Influenza antibodies from hosts with preexisting immunity showed broader cross-reactivity than those without preexisting immunity. Repeated influenza exposures imprinted both antibody quantity and antibody quality. To improve vaccine effectiveness, vaccine strain selection needs to consider the preexisting immunity in humans.
Keywords: seasonal influenza vaccine, vaccine strain selection, antibody cross-reactivity, antibody avidity, repeated influenza exposure
Abstract
Background
Reduced seasonal influenza vaccine effectiveness (VE) was observed in individuals who received repeated annual vaccinations. Preexisting influenza antibody levels were also found inversely correlated with postvaccination titers. These reports suggest that preexisting immunity may affect contemporary seasonal vaccine performance.
Methods
Influenza A/H3 specific cross-reactivity of postvaccination sera from humans with or without preexisting immunity was assessed by hemagglutination inhibition (HAI) assay. Ferret antisera induced by repeated H3 exposures were also subjected to HAI, antibody affinity, and antibody avidity analyses.
Results
Human postvaccination sera derived from subjects with or without preexisting immunity showed different cross-reactivity against H3 variant viruses. Similarly, the breadth of cross-reactive ferret antibodies induced by repeated H3 exposures was also broadened. Antigenic differences between H3 viruses characterized by ferret antisera became smaller as the number of exposures increased. Although repeated H3 exposures induced “original antigenic sin” phenomena in HAI titers against later exposed viruses, resultant ferret antibodies showed gradually enhanced avidity for different H3/hemagglutinin. Increased antibody avidity was found to be inversely correlated with decreased antigenic differences among H3 viruses characterized.
Conclusions
Our results suggest that repeated H3 exposures imprinted not only antibody quantity but also antibody quality. The “naive” ferret model currently used for vaccine strain selection does not recapitulate the complexity of human preexisting immunity. Vaccine strains identified hereby may not provide coverage sufficient for those who were frequently infected and/or vaccinated, leading to the reduced VE observed.
(See the Editorial Commentary by Treanor on pages 1533–4.)
Annual vaccination is the main preventive strategy for control of seasonal influenza, the recommendation of which was first introduced in 1960 for older adults and immunocompromised individuals at high risk for severe influenza-like illness [1]. Since 2010, the annual vaccination policy has been expanded to include all healthy persons aged ≥6 months in the United States [2], and a similar policy has also been adopted by many countries worldwide. Thus, a child born after 2010 would expect to be vaccinated ≥70 times during an average 75-year life expectancy. The annual vaccination policy is made largely due to virus evolution resulting in frequent antigenic drift or shift, which in turn requires seasonal vaccine strains to be updated yearly to match with circulating viruses. Even in seasons without vaccine strain changes, concerns of vaccine-induced immunity waning also necessitate revaccination [2]. This allows individuals with frequent vaccinations to develop influenza antibody repertoires that are readily recalled on later exposure to antigenically similar or related viruses. However, recent epidemiologic studies have reported that lower vaccine effectiveness (VE) was observed in individuals with repeated annual vaccination than those who were not vaccinated in previous season(s) [3–10]. A potential negative effect of prior vaccination was found more pronounced for H3N2-specific VE [3, 4, 6, 9]. These reports suggest that seasonal vaccine-induced protection may be dampened in frequent vaccinees, prompting questions about the benefits of annual vaccination policy.
Annual vaccine strain evaluation and selection organized by the World Health Organization (WHO) is key to controlling seasonal vaccine performance. In this complex process, standard antisera raised in seronegative ferrets infected with representative influenza strains are used to antigenically characterize circulating viruses and play a decisive role in identifying final vaccine strains [11–13]. However, influenza-specific immunity in humans is greatly shaped by previous exposures including natural infections and annual vaccinations, the complexity of which cannot be recapitulated by seronegative ferrets infected with a single influenza strain [12, 14–21]. Antigenic distances derived by a seronegative ferret model have been found to correlate poorly with both H1- and H3-specific VE in humans [20, 22, 23]. This fundamental difference is one of the major factors responsible for the 2014–2015 Northern Hemisphere vaccine strain mismatch and poor vaccine performance [12]. Even in humans, preexisting immunity is also highly variable due to age and infection/vaccination histories [14–19, 24]. In vaccine trials or VE studies, however, the preexisting background of recruited subjects is seldom explicitly determined or analyzed. This is also a problem for annual vaccine strain selection, in which anonymous postvaccination sera provided by courtesy of international government/industry partnerships are used for human serology [12, 18].
In this study, we demonstrated that repeated H3 exposures imprinted both antibody quantity and antibody quality, thus significantly affecting virus antigenic characterization. To improve seasonal vaccine performance, we must take into account of human preexisting immunity during vaccine strain selection.
METHODS
Viruses
All H3N2 viruses were propagated in 9- to 10-day old embryonated eggs, including (1) clade 1 A/Uruguay/716/2007X175C (175C); (2) clade 3C.1 A/Texas/50/2012 (TX/50); (3) clade 3C.2a A/Hong Kong/4801/2014 (HK/4801), A/Singapore/KK934/2014 (SGPKK934), A/Fiji/2/2015 (Fiji/2), A/South Australia/09/2015 (SA/09), A/South Australia/21/2015 (SA/21), A/Victoria/503/2015 (Vic/503), A/Brisbane/47/2015 (Bris/47), and A/Brisbane/82/2015 (Bris/82); (4) clade 3C.3 A/New York/39/2012 (NY/39); (5) clade 3C.3a A/Switzerland/9715293/2013 (SWZ/13), A/North Carolina/13/2014 (NC/13), and A/Palau/6759/2014 (PA/14); (6) clade 3C.3b A/Victoria/511/2015 (Vic/511).
Human Sera
Archived human sera were obtained from healthy subjects administered the 2015–2016 egg-based inactivated, nonadjuvanted, trivalent or quadrivalent influenza vaccines and analyzed anonymously as part of a public health, nonresearch, regulatory activity in support of WHO annual influenza vaccine strain selection, which is exempt from human subjects review. Postvaccination sera from children (8–30 months of age) and adults (28–74 years of age) showing high seroconversion (≥16-fold rise in postvaccination hemagglutination inhibition [HAI] titers against the 2015–2016 H3 vaccine prototype virus– SWZ/13) were selected to assess the HAI cross-reactivity against a panel of previous and recent H3N2 viruses.
Ferret Infection Experiments
Seronegative male ferrets (Triple F Farm) at 15–16 weeks old were anesthetized with ketamine/xylazine mixture followed by intranasal inoculation with H3N2 viruses individually or sequentially at 2-week intervals: (1) HK/4801 only (HK); (2) SWZ/13 followed by HK/4801 (SWZ/HK); (3) TX/50 followed by SWZ/13 and then HK/4801 (TX/SWZ/HK); and (4) 175C followed by TX/50, SWZ/13, and then HK/4801 (175C/TX/SWZ/HK). Blood was collected at 14 days postinfection immediately before each new inoculation. All the procedures were carried out in accordance with the protocol approved by the Institutional Animal Care and Use Committee of the Center for Biologics Evaluation and Research, US Food and Drug Administration.
HAI Assay
The HAI assays were performed using 8 hemagglutinin (HA) units/50 μL of stock viruses and 0.75% guinea pig erythrocytes in the presence of 20 nM oseltamivir for H3N2 viruses, or 0.5% turkey erythrocytes for H1N1 and type B viruses as described previously [12, 18]. Following treatment with receptor-destroying enzyme (Denka-Seiken), prevaccination and postvaccination sera were serially 2-fold diluted for HAI testing. HAI titers represent the reciprocal of the highest serum dilution that yielded a complete HA inhibition. A titer 5 was assigned if no inhibition was observed at the starting 1:10 serum dilution. HAI geometric mean titers (GMTs) were calculated.
Microneutralization Assay
An enzyme-linked immunosorbent assay (ELISA)–based microneutralization (MN) assay was performed as previously described [25, 26]. Receptor-destroying enzyme–treated sera were incubated with one hundred 50% tissue culture infectious dose at 37°C, 5% carbon dioxide for 1 hour, and then were added to Madin-Darby canine kidney–SIAT1 cells (Sigma). After overnight incubation, infected cells were detected using influenza A nucleoprotein-specific monoclonal antibodies (Millipore). MN titers represent the reciprocal of the highest serum dilution resulting in ≥50% neutralization.
Ferret Immunoglobulin G Purification
Immunoglobulin G (IgG) antibodies in ferret antisera were purified using the NAb™ Protein A Plus Spin Kit (ThermoFisher). Total amount of purified ferret IgG was quantitated using reducing agent-compatible Pierce microplate bicinchoninic acid protein assay kit (ThermoFisher).
Anti-HA ELISA and Avidity Assays
Nunc MaxiSorp microtiter plates (ThermoFisher) were coated with 0.2 µg/mL of recombinant HA (Protein Sciences) in pH 9.5 bicarbonate/carbonate coating buffer at 4°C overnight followed by 2 hours’ room temperature blocking in blocking buffer (pH 7.4 phosphate-buffered saline [PBS] containing 1% bovine serum albumin [BSA] and 5% sucrose). Ferret antisera or purified antibodies were serially diluted in assay buffer (pH 7.4 PBS containing 0.1% BSA and 0.05% Tween 20) and incubated on blocked plates at 37°C for 90 minutes. For avidity assays, the plates were washed and overlaid with 100 µL/well of 4 M urea for 15 minutes [27]. After thorough washing, the plates were reblocked in blocking buffer for another hour. Bound antibody was detected using peroxidase-conjugated goat antiferret IgG (Abcam) followed by 1-Step Ultra TMB-ELISA substrate (ThermoFisher). Absorbance values were measured at 450 nm in a Victor V multilabel reader (PerkinElmer). Avidity index was calculated based on the area of the entire antibody titration curve as previously reported [28].
Antigenic Cartography and Correlation Coefficient Analysis
The 2-dimensional antigenic maps with multidimensional scaling were constructed based on human or ferret HAI titers using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap) with each horizontal or vertical gridline representing 1 antigenic unit distance corresponding to a 2-fold difference in HAI titers [12, 29, 30]. The correlation coefficient for any 2 sets of antigenic distances derived from antigenic cartography was also determined with 1 indicating a perfect positive correlation and −1 denoting a perfect negative correlation, respectively [18].
