Due to the rapid evolution of the influenza A virus, vaccines require continuous strain updates. Additionally, the platform used to deliver the vaccine can have an impact on the breadth of protection. Currently, there are various vaccine platforms available to prevent influenza A virus infection in swine, and we experimentally tested two: adjuvanted-whole inactivated virus and live-attenuated virus. When challenged with an antigenically distinct virus, adjuvanted-whole inactivated virus provided partial protection, while live-attenuated virus provided effective protection. Additional strategies are required to broaden the protective properties of inactivated virus vaccines, given the dynamic antigenic landscape of cocirculating strains in North America, whereas live-attenuated vaccines may require less frequent strain updates, based on demonstrated cross-protection. Enhancing vaccine efficacy to control influenza infections in swine will help reduce the impact they have on swine production and reduce the risk of swine-to-human transmission.
KEYWORDS: influenza, live-attenuated, swine, vaccine
ABSTRACT
Influenza A viruses in swine (IAV-S) circulating in the United States of America are phylogenetically and antigenically distinct. A human H3 hemagglutinin (HA) was introduced into the IAV-S gene pool in the late 1990s, sustained continued circulation, and evolved into five monophyletic genetic clades, H3 clades IV-A to -E, after 2009. Across these phylogenetic clades, distinct antigenic clusters were identified, with three clusters (cyan, red, and green antigenic cluster) among the most frequently detected antigenic phenotypes (Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, et al. J Virol 90:8266–8280, 2016, https://doi.org/10.1128/JVI.01002-16). Although it was demonstrated that antigenic diversity of H3N2 IAV-S was associated with changes at a few amino acid positions in the head of the HA, the implications of this diversity for vaccine efficacy were not tested. Using antigenically representative H3N2 viruses, we compared whole inactivated virus (WIV) and live-attenuated influenza virus (LAIV) vaccines for protection against challenge with antigenically distinct H3N2 viruses in pigs. WIV provided partial protection against antigenically distinct viruses but did not prevent virus replication in the upper respiratory tract. In contrast, LAIV provided complete protection from disease and virus was not detected after challenge with antigenically distinct viruses.
IMPORTANCE Due to the rapid evolution of the influenza A virus, vaccines require continuous strain updates. Additionally, the platform used to deliver the vaccine can have an impact on the breadth of protection. Currently, there are various vaccine platforms available to prevent influenza A virus infection in swine, and we experimentally tested two: adjuvanted-whole inactivated virus and live-attenuated virus. When challenged with an antigenically distinct virus, adjuvanted-whole inactivated virus provided partial protection, while live-attenuated virus provided effective protection. Additional strategies are required to broaden the protective properties of inactivated virus vaccines, given the dynamic antigenic landscape of cocirculating strains in North America, whereas live-attenuated vaccines may require less frequent strain updates, based on demonstrated cross-protection. Enhancing vaccine efficacy to control influenza infections in swine will help reduce the impact they have on swine production and reduce the risk of swine-to-human transmission.
INTRODUCTION
Influenza A virus (IAV) is an enveloped virus comprised of 8 negative-sense RNA segments that can encode up to 15 proteins (1–7). Most vaccine efforts target the production of neutralizing antibodies against hemagglutinin (HA), a glycoprotein on the virion's surface that mediates virus entry into susceptible cells (8). Influenza A viruses in swine (IAV-S) of the H1N1, H1N2, and H3N2 subtypes are endemic in swine in North America (9–11), and antigenically distinct strains cocirculate (10, 12–14). Expansion of the IAV-S genetic pool was observed after the introduction into swine of a triple-reassortant internal gene (TRIG) constellation in the late 1990s with a human-origin H3 HA (15). The H3 phylogenetic clade IV emerged in North America in the mid-2000s as an evolutionary branch of clade III TRIG viruses introduced in the 1990s (11, 16) and has continued to increase in diversity. Another human H3 HA, which emerged in the swine population in 2012 and has persisted, is phylogenetically and antigenically distinct from contemporary swine H3 viruses (17), further extending the diversity of an already complex viral ecosystem. In this context of cocirculation of antigenically diverse viruses, it is important to have a better understanding of the breadth of cross-reactions among contemporary antigenically distinct IAV-S viruses.
In contrast to the concerted, global effort that is carried out to select vaccine strains for human seasonal IAV, there are no standardized systems for strain recommendations for IAV-S, although there are several vaccines licensed and available, primarily in the United States of America (18–20). In addition to vaccine composition, the platform and method of vaccine delivery can have a significant impact on protection. Currently licensed technologies available for use in pigs in the United States include whole-inactivated virus (WIV), either commercial or prepared as an autogenous vaccine (18), an HA subunit vaccine that is delivered as RNA in an alphavirus vector (21–23), and a live-attenuated influenza virus (LAIV) vaccine platform (24, 25). Manufacturers are not required to disclose the strain names used in the vaccines on the label or product insert, although one manufacturer reports inclusion of two clade IV H3N2 strains (19), and another manufacturer reports inclusion of clade I and clade IV H3N2 components (20).
Important advances have been made in predicting the antigenic phenotype of H3 IAV based on the genetics of the HA (26–29), and based on the number of predicted antigenic phenotypes in current H3 HA, it is unlikely that a single representative of clade IV would protect against all clade IV H3 viruses in WIV formulations. Recent work on antigenic characterization of contemporary swine H3 viruses in the United States using antigenic cartography determined that, since 2009, there have been four predominant antigenic clusters, defined as cyan, red, and green and a recent human-like antigenic cluster (12, 13, 17). In this study, we tested the efficacies of WIV and LAIV containing H3N2 clade IV viruses of one antigenic cluster to protect against challenge with contemporary viruses of another antigenic cluster. We observed that as few as two amino acid differences in the HA1 of clade IV viruses were sufficient to reduce the efficacy of the WIV platform. More notably, the LAIV provided complete protection even in the presence of antigenic mismatch. These results have significant implications for the use of influenza vaccines in swine in the United States.
RESULTS
Viruses selected by antigenic motif demonstrated loss in HI cross-reactivity.
The four virus strains used in these studies included A/turkey/Ohio/313053/2004 H3N2 (OH/04), A/swine/Indiana/A01260254/2013 H3N2 (IN/13), A/swine/Iowa/A01480656/2014 H3N2 (IA/14), and A/swine/New York/A01104005/2011 H3N2 (NY/11) (study design shown in Table 1). Prior work has characterized the HA antigenic phenotype of OH/04 as belonging to the cyan antigenic cluster and the IN/13 and NY/11 strains to the red antigenic cluster. Sequence analysis predicts that the IA/14 virus belongs to the green antigenic cluster (12, 13, 30). Cross-hemagglutination inhibition (cross-HI) assays were employed to confirm the antigenic properties of the selected viruses. There were ≥2-fold-dilution reductions in HI activity across viruses from different antigenic clusters compared to the homologous HI titers (Table 2).
TABLE 1.
Animal study designa
| Study | Groupb | Vaccine platform (antigenic cluster) | Challenge virus (antigenic cluster) | No. of pigs |
|---|---|---|---|---|
| 1 | NV-NC | No vaccine | No challenge | 8 |
| NV-IN/13 | No vaccine | IN/13 (red) | 8 | |
| OH/04WIV-IN/13 | OH/04WIV (cyan) | IN/13 (red) | 8 | |
| OH/04LAIV-IN/13 | OH/04LAIV (cyan) | IN/13 (red) | 8 | |
| 2 | NV-NC | No vaccine | No challenge | 5 |
| NV-IA/14 | No vaccine | IA/14 (green) | 10 | |
| IA/14WIV-IA/14 | IA/14WIV (green) | IA/14 (green) | 10 | |
| IA/14LAIV-IA/14 | IA/14LAIV (green) | IA/14 (green) | 10 | |
| NV-NY/11 | No vaccine | NY/11 (red) | 9 | |
| IA/14WIV-NY/11 | IA/14WIV (green) | NY/11 (red) | 10 | |
| IA/14LAIV-NY/11 | IA/14LAIV (green) | NY/11 (red) | 10 |
NV, nonvaccinated; NC, nonchallenged; WIV, adjuvanted-whole inactivated virus vaccine; LAIV, live-attenuated influenza A virus vaccine.
The vaccine and challenge strains used in each group are indicated.
TABLE 2.
Cross-HI titers reported as reciprocal GMTsa
| Virus (clade [antigenic cluster])b | Cross-HI GMT (no. of HA1 aa differencesc) for antiserum against indicated virus in study: |
|||
|---|---|---|---|---|
| 1 |
2 |
|||
| OH/04 | IN/13 | IA/14 | NY/11 | |
| OH/04 (IV [cyan]) | 453 | 61 (18) | 14 (17) | 320 (15) |
| IN/13 (IV-A [red]) | 160 (18) | 640 | 160 (3) | 2,560 (3) |
| IA/14 (IV-A [green]) | 57 (17) | 160 (3) | 1,810 | 320 (2) |
| NY/11 (IV-A [red]) | 57 (15) | 485 (3) | 160 (2) | 1,280 |
Paired antisera were generated in swine by delivering two doses of adjuvanted-whole inactivated virus. HI, hemagglutination inhibition; GMT, geometric mean titer.
Phylogenetic and antigenic analysis (color name refers to antigenic clusters) was performed previously by Abente et al. (13).
Number of amino acid (aa) differences out of 329 total amino acids in HA1.
LAIV vaccination protected against antigenically distinct strains.
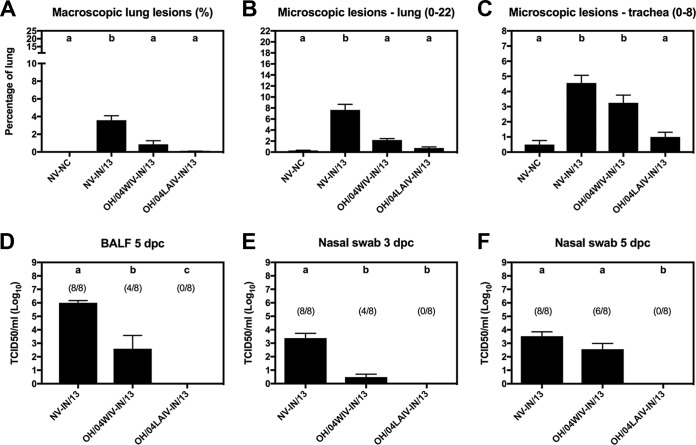
Nonvaccinated (NV) pigs challenged with IN/13 (NV-IN/13) in study 1 had mild lung lesions but high viral titers in bronchoalveolar lavage fluid (BALF) and shed virus in the nasal cavity at 3 and 5 days postchallenge (dpc) (Fig. 1A to F). In vaccinated pigs, protection was defined as statistically significant reductions in lung lesions and virus titers detected in BALF and nasal swab samples compared to the results for the nonvaccinated group. Pigs vaccinated with OH/04 in the WIV platform (OH/04WIV) (cyan) and challenged with IN/13 (red) (OH/04WIV-IN/13) showed significantly reduced lesions of the lungs but no significant reduction in microscopic lesions of the trachea compared to those in the NV-IN/13 control group (Fig. 1A to C). Consistent with these findings and compared to the results for the NV-IN/13 pigs, the OH/04WIV-vaccinated pigs showed no significant reduction in virus titers from nasal swabs at 5 dpc, although they did show significantly reduced viral titers in nasal swabs at 3 dpc and in BALF at 5 dpc (Fig. 1E and F). Notably, pigs vaccinated with OH/04 in the LAIV platform (OH/04LAIV) and challenged with IN/13 (OH/04LAIV-IN/13) had no detectable macroscopic or microscopic lesions in the lungs, minimal detectable lesions in the trachea, and no detectable virus in BALF or in nasal swabs at 3 or 5 dpc (Fig. 1A to F).
FIG 1.
Protection against challenge strains in pigs vaccinated with whole inactivated virus (WIV) or live-attenuated influenza virus (LAIV) in study 1. (A to C) Lung and trachea lesions were evaluated at 5 dpc. (D to F) Viral titers were measured in bronchoalveolar lavage fluid (BALF) at 5 dpc (D) and in nasal swab samples at 3 and 5 dpc (E, F); the number of pigs with a positive virus titer/total number of pigs is indicated above each bar. Bars are labeled with the vaccine and challenge strain used for each group of pigs. Data are presented as mean values ± standard errors of the means. Different lowercase letters within each graph indicate statistically significant differences (P ≤ 0.05). NV, not vaccinated; NC, not challenged.
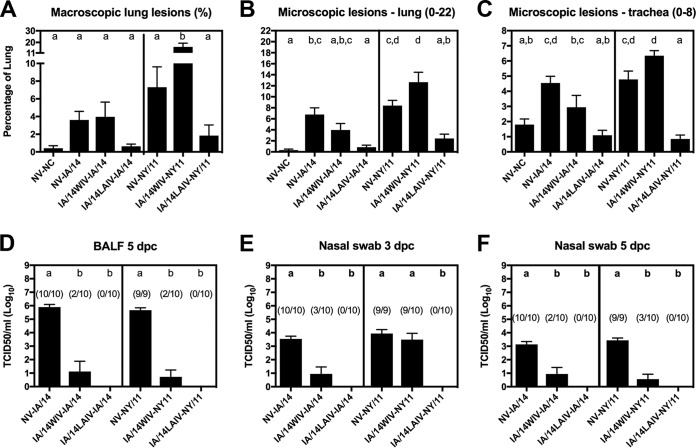
In study 2, the groups of NV-IA/14 and NV-NY/11 pigs developed mild-to-moderate lung lesions with high viral loads in the lungs at 5 dpc and in nasal swabs at 3 and 5 dpc (Fig. 2A to F). Pigs vaccinated with IA/14WIV and challenged with IA/14 showed significantly reduced virus titers in the lungs and in nasal swabs, although lung lesions were not reduced compared to those in the NV-IA/14 control group (Fig. 2A to F). Compared to the NV-NY/11 group, a significant increase in lung macroscopic lesions and a trend toward higher levels of microscopic lesions in the lung and trachea was observed in pigs vaccinated with IA/14WIV (green) and challenged with NY/11 (red) in the antigenically mismatched WIV challenge group. The IA/14WIV-NY/11 group had significantly reduced viral titers in the lung and in nasal swabs at 5 dpc (but not at 3 dpc) compared to the results for the NV-NY/11 control group (Fig. 2D to F). Consistent with the results from study 1, pigs vaccinated with the IA/14LAIV showed significant protection against challenge with either the antigenically matched IA/14 or mismatched NY/11 virus. The IA/14LAIV-vaccinated pigs showed no detectable virus in BALF or nasal swabs irrespective of the challenge virus used. More importantly, the lungs of pigs in the IA/14LAIV groups showed lung lesion scores indistinguishable from those in the negative-control pigs (not vaccinated and not challenged [NV-NC]), indicating efficacious protection after challenge (Fig. 2A to C).
FIG 2.
Protection against challenge strains in pigs vaccinated with whole inactivated virus (WIV) or live-attenuated influenza virus (LAIV) in study 2. (A to C) Lung and trachea lesions were evaluated at 5 dpc. (D to F) Viral titers were measured in bronchoalveolar lavage fluid (BALF) at 5 dpc (D) and in nasal swab samples at 3 and 5 dpc (E, F); the number of pigs with a positive virus titer/total number of pigs is indicated above each bar. Bars are labeled with the vaccine and challenge strain used for each group of pigs. Data are presented as mean values ± standard errors of the means. Different lowercase letters within each graph indicate statistically significant differences (P ≤ 0.05). NV, not vaccinated; NC, not challenged.
IAV-specific systemic antibodies were not predictive of protection.
The serum HI antibody responses in pigs varied depending on the vaccine platform used (Fig. 3A). In study 1, the HI reciprocal geometric mean titers (GMT) prior to challenge in the OH/04WIV- and the OH/04LAIV-vaccinated groups were 1,280 and 247, respectively, against the OH/04 virus (Fig. 3A, left, white bars). Reduced HI cross-reactivity was observed against the antigenically mismatched IN/13 virus, with a reciprocal HI titer of 320 in the OH/04WIV-vaccinated group and a reciprocal HI titer of 40 in the OH/04LAIV-vaccinated group (Fig. 3A, right, white bars). At 5 dpc with the IN/13 virus, no significant effect on the HI titers against the OH/04 virus was observed regardless of the vaccine platform (Fig. 3A, left, gray bars). A modest boost was observed against the IN/13 virus at 5 dpc in the OH/04WIV-vaccinated group, with a reciprocal HI titer of 538 observed, and a >2-fold increase with a reciprocal HI titer of 174 was observed in the OH/04LAIV-vaccinated group (Fig. 3A, right, gray bars).
FIG 3.
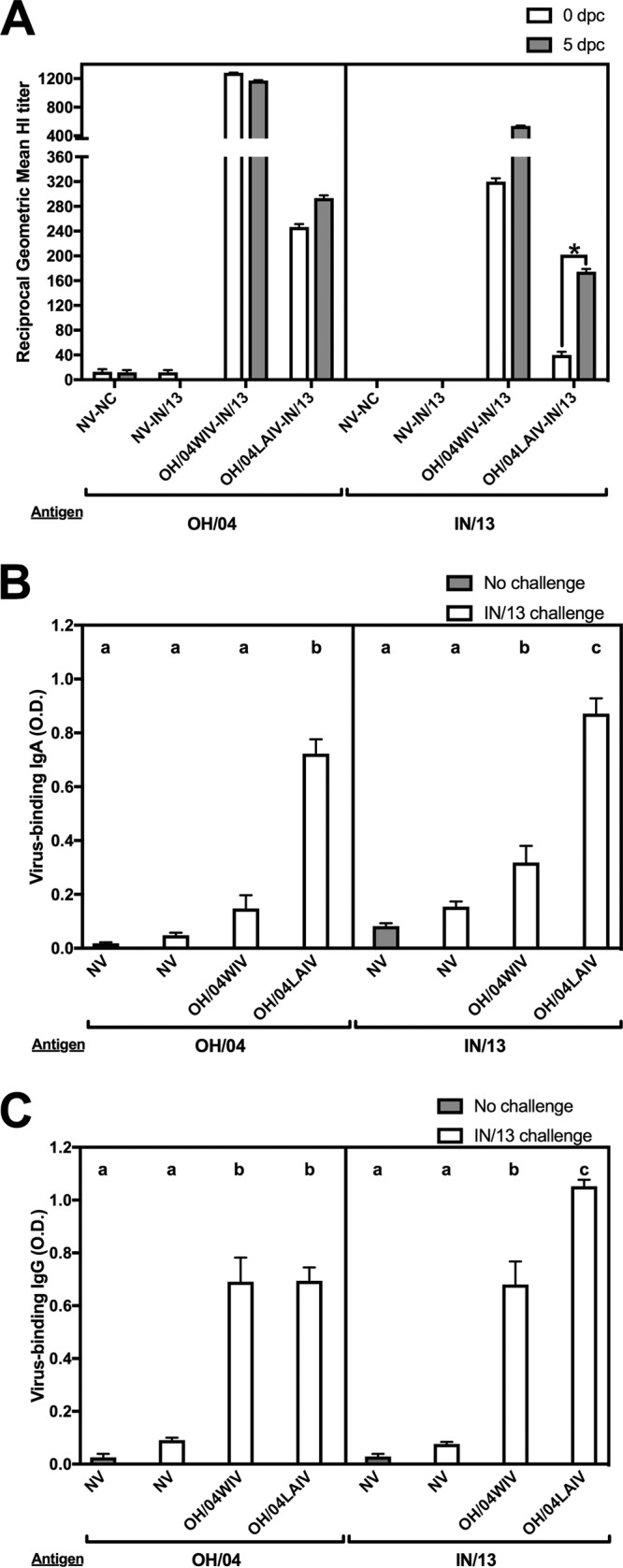
Antibody responses induced in pigs by whole inactivated virus (WIV) and live-attenuated influenza virus (LAIV) vaccines in study 1. (A) Serum hemagglutination inhibition (HI) geometric mean titers against OH/04 and IN/13 at 0 and 5 dpc. *, HI titers differed by more than two serial dilutions between 0 and 5 dpc. (B, C) Whole-virus IgA (B) and IgG (C) assays were performed using BALF from 5 dpc with OH/04 and IN/13 as antigens. Bars are labeled with the vaccine and challenge strain used in each group of pigs. Data are presented as mean optical densities (O.D.) or geometric mean titers ± standard errors of the means. Different lowercase letters within the same graph indicate statistically significant differences (P ≤ 0.05). NV, not vaccinated; NC, not challenged.
Pigs that received the OH/04WIV vaccine had lower levels of IgA-specific antibodies against OH/04 and IN/13 in BALF at 5 dpc, while the OH/04LAIV-vaccinated pigs had high levels of IgA-specific antibodies against both antigens (Fig. 3B). The OH/04WIV and the OH/04LAIV groups had robust IgG-specific antibodies in BALF against both OH/04 and IN/13, but the IgG levels against IN/13 were significantly higher for the OH/04LAIV group than for the OH/04WIV group (Fig. 3C).
In study 2, at 0 dpc, the reciprocal HI GMTs of the two IA/14WIV-vaccinated groups were 43 and 37, while those of IA/14LAIV-vaccinated pigs were 99 and 106 (Fig. 4A, left, white bars). Prior to challenge, the cross-HI activity against the antigenically mismatched NY/11 virus was ≤20 in all vaccinated groups (Fig. 4A, right, white bars). At 5 dpc, HI titers against the matched IA/14 challenge virus showed no significant changes (Fig. 4A, left, gray bars). In contrast, significant HI GMT boosts against IA/14 (from 37 to 279) and NY/11 (from <20 to 121) were observed in IA/14WIV-vaccinated pigs after challenge with NY/11 virus (Fig. 4A). Modest HI GMT boosts were seen against IA/14 (from 106 to 171) and against NY/11 (from <20 to 37) in the IA/14LAIV-vaccinated group after challenge with NY/11 virus.
FIG 4.
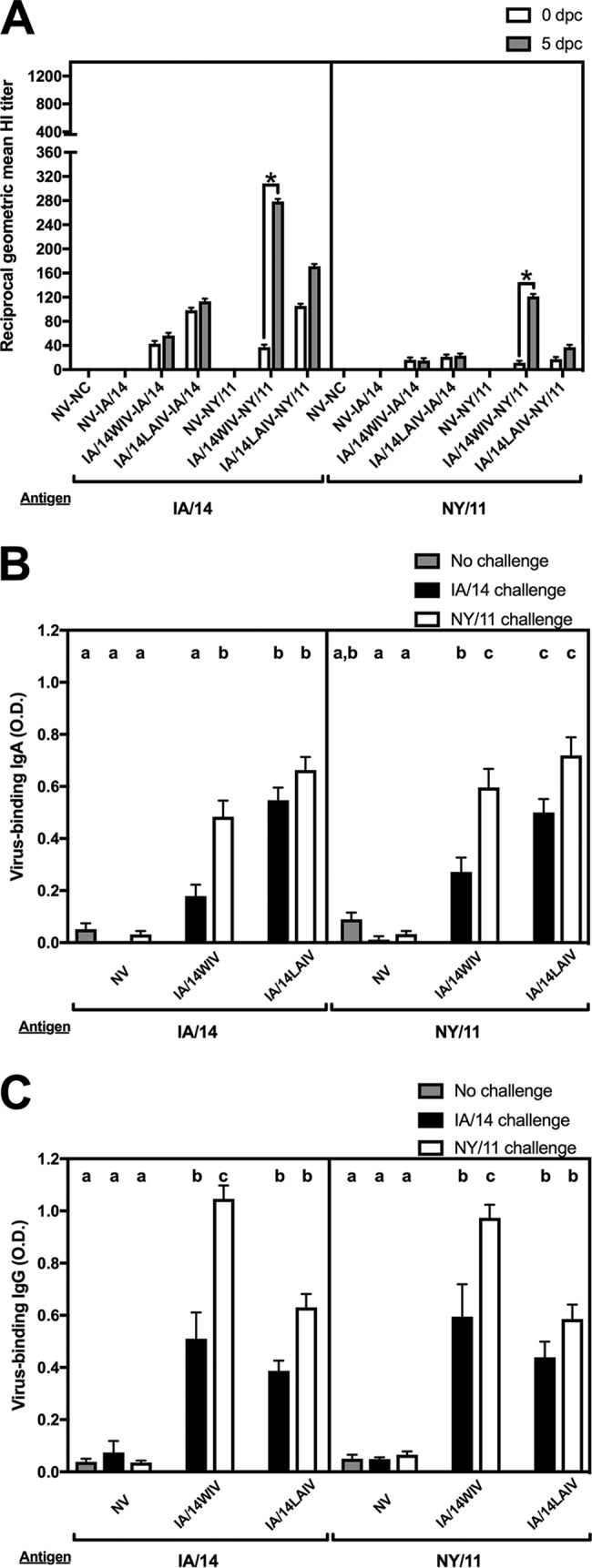
Antibody responses induced in pigs by whole inactivated virus (WIV) and live-attenuated influenza virus (LAIV) vaccines in study 2. (A) Serum hemagglutination inhibition (HI) geometric mean titers against IA/14 and NY/11 at 0 and 5 dpc. *, HI titers differed by more than two serial dilutions between 0 and 5 dpc. (B, C) Whole-virus IgA (B) and IgG (C) assays were performed using BALF from 5 dpc with IA/14 and NY/11 as antigens. Bars are labeled with the vaccine and challenge strain used in each group of pigs. Data are presented as mean optical densities (O.D.) or geometric mean titers ± standard errors of the means. Different lowercase letters within the same graph indicate statistically significant differences (P ≤ 0.05). NV, not vaccinated; NC, not challenged.
At 5 dpc, IA/14- and NY/11-specific IgA antibodies were relatively low in the BALF samples obtained from pigs in the IA/14WIV-IA/14 group compared to the results for the IA/14WIV-NY/11 group (Fig. 4B). After challenge, the IA/14LAIV-vaccinated pigs had robust levels of IgA-specific antibodies against both antigens, independent of challenge virus (Fig. 4B).
The IA/14WIV and IA/14LAIV groups had robust IgG-specific antibodies against both IA/14 and NY/11 regardless of the challenge strain used (Fig. 4C), which were particularly significant in the IA/14WIV group after challenge with the NY/11 virus. Overall, challenge with the NY/11 virus led to significant expression of virus-specific IgG regardless of whether the pigs received the WIV or LAIV (Fig. 4C).
We assessed the neuraminidase (NA) inhibition (NI) titers from a subset of vaccinated pigs to examine the NI antibody responses for the two different vaccine platforms (Table 3). All vaccinated pigs, regardless of platform, were vaccinated with an antigen that contained the OH/04 NA, and we only detected anti-OH/04 NI titers in pigs that received the LAIV.
TABLE 3.
NI titers in sera from a subset of pigs at 0 days postchallenge
| Study | Groupa | NI titer against NA of indicated virusb |
||
|---|---|---|---|---|
| OH/04 | IA/14 (n = 15 NA aa differences)c | NY/11 (n = 21 NA aa differences)c | ||
| 1 | NV-IN/13 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 |
| OH/04WIV-IN/13 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 | |
| OH/04LAIV-IN/13 | 10, 10, 80, 40, 20 | 10, 20, 40, 160, 10 | <10, <10, <10, 10, <10 | |
| 2 | NV-IA/14 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 |
| IA/14WIV-IA/14 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 | |
| IA/14LAIV-IA/14 | 20, 40, <10, 20, 20 | 20, 20, <10, 20, 20 | <10, <10, <10, <10, <10 | |
| NV-NY/11 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 | |
| IA/14WIV-NY/11 | 10, <10, <10, <10, <10 | 10, <10, <10, <10, <10 | <10, <10, <10, <10, <10 | |
| IA/14LAIV-NY/11 | 20, 40, 10, 10, 40 | <10, 10, <10, <10, 40 | <10, 10, <10, <10, <10 | |
| Serum paneld | OH/04WIV | 80, 20 | ||
The vaccine and challenge strains used in each group are defined in Table 1. WIV and LAIV contain the NA of OH/04. NV, nonvaccinated; WIV, adjuvanted-whole inactivated virus vaccine; LAIV, live-attenuated influenza A virus vaccine.
A titer of ≥20 was considered positive. Boldface indicates positive titers. NI, neuraminidase inhibition; NA, neuraminidase.
Number of amino acid (aa) differences out of 469 total amino acids in the NA compared to the sequence of OH/04 NA.
OH/04 antisera generated previously used as part of a panel of sera to generate antigenic maps of H3 IAV-S (13).
DISCUSSION
Several studies have shed light on the antigenic diversity of IAV-S cocirculating in the United States. Antigenic analyses of contemporary H3 IAV-S from the clade IV lineage have revealed at least three major antigenic clusters in the United States (12, 31). The impact on vaccine efficacy due to this antigenic diversity had yet to be explored, and we were particularly interested in examining the levels of cross-protection conferred across antigenically distinct viruses within the H3 IAV-S subtype from the phylogenetic clade IV. We tested two vaccine platforms, an adjuvanted WIV and an LAIV, to assess their efficacies of protection.
In study 1, both vaccine platforms with a cyan-antigenic-cluster virus provided significant protection against a red-antigenic-cluster challenge virus. OH/04, a well-characterized virus that has been used previously in vaccine studies (32, 33) and characterized antigenically to be within the cyan antigenic cluster (13), was chosen as the vaccine antigen. IN/13, a contemporary virus that was recently examined in an animal study (30), contains a red antigenic cluster motif, and it was selected as the challenge strain. The OH/04LAIV provided efficacious protection, despite the antigenic difference with the challenge strain. The OH/04WIV vaccine provided partial protection, which may have been associated with high HI antibody titers. However, infectious virus was still detected in nasal swabs in the OH/04WIV-vaccinated group, suggesting that the WIV vaccine may not be able to block challenge virus transmission, especially to naive contacts, although additional animal studies are required to demonstrate transmission to contacts.
In study 2, pigs were vaccinated with a virus expressing an HA belonging to the green antigenic cluster (IA/14) and challenged with a virus with a HA from the red antigenic cluster (NY/11). Despite high homology between the HA proteins of IA/14 and NY/11, differing only at positions 145 and 289 in the HA1 region, these viruses were markedly distinct antigenically (Table 2). In agreement with the results from study 1, the IA/14LAIV provided complete protection against challenge with the antigenically distinct NY/11 virus. Efficacious protection was not observed in the IA/14WIV-mismatched NY/11 challenge group, which ultimately led to enhanced lung lesions and detectable levels of viral shedding in the upper respiratory tract. Vaccine-associated enhanced respiratory disease (VAERD) has been observed previously in WIV vaccine animal studies when pigs were challenged with an antigenically distinct virus from either the H1 or H3 subtype (34, 35). Our findings are consistent with previous descriptions of VAERD and warrant further investigation, particularly due to the cocirculation of antigenically distinct viruses in the swine population and the use of WIV in the United States.
It must be noted that there were important differences between studies 1 and 2. The age at which pigs were administered the first dose of the vaccine was different. Pigs in study 1 were 3 weeks old when they received the first dose of the vaccine, while pigs in study 2 were 9 weeks old at the time of first vaccination. There is evidence that priming pigs at 4 weeks of age results in higher HI titers against the antigenically matched strains than priming at 9 weeks of age (36), consistent with our finding that pigs in study 1 developed higher HI titers than pigs in study 2. Despite the differences between the two studies presented here, there is a consistent pattern that the LAIV conferred efficient protection against antigenically matched and mismatched strains. In contrast, WIV provided only partial protection against mismatched strains and was associated with the risk of enhanced disease. This observation is consistent with other findings that have examined the vaccine efficacies of WIV and LAIV in pigs in side-by-side comparisons (33, 37, 38). Of interest, an LAIV distinct from the one used in the studies presented here has recently been approved for use in swine in the United States (24, 25, 39) and may provide broader protection, but the H3 strain in the commercial LAIV is of a different phylogenetic clade and further study is required to speculate on its capacity for protection from the contemporary strains tested here.
In the face of the vast antigenic diversity of IAV, not only in swine but also in humans and avian species, efforts have been focused on exploring mechanisms to potentiate immune responses to vaccine antigens to induce broader protection. Some approaches include testing different adjuvants to target specific pathways of the immune response (40–42), and others have examined various prime-boost strategies (43–49). One salient observation from our studies was a detectable back-boost in pigs that were vaccinated with one antigenic phenotype and challenged with an antigenically distinct virus. A significant boost in HI-specific antibodies against the vaccine was observed in the IA/14 WIV group after challenge with NY/11, which suggests that a heterologous prime-boost regime with WIV and LAIV could help broaden the antibody repertoire and warrants further investigation.
In recent years, there has been a collective call to produce a “universal” flu vaccine that can provide broad protection across multiple subtypes (50). Many strategies are being pursued in an effort to improve the protective quality of current vaccines, and one approach includes specifically targeting NA-based immunity (51). In humans, there is evidence that IAV infection produces broadly cross-reactive NA antibodies, in contrast to minimal NA antibody induction in response to nonadjuvanted split-virion vaccine (52). Here, homologous NA antibodies were only induced by the LAIV, which mimics a live exposure. As increasing evidence is reported for a role of NA antibodies in providing protection in humans (53), additional studies are required to better understand the NA antigenic diversity of IAV-S and its implications for vaccine design.
This study has two important limitations. First, we did not compare the T cell response across both vaccine platforms. It is well documented that robust CD4 and CD8 memory T cell responses contribute to heterologous protection (54–56). Previous work in pigs found that LAIV can prime a T cell response in naive pigs (57–59), although when comparing WIV and LAIV, a moderate T cell response was observed in adjuvanted-WIV-treated pigs as opposed to a more robust response in LAIV-treated pigs (35, 58). Second, we assessed functional HA and NA antibodies and did not quantitate the total antibody response to either protein. Nonneutralizing antibodies have been shown to mediate protection and virus clearance indirectly through complement- and antibody-dependent cell-mediated cytotoxicity (60–68). Future work examining these two components of the immune response may help to elucidate the mechanistic factors that result in LAIV providing broader protection.
Better vaccines can help reduce the burden of disease caused by IAV-S on swine production systems and reduce the viruses' chances of two-way interspecies transmission between swine and humans. As IAV-S continues to diversify phylogenetically and antigenically, improved intervention strategies are required. Consistent with our findings, there is evidence that LAIVs can provide broader protection, although factors such as multivalent killed vaccines, adjuvants, and different prime-boost regimens should be explored to identify additional effective approaches.
MATERIALS AND METHODS
Viruses and vaccine preparations.
The A/turkey/Ohio/313053/2004 H3N2 LAIV (OH/04LAIV) with temperature-sensitive and epitope tag-attenuating mutations was previously described (32, 69). In study 1, the OH/04LAIV was administered intranasally (i.n.) at 1 × 106 50% tissue culture infective dose (TCID50) per ml. In study 2, the IA/14LAIV was produced by reverse genetics using the HA gene segment of A/swine/Iowa/A01480656/2014 H3N2 (IA/14) in the background of the OH/04LAIV virus (1 + 7 reassortant) and given i.n. at 1 × 105 TCID50/ml. In order to compare them side by side, the WIV vaccines were prepared from the LAIV of each respective study, either the OH/04LAIV (OH/04WIV) or the IA/14LAIV (IA/14WIV), by UV inactivation and emulsion in a commercial oil-in-water adjuvant (1:5 ratio, Emulsigen D; MVP Laboratories, Inc., Ralston, NE) at 128 hemagglutinating units (HAU)/dose.
The clade IV H3N2 viruses used for challenge were propagated in Madin-Darby canine kidney (MDCK) cells and corresponded to the following field isolates: IA/14 (described above), A/swine/Indiana/A01260254/2013 H3N2 (IN/13), and A/swine/New York/A01104005/2011 H3N2 (NY/11).
Virus antigenic characterization.
The OH/04 HA gene encodes the antigenic motif NHNNYR (amino acids at positions 145, 155, 156, 158, 159, and 189 in the HA's globular head) and belongs to the cyan antigenic cluster. The antigenic motif is known to modulate the antigenic phenotype of a virus as it relates to antibody recognition and hemagglutination inhibition (HI) activity (13). The antigenic motif of the IA/14 HA protein is KYNNYK (green antigenic cluster), and the HA proteins of IN/13 and NY/11 contain the antigenic motif NYNNYK (red antigenic cluster), based on prior findings (12, 13, 30). The cross-HI assays whose results are shown in Table 2 were performed with a subset of a reference antiserum panel described previously (12). Prior to HI testing, sera were treated with receptor-destroying enzyme (Sigma-Aldrich, St. Louis, MO, USA), heat inactivated at 56°C for 30 min, and adsorbed with 50% turkey red blood cells (RBC) to remove nonspecific inhibitors of hemagglutination. Serial 2-fold dilutions starting at 1:10 were tested for the ability to inhibit the agglutination of 0.5% turkey RBC with 8 HAU of OH/04, IN/13, IA/14, NY/11, and reference viruses.
Animal study design.
All pigs were cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center. Pigs were treated with ceftiofur (crystalline free acid) and tulathromycin (Zoetis Animal Health, Florham Park, NJ) and were seronegative to IAV antibodies by a commercial enzyme-linked immunosorbent assay (ELISA) kit (swine influenza virus antibody test; IDEXX, Westbrook, ME) prior to the start of the study. Two animal studies were conducted to examine protection against heterologous challenge with antigenically distinct H3N2 viruses (Table 1). Both studies employed two different vaccine platforms, WIV and LAIV (described above).
Due to pig availability and coordination with an unrelated study, pig ages varied in the two studies. In study 1, 32 3-week-old, cross-bred pigs were divided into 4 groups (Table 1). Pigs in the WIV group were vaccinated intramuscularly and pigs in the LAIV group were vaccinated intranasally with 2 ml of each respective vaccine at 3 and 5 weeks of age (35 and 21 days prior to challenge, respectively). To further explore the extent of protection between antigenic clusters in study 2, 44 9-week-old pigs were divided into 7 groups (Table 1). Similarly, pigs in the WIV groups were vaccinated intramuscularly and pigs in the LAIV groups were vaccinated intranasally with 2 ml of each vaccine at 9 and 11 weeks of age (36 and 19 days prior to challenge, respectively).
On day 0, 3 ml of 1 × 105 TCID50/ml of challenge virus, either IN/13, NY/11, or IA/14, was delivered to pigs intratracheally (2 ml) and intranasally (1 ml). Pigs were challenged under anesthesia, using an intramuscular injection of a ketamine (8 mg/kg of body weight; Phoenix, St. Joseph, MO), xylazine (4 mg/kg; Lloyd Inc., Shenandoah, IA), and Telazol (tiletamine HCl and zolazepam HCl, 6 mg/kg; Zoetis Animal Health, Florham Park, NJ) cocktail. Nasal swabs (Fisherbrand Dacron swabs, Fisher Scientific, Pittsburg, PA) were collected on 0, 3, and 5 days postchallenge (dpc) and used for virus isolation as previously described (37). Pigs were humanely euthanized with a lethal dose of pentobarbital (Fatal Plus; Vortech Pharmaceuticals, Dearborn, MI) at 5 dpc. Bronchoalveolar lavage fluids (BALF) were collected in minimal essential medium (MEM).
Antibody responses.
HI assays were performed with 0.5% turkey RBC according to standard techniques (70). Sera were treated with receptor-destroying enzyme as described above in “LAIV vaccination protected against antigenically distinct strains.” Results are reported as geometric mean antibody titers (GMT).
Isotype-specific (IgA and IgG) ELISAs were performed using OH/04, IN/13, IA/14, and NY/11 as antigens after BALF samples were treated with 10 mM dithiothreitol (DTT; Sigma-Aldrich, St. Louis, MO) and diluted 1:4, as previously described (37). Results are reported as average optical density (OD) values of duplicate wells for each sample.
Neuraminidase-inhibiting (NI) antibodies were assessed by enzyme-linked lectin assay (ELLA) as described by Couzens et al. (71). Briefly, fetuin-coated plates were incubated for 18 to 20 h at 37°C with serial dilutions of postvaccination sera (0 dpc) and H9N2 reassortant viruses containing the NA of A/turkey/Ohio/313053/2004, A/swine/New York/A01104005/2011, or A/swine/Iowa/A01480656/2014. Following incubation, plates were washed 6 times with PBS–0.05% Tween 20 and then incubated for 2 h at room temperature with horseradish peroxidase-conjugated peanut agglutinin (HRP-PNA) (Sigma-Aldrich, St. Louis, MO). Excess HRP-PNA was removed by washing. TMB (3,3′,5,5′-tetramethylbenzidine) substrate (KPL Laboratories, Gaithersburg, MD) was added and the reaction stopped following 10 min of incubation at room temperature. Plates were read at 650 nm, and the resulting OD values were used to calculate the percentage of inhibition. The inverse of the dilution that gave a 50% inhibition of NA activity was considered the NI titer.
Pathological examination and virus detection.
At necropsy, the percentage of lung surface affected with pneumonia was calculated as previously described (72, 73). Right cardiac or affected lung lobes and trachea were collected in formalin, routinely processed, and stained with hematoxylin and eosin. Microscopic lesions in lung and trachea were scored according to previously described parameters (34), and individual composite scores were computed for each pig. Virus isolation-positive nasal swabs and BALF samples were titrated in MDCK cells as previously described (37, 74), and virus titers (TCID50/ml) were calculated for each sample by the method of Reed and Muench (75). The limit of detection for virus titration was 1.78 × 102 TCID50/ml, and any value below this limit was calculated as 0.
Statistical analysis.
Results were analyzed by analysis of variance (ANOVA), with a P value of ≤0.05 considered significant (Prism software; GraphPad, La Jolla, CA), and variables with significant effects by treatment group were subjected to pairwise mean comparisons using the Tukey-Kramer test.
ACKNOWLEDGMENTS
We thank Michelle Harland, Gwen Nordholm, Steven Kellner, and Sarah Shore for technical assistance and Jason Huegel, Justin Miller, Aaron Hebeisen, and Keiko Sampson for assistance with animal studies.
D.R.P. acknowledges that the LAIV technology described in the paper is covered by U.S. patent 8,475,807 B2 and other pending patent applications.
This study was supported by USDA-ARS and by an NIH-National Institute of Allergy and Infectious Diseases (NIAID) interagency agreement (AAI14006-001-00004) associated with the Center of Research in Influenza Pathogenesis, an NIAID-funded Center of Excellence in Influenza Research and Surveillance (grant number HHSN272201400008C). E.J.A. and B.S.K. were supported in part by an appointment to the ARS-USDA Research Participation Program administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the United States Department of Energy and USDA under contract number DE-AC05-06OR23100.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Government.
REFERENCES
- 1.Palese P. 1977. The genes of influenza virus. Cell 10:1–10. doi: 10.1016/0092-8674(77)90133-7. [DOI] [PubMed] [Google Scholar]
- 2.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 4.Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. 2013. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol 87:2455–2462. doi: 10.1128/JVI.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P. 2009. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol 83:8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb RA, Lai CJ. 1980. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell 21:475–485. doi: 10.1016/0092-8674(80)90484-5. [DOI] [PubMed] [Google Scholar]
- 7.Lamb RA, Lai CJ, Choppin PW. 1981. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc Natl Acad Sci U S A 78:4170–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 9.Kitikoon P, Nelson MI, Killian ML, Anderson TK, Koster L, Culhane MR, Vincent AL. 2013. Genotype patterns of contemporary reassorted H3N2 virus in U.S. swine. J Gen Virol 94:1236–1241. doi: 10.1099/vir.0.051839-0. [DOI] [PubMed] [Google Scholar]
- 10.Lorusso A, Vincent AL, Harland ML, Alt D, Bayles DO, Swenson SL, Gramer MR, Russell CA, Smith DJ, Lager KM, Lewis NS. 2011. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J Gen Virol 92:919–930. doi: 10.1099/vir.0.027557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG. 2000. Evolution of swine H3N2 influenza viruses in the United States. J Virol 74:8243–8251. doi: 10.1128/JVI.74.18.8243-8251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. 2014. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J Virol 88:4752–4763. doi: 10.1128/JVI.03805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, Skepner E, Anderson TK, Rajao DS, Perez DR, Vincent AL. 2016. The molecular determinants of antibody recognition and antigenic drift in the H3 hemagglutinin of swine influenza A virus. J Virol 90:8266–8280. doi: 10.1128/JVI.01002-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, Burke DF, Dudas G, Fonville JM, Fouchier RA, Kellam P, Koel BF, Lemey P, Nguyen T, Nuansrichy B, Peiris JM, Saito T, Simon G, Skepner E, Takemae N, ESNIP3 Consortium, Webby RJ, Van Reeth K, Brookes SM, Larsen L, Watson SJ, Brown IH, Vincent AL. 2016. The global antigenic diversity of swine influenza A viruses. Elife 5:e12217. doi: 10.7554/eLife.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73:8851–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsen CW, Karasin AI, Carman S, Li Y, Bastien N, Ojkic D, Alves D, Charbonneau G, Henning BM, Low DE, Burton L, Broukhanski G. 2006. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg Infect Dis 12:1132–1135. doi: 10.3201/eid1207.060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajao DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, Perez DR, Sutton TC, Zhang J, Vincent AL. 2015. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J Virol 89:11213–11222. doi: 10.1128/JVI.01675-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandbulte MR, Spickler AR, Zaabel PK, Roth JA. 2015. Optimal use of vaccines for control of influenza A virus in swine. Vaccines (Basel) 3:22–73. doi: 10.3390/vaccines3010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anonymous. 2017. Zoetis. FluSure XP. Zoetis, Florham Park, NJ: https://www.zoetisus.com/products/pork/flusure-xp/index.aspx Accessed 20 June 2018. [Google Scholar]
- 20.Anonymous. MaxiVac Excell 5.0. Product bulletin. Merck Animal Health, Summit, NJ. [Google Scholar]
- 21.Vander Veen RL, Harris DL, Kamrud KI. 2012. Alphavirus replicon vaccines. Anim Health Res Rev 13:1–9. doi: 10.1017/S1466252312000011. [DOI] [PubMed] [Google Scholar]
- 22.Vander Veen RL, Loynachan AT, Mogler MA, Russell BJ, Harris DL, Kamrud KI. 2012. Safety, immunogenicity, and efficacy of an alphavirus replicon-based swine influenza virus hemagglutinin vaccine. Vaccine 30:1944–1950. doi: 10.1016/j.vaccine.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Anonymous. 2017. Issuance of licenses. Center for Veterinary Biologics notice no. 17-01. Center for Veterinary Biologics, USDA, Ames, IA: https://www.aphis.usda.gov/animal_health/vet_biologics/publications/notice_17_01.pdf. [Google Scholar]
- 24.Anonymous. 2017. Issuance of licenses. Center for Veterinary Biologics notice no. 17-09. Center for Veterinary Biologics, USDA, Ames, IA: https://www.aphis.usda.gov/animal_health/vet_biologics/publications/notice_17_09.pdf. [Google Scholar]
- 25.Genzow M, Goodell C, Kaiser TJ, Johnson W, Eichmeyer M. 2018. Live-attenuated influenza virus vaccine reduces virus shedding of newborn piglets in the presence of maternal antibody. Influenza Other Respir Viruses 12:353–359. doi: 10.1111/irv.12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volz EM, Koelle K, Bedford T. 2013. Viral phylodynamics. PLoS Comput Biol 9:e1002947. doi: 10.1371/journal.pