rabbit | https://www.sigmaaldrich.com/catalog/product/sigma/duo92101 |
| DUO92102 | Duolink® In Situ Orange starter kit Mouse/Rabbit | https://www.sigmaaldrich.com/catalog/product/sigma/duo92102 |
| DUO92103 | Duolink® In Situ Red starter kit Mouse/Goat | https://www.sigmaaldrich.com/catalog/product/sigma/duo92103 |
| DUO92104 | Duolink® In Situ Orange starter kit Mouse/Goat | https://www.sigmaaldrich.com/catalog/product/sigma/duo92104 |
| DUO92105 | Duolink® In Situ Red starter kit Goat/Rabbit | https://www.sigmaaldrich.com/catalog/product/sigma/duo92105 |
| DUO92106 | Duolink® In Situ Orange starter kit Goat/Rabbit | https://www.sigmaaldrich.com/catalog/product/sigma/duo92106 |
Cell preparation
-
1
Plate 50 μl cells per well (1–5×105 cells depending on the cell size) in a ibidi μ-angiogenesis plate.
Ibidi μ-angiogenesis plate is a convenient format to do PLA because it allows to visualize the cells in one focal plane. Furthermore, it makes the assay cost-effective, requiring only 10–15 μl of cell suspension and antibody mixtures per well.If cells need to first be stimulated, the plate can be coated with the antibody or other reagents at the appropriate concentration. Plates can be washed before applying the cells. If stimulation is not needed after applying the cells onto the wells incubate the cells in 5% CO2 incubator for 20 min to attach the cell into the wells. -
2
Aspirate off media and fix cells with 4% PFA in PBS for 20 minutes at room temperature (RT).
A low speed aspirator would work best but pipette aspiration would also work to decant the solution. -
3
Aspirate off PFA and wash twice with PBS.
-
4
Stain cell membrane with 5 μg/ml WGA (Stock conc. 1 mg/ml) in PBS and incubate at RT for 5 min at dark.
From this step, the cells need to operate in dark. Staining with WGA (Wheat germ agglutinin) is optional but better to determine specific dots within the cell. WGA is a lectin and Alexa fluor® 488 conjugated form is widely used to stain plasma membrane to visualize by fluorescent microscopy (excitation/emission maxima~495/519 nm). It is useful to detect number of specific dots and intensity per cell after acquisition of images by image analysis softwares. -
5
Aspirate off PBS and add 50 μl of ice-cold 100% methanol to cells. Incubate at -20 °C for 15–30 minutes to permeabilize.
Although methanol works perfectly to permeabilize many cells/cell lines to stain with different antibody combinations, it can be changed depending on the cells or antibody combinations. -
6
Wash cells twice with PBS.
-
7
Block cells with Duolink® block solution for 1 hr at RT.
Antibody staining
-
8
Dilute primary antibodies in Duolink® antibody dilution buffer and stain overnight at 4°C.
The dilution of the antibody is usually the recommended dilution for immunohistochemistry or immunofluorescence applications. Optional: For negative control, at this point cells can be stained with antibody of different species. For instance, in case of mouse/rabbit pair, cells can be incubated with anti-goat antibody. -
9
Wash twice with large volume of 5% BSA in PBS for 10 minutes each.
-
10
While washing prepare the PLA probes (or antibody) according to the following procedure:
15 μl total volume per reaction: 3 μl PLUS antibody + 3 μl MINUS antibody + 9 μl Duolink® ab dilution buffer.
-
Mix and let stand for 20 min at RT.
It is better to prepare a master mixture for multiple samples and mix well before applying to the wells.
-
11
Add 15 μl of secondary antibody mix to each sample and incubate at 37 °C for 1 hr.
Ligation and Amplification
-
12
Gently aspirate the secondary antibody mix and wash slides in 1× buffer A 2 × 5 min twice for 5 minutes.
-
13
Prepare Ligation mix according to the following procedure:
-
15 μl total volume per reaction: 3 μl (5×) ligation stock + 11.63 μl distilled water + 0.375 μl ligase.