Statistical Analysis
HA-specific ferret IgG binding affinity was analyzed using nonlinear regression curve fit (Prism 6.02, GraphPad). P ˂ .05 determined by 2-way analysis of variance was considered statistically significant.
RESULTS
Preexisting Immunity Affected Human H3-Specific HAI Cross-reactivity
Because of lack of infection/vaccination records, selected pediatric or adult postvaccination sera were grouped as previously reported [12, 14, 15, 18]: (1) undetectable H3-specific preexisting immunity (prevaccination HAI titer of <40 against SWZ/13), and (2) detectable H3-specific preexisting immunity (prevaccination HAI titer of ≥40 against SWZ/13). Compared to the pediatric or adult group with undetectable preexisting immunity, the corresponding age group with detectable preexisting immunity responded more evenly to all H3 viruses tested (Figure 1A and 1B). When these same postvaccination HAI titers were visualized using antigenic cartography (Figure 1C–F), interestingly, the groups with detectable preexisting immunity, regardless of ages, had difficulties in distinguishing different H3 clades as compared to the groups with undetectable preexisting immunity (Figure 1C vs 1D and Figure 1E vs 1F). For instance, clade 3C.2a (green) and clade 3C.3a (red) viruses were well separated in the antigenic map derived from pediatric postvaccination sera with undetectable preexisting immunity, indicative of distinct antigenicity (Figure 1C). However, in the antigenic map derived from pediatric postvaccination sera with detectable preexisting immunity, these 2 clades tended to cluster together and were not distinctly separated (Figure 1D). Similar phenomena were observed in the maps derived from adult postvaccination sera with and without detectable preexisting immunity (Figure 1E vs 1F). In particular, clade 3C.2a and clade 3C.3a viruses became completely indistinguishable in the adult map with detectable preexisting immunity (Figure 1F). The smaller antigenic distances in pediatric or adult map with detectable preexisting immunity indicated smaller antigenic differences among H3 viruses characterized (1.2763 vs 1.4629 and 0.8339 vs 1.0340 in children and adults with undetectable preexisting immunity, respectively; Figure 1G). Correlation coefficient analysis also showed that the maps with detectable preexisting immunity correlated poorly with those with undetectable preexisting immunity (Figure 1G). These results indicated that the postvaccination sera from the subjects with detectable preexisting immunity had different cross-reactivity toward H3N2 variants from those with undetectable preexisting immunity.
Figure 1.
Different antigenic patterns of human postvaccination sera with or without detectable H3-specific preexisting immunity. A and B, Normalized hemagglutination inhibition (HAI) geometric mean titers (GMTs) in pediatric (A) or adult (B) subjects with or without detectable H3-specific preexisting immunity. Postvaccination HAI GMTs were normalized to vaccine strain A/Hong Kong/4801/2014 (HK/4801)-specific GMTs in the same subpopulation without detectable preexisting immunity. C–F, Antigenic maps constructed using postvaccination HAI titers from the same pediatric (C and D) or adult (E and F) subjects with or without detectable H3-specific preexisting immunity. Each gridline (horizontal and vertical) in the antigenic maps represents 1 antigenic unit distance corresponding to a 2-fold difference in HAI titers. G, Antigenic distances (average ± standard error of the mean [SEM]) of H3 viruses determined in each antigenic map and correlation analysis within the same population. Correlation coefficients of 1, −1, and 0 represent a perfect positive correlation, a perfect negative correlation, and no correlation, respectively. Both pediatric and adult participants were vaccinated with the 2015/16 egg-based inactivated, nonadjuvanted, trivalent or quadrivalent vaccines. Egg-grown H3 viruses in the testing panel included clade 1, A/Uruguay/716/2007X175C (175C); clade 3C.1, A/Texas/50/2012 (TX/50); clade 3C.2a, HK/4801, A/Singapore/KK934/2014 (SGPKK934), A/Fiji/2/2015 (Fiji/2), A/South Australia/09/2015 (SA/09), A/South Australia/21/2015 (SA/21), A/Victoria/503/2015 (Vic/503), A/Brisbane/47/2015 (Bris/47), and A/Brisbane/82/2015 (Bris/82); clade 3C.3, A/New York/39/2012 (NY/39); clade 3C.3a, A/Switzerland/9715293/2013 (SWZ/13), A/North Carolina/13/2014 (NC/13), and A/Palau/6759/2014 (PA/14); clade 3C.3b, A/Victoria/511/2015 (Vic/511).
Prior H3 Exposures Affected Both Quantity and Quality of Ferret Antibodies
We then investigated how prior H3 exposures may affect antibody development in a ferret model. Seronegative ferrets were either exposed to a single HK/4801 (clade 3C.2a) infection or sequentially infected with antigenically drifted 175C (clade 1), TX/50 (clade 3C.1), or SWZ/13 (clade 3C.3a) followed by HK/4801. As expected, ferret antisera raised from single HK/4801 infection showed much higher HAI titers against the homologous virus (GMT = 557) than drifted H3N2 variants (Figure 2A and Supplementary Table 1). In contrast, ferret antisera derived from sequential H3N2 infections showed better HAI responses toward first encountered H3N2 virus than later exposed HK/4801 (Figure 2A–D). For instance, ferret 175C/TX/SWZ/HK antisera exhibited higher HAI response toward the first exposed 175C virus (GMT = 226), but much lower titers against later infected HK/4801 (GMT = 80) (Figure 2D), indicating a typical “original antigenic sin” (OAS) [31]. Yet the OAS phenomena were less pronounced in ferret MN titers against the same set of viruses (Supplementary Figure 1).
Figure 2.
H3 virus-specific hemagglutination inhibition (HAI) responses in ferrets with different exposure histories. Sera from ferrets infected with A/Hong Kong/4801/2014 (HK/4801) only (HK; A), or A/Switzerland/9715293/2013 (SWZ/13) followed by HK/4801 (SWZ/HK; B), or A/Texas/50/2012 (TX/50) followed by SWZ/13 and then HK/4801 (TX/SWZ/HK; C), or A/Uruguay/716/2007X175C (175C) followed by TX/50, SWZ/13, and then HK/4801 (175C/TX/SWZ/HK; D) were assessed for virus-specific HAI titers. Individual ferret titers with geometric mean titers (GMTs) (bar graphs) are shown.
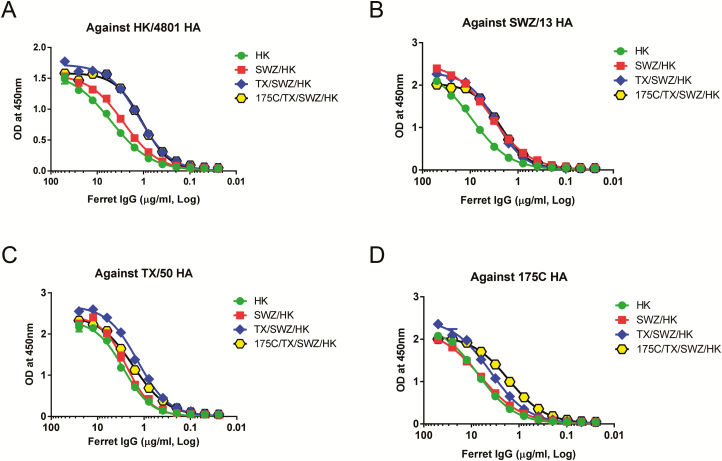
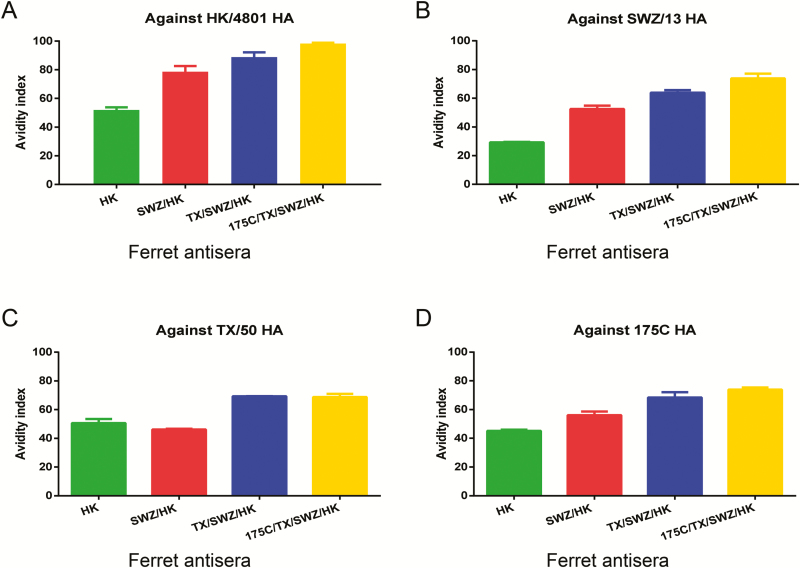
Despite lower HAI titers against later exposed viruses, ferret IgG purified from antisera with repeated H3 exposures in general exhibited enhanced binding affinity for both early and late exposed HA antigens as compared to ferret IgG elicited by single HK/4801 infection (Figure 3A–D). Using urea as the chaotrope, we then measured antibody avidity which detects the total strength of multivalent interactions between antibody and antigen, instead of a specific interaction between one antigenic epitope and one antibody binding site detected by antibody affinity. As shown in Figure 4, antibodies induced by single HK/4801 infection had the lowest avidity index not only against the homologous HA but also against drifted H3 HA. In contrast, repeated H3 infections gradually enhanced the avidity of the resulting ferret antibodies; for example, ferret 175C/TX/SWZ/HK antisera had the highest avidity index against all 4 H3 HA antigens tested (Figure 4A–D). Apparently, the more prior H3 exposures, the higher H3-specific antibody avidity.
Figure 3.
H3-specific immunoglobulin G (IgG) affinity in ferrets with different exposure histories. Sera were collected from ferrets infected with HK (A), SWZ/HK (B), TX/SWZ/HK (C), or 175C/TX/SWZ/HK (D). Purified ferret IgG was assessed and analyzed for hemagglutinin-specific binding affinity using nonlinear regression (curve fit). Please see the Figure 2 legend for descriptions of influenza virus strains. Abbreviations: HA, hemagglutinin; IgG, immunoglobulin G; OD, optical density.
Figure 4.
H3-specific immunoglobulin G (IgG) avidity in ferrets with different exposure histories. Sera were collected from ferrets infected with HK (A), SWZ/HK (B), TX/SWZ/HK (C), or 175C/TX/SWZ/HK (D). Hemagglutinin-specific ferret IgG avidity was determined in the presence of 4M urea. Please see the Figure 2 legend for descriptions of influenza virus strains.
These results suggest that prior H3 exposures not only affect the quantity but also the quality of ferret HA-specific antibodies, and the impact seems to depend on the number of previous exposures.
Repeated Prior H3 Exposures Expanded Ferret HAI Cross-reactivity
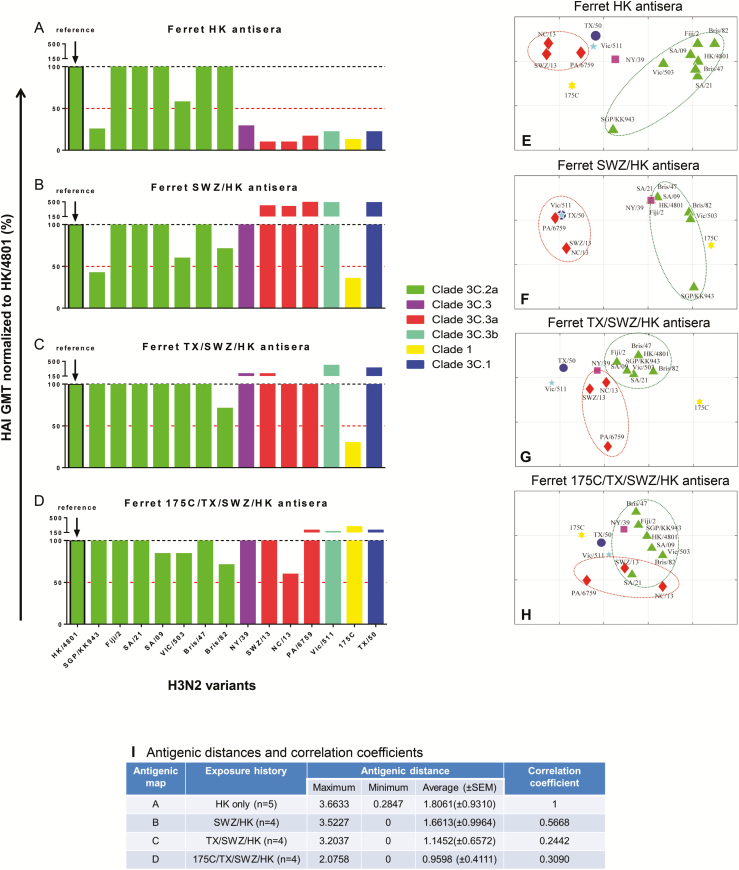
As expected, ferret antisera raised from single infection mainly cross-reacted with homologous viruses (Figure 5A and Supplementary Figure 2). When ferrets were sequentially infected with SWZ/13 followed by HK/4801, the resulting SWZ/HK antisera had HAI cross-reactivity extend to clade 3C.1, 3C.3, 3C.3a, and 3C.3b viruses by showing >50% rise in HK/4801-specific HAI GMT (Figure 5B). Similarly, TX/SWZ/HK antisera induced by sequential TX/50, SWZ/13, and HK/4801 exposures broadened HAI cross-reactivity to clade 3C.1, 3C.3, 3C.3a, and 3C.3b viruses, including the 2 clade 3C.2a viruses, SGP/KK943 and VIC/503 (Figure 5C), in which HK and SWZ/HK antisera showed ≥50% reduction in HAI GMT (Figure 5A and 5B). When ferrets had been sequentially exposed to 175C, TX/50, and SWZ/13 followed by HK/4801, the resulting 175C/TX/SWZ/HK antisera expanded the HAI cross-reactivity to all 6 H3N2 clades tested including clade 1 virus (Figure 5D).
Figure 5.
H3 antigenic patterns determined by ferret antisera with different exposure histories. A–D, Normalized H3-specific ferret cross-reactive hemagglutination inhibition (HAI) geometric mean titers (GMTs). Egg-grown H3 viruses in the testing panel included clade 1, A/Uruguay/716/2007X175C (175C); clade 3C.1, A/Texas/50/2012 (TX/50); clade 3C.2a, A/Hong Kong/4801/2014 (HK/4801), A/Singapore/KK934/2014 (SGPKK934), A/Fiji/2/2015 (Fiji/2), A/South Australia/09/2015 (SA/09), A/South Australia/21/2015 (SA/21), A/Victoria/503/2015 (Vic/503), A/Brisbane/47/2015 (Bris/47), and A/Brisbane/82/2015 (Bris/82); clade 3C.3, A/New York/39/2012 (NY/39); clade 3C.3a, A/Switzerland/9715293/2013 (SWZ/13), A/North Carolina/13/2014 (NC/13), and A/Palau/6759/2014 (PA/14); clade 3C.3b, A/Victoria/511/2015 (Vic/511). Sera were collected from ferrets infected with HK (A), SWZ/HK (B), TX/SWZ/HK (C), or 175C/TX/SWZ/HK (D). (Please see the Figure 2 legend for descriptions of influenza virus strains.) Ferret HAI GMTs specific for individual H3 viruses are normalized to HK/4801-specific GMT with the red dotted lines indicating 50% response. E–H, Antigenic maps constructed using H3-specific HAI titers of ferret antisera. Each gridline (horizontal and vertical) in the antigenic maps represents 1 antigenic unit distance corresponding to a 2-fold difference in HAI titers. I, Antigenic distances (with standard error of the mean [SEM]) of H3 viruses determined in each ferret antigenic map and correlation analysis relative to ferret HK antisera-derived antigenic map. Correlation coefficients of 1, −1, and 0 represent a perfect positive correlation, a perfect negative correlation, and no correlation, respectively.
When these same ferret HAI titers were plotted using antigenic cartography, it became obvious that different H3N2 clades were less distinguishable by antisera raised from ferrets with increased exposure histories (Figure 5E–H). For example, clade 3C.2a (green) and clade 3C.3a (red) were well separated in the map derived from ferret antisera raised after single HK/4801 infection (Figure 5E). However, the antigenic differences between clade 3C.2a and clade 3C.3a viruses became smaller in the map derived from the TX/SWZ/HK antisera (Figure 5G). Eventually clade 3C.2a and 3C.3a groups clustered together and became antigenically indistinguishable as determined by the 175C/TX/SWZ/HK antisera (Figure 5H). The average antigenic distances in individual ferret maps decreased when the number of previous H3N2 infections increased (Figure 5I).
The correlation analysis showed that the antigenic maps derived by ferret antisera with previous H3N2 infections correlated poorly with the map derived by ferret antisera with single HK/4801 infection: more prior H3N2 exposures, smaller correlation coefficients (Figure 5I).
Ferret Antibody Avidities Inversely Correlated With Antigenic Distances of H3N2 Viruses Characterized
We then correlated the avidity indexes of individual ferret antisera in Figure 4 with the average antigenic distance derived by corresponding ferret antisera in Figure 5I. It showed that reduced average antigenic distances inversely correlated with increased antibody avidities in ferrets with multiple prior H3N2 exposures (R2 = 0.5765, P = .0006): higher antibody avidity and shorter antigenic distances (Figure 6). These results suggest that repeated H3N2 exposures enhance antibody avidity, thus affecting virus antigenic characterization.
Figure 6.
Correlation between immunoglobulin G (IgG) avidities and antigenic distances determined by ferret antisera with different exposure histories. H3-specific IgG avidities of ferret antisera determined in Figure 4 were correlated with average antigenic distances determined by corresponding ferret antisera in Figure 5I using linear regression. Please see the Figure 2 legend for descriptions of influenza virus strains.
DISCUSSION
Current vaccine strain selection uses seronegative ferret model without influenza-specific preexisting immunity to detect epidemic viruses that are antigenically different from vaccine strains [11–13]. However, accumulated evidence indicates that early life exposure to influenza can leave an imprint on human antibody repertoires, and resulted residual protection may last a lifetime [15, 16, 21, 31–35]. Intensified global connectivity helps to spread antigenically drifted influenza strains [36]. Repeated annual vaccination also contributes to widespread influenza preexisting immunity in humans. Virtually all humans have been exposed—asymptomatically or symptomatically—to influenza, and there exist no such “influenza-naive” persons except newborns. As shown in this study, the H3 viruses representing different genetic clades were “seen” as antigenically related by human postvaccination sera with detectable H3 preexisting immunity, rather than antigenically distinguishable by those with undetectable H3 preexisting immunity. This difference is not due to poor vaccination response as all human postvaccination sera used in this study had ≥16-fold seroconversion after seasonal vaccination. Rather, our results confirm that the background of previous exposures—whether by natural infections or annual vaccinations—can significantly change human postvaccination antibody cross-reactivity [18].
Similarly, preexisting immunity also can influence virus antigenic characterizations in ferrets. Clade 3C.2a and clade 3C.3a viruses were antigenically well separated by ferret antisera stimulated by single H3 infection, whereas their antigenic difference diminished as determined by ferret antisera with increased prior H3 exposures. This is because repeated H3 infections extended the cross-reactivity of resultant ferret antibodies from clade 1 to clade 3C.X, in contrast to strain-specific antibodies elicited by single H3 infection. Consequently, clade 3C.2a and clade 3C.3a viruses were no longer “seen” antigenically distinct by ferret antisera with repeated H3 exposures.
The broadened breath of ferret antibody cross-reactivity apparently occurred at the expense of absolute HAI titers against more recent viruses—an OAS phenomenon that has traditionally been considered detrimental to contemporary vaccine performance [31–34]. Human preexisting influenza-specific antibody levels have been found to inversely correlate with postvaccination titers [35–37]. In individuals with high prevaccination antibody titers, vaccination is found to primarily boost preexisting influenza-specific repertoires rather than to induce de novo antibody clonotypes [37]. Many preexisting antibodies are cross-reactive and can help to restrict subsequent infections caused by antigenic variants of influenza [37, 38]. Correspondingly, a suppressed de novo antibody response specific for later exposed virus may occur. In this study, we observed that repeated H3 infections induced more pronounced OAS phenomena in ferret HAI titers than in ferret MN titers. This could be because some preexisting antibodies with broad cross-reactivity have no HAI activity [37].
Our current study also suggests that repeated H3 exposures resulted in improved quality of recall antibodies, which might be partially via OAS induction. Antibodies from ferrets primed with 175C may target HA antigenic sites that are also conserved by later H3 strains. When the same ferrets were repeatedly infected with evolved but still antigenically related TX/50, SWZ/13, and HK/4801, preexisting cross-reactive antibodies responded to limit viral infections, leading to reduced HAI titers against recall viruses. This may also give opportunities to B cells specific for other antigenic sites to expand [39]. Hence, the resulting pool of ferret antibodies had diversified specificities against a wide range of H3 viruses. Because individual viruses have different antigenic sites to play a dominant role in immune system [40], repeated exposures to different H3 viruses may not necessarily increase the binding affinity of resultant antibodies for a specific antigenic determinant. However, their avidity—the multivalent binding capacity for all antigenic sites—was enhanced along with increased virus encounters, thus resulting in less distinguishable antigenic differences between H3 variants characterized.
Taken together, our study shows that repeated influenza exposures imprinted not only antibody quantity but also antibody quality. The naive ferret model currently used for vaccine strain selection does not reflect the complexity of human exposure history. Whether viruses appear antigenically identical or drift to naive ferrets may not hold true for individuals with frequent exposures. This could lead to misidentifying a strain that indeed is antigenic drift to repeatedly vaccinated individuals [20]. Vaccine strains selected hereby may not provide coverage sufficient for those with frequent vaccinations/infections, resulting in the compromised VE observed. To improve seasonal vaccine performance, we should take the widespread human preexisting immunity into consideration during vaccine strain selection, and select vaccine strains that are optimal for populations with different immune backgrounds.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. H. X. conceived the ideas and designed the study. M. K., P. R., and H. X. conducted the ferret infection experiments. M. K., Z. Y., and H. X. performed the HAI assays. M. K. conducted antibody quality experiments. L. L. and X.-F. W. generated the antigenic maps and did correlation coefficient analyses. H. X. analyzed the data and performed the statistical analyses. H. X. and M. K. wrote the manuscript.
Acknowledgments. The authors sincerely appreciate Dr Xiyan Xu of the US Centers for Diseases Control and Prevention for providing influenza viruses.
Disclaimer. The findings and conclusions in this article have not been formally disseminated by the US Food and Drug Administration (FDA) and should not be construed to represent any agency determination or policy.
Financial support. This project was supported by the intramural research fund of the Center for Biologics Evaluation and Research, FDA. X.-F. W. and L. L. were supported by the National Institutes of Health (grant number R01AI116744 to X.-F. W.).
Potential conflicts of interest. All authors: No potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Burney LE. Influenza immunization: statement. Public Health Rep 1960; 75:944. [PMC free article] [PubMed] [Google Scholar]
- 2. Fiore AE, Uyeki TM, Broder K, et al. ; Centers for Disease Control and Prevention (CDC) Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Recomm Rep 2010; 59:1–62. [PubMed] [Google Scholar]
- 3. McLean HQ, Thompson MG, Sundaram ME, et al. Impact of repeated vaccination on vaccine effectiveness against influenza A(H3N2) and B during 8 seasons. Clin Infect Dis 2014; 59:1375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohmit SE, Thompson MG, Petrie JG, et al. Influenza vaccine effectiveness in the 2011–2012 season: protection against each circulating virus and the effect of prior vaccination on estimates. Clin Infect Dis 2014; 58:319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ohmit SE, Petrie JG, Malosh RE, Fry AM, Thompson MG, Monto AS. Influenza vaccine effectiveness in households with children during the 2012–2013 season: assessments of prior vaccination and serologic susceptibility. J Infect Dis 2015; 211:1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Skowronski DM, Chambers C, Sabaiduc S, et al. A perfect storm: impact of genomic variation and serial vaccination on low influenza vaccine effectiveness during the 2014–2015 season. Clin Infect Dis 2016; 63:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rondy M, Launay O, Castilla J, et al. ; InNHOVE/I-MOVE+Working Group Repeated seasonal influenza vaccination among elderly in Europe: effects on laboratory confirmed hospitalised influenza. Vaccine 2017; 35:4298–306. [DOI] [PubMed] [Google Scholar]
- 8. Saito N, Komori K, Suzuki M, et al. Negative impact of prior influenza vaccination on current influenza vaccination among people infected and not infected in prior season: a test-negative case-control study in Japan. Vaccine 2017; 35:687–93. [DOI] [PubMed] [Google Scholar]
- 9. Belongia EA, Skowronski DM, McLean HQ, Chambers C, Sundaram ME, De Serres G. Repeated annual influenza vaccination and vaccine effectiveness: review of evidence. Expert Rev Vaccines 2017; 16:1–14. [DOI] [PubMed] [Google Scholar]
- 10. Ferdinands JM, Fry AM, Reynolds S, et al. Intraseason waning of influenza vaccine protection: evidence from the US Influenza Vaccine Effectiveness Network, 2011-12 through 2014-15. Clin Infect Dis 2017; 64:544–50. [DOI] [PubMed] [Google Scholar]
- 11. Russell CA, Jones TC, Barr IG, et al. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine 2008; 26:D31–4. [DOI] [PubMed] [Google Scholar]
- 12. Xie H, Wan XF, Ye Z, et al. H3N2 mismatch of 2014-15 Northern Hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Sci Rep 2015; 5:15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stöhr K, Bucher D, Colgate T, Wood J. Influenza virus surveillance, vaccine strain selection, and manufacture. Methods Mol Biol 2012; 865:147–62. [DOI] [PubMed] [Google Scholar]
- 14. Xie H, Jing X, Li X, et al. Immunogenicity and cross-reactivity of 2009–2010 inactivated seasonal influenza vaccine in US adults and elderly. PLoS One 2011; 6:e16650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xie H, Li X, Gao J, et al. Revisiting the 1976 “swine flu” vaccine clinical trials: cross-reactive hemagglutinin and neuraminidase antibodies and their role in protection against the 2009 H1N1 pandemic virus in mice. Clin Infect Dis 2011; 53:1179–87. [DOI] [PubMed] [Google Scholar]
- 16. Li Y, Myers JL, Bostick DL, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med 2013; 210:1493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hensley SE. Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Curr Opin Virol 2014; 8:85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie H, Li L, Ye Z, et al. Differential effects of prior influenza exposures on H3N2 cross-reactivity of human postvaccination sera. Clin Infect Dis 2017; 65:259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cobey S, Hensley SE. Immune history and influenza virus susceptibility. Curr Opin Virol 2017; 22:105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gupta V, Earl DJ, Deem MW. Quantifying influenza vaccine efficacy and antigenic distance. Vaccine 2006; 24:3881–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Linderman SL, Chambers BS, Zost SJ, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc Natl Acad Sci U S A 2014; 111:15798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X, Deem MW. Influenza evolution and H3N2 vaccine effectiveness, with application to the 2014/2015 season. Protein Eng Des Sel 2016; 29:309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pan K, Subieta KC, Deem MW. A novel sequence-based antigenic distance measure for H1N1, with application to vaccine effectiveness and the selection of vaccine strains. Protein Eng Des Sel 2011; 24:291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herati RS, Muselman A, Vella L, et al. Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells. Sci Immunol 2017; 2. doi:10.1126/sciimmunol.aag2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levine MZ, Martin JM, Gross FL, et al. Neutralizing antibody responses to antigenically drifted influenza A(H3N2) viruses among children and adolescents following 2014–2015 inactivated and live attenuated influenza vaccination. Clin Vaccine Immunol 2016; 23:831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. World Health Organization Global Influenza Surveillance Network. Serological diagnosis of influenza by haemagglutination inhibition testing. Available at: http://www.who.int/influenza/gisrs_laboratory/manual_diagnosis_surveillance_ influenza/en/. Accessed 20 February 2018. [Google Scholar]
- 27. Klinman DM, Xie H, Little SF, Currie D, Ivins BE. CpG oligonucleotides improve the protective immune response induced by the anthrax vaccination of rhesus macaques. Vaccine 2004; 22:2881–6. [DOI] [PubMed] [Google Scholar]
- 28. Perciani CT, Peixoto PS, Dias WO, Kubrusly FS, Tanizaki MM. Improved method to calculate the antibody avidity index. J Clin Lab Anal 2007; 21:201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cai Z, Zhang T, Wan XF. A computational framework for influenza antigenic cartography. PLoS Comput Biol 2010; 6:e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cai Z, Zhang T, Wan XF. Antigenic distance measurements for seasonal influenza vaccine selection. Vaccine 2012; 30:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Francis T., Jr On the doctrine of original antigenic sin. Proc Am Philos Soc 1960; 104:572–8. [Google Scholar]
- 32. Kim JH, Skountzou I, Compans R, Jacob J. Original antigenic sin responses to influenza viruses. J Immunol 2009; 183:3294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choi YS, Baek YH, Kang W, et al. Reduced antibody responses to the pandemic (H1N1) 2009 vaccine after recent seasonal influenza vaccination. Clin Vaccine Immunol 2011; 18:1519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monto AS, Malosh RE, Petrie JG, Martin ET. The doctrine of original antigenic sin: separating good from evil. J Infect Dis 2017; 215:1782–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sasaki S, He XS, Holmes TH, et al. Influence of prior influenza vaccination on antibody and B-cell responses. PLoS One 2008; 3:e2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andrews SF, Huang Y, Kaur K, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med 2015; 7:316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J, Boutz DR, Chromikova V, et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med 2016; 22:1456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Linderman SL, Hensley SE. Antibodies with ‘original antigenic sin’ properties are valuable components of secondary immune responses to influenza viruses. PLoS Pathog 2016; 12:e1005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Angeletti D, Gibbs JS, Angel M, et al. Defining B cell immunodominance to viruses. Nat Immunol 2017; 18:456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muñoz ET, Deem MW. Epitope analysis for influenza vaccine design. Vaccine 2005; 23:1144–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.