Inflammatory responses require mobilization of innate immune cells from the bone marrow. The functionality of this process depends on the state of the bone marrow microenvironment.
KEYWORDS: Fra-2/AP-1, osteopontin, bone niche, systemic inflammation, lung injury, Fra-2, osteoblasts
ABSTRACT
Inflammatory responses require mobilization of innate immune cells from the bone marrow. The functionality of this process depends on the state of the bone marrow microenvironment. We therefore hypothesized that molecular changes in osteoblasts, which are essential stromal cells of the bone marrow microenvironment, influence the inflammatory response. Here, we show that osteoblast-specific expression of the AP-1 transcription factor Fra-2 (Fra-2Ob-tet) induced a systemic inflammatory state with infiltration of neutrophils and proinflammatory macrophages into the spleen and liver as well as increased levels of proinflammatory cytokines, such as interleukin-1β (IL-1β), IL-6, and granulocyte-macrophage colony-stimulating factor (GM-CSF). By in vivo inhibition of osteopontin (OPN) in Fra-2Ob-tet mice, we demonstrated that this process was dependent on OPN expression, which mediates alterations of the bone marrow niche. OPN expression was transcriptionally enhanced by Fra-2 and stimulated mesenchymal stem cell (MSC) expansion. Furthermore, in a murine lung injury model, Fra-2Ob-tet mice showed increased inflammatory responses and more severe disease features via an enhanced and sustained inflammatory response to lipopolysaccharide (LPS). Our findings demonstrate for the first time that molecular changes in osteoblasts influence the susceptibility to inflammation by altering evasion of innate immune cells from the bone marrow space.
INTRODUCTION
Bone is an endocrine organ that is able to regulate mineral metabolism, glucose metabolism, and immune activation (1, 2). Of interest is that the stromal cells of the bone interact with the immune system, which is mediated by several soluble factors and cell-cell contacts (3, 4). It is known that bone marrow stromal cells control the state and function of hematopoietic stem cells (HSCs) and hematopoietic stem progenitor cells (HSPCs) (5), thereby affecting immune cell development. In particular, mesenchymal stem cells (MSCs) in the bone marrow were found to affect the maintenance of various lymphocytes (3, 6), which in turn control MSC mobilization from the bone marrow to distant locations, contributing to tissue repair (7). In addition, abundant studies also indicate that MSCs affect the activation of innate immune cells such as macrophages, monocytes, and dendritic cells (DCs) both in vivo and in vitro (6, 8). These observations suggest that molecular changes in the stromal cell compartment of bone may affect susceptibility to inflammation. The mechanism by which stromal cells may influence inflammation, however, remains incompletely described to date. Fra-2, a Fos member of the AP-1 transcription factor family, is an attractive candidate linking bone physiology to inflammation. Constitutive Fra-2 overexpression was linked to fibrosis and inflammation in skin and lung (9, 10). Moreover, Fra-2 is a master regulator of bone homeostasis regulating osteoclasts and osteoblasts (11, 12). Importantly, Fra-2 controls osteoblast differentiation and activity by transcriptional regulation of type 1 collagen alpha 2 (COL1A2) and osteocalcin (OCN) gene expression (12). Fra-2 expression in osteoblasts could also regulate glucose metabolism via an adiponectin- and OCN-dependent mechanism, linking bone physiology to metabolism (2) Considering the intense relationship between glucose metabolism and immune cell activation (2, 13, 14), we hypothesized that Fra-2 expression in osteoblasts might also influence inflammatory responses.
For instance, stromal cell-derived mediators may be instrumental in inducing proinflammatory changes in the immune system. Osteopontin (OPN), for instance, is a cytokine that influences both the immune response and bone remodelling (15–17). In bone marrow, OPN can be expressed by stromal cells and is recognized as a negative regulator of HSC homing and proliferation (18, 19). Additionally, OPN promotes MSC differentiation into osteoblasts via its interaction with integrin (20). Functionally, OPN was shown to stimulate MSC migration and attachment to fracture sites in vivo (21). Furthermore, OPN induces monocyte/macrophage chemotaxis, spreading, and activation (22, 23). Mice with OPN deficiency display reduced neutrophil recruitment and migration (24). Physiologically, it has been shown that OPN neutralization attenuates a variety of inflammation-related disorders such as sepsis-induced acute lung injury (25), rheumatoid arthritis (26), and obesity-induced inflammation (27).
In this study, we show that specific expression of Fra-2 in osteoblasts (Fra-2Ob-tet) induces an inflammatory state by a profound upregulation of OPN. Furthermore, we show the clinical relevance of this process in a lipopolysaccharide (LPS)-induced lung injury model. Fra-2 expression in osteoblasts exacerbated lung injury via an enhanced and sustained inflammatory response to LPS.
RESULTS
Fra-2 expression in osteoblasts leads to MSC expansion and molecular changes in the bone marrow niche.
Fra-2 was previously shown to be essential for osteoblast differentiation and activity. Therefore, we hypothesized that overexpression of Fra-2 in osteoblasts also regulates osteoprogenitor cells such as mesenchymal stem cells (MSCs) and thereby alters the hematopoietic niche in the bone marrow. To test this hypothesis, bone marrow of mice expressing Fra-2 under the control of the osterix promoter (Fra-2Ob-tet) was analyzed at 10 weeks of age. These mutant mice were previously shown to overexpress specifically Fra-2 in the osteoblastic lineages (2). When Fra-2 expression was assessed in different tissues, including the fat, liver, lung, spleen, brain, bone marrow, and long bones, from wild-type and Fra-2Ob-tet mice, we could confirm the specifically increased expression of Fra-2 in bone and bone marrow from Fra-2Ob-tet mice (Fig. 1A). Moreover, Fra-2 expression was increased only in osteoblasts differentiated from Fra-2Ob-tet mice MSC and not in the precursor cells or in adipocytes differentiated from MSCs (Fig. 1A). When assessing MSCs, identified as CD45− Ter119− Sca-1+cells by flow cytometry, a significant increase in bone marrow from Fra-2Ob-tet mice compared to that from littermate controls was observed (Fig. 1B). In accordance, expression of the kit ligand (kit-l) gene, a marker of MSCs, was significantly increased in bone marrow from Fra-2Ob-tet mice (Fig. 1D). Conversely, the expression of sdf-1, a factor critically involved in regulating the HSC niche (5), was reduced in the bone marrow of Fra-2Ob-tet mice, while no difference in expression was found in bone tissue deprived of bone marrow (Fig. 1C and D and data not shown). Similarly, tie-2 and jag-1 expression was reduced in Fra-2Ob-tet bone marrow (Fig. 1D), while mRNA levels of other bone marrow niche markers, such as vegfr2/3, ang-1, cxcr4, il-7, vcam-1, Notch-1, and HES-1, were comparable in mutant and control mice (Fig. 1D). These data suggest that Fra-2 expression in osteoblasts induces MSC expansion and molecular changes in the bone marrow.
FIG 1.
Fra-2 expression in osteoblasts induces mesenchymal stem cell alterations. (A) Quantitative PCR analyses of Fra-2 in fat pads, livers, lungs, spleens, brains, bone marrow, and long bones isolated from Fra-2Ob-tet and littermate mice (n = 6), as well as in osteoblast precursors (OBP)-, osteoblast (Ob)-, and adipocyte (Adip)-derived MSCs from Fra-2Ob-tet and littermate mice (n = 4). (B) FACS quantification of mesenchymal stem cells (Ter119− CD45− Sca-1+) in bone marrow from 10-week-old Fra-2Ob-tet and littermate mice (n = 8). (C) Immunofluorescence staining for SDF-1 in the tibiae of 10-week-old Fra-2Ob-tet and littermate mice (magnification, ×100). Arrows indicate SDF-1+ cells. (D) Real-time PCR analyses of bone niche genes in flushed bone marrow from 10-week-old Fra-2Ob-tet and littermate mice (n = 6). Significant differences are indicated as follows: *, P < 0.05; **, P < 0.01.
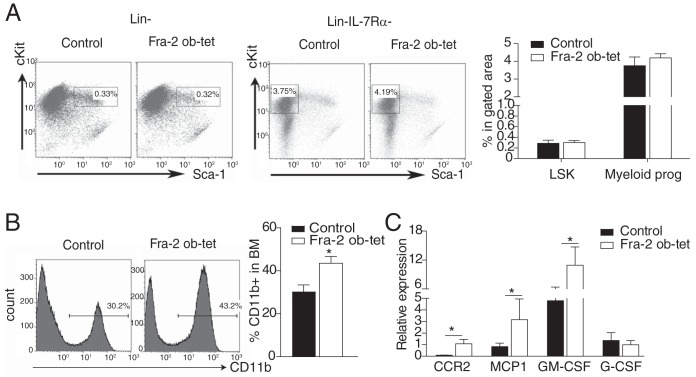
Next, we examined whether hematopoietic stem cells (HSCs) and HSC subpopulations were affected in Fra-2Ob-tet mice. The relative and absolute numbers of HSCs, defined as Lin− Sca-1+ c-Kit+ (LSK) cells, were not significantly altered in Fra-2Ob-tet mice (Fig. 2A and Table 1). Moreover, the myeloid progenitor populations were also comparable in the bone marrow of the two groups (Fig. 2A and Table 1). Accordingly, there was no significant difference in the percentage or absolute number of common lymphoid progenitors (CLP) between mutant and control bone marrow (Table 1 and data not shown), suggesting that the reduced bone marrow space in Fra-2Ob-tet mice (2) was not sufficient to affect the early hematopoiesis process. However, we found an increase in CD11b+ monocytes in the bone marrow of Fra-2Ob-tet mice (Fig. 2B) together with increased mRNA expression of the ccr2, mcp-1, and granulocyte-macrophage colony-stimulating factor (GM-CSF) genes in Fra-2Ob-tet mice (Fig. 2C), which could reflect a proinflammatory milieu in the bone marrow space.
FIG 2.
Fra-2 expression in osteoblasts induces myeloid cell alterations. (A) FACS dot plots and quantification of hematopoietic stem cells (LSK, Lin− Sca1+ c-Kit+) and myeloid progenitor cells (Lin− IL-7Rα− Sca-1− c-Kit+) in bone marrow from 10-week-old Fra-2Ob-tet and littermate mice (n = 6 to 8). (B) FACS dot plots and quantification of CD11b+ cells in bone marrow from 10-week-old Fra-2Ob-tet and littermate mice (n = 8). (C) Real-time PCR analyses of the ccr2, mcp-1, GM-CSF, and G-CSF genes in bone marrow from 10-week-old Fra-2Ob-tet and littermate mice (n = 10). Data represent the mean values ± SEM. *, P < 0.05 (unpaired t test).
TABLE 1.
Absolute numbers of HSCs and myeloid cells in the bone marrow and spleens from wild-type and Fra-2Ob-tet mice
| Cell type (unit) | No. (mean ± SEM) in: |
P valuea | |
|---|---|---|---|
| Control mice (n = 4) | Fra-2Ob-tet mice (n = 6) | ||
| LSK cells (×104/whole bone) | 5.03 ± 0.66 | 3.64 ± 0.33 | 0.13 |
| Myeloid progenitors (×105/whole bone) | 1.09 ± 0.08 | 1.98 ± 0.37 | 0.07 |
| Common lymphoid progenitors (×104/whole bone) | 0.19 ± 0.02 | 0.14 ± 0.03 | 0.46 |
| Neutrophils (×106/spleen) | 1.99 ± 0.30 | 2.71 ± 0.15 | 0.04* |
| Eosinophils (×106/spleen) | 0.98 ± 0.23 | 1.35 ± 0.28 | 0.21 |
| Macrophages (×106/spleen) | 1.27 ± 0.21 | 1.58 ± 0.13 | 0.26 |
| MI macrophages (×106/spleen) | 0.94 ± 0.12 | 1.63 ± 0.19 | 0.04* |
| MII macrophages (×105/spleen) | 3.06 ± 0.76 | 2.27 ± 0.35 | 0.44 |
*, P < 0.05.
Osteoblast-specific Fra-2 expression induces systemic inflammation.
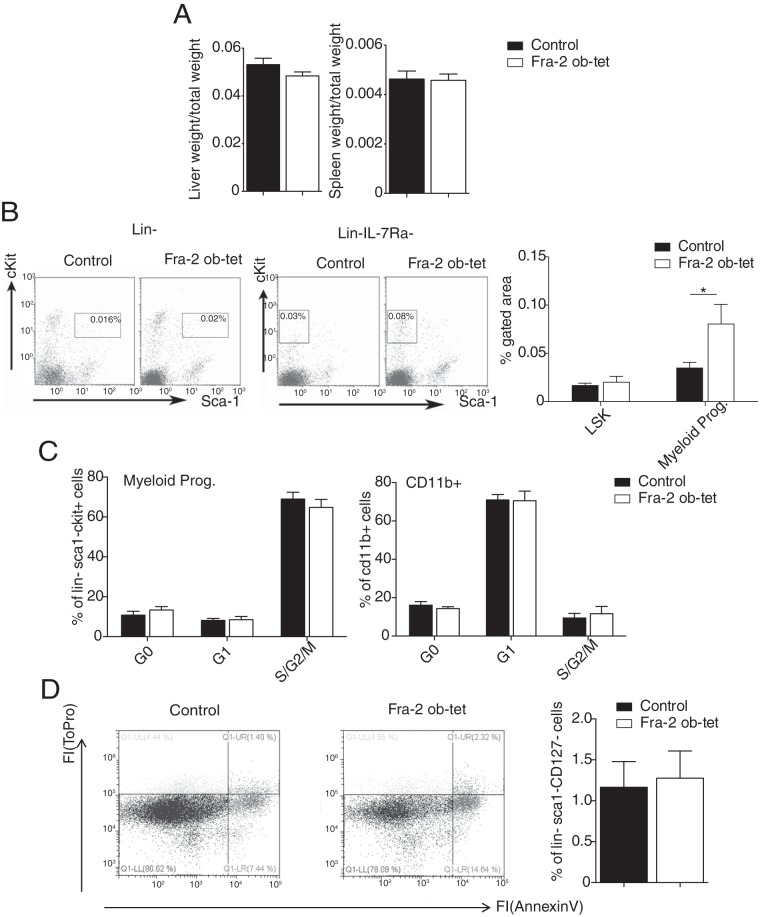
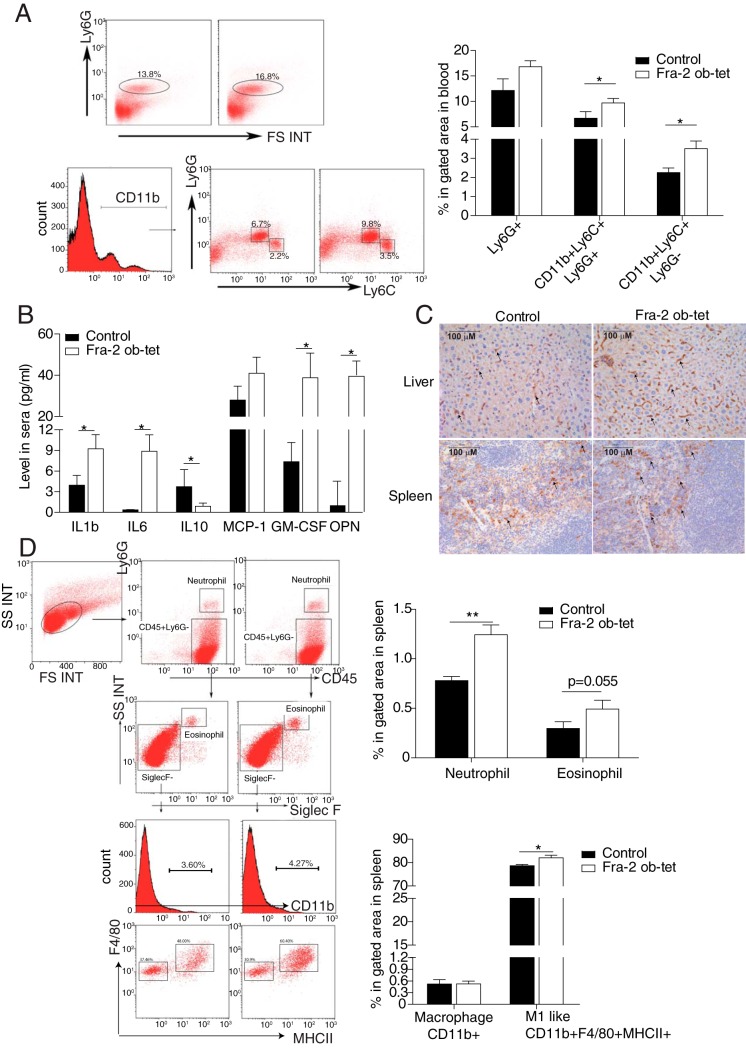
The increased number of CD11b+ myeloid cells in Fra-2Ob-tet bone marrow prompted us to examine whether Fra-2Ob-tet mice also display signs of systemic inflammation. First, we measured spleen and liver weights, but we could not detect any difference between wild-type and mutant mice (Fig. 3A). However, peripheral blood analysis revealed a 2.5-fold increase in myeloid progenitor cells in Fra-2Ob-tet mice compared to littermate controls (Fig. 3B). Cell cycle and apoptosis analyses revealed no alteration in blood myeloid progenitor cell proliferation or apoptosis (Fig. 3C and D), suggesting an increased myeloid progenitor cell migration from the bone marrow into the circulation. Furthermore, monocyte numbers were higher in the circulation of Fra-2Ob-tet mice than in that of littermate controls (Table 2). More-detailed characterization of the circulating cell populations revealed that the number of inflammatory monocytes, identified as CD11b+ Ly6C+ Ly6G− cells, was significantly increased in Fra-2Ob-tet mice (Fig. 4A). In accordance, Fra-2Ob-tet mice had higher levels of proinflammatory cytokines interleukin-1β (IL-1β), IL-6, and GM-CSF in serum and lower levels of the anti-inflammatory cytokine IL-10 in serum (Fig. 4B). Interestingly, the levels of serum OPN were dramatically increased (∼30- to 40-fold) in Fra-2Ob-tet mice compared to controls (Fig. 4B).
FIG 3.
Fra-2 expression in osteoblasts increases circulant myeloid progenitor cell numbers. (A) Liver and spleen weights per body weight from Fra-2Ob-tet and littermate mice (n = 6). (B) FACS dot plots and quantification of hematopoietic stem cells (LSK, Lin− Sca1+ c-Kit+) and myeloid progenitors (Lin− IL-17Ra− Sca1− c-Kit+) in 10-week-old Fra-2Ob-tet and littermate mouse blood (n = 4 to 8). (C) Quantification of myeloid progenitor and CD11b+ cell proliferation in blood from Fra-2Ob-tet and littermate mice (n = 6). (D) FACS dot plots and quantification of myeloid progenitor apoptosis in blood from Fra-2Ob-tet and littermate mice (n = 6). Data represent the mean values ± SEM. *, P < 0.05 (by unpaired t test).
TABLE 2.
Peripheral blood cell profiles of wild-type and Fra-2Ob-tet mice
| Lineage (unit)a | Value (mean ± SEM)b in: |
|
|---|---|---|
| Control mice (n = 4) | Fra-2Ob-tet mice (n = 3) | |
| WBC (×103/μl) | 7.98 ± 0.67 | 8.98 ± 1.11 |
| Neutrophils (%) | 7.13 ± 0.53 | 8.3 ± 0.15 |
| Lymphocytes (%) | 82.90 ± 5.74 | 66.10 ± 8.4 |
| Monocytes (%) | 0.97 ± 0.05 | 1.47 ± 0.18* |
| RBCs (×103/μl) | 7.03 ± 1.17 | 6.36 ± 0.97 |
| Platelets (×103/μl) | 1,442 ± 86.03 | 812.0 ± 151.4** |
WBC, indicates white blood cell; RBC, red blood cell.
*, P < 0.05; **, P < 0.01.
FIG 4.
Mice expressing Fra-2 in osteoblasts develop a systemic inflammation. (A) Representative flow cytometry plots of neutrophil or monocyte populations in the blood from 10-week-old Fra-2Ob-tet and littermate mice. Quantification of neutrophils (Ly6G+), inflammatory monocytes (CD11b+ Ly6C+ Ly6G−), and granulocytes (CD11b+ Ly6C+ Ly6G+) in Fra-2Ob-tet and littermate blood is shown (n = 6). (B) Levels of cytokines and chemokines in serum from 10-week-old Fra-2Ob-tet and littermate mice (n = 10). (C) Immunohistochemistry staining for F4/80 monocytes/macrophages in spleens and livers from 10-week-old Fra-2Ob-tet and littermate mice (magnification, ×20). (D) Quantification of neutrophils (Ly6G+), eosinophils (CD45+ Ly6G− SiglecF+), macrophages (CD45+ Ly6G− SiglecF− F4/80+ CD11bHi), and proinflammatory (MI-like) macrophages (Ly6G− SiglecF− F4/80+ CD11bHi MHCII+) in spleens from 10-week-old Fra-2Ob-tet and littermate mice (n = 6). Data represent the mean values ± SEM. *, P < 0.05; **, P < 0.01 (by unpaired t test).
Increased circulating proinflammatory monocytes in Fra-2Ob-tet could suggest the presence of immune cell infiltrates in other organs. Interestingly, we observed an increase of F4/80+ cell populations in the spleens and livers of Fra-2Ob-tet mice (Fig. 4C). Neutrophil, eosinophil, and macrophage numbers in the spleen were further quantified by flow cytometry. Neutrophils, identified as CD45+ Ly6G+ cells, were drastically increased in Fra-2Ob-tet spleens (Fig. 4D and Table 1), whereas no significant difference in eosinophil numbers was observed (Fig. 4D and Table 1). Despite a lack of change in the total number of macrophages in the spleens of mutant mice, significantly increased percentages and absolute numbers of proinflammatory (M1-like) macrophages was found in Fra-2Ob-tet mice (Fig. 4D and Table 1). These data indicate that osteoblast-specific expression of Fra-2 induces systemic inflammation, as shown by increased proinflammatory cytokines in the circulation and monocyte/macrophage and neutrophil infiltration in peripheral organs.
Fra-2 expression in osteoblasts regulates OPN expression, which induces MSC expansion.
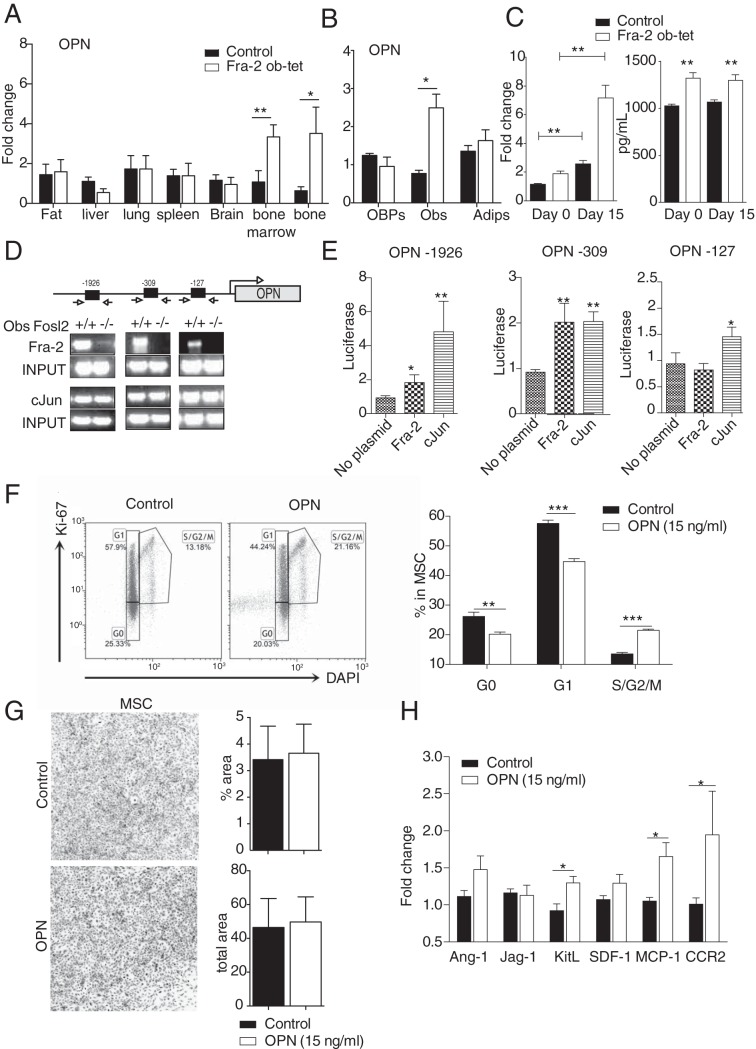
The strong increase in the serum OPN levels (Fig. 4B) prompted us to further analyze OPN mRNA levels in the different organs and in MSCs. Interestingly, OPN mRNA levels followed the same expression pattern as those for Fra-2, with no difference in fat pad, liver, spleen, lung, or brain tissues between mutants and controls but upregulation in the long bones and the bone marrow from the mutant mice as well as in osteoblasts differentiated from mutant MSCs (Fig. 5A and B), suggesting that Fra-2 controls opn expression and secretion by stromal cells. Moreover, opn expression was analyzed in osteoblast cultures isolated from Fra-2Ob-tet and littermate calvariae. Compared to control osteoblasts, Fra-2Ob-tet osteoblasts expressed increased levels of opn mRNA already in progenitor cells at day 0 of culture. Moreover, opn mRNA levels were further increased in mutant osteoblasts at day 15 of differentiation (Fig. 5C). Accordingly, the level of OPN protein was significantly increased in the supernatants of osteoblast cultures derived from Fra-2Ob-tet mice at both days 0 and 15 of differentiation (Fig. 5C). These data demonstrate that Fra-2 regulates OPN expression and production in osteoblasts.
FIG 5.
Fra-2 transcriptionally induces osteopontin (OPN) expression, which regulates mesenchymal stem cell (MSC) expansion. (A) Quantitative PCR analyses of OPN in fat pads, livers, lungs, spleens, brains, bone marrow, and long bones isolated from Fra-2Ob-tet and littermate mice (n = 6). (B) Quantitative PCR analyses of OPN in mesenchymal stem cells and derived osteoblasts or adipocytes isolated from 10-week-old Fra-2Ob-tet and littermate mice (n = 4). (C) OPN mRNA and protein levels in calvaria-derived osteoblast culture and supernatant isolated from Fra-2Ob-tet and littermate mice at days 0 and 15 postdifferentiation in vitro (n = 6). (D) ChIP for the OPN promoter. Arrows indicate primers amplifying fragments of the promoter. Chromatin of the indicated genotypes was immunoprecipitated with AP-1 antibodies. Endpoint PCR-amplified fragments are shown (n = 3). (E) OPN-luc reporter assay for the OPN promoter fragments in the presence of Fra-2- or c-Jun-expressing vectors (n = 3). (F) FACS dot plots and quantification of MSC proliferation after OPN (15 ng/ml) treatment for 48 h in vitro (n = 6). (G) CFU-MSC pictures and quantification after OPN (15 ng/ml) treatment for 48 h in vitro (n = 6). (H) RT-PCR analyses of ang-1, jag-1, sdf-1, kit-l, mcp-1 and ccr2 gene expression in MSCs (Ter119− CD45− Sca-1+) after OPN (15 ng/ml) treatment for 48 h (n = 6). Data represent the mean values ± SEM. *, P < 0.05; **, P < 0.01 (by unpaired t test). In vitro experiments were performed 3 times in triplicate for each sample.
To further examine how Fra-2 controls opn expression, the opn promoter was analyzed for possible AP-1 binding sites. Three putative nonconsensus binding regions for AP-1 protein were found at bp −127, −309, and −1926 after the opn start site (Fig. 5D). Using chromatin immunoprecipitation (ChIP) with an anti-Fra-2 antibody in Fra-2 control and knockout primary osteoblasts, we observed that Fra-2 bound the opn promoter at these 3 sites (Fig. 5D). To further characterize the specificity and potential dimerizing partners of Fra-2 on the opn promoter, ChIP was performed for the c-Jun transcription factor or IgG as a control. We observed binding of c-Jun to the opn promoter in control and Fra-2 knockout cells, whereas IgG showed no binding (Fig. 5D and data not shown), suggesting that c-Jun could likely regulate opn transcription in the presence or absence of Fra-2. Next, luciferase assays were performed using 3 clones of opn promoter regions in the presence of Fra-2- or c-Jun-overexpressing plasmids. As shown in Fig. 5E, Fra-2 could enhance luciferase activity at positions −1926 and −309 on the opn promoter, whereas c-Jun could activate its expression at all 3 sites. Altogether, these data demonstrate that Fra-2 can regulate opn at the transcription level in osteoblasts.
It is well known that Fra-2 regulates osteoblast maturation (2) (12); however, here we show that Fra-2 also controls MSC numbers (Fig. 1B). Therefore, we tested whether OPN production regulated by Fra-2 can impact MSC proliferation. To do so, MSCs were stimulated with OPN at 15 ng/ml, a concentration found in Fra-2Ob-tet osteoblast culture supernatants. At 48 h after OPN stimulation, the cell cycle profile of MSCs showed a lower percentage of cells in G0 and G1 and a higher percentage of cells in S/G2/M phase (Fig. 5F) but no difference in CFU formation (Fig. 5G). Importantly, gene profiling performed in MSCs with or without OPN stimulation revealed higher mRNA levels of kit-l, mcp-1, and ccr2 but no differences in ang-1, jag-1, and sdf-1 after OPN treatment (Fig. 5H). Collectively, these data suggest that OPN, transcriptionally controlled by Fra-2 in osteoblasts, regulates MSC expansion and MSC gene expression.
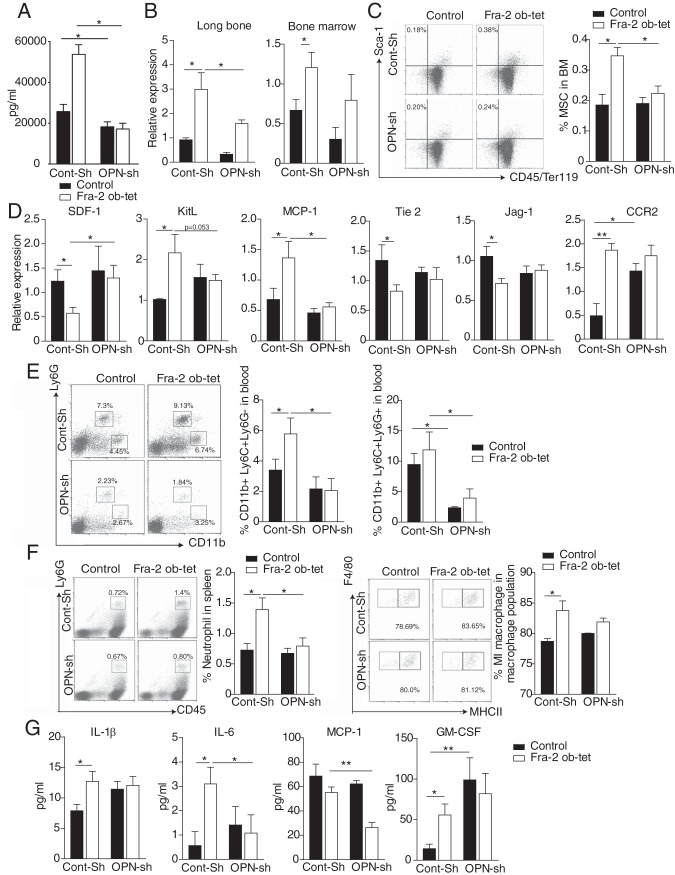
Inhibition of OPN in Fra-2Ob-tet mice rescues the bone marrow niche changes and the inflammatory phenotype.
Next, we investigated whether the changes in the bone marrow niche and the systemic inflammatory phenotype were due to increased OPN levels in Fra-2Ob-tet mice. To achieve knockdown of OPN, hydrodynamic intravenous injection of 20 μg opn short hairpin RNA (shRNA) (OPN-Sh) or scramble shRNA (Cont-Sh) was performed. As shown in Fig. 6A, opn shRNA injection efficiently reduced OPN levels in sera from wild-type and Fra-2Ob-tet mice compared to those from Cont-Sh-injected mice. Furthermore, opn shRNA injection led to a significant decrease of opn gene expression in both the bone marrow and long bones of Fra-2Ob-tet mice (Fig. 6B). Interestingly, the percentage of MSCs (CD45− Ter119− Sca-1+ cells) was rescued in Fra-2Ob-tet bone marrow after opn shRNA injection compared to that in vehicle-injected mice (Fig. 6C). Additionally, decreased OPN in Fra-2Ob-tet mice completely rescued mRNA levels of tie-2, jag-1, ccr2, sdf-1, kit-l, and mcp-1 in bone marrow from Fra-2Ob-tet mice (Fig. 6D). Only GM-CSF mRNA levels remained increased in Fra-2Ob-tet bone marrow despite the injection of opn shRNA (Fig. 6D). These data indicate that MSC expansion and alterations in the expression of bone marrow niche genes in Fra-2Ob-tet bone marrow were OPN dependent.
FIG 6.
Osteopontin depletion in Fra-2Ob-tet mice rescues their bone niche and their systemic inflammation phenotypes. (A) OPN levels in sera of Fra-2Ob-tet and littermate mice injected with control (Cont-Sh) or OPN (OPN-Sh) shRNA by hydrodynamic injection (n = 6). (B) Quantitative PCR analyses of OPN in long bones (without bone marrow) and bone marrow of Fra-2Ob-tet and littermate mice injected with control (Cont-Sh) or OPN (OPN-Sh) shRNA by hydrodynamic injection (n = 6). (C) FACS dot plots and quantification of mesenchymal stem cells (Ter119− CD45− Sca-1+) in Fra-2Ob-tet and littermate mice injected with control (Cont-Sh) or OPN (OPN-Sh) shRNA (n = 6). (D) Quantitative PCR analyses of bone niche markers in flushed bone marrow from Fra-2Ob-tet and littermate mice injected with control (Cont-Sh) or OPN (OPN-Sh) shRNA (n = 6). (E) FACS dot plots and quantification of inflammatory monocytes (CD11b+ Ly6C+ Ly6G−) or granulocytes (CD11b+ Ly6C+ Ly6G+) in blood from Fra-2Ob-tet and littermate mice injected with control (Cont-Sh) or OPN (OPN-Sh) shRNA (n = 6). (F) FACS dot plots and quantification of neutrophils and proinflammatory (MI-like) macrophages in spleens from Fra-2Ob-tet and littermate mice injected with control (Cont-Sh) or OPN (OPN-Sh) shRNA (n = 6). (G) Inflammatory cytokine and chemokine levels in sera from Fra-2Ob-tet and littermate control mice injected with control (Cont-Sh) or OPN (OPN-Sh) shRNA (n = 6). Data represent the mean values ± SEM. *, P < 0.05; **, P < 0.01 (by ANOVA).
Interestingly, opn shRNA injection also fully rescued the elevation of CD11b+ Ly6C+ Ly6G− and CD11b+ Ly6C+ Ly6G+ populations in the blood of Fra-2Ob-tet mice (Fig. 6E). In addition, flow cytometry analysis indicated a similar frequency of neutrophil and proinflammatory macrophages in the spleens from mutant and control mice treated with opn shRNA (Fig. 6F). Moreover, the levels of the proinflammatory cytokines IL-1β and IL-6 were comparable in mutant and control mice after injection with OPN-Sh (Fig. 6G). These data suggest that increased OPN expression in Fra-2Ob-tet osteoblasts contributes to the inflammatory phenotype of Fra-2Ob-tet mice.
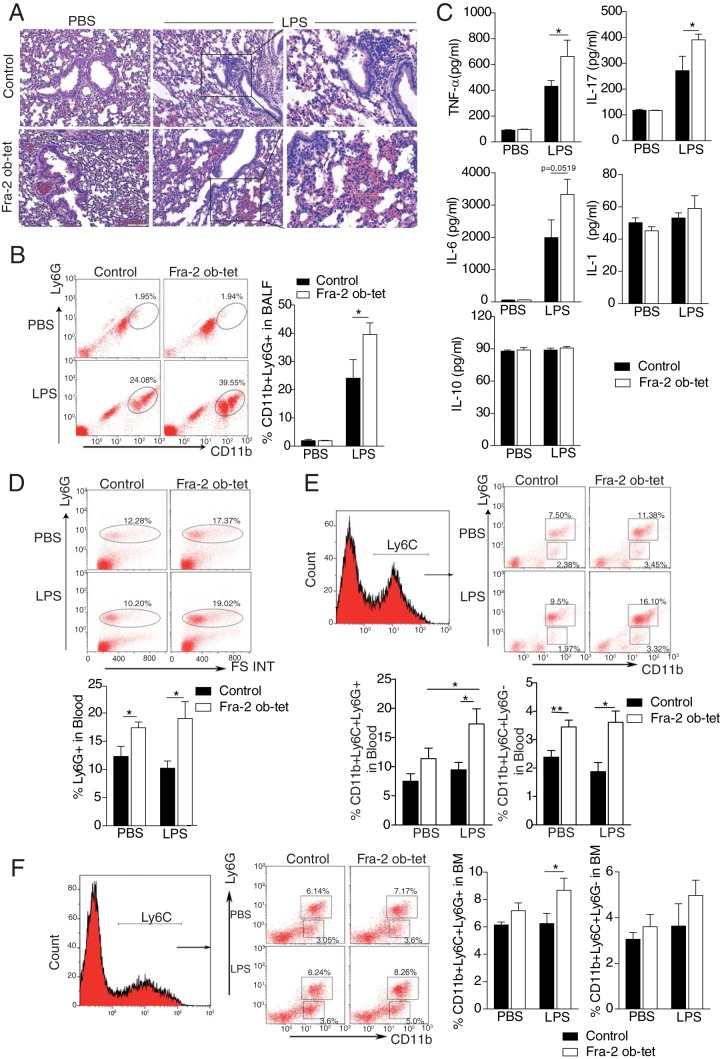
Osteoblast-specific Fra-2 expression exacerbates acute lung injury and enhances the recruitment of neutrophils to the lung.
To examine whether osteoblast-specific Fra-2 expression might have an impact on disease development, we exposed Fra-2Ob-tet mice to an LPS-induced acute lung injury model. LPS instilled intratracheally (i.t.) has been reported to induce monocyte, especially neutrophil, recruitment and vascular leakage into the lungs (28). Compared to that for the vehicle group, LPS treatment led to a prominent increase in the inflammatory cell infiltration into the lungs of Fra-2Ob-tet and control mice (Fig. 7A). However, in accordance with the basal proinflammatory phenotype, Fra-2Ob-tet mice exhibited a more severe lung injury than controls (Fig. 7A), which was associated with increased recruitment of neutrophils (CD11b+ Ly6G+) to the bronchiolar lavage fluid (BALF) (Fig. 7B). The levels of proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and IL-17 were increased in the BALF of Fra-2Ob-tet mice compared to littermate controls (Fig. 7C). Under LPS exposure, Fra-2Ob-tet mice also had markedly higher IL-6 levels in BALF than controls (Fig. 7C). In contrast, the levels of other cytokines, such as IL-2, IL-4, gamma interferon (IFN-γ), IL-5, GM-CSF, IL-1β, and IL-10, were identical in mutant and control mice after LPS treatment (Fig. 7C and data not shown). To further investigate whether LPS treatment can enhance the myeloid cell expansion in Fra-2Ob-tet mice, Ly6G+, CD11b+ Ly6C+ Ly6G−, and CD11b+ Ly6C+ Ly6G+ cells in blood and bone marrow were evaluated. The percentages of Ly6G+ and CD11b+ Ly6C+ Ly6G− cells in blood were higher in Fra-2Ob-tet mice than in control mice but were not further enhanced by LPS (Fig. 7D and E). In contrast, the percentages of CD11b+ Ly6C+ Ly6G+ cells were higher in blood of Fra-2Ob-tet mice (Fig. 7E), in accordance with the observed increase of these cells in bone marrow (Fig. 7F). Altogether, these data indicate that Fra-2 expression in osteoblasts can regulate inflammatory responses in LPS-induced acute lung injury by controlling the accumulation of inflammatory cells in bone marrow and in circulation, as well as the neutrophil infiltration in the target tissue.
FIG 7.
Fra-2Ob-tet mice show an acute lung injury exacerbation. (A) Hematoxylin and eosin (H&E) staining in Fra-2Ob-tet and littermate lungs at 24 h after PBS or LPS treatment (n = 3). (B) FACS dot plots and quantification of infiltrating neutrophils (Ly6G+ CD11b+) in the bronchiolar lavage fluid (BALF) of Fra-2Ob-tet and littermate mice at 24 h after PBS or LPS treatment (n = 6). (C) Cytokine levels in BALF of Fra-2Ob-tet and littermate lungs at 24 h after PBS or LPS treatment (n = 6). (D and E) Representative flow cytometry plots of neutrophils (Ly6G+) or inflammatory monocytes (CD11b+ Ly6C+ Ly6G−) and granulocytes (CD11b+ Ly6C+ Ly6G+) in blood of Fra-2Ob-tet and littermate mice at 24 h after PBS or LPS treatment (n = 6). (F) Representative flow cytometry plots indicating neutrophils or inflammatory monocytes (CD11b+ Ly6C+ Ly6G−) and granulocytes (CD11b+ Ly6C+ Ly6G+) in bone marrow from Fra-2Ob-tet and littermate mice at 24 h after PBS or LPS treatment (n = 6). Data represent the mean values ± SEM. *, P < 0.05; **, P < 0.01 (by ANOVA).
DISCUSSION
In this study, we demonstrate that Fra-2 expression in osteoblasts can control the bone marrow microenvironment, thereby regulating myeloid cell mobilization into the circulation and subsequently triggering a systemic inflammatory phenotype. Mechanistically, opn expression is transcriptionally enhanced by Fra-2, thereby stimulating mesenchymal stem cell (MSC) expansion. OPN was essential for the systemic inflammation observed in Fra-2Ob-tet mice. Using a model of acute lung injury by LPS instillation, we showed that myeloid cells were rapidly recruited to the injured tissue in Fra-2Ob-tet mice, leading to an exacerbation of lung inflammation.
MSCs are multipotent stromal cells of the bone marrow which are essential for the maintenance of bone homeostasis through their ability to differentiate into chondrocytes, adipocytes, and osteoblasts (29). Previously, Fra-2 was shown to be crucial for the maturation and function of the bone-forming osteoblasts (12). In this study, we showed that Fra-2 expression in osteoblasts regulates proliferation of MSCs, controlling their expansion. We could observe that Fra-2Ob-tet osteoblasts secrete large amounts of OPN. It is possible that the increased local OPN level caused by Fra-2 contributes to the enhanced osteoblastogenesis previously described in Fra-2Ob-tet mice (2). Chen et al. had previously shown that OPN induces MSC osteoblastogenesis and that its absence increases adipogenesis (20). However, the OPN effect is covered by the cell autonomous activation of osteoblastogenesis by Fra-2 (12). It is not excluded, however, that bone marrow cells other than osteoblasts also secrete OPN. Here we show that OPN can polarize MSCs toward an inflammatory phenotype, as examined by expression of mcp-1 and other genes. In accordance, previous studies have also shown that alterations in MSCs could be associated with a distinct inflammatory secretome (30, 31). For example, MSCs from Gaucher disease patients have increased COX-2, prostaglandin E2, interleukin-8, and CCL2 production, which is responsible for systemic inflammation (31). Our study suggests that the increased expression of ccr2 and mcp-1 in MSCs from Fra-2Ob-tet mice could contribute to the inflammatory phenotype. Moreover, multiple bone marrow niche gene such as the angiopoietin-1 gene (ang-1)/tie-2, jag-1, the kit ligand (also known as stem cell factor) gene (kit-l), and sdf-1 were also found to be differentially expressed in Fra-2Ob-tet bone marrow. ang-1 expressed by osteoblasts could interact with tie-2 on hematopoietic stem cells (HSCs) or endothelial cells, enhancing the ability of HSCs to become quiescent (32). On the other hand, jag-1 expressed by osteoblasts and bone marrow stromal cells could bind Notch, leading to HSC expansion (32, 33). Surprisingly, although the expression of the jag-1 gene was altered in bone marrow of Fra-2Ob-tet mice, no difference in Notch-1 and HES-1 or HSC number was observed, suggesting that osteoblasts have a minor role in HSC maintenance. In accordance, HSC differentiation toward the lymphoid or myeloid progenitors remain identical in mutant and control mice. However, the percentage of myeloid cells was strongly increased in peripheral blood of mutant mice, indicating a higher capacity to mobilize into the circulation. This likely explains the inflammatory cell infiltration in the spleens and livers of Fra-2Ob-tet mice.
There are at least two possible pathways to mobilize myeloid progenitors and monocytes and release these cells into the circulation. One mechanism is through the SDF-1/CXCR4 axis, which has been implicated in the regulation of HSCs and leukemic hematopoietic cell migration in acute myeloid leukemia (AML) (32, 33). Indeed, depletion of SDF-1 from perivascular stromal cells could induce HSC migration into the circulation (34). Moreover, CXCR4 antagonists is used in human clinical trials to facilitate release of stem cells from the bone marrow (35, 36). In fact, we detected significantly decreased SDF-1 expression at both the mRNA and protein levels, suggesting that SDF-1 might contribute to the inflammatory phenotype observed in Fra-2Ob-tet mice. Further investigations will determine the role of the SDF-1/CXCR4 axis in this model. Another possible mechanism is through the monocyte chemoattractant protein 1 (MCP-1)/CCR2 pathway. It was shown that depletion of MCP-1 in MSCs, CAR cells, and osteoblasts can significantly reduce the number of circulating Ly6Chi inflammatory monocytes after injection of LPS at low concentrations (37, 38). Furthermore, its receptor, CCR2, was shown to mediate the release of mature monocytes from the bone marrow. Accordingly, CCR2−/− mice have decreased numbers of “inflammatory” (Ly6Chi) monocytes in the blood and fail to expand these monocytes in response to inflammation or infection (39, 40). In our study, when Fra-2 was specifically expressed in osteoblasts, both the mcp-1 and ccr2 genes were drastically increased in bone marrow, which could subsequently augment the number of inflammatory monocytes in circulation and in peripheral tissues.
The interaction between the bone marrow environment and peripheral inflammation is an emerging field of interest. For example, systemic inflammation caused by adjuvant challenge or sepsis has been shown to rapidly lead to accumulation of B cells in the bone marrow (41). Upon LPS treatment, bone marrow stromal cells could enhance myeloid cell development and progenitor cell maintenance in humans (42). Here, we present a new mechanism by which Fra-2 expression in osteoblasts induces systemic inflammation, which is strongly increased upon an additional inflammatory trigger. The high levels of G-CSF and GM-CSF in Fra-2 mutant mice could potentially affect the migration of neutrophils. Moreover, one could hypothesize that the increase of GM-CSF levels after OPN knockdown is a feedback response to maintain cell survival, since OPN is induced in hematopoietic cells by GM-CSF signaling and OPN and GM-CSF could synergize to promote survival of IL-3-dependent mouse bone marrow cells (43). However, the exact role of the interaction of OPN, GM-CSF, and G-CSF in this model needs to be further addressed. Interestingly, Eferl et al. have previously demonstrated that broad overexpression of Fra-2 leads to pulmonary fibrosis, followed by release of profibrogenic and inflammatory factors, including an upregulation of OPN (9). The latter is known for its function as a bridge between bone and blood. However, most studies to date have focused on the impact of OPN on HSCs (18). Of interest is that we have demonstrated that OPN, transcriptionally regulated by Fra-2 in osteoblastic cells, regulates the expression of mcp-1 and ccr2 in MSCs, providing a novel pathway of inflammatory cell trafficking. In summary, our findings highlight the role of osteoblasts in influencing inflammatory responses and key molecular mechanisms such as the Fra-2/OPN axis in this process.
MATERIALS AND METHODS
Mice.
Osteoblast-specific expression of Fra-2 mice (Fra-2Ob-tet) was generated as described earlier (2). Briefly, tet-switchable Fosl2 allele mice were crossed with Osx-tTA-Cre mice for generating Fra-2Ob-tet mice. In the absence of doxycycline, these mice express Fra-2 while osterix is expressed. All mice were maintained on a mixed C57BL6/129 background, and littermates (Fra-2 wild-type osterix Cre transgenic and Fra-2 knock-in osterix Cre wild type) were used as controls. All mice were bred and maintained without doxycycline to ensure Fra-2 expression as early as embryonic development. Genotyping was performed by PCR analyses of genomic DNA from tail biopsy specimens. All mouse experiments were performed in accordance with local and institutional regulations. Male mice were analyzed at 10 weeks of age.
For the peripheral blood cell profile, 200 μl of blood was collected by retro-orbital puncture using EDTA-coated capillaries. Blood samples were diluted with phosphate-buffered saline (PBS) (1:1), transferred into 1.5-ml Eppendorf tubes, and then analyzed with an Advia 120 hematology system.
To decrease osteopontin (OPN) expression in vivo, hydrodynamic intravenous injection was performed with 1 ml of OPN shRNA (20 μg/mouse) (Origene). At 72 h after OPN shRNA injection, mice were sacrificed and analyzed. The control groups were injected with scramble shRNA (20 μg/mouse; Origene).
For the murine model of LPS-induced acute lung injury, 8-week-old mice were anesthetized with ketamine (125 mg/kg) and xylazine (8 mg/kg) and challenged with intratracheal instillation of sterile PBS or 40 μl of LPS (1 mg/ml; E. coli 0111:B4 [Sigma]). After challenge, the incision was sutured using surgical silk, and 24 h later, mice were sacrificed to collect tissues for analyses. Lungs were dissected and fixed in 4% formaldehyde for histopathology. Bronchiolar lavage fluid (BALF) was collected and then centrifuged at 800 × g. The supernatant was collected for cytokine/chemokine measurement. BALF cells were isolated for fluorescence-activated cell sorter (FACS) analysis.
Histology and immunofluorescence staining. (i) Histology.
Freshly isolated spleen, liver, and lung were fixed in 4% formaldehyde overnight at 4°C and embedded into paraffin. Long bones were decalcified in EDTA until the bones became flexible before embedding into paraffin. Tissue sections (5 μm) were deparaffinized with xylene and then rehydrated with 100%, 95%, and 80% ethanol, followed by staining with hematoxylin and eosin (H&E) (44).
(ii) Immunohistochemistry an immunofluorescence staining.
Immunohistochemistry staining for F4/80 (Abcam) expression in spleen and liver was performed using proteinase K as an unmasking solution, and then the manufacturer's instructions for the Vectastain kit (Vector Laboratories, Inc., Burlingame, CA) were followed. For immunofluorescence staining, primary SDF-1 antibodies were applied to the long-bone slides overnight at 4°C, followed by incubation with secondary antibodies for 1 h at room temperature with repeated washes in between. Slides were mounted with Fluoroshield with DAPI (4′,6′-diamidino-2-phenylindole) (Sigma), and images were acquired in a Nikon Eclipse 80i microscope.
For the primary antibodies, rabbit anti-F4/80 (eBioscience; 1/400) and rabbit anti-mouse SDF-1 (LSBio; 1/100) were used. For the secondary antibodies, eFluor594-conjugated goat anti-rabbit IgG (Invitrogen; 1/1,000), and fluorescein isothiocyanate (FITC)-conjugated goat anti-rat IgG (Thermo Fisher; 1/1,500) were used.
Flow cytometry.
Bone marrow cells were isolated by flushing the long bones with PBS. Splenocytes were obtained by crushing the spleen, which then was gently filtered through a 70-μm cell strainer (BD). Blood cells were obtained by intracardiac puncture. After red blood cell lysis, mesenchymal cells were analyzed using allophycocyanin (APC)-eFluor780-conjugated CD45, Ter-119, and phycoerythrin (PE)-Cy7-conjugated Sca-1 (all from eBioscience). Hematopoietic stem cell (HSCs) were analyzed after exclusion of lineage (Lin)-positive cells using a cocktail of biotin-conjugated Gr1, B220, CD3ε, CD11b, and Ter-119 and after labeling with PE-Cy7-conjugated Sca-1 and PE-conjugated c-Kit. HSCs were defined as Lin− Sca-1+ c-Kit+ (LSK) cells. For myeloid progenitor cell analysis, cells were stained with a cocktail of biotin-conjugated Gr1, B220, CD3ε, CD11b, and Ter-119, PE-Cy7-conjugated Sca-1, PE-conjugated c-Kit, and PE-Cy5-conjugated IL-7Rα. Myeloid progenitors were defined as Lin− IL-7Rα− Sca-1− c-Kit+ cells. Macrophages/monocytes were analyzed using FITC-conjugated Ly6G, PE-conjugated CD11b, and APC-conjugated Ly6C (BD). Splenocytes were stained with peridinin chlorophyll protein (PerCP) Cy5.5-conjugated Ly6G, APC-eFluor780-conjugated CD45, PE-conjugated Siglec F, APC-conjugated F4/80, FITC-conjugated CD11b, and Pacific blue-conjugated major histocompatibility complex class II (MHCII). After washing, the labeled cells were analyzed on a Gallios flow cytometer (Beckman Coulter, Inc.). Absolute numbers were obtained by using counting beads (6602796; Beckman Coulter, Inc.) as instructed by the manufacturer.
Quantitative RT-PCR.
RNA from bone marrow or flushed long bones was isolated using TRIzol (Invitrogen) or peqGOLDRNAPure reagent (PeQlab) according to the manufacturer's instructions. RNA was digested with DNase I and reverse transcribed into cDNA using an oligo(dT) primer. Quantitative real-time PCR (RT-PCR) was performed using SYBR green I-dTTP (Eurogentec). Samples were analyzed in duplicate, and β-actin and hypoxanthine phosphoribosyltransferase (HPRT) were used to normalize the RNA content of samples. Primer sequences are listed in Table 3.
TABLE 3.
Primer sequences for quantitative RT-PCR analyses
| Target | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| Ang-1 | GAAGCAACAACTGGAGCTCATG | TCCTCCCTTTAGCAAAACACCTT |
| Jag-1 | AACGACCGTAATCGCATCGT | TATCAGGTTGAATAGTGTCATTACTGGAA |
| CXCR4 | GTGTAAGGCTGTCCATATCATC | GACAGCTTAGAGATGATGATGC |
| SDF-1 | GAGCCAACGTCAAGCATCTG | CGGGTCAATGCACACTTGT |
| Tie-2 | GAACTGAGGACGCTTCCACATTC | TCAGAAACGCCAACAGCACGGT |
| IL-7 | CAGGAACTGATAGTAATTGCCCG | CTTCAACTTGCGAGCAGCACGA |
| VCAM-1 | GCTATGAGGATGGAAGACTCTGG | ACTTGTGCAGCCACCTGAGATC |
| KitL | GATTCCAGAGTCAGTGTCACAA | TTGTCAGTTCAGAATGTTCCCG |
| VEGFR2 | CATCACCGAGAACAAGAACA | CATTGATCTTTGCCTCACAG |
| VEGFR3 | GGAAGGCTCTGAAGATAAAGG | CTTTATCTTCAGAGCCTTCCAC |
| TNF-α | GGAGGCAACAAGGTAGAGAGG | CACAGCCTTCCTCACAGAGC |
| IL-1α | CAG GCA GGC AGT ATC ACT CA | AGG TGC TCA TGT CCT CAT CC |
| IL-6 | TGTGCAATGGCAATTCTGAT | TCCAGTTTGGTAGCATCCATC |
| G-CSF | CCTCTGGTGCCATTCTCTACTA | GGAGACCACGGTAAGAGATGAT |
| GM-CSF | ACACAGCCCTGGGAGCAT | CTGGCCGGTCTCACTCCT |
| MCP-1 | GCTACAAGAGGATCACCAGCAG | GTCTGGACCCATTCCTTCTTGG |
| CCR2 | GCTGTGTTTGCCTCTCTACCAG | CAAGTAGAGGCAGGATCAGGCT |
| OPN | AAGCAGACACTTTCACTCCAATCG | TGACCTCAGTCCATAAGCCAAGC |
| β-Actin | TGTCCACCTTCCAGCAGATGT | AGCTCAGTAACAGTCCGCCTAGA |
ChIP and luciferase assay.
Chromatin immunoprecipitation (ChIP) was performed according to a standard protocol with antibodies against Fra-2 and c-Jun (Santa Cruz Biotechnology) (11). The specific binding of Fra-2 antibody was verified in parallel using control and Fra-2-deficient osteoblasts in 3 independent experiments.
The osteopontin promoter-reporter vectors were generated using the pGL4.23[luc2/minP] vector following the kit instructions (Promega). HEK293 cells were plated in 24-well dishes, and 1.5 mg of the luciferase reporter construct, 0.2 mg of the Renilla internal control (pHRG-tk; Promega), and 0.2 to 1 mg of each AP-1 expression vector were cotransfected using Lipofectamine (Invitrogen). The experiments were performed 3 times, with triplicate sample each time. Luciferase activity was quantified using the dual-luciferase kit (Promega) following the manufacturer's instructions.
Multiplex cytokines and ELISA measurement.
Chemokines and cytokines were measured using the mouse Th1/Th2 10plex FlowCytomix Multiplex kit (eBioscience; catalog number BMS820FF) according to the manufacturer's instructions. Osteopontin, MCP-1, and GM-CSF levels in serum were determined using the DuoSet enzyme-linked immunosorbent assay (ELISA) kit (R&D).
In vitro experiments. (i) Osteoblast culture.
Calvariae were sequentially digested for 30 min in modified Eagle's medium type α (α-MEM) containing 0.1% collagenase and 0.2% dispase. Cells isolated in fractions 2 and 3 were combined as an osteoblastic cell population, expanded for 2 days in α-MEM, with 10% fetal calf serum (FCS), and plated at a density of 5 × 105 cells/well. The medium was supplemented with 10 mM β-glycerophosphate and 50 μg/ml ascorbic acid for osteoblast cultures. After 2 weeks of culture, RNAs were extracted from osteoblasts. To control cell differentiation, Alizarin Red (Sigma-Aldrich) staining was performed for osteoblast cultures (data not shown).
(ii) Bone marrow-derived MSC culture. Bone marrow cells were harvested by flushing the tibiae and femurs from mutant and littermate control mice. The cell suspension was filtered through a 70-mm filter. Bone marrow cells (25 × 106 cells/ml) were cultured in 6-well plates with 2 ml of complete Dulbecco modified Eagle medium (DMEM) at 37°C with 5% CO2 in a humidified chamber. After 3 h, nonadherent cells were removed and replaced with fresh complete medium. The medium was changed daily until the cells became confluent (90%). At passage 6, the cells were stained with antibodies against APC-conjugated Ter119, APC-conjugated CD45, and PE-Cy7-conjugated Sca-1. Ter119− CD45− Sca-1+ cells were identified as MSCs. The MSCs were then exposed to medium supplemented with 10 mM β-glycerophosphate and 50 μg/ml ascorbic acid for osteoblast differentiation (7 days) or to medium supplemented with 5 mM isobutylmethylxanthine, 60 mM indomethacin, 100 nM dexamethasone, and 10 mg/ml insulin for adipocyte differentiation.
For CFU analysis, 1 × 105 murine MSCs were plated into 6-well plate and incubated with α-MEM containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin, with or without osteopontin (15 ng/ml), for 48 h. Cells where fixed and stained in 0.5% crystal violet in methanol for 10 min at room temperature. The total area of crystal violet-positive CFU was calculated by automatic particle counting using the ImageJ software. In short, pictures are converted to binary black-and-white images, and a threshold range is manually set to distinguish objects of interest from the background. Particles with sizes of 0.1 to 150 pixels2 are then counted by particle analysis.
To determine the effect of osteopontin on MSC proliferation, 1 × 105 cells were plated into 6-well plates and incubated with α-MEM medium containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin, with or without osteopontin (15 ng/ml), for 48 h. mRNAs were extracted as described before for RT-PCR analyses. For cell cycle analyses, the cells were fixed using the Foxp3 staining buffer set (eBioscience), stained with eFluor660-conjugated Ki-67 (eBioscience), and resuspended in 1 mg/ml of DAPI.
Statistics.
All data are presented as mean ± standard error of the mean (SEM). The statistical significances were determined by Student's t test for single comparisons or by one-way analysis of variance (ANOVA) for multiple comparisons using GraphPad Prism software. Significant differences are indicated by for P values of <0.05 and <0.01.
ACKNOWLEDGMENTS
We thank Wolfgang Baum for assistance with animal experiments and Christine Zech for excellent technical assistance. We thank Alvaro Ucero for critical comments that helped to improve the manuscript.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG-CRC1181-A01, Emmy Noether BO3811/1-1, SPP2084, DFG-BO3811/5, and DFG-BO3811/6), IZKF-Projekt D23, and the Sichuan Province International Cooperative Foundation (grant 2017HH0109).
We declare no conflict of interest.
REFERENCES
- 1.Karsenty G, Ferron M. 2012. The contribution of bone to whole-organism physiology. Nature 481:314–320. doi: 10.1038/nature10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozec A, Bakiri L, Jimenez M, Rosen ED, Catala-Lehnen P, Schinke T, Schett G, Amling M, Wagner EF. 2013. Osteoblast-specific expression of Fra-2/AP-1 controls adiponectin and osteocalcin expression and affects metabolism. J Cell Sci 126:5432–5440. doi: 10.1242/jcs.134510. [DOI] [PubMed] [Google Scholar]
- 3.Bocelli-Tyndall C, Bracci L, Schaeren S, Feder-Mengus C, Barbero A, Tyndall A, Spagnoli GC. 2009. Human bone marrow mesenchymal stem cells and chondrocytes promote and/or suppress the in vitro proliferation of lymphocytes stimulated by interleukins 2, 7 and 15. Ann Rheum Dis 68:1352–1359. doi: 10.1136/ard.2008.094003. [DOI] [PubMed] [Google Scholar]
- 4.Pistoia V, Raffaghello L. 2014. Unveiling the role of TNF-alpha in mesenchymal stromal cell-mediated immunosuppression. Eur J Immunol 44:352–356. doi: 10.1002/eji.201344372. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama T, Nagasawa T. 2012. Bone marrow niches for hematopoietic stem cells and immune cells. Inflamm Allergy Drug Targets 11:201–206. doi: 10.2174/187152812800392689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovach TK, Dighe AS, Lobo PI, Cui Q. 2015. Interactions between MSCs and immune cells: implications for bone healing. J Immunol Res 2015:752510. doi: 10.1155/2015/752510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Chen X, Cao W, Shi Y. 2014. Plasticity of mesenchymal stem cells in immunomodulation: pathological and therapeutic implications. Nat Immunol 15:1009–1016. doi: 10.1038/ni.3002. [DOI] [PubMed] [Google Scholar]
- 8.Nemeth K, Leelahavanichkul A, Yuen PS, Mayer B, Parmelee A, Doi K, Robey PG, Leelahavanichkul K, Koller BH, Brown JM, Hu X, Jelinek I, Star RA, Mezey E. 2009. Bone marrow stromal cells attenuate sepsis via prostaglandin E(2)-dependent reprogramming of host macrophages to increase their interleukin-10 production. Nat Med 15:42–49. doi: 10.1038/nm.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eferl R, Hasselblatt P, Rath M, Popper H, Zenz R, Komnenovic V, Idarraga MH, Kenner L, Wagner EF. 2008. Development of pulmonary fibrosis through a pathway involving the transcription factor Fra-2/AP-1. Proc Natl Acad Sci U S A 105:10525–10530. doi: 10.1073/pnas.0801414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reich N, Maurer B, Akhmetshina A, Venalis P, Dees C, Zerr P, Palumbo K, Zwerina J, Nevskaya T, Gay S, Distler O, Schett G, Distler JH. 2010. The transcription factor Fra-2 regulates the production of extracellular matrix in systemic sclerosis. Arthritis Rheum 62:280–290. doi: 10.1002/art.25056. [DOI] [PubMed] [Google Scholar]
- 11.Bozec A, Bakiri L, Hoebertz A, Eferl R, Schilling AF, Komnenovic V, Scheuch H, Priemel M, Stewart CL, Amling M, Wagner EF. 2008. Osteoclast size is controlled by Fra-2 through LIF/LIF-receptor signalling and hypoxia. Nature 454:221–225. doi: 10.1038/nature07019. [DOI] [PubMed] [Google Scholar]
- 12.Bozec A, Bakiri L, Jimenez M, Schinke T, Amling M, Wagner EF. 2010. Fra-2/AP-1 controls bone formation by regulating osteoblast differentiation and collagen production. J Cell Biol 190:1093–1106. doi: 10.1083/jcb.201002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobsen MJ, Mentzel CM, Olesen AS, Huby T, Jorgensen CB, Barres R, Fredholm M, Simar D. 2016. Altered methylation Profile of lymphocytes is concordant with perturbation of lipid metabolism and inflammatory response in obesity. J Diabetes Res 2016:8539057. doi: 10.1155/2016/8539057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zietek T, Rath E. 2016. Inflammation meets metabolic disease: gut feeling mediated by GLP-1. Front Immunol 7:154. doi: 10.3389/fimmu.2016.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Regan A, Berman JS. 2000. Osteopontin: a key cytokine in cell-mediated and granulomatous inflammation. Int J Exp Pathol 81:373–390. doi: 10.1046/j.1365-2613.2000.00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Regan AW, Chupp GL, Lowry JA, Goetschkes M, Mulligan N, Berman JS. 1999. Osteopontin is associated with T cells in sarcoid granulomas and has T cell adhesive and cytokine-like properties in vitro. J Immunol 162:1024–1031. [PubMed] [Google Scholar]
- 17.McKee MD, Nanci A. 1996. Osteopontin: an interfacial extracellular matrix protein in mineralized tissues. Connect Tissue Res 35:197–205. doi: 10.3109/03008209609029192. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson SK, Johnston HM, Whitty GA, Williams B, Webb RJ, Denhardt DT, Bertoncello I, Bendall LJ, Simmons PJ, Haylock DN. 2005. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood 106:1232–1239. doi: 10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- 19.Stier S, Ko Y, Forkert R, Lutz C, Neuhaus T, Grunewald E, Cheng T, Dombkowski D, Calvi LM, Rittling SR, Scadden DT. 2005. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med 201:1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q, Shou P, Zhang L, Xu C, Zheng C, Han Y, Li W, Huang Y, Zhang X, Shao C, Roberts AI, Rabson AB, Ren G, Zhang Y, Wang Y, Denhardt DT, Shi Y. 2014. An osteopontin-integrin interaction plays a critical role in directing adipogenesis and osteogenesis by mesenchymal stem cells. Stem Cells 32:327–337. doi: 10.1002/stem.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin H, Zygourakis K, Farach-Carson MC, Yaszemski MJ, Mikos AG. 2004. Attachment, proliferation, and migration of marrow stromal osteoblasts cultured on biomimetic hydrogels modified with an osteopontin-derived peptide. Biomaterials 25:895–906. doi: 10.1016/S0142-9612(03)00602-1. [DOI] [PubMed] [Google Scholar]
- 22.Zhu B, Suzuki K, Goldberg HA, Rittling SR, Denhardt DT, McCulloch CA, Sodek J. 2004. Osteopontin modulates CD44-dependent chemotaxis of peritoneal macrophages through G-protein-coupled receptors: evidence of a role for an intracellular form of osteopontin. J Cell Physiol 198:155–167. doi: 10.1002/jcp.10394. [DOI] [PubMed] [Google Scholar]
- 23.Weber GF, Zawaideh S, Hikita S, Kumar VA, Cantor H, Ashkar S. 2002. Phosphorylation-dependent interaction of osteopontin with its receptors regulates macrophage migration and activation. J Leukoc Biol 72:752–761. [PubMed] [Google Scholar]
- 24.Koh A, da Silva AP, Bansal AK, Bansal M, Sun C, Lee H, Glogauer M, Sodek J, Zohar R. 2007. Role of osteopontin in neutrophil function. Immunology 122:466–475. doi: 10.1111/j.1365-2567.2007.02682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano Y, Aziz M, Yang WL, Wang Z, Zhou M, Ochani M, Khader A, Wang P. 2015. Neutralization of osteopontin attenuates neutrophil migration in sepsis-induced acute lung injury. Crit Care 19:53. doi: 10.1186/s13054-015-0782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yumoto K, Ishijima M, Rittling SR, Tsuji K, Tsuchiya Y, Kon S, Nifuji A, Uede T, Denhardt DT, Noda M. 2002. Osteopontin deficiency protects joints against destruction in anti-type II collagen antibody-induced arthritis in mice. Proc Natl Acad Sci U S A 99:4556–4561. doi: 10.1073/pnas.052523599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiefer FW, Zeyda M, Gollinger K, Pfau B, Neuhofer A, Weichhart T, Saemann MD, Geyeregger R, Schlederer M, Kenner L, Stulnig TM. 2010. Neutralization of osteopontin inhibits obesity-induced inflammation and insulin resistance. Diabetes 59:935–946. doi: 10.2337/db09-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dagvadorj J, Shimada K, Chen S, Jones HD, Tumurkhuu G, Zhang W, Wawrowsky KA, Crother TR, Arditi M. 2015. Lipopolysaccharide induces alveolar macrophage necrosis via CD14 and the P2X7 receptor leading to interleukin-1alpha release. Immunity 42:640–653. doi: 10.1016/j.immuni.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glenn JD, Whartenby KA. 2014. Mesenchymal stem cells: emerging mechanisms of immunomodulation and therapy. World J Stem Cells 6:526–539. doi: 10.4252/wjsc.v6.i5.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranganath SH, Tong Z, Levy O, Martyn K, Karp JM, Inamdar MS. 2016. Controlled inhibition of the mesenchymal stromal cell pro-inflammatory secretome via microparticle engineering. Stem Cell Rep 6:926–939. doi: 10.1016/j.stemcr.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campeau PM, Rafei M, Boivin MN, Sun Y, Grabowski GA, Galipeau J. 2009. Characterization of Gaucher disease bone marrow mesenchymal stromal cells reveals an altered inflammatory secretome. Blood 114:3181–3190. doi: 10.1182/blood-2009-02-205708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison SJ, Scadden DT. 2014. The bone marrow niche for haematopoietic stem cells. Nature 505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peled A, Tavor S. 2013. Role of CXCR4 in the pathogenesis of acute myeloid leukemia. Theranostics 3:34–39. doi: 10.7150/thno.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding L, Morrison SJ. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broxmeyer HE, Orschell CM, Clapp DW, Hangoc G, Cooper S, Plett PA, Liles WC, Li X, Graham-Evans B, Campbell TB, Calandra G, Bridger G, Dale DC, Srour EF. 2005. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med 201:1307–1318. doi: 10.1084/jem.20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abraham M, Biyder K, Begin M, Wald H, Weiss ID, Galun E, Nagler A, Peled A. 2007. Enhanced unique pattern of hematopoietic cell mobilization induced by the CXCR4 antagonist 4F-benzoyl-TN14003. Stem Cells 25:2158–2166. doi: 10.1634/stemcells.2007-0161. [DOI] [PubMed] [Google Scholar]
- 37.Shi C, Jia T, Mendez-Ferrer S, Hohl TM, Serbina NV, Lipuma L, Leiner I, Li MO, Frenette PS, Pamer EG. 2011. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity 34:590–601. doi: 10.1016/j.immuni.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugiyama T, Nagasawa T. 2011. Emergency evacuation! Hematopoietic niches induce cell exit in infection. Immunity 34:463–465. doi: 10.1016/j.immuni.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Serbina NV, Pamer EG. 2006. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol 7:311–317. [DOI] [PubMed] [Google Scholar]
- 40.Tsou CL, Peters W, Si Y, Slaymaker S, Aslanian AM, Weisberg SP, Mack M, Charo IF. 2007. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest 117:902–909. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreau JM, Berger A, Nelles ME, Mielnik M, Furlonger C, Cen SY, Besla R, Robbins CS, Paige CJ. 2015. Inflammation rapidly reorganizes mouse bone marrow B cells and their environment in conjunction with early IgM responses. Blood 126:1184–1192. doi: 10.1182/blood-2015-03-635805. [DOI] [PubMed] [Google Scholar]
- 42.Ziegler P, Boettcher S, Takizawa H, Manz MG, Brummendorf TH. 2016. LPS-stimulated human bone marrow stroma cells support myeloid cell development and progenitor cell maintenance. Ann Hematol 95:173–178. doi: 10.1007/s00277-015-2550-5. [DOI] [PubMed] [Google Scholar]
- 43.Lin YH, Huang CJ, Chao JR, Chen ST, Lee SF, Yen JJ, Yang-Yen HF. 2000. Coupling of osteopontin and its cell surface receptor CD44 to the cell survival response elicited by interleukin-3 or granulocyte-macrophage colony-stimulating factor. Mol Cell Biol 20:2734–2742. doi: 10.1128/MCB.20.8.2734-2742.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bozec A, Hannemann N. 2016. Mechanism of regulation of adipocyte numbers in adult organisms through differentiation and apoptosis homeostasis. J Vis Exp doi: 10.3791/53822. [DOI] [PMC free article] [PubMed] [Google Scholar]