It is better to prepare master mixture for multiple samples and mix well before applying to the wells.
-
-
14
Add 15 μl of ligation mix to each sample and incubate at 37°C for 30 min.
-
15
Wash slides twice with 1× buffer A each for 2 min.
-
15
Prepare amplification mix according to the following procedure:
-
15 μl total volume per reaction: 3 μl (5×) amplification stock + 11.81 μl distilled water + 0.1875 μl ligase.
It is better to prepare a master mixture for multiple samples and mix well before applying to the wells.
-
-
17
Add 15 μl of amplification mix to each sample and incubate for 100 min at 37°C.
-
18
Aspirate off amplification mix and wash twice with 1× buffer B each for 10 min.
-
19
Wash slides once with 0.01× buffer B for 1 min.
-
20
Aspirate buffer B and then mount Prolong Gold mounting medium with DAPI.
The images can be taken instantly, or the plates can be stored at 4°C in the dark until images are taken. The plates should be good for up to 4 days.
Image Analysis
For fluorescence applications, store slides in the dark at 4°C after mounting with Duolink® Mounting Media with DAPI for up to 4 days.
The fluorescent images can be analyzed by Blobfinder (http://www.cb.uu.se/~amin/BlobFinder/) or Image-Pro Premier (http://www.mediacy.com/imagepropremier) program.
Images should be produced with the same acquisition parameters between experimental and control samples.
COMMENTARY
Background Information
Detection of protein-protein interactions is one of the major objectives in studies of cell biology. It has been estimated that over 80% of proteins function in complexes to elicit biological responses (Berggård et al., 2007). Therefore, it is crucial to investigate protein-protein interactions to understand mechanisms underlying cellular functions.
Co-immunoprecipitation (Co-IP) is the gold standard to identify protein-protein interactions, where an antibody to one protein is used to immunoprecipitate it and its partners in a complex, which are typically detect by immunoblotting with antibodies to proteins of interest (Ni et al., 2016; Lee, 2007). However, Co-IP may require large amounts of the proteins of interest, is not well suited to observe interactions in nuclear lysates and reveals protein associations in a complex but not necessarily direct interaction.
Tandem affinity purification (TAP) provides high throughput screening of the binding partners of a particular protein (Gould et al., 2004). In this method, the protein of interest needs to be tagged with an epitope and affinity purification of the complex performed, followed by mass spectrometry (MS). However, the protein purification procedures can detect non-specific proteins that can give false positives. Moreover, the tag can interfere with protein-protein interactions, resulting in failure to detect transient interactions (Collins et al., 2007).
Yeast two-hybrid assays allow detection of interacting proteins based on transcriptional activity (Phizicky and Fields, 1995). The assay is performed with genetically modified yeast strains, in which the interaction of two proteins leads to transcription of a reporter gene. However, this method is prone to false positives, revealing interactions that don’t occur under physiological condition.
FRET (Fluorescence Resonance Energy Transfer) is an advanced technology that analyzes interactions between two fluorescently-tagged proteins, such as cyan fluorescent protein (CFP) and the yellow fluorescent protein (YFP) (Sekar and Periasamy, 2003; Kremers et al., 2006). If the two fluorescent proteins are in close proximity (<10 nm), the absorbed energy of the light-excited CFP transfers to the YFP, which then emits light at a higher wavelength. The major limitation of FRET is the low signal-to-noise ratio associated with imaging. Furthermore, fluorescent proteins are sensitive to changes in the local environment such pH, ion concentrations, oxidation, and temperature (Leavesley and Rich, 2016).
In situ PLA offers an advanced method to detect protein interactions in cells or tissues as long as suitable antibodies are available. The method depends on the recognition of target molecules in close proximity (<40 nm) by pairs of affinity probes, giving rise to an amplifiable detection signal. Because the assay detects the amplified DNA as dots, a few interacting molecules can produce a very strong, robust, and visible signal, making the assay highly sensitive. Duolink® PLA cannot be used for live cells because permeabilization is necessary for the antibodies and probes to find their targets. Furthermore, some PLA reagents like enzymes or DNA oligonucleotides might destroy active mechanisms of cells. However, it provides a highly sensitive and quick assay of protein interactions without any manipulation of cells.
Critical Parameters
Use appropriate primary antibodies with conditions for optimal performance that include sample-processing parameters such as fixation and permeabilization etc.
Include both technical and if possible, positive or negative controls in experiment to properly evaluate the results.
Carry out the incubation steps in a humidity chamber to avoid dryness of the samples.
Completely remove the wash solutions for optimal antibody binding, efficient ligation and amplification.
Perform all steps at the appropriate temperatures and incubation times for best results, in particular the enzymatic steps (ligation and amplification).
For detection of low-abundance proteins, extended amplification times may be required.
Perform washes in ample wash buffer and ensure the samples are fully covered. Perform washes at room temperature.
Keep enzymes on ice while in use. Make sure other reagents are completely thawed (e.g., ligation buffer and amplification buffer) and mixed prior to use.
Troubleshooting
A. High Background Signal
-
Blocking:
Bring the blocking solution to room temperature before applying to the cells.
Ensure entire sample is covered in blocking reagent.
Increase incubation time for blocking.
Dilute the primary antibodies and PLA probes in the antibody diluent, which contain blocking agents optimized for Duolink® PLA.
-
Primary antibodies:
Titrate each primary antibody in different dilution to determine optimum binding without background.
Try an alternative antibody from different clone or vendor
-
Washing:
Increase the number of washes and wash times
Use appropriate wash buffer A and B when specified. If made in the laboratory, filter the buffer with 0.45 μm filter.
Use freshly prepared buffer.
-
Incubation:
Make sure to incubate samples in a good humidity chamber to avoid dryness of the cells.
B. Low or lack of positive signal
-
Cell density:
Perform the PLA in 50–70% confluence, which is optimal for efficient reagent penetration. Over confluent cells give lower signal.
-
Primary antibodies:
Standardize the fixation and permeabilization conditions optimal for primary antibodies.
-
Incubation temperatures:
Perform all steps at the appropriate temperatures, in particular the blocking and the enzymatic steps (ligation and amplification).
-
Use of wash buffers:
Bring wash buffers to room temperature before use.
Use appropriate wash buffers A and B when specified.
After every washing make sure to completely remove the wash buffer before addition of ligation and amplification reagents.
Adjust the amplification time for low-abundance interactions (can go upto overnight at 37°C).
-
Reagent storage and activity:
Make sure that the ligase and polymerase are active and stored at -20°C.
-
Filter used for acquisition:
Make sure to use appropriate filters to acquire the images in microscope depending on the kit used.
C. Poor imaging or signal coalescence
-
Primary Antibodies:
Use standardized concentration of primary antibodies as high concentration cause signal coalescence.
-
Amplification duration:
Follow recommended amplification times as extended amplification duration can cause signal coalescence.
-
Image capture:
Over-exposure during image capture can result signal coalescence. Use appropriate setting during image capture.
Understanding Results
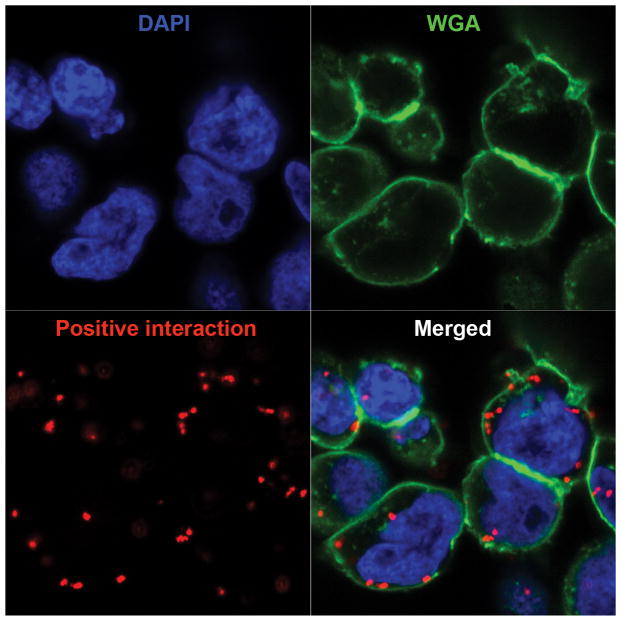
Figure-2 shows a typical result of the PLA assay in Jurkat cells. Jurkat cells were stimulated with anti-CD3/CD28 for 15 min, fixed with 4% PFA followed by permeabilization with methanol. The cells were stained with rabbit anti-human ZAP-70 and mouse anti-human TCR-zeta antibody. Alexa Fluor 488 (green)-conjugated wheat germ agglutinin (WGA) was used to stain plasma membrane and DAPI is for nucleus.
Figure-2. ZAP-70 and TCR zeta interaction in Jurkat cells.
Confocal images of in situ PLA using rabbit anti-ZAP-70 and mouse anti-TCR zeta antibody in Jurkat cells that had been stimulated with anti-CD3/CD28 for 15 min.
Time Considerations
The PLA assay requires ~ 2 days. Once developed the plate can be kept in dark at 4°C for up to 4 days until images collected by the fluorescence microscope.
Significance Statement.
Detection of protein-protein interactions in cells or tissues at endogenous expression levels.
Acknowledgments
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Footnotes
- Different secondary antibody combinations and ordering information: https://www.sigmaaldrich.com/life-science/molecular-biology/molecular-biology-products.html?TablePage=112232138
- Step by step guide to analyse image using Blobfinder. https://www.sciencedirect.com/science/article/pii/S0169260708002071
LITERATURE CITED
- Berggård T, Linse S, James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- Braun P, Gingras AC. History of protein-protein interactions: from egg-white to complex networks. Proteomics. 2012;12:1478–1498. doi: 10.1002/pmic.201100563. [DOI] [PubMed] [Google Scholar]
- Cheng P-C. In: The Contrast Formation in Optical Microscopy Handbook of Biological Confocal Microscopy. Pawley JB, editor. Boston, MA: Springer US; 2006. pp. 162–206. [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Edidin M. Fluorescence resonance energy transfer: techniques for measuring molecular conformation and molecular proximity. Curr Protoc Immunol. 2003;Chapter 18(Unit 18.10) doi: 10.1002/0471142735.im1810s52. [DOI] [PubMed] [Google Scholar]
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Gould KL, Ren L, Feoktistova AS, Jennings JL, Link AJ. Tandem affinity purification and identification of protein complex components. Methods. 2004;33:239–244. doi: 10.1016/j.ymeth.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Gullberg M, Gústafsdóttir SM, Schallmeiner E, Jarvius J, Bjarnegård M, Betsholtz C, Landegren U, Fredriksson S. Cytokine detection by antibody-based proximity ligation. Proc Natl Acad Sci U S A. 2004;101:8420–8424. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers GJ, Goedhart J, van Munster EB, Gadella TW. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry. 2006;45:6570–6580. doi: 10.1021/bi0516273. [DOI] [PubMed] [Google Scholar]
- Leavesley SJ, Rich TC. Overcoming limitations of FRET measurements. Cytometry A. 2016;89:325–327. doi: 10.1002/cyto.a.22851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Coimmunoprecipitation assay. Methods Mol Biol. 2007;362:401–406. doi: 10.1007/978-1-59745-257-1_31. [DOI] [PubMed] [Google Scholar]
- Ni D, Xu P, Gallagher S. Immunoblotting and Immunodetection. Curr Protoc Mol Biol. 2016;114:10.8.1–10.8.37. doi: 10.1002/0471142727.mb1008s114. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar RB, Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol. 2003;160:629–633. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Zheng J. In: Spectroscopy-Based Quantitative Fluorescence Resonance Energy Transfer Analysis Ion Channels: Methods and Protocols. Stockand JD, Shapiro MS, editors. Totowa, NJ: Humana Press; 2006. pp. 65–77. [DOI] [PubMed] [Google Scholar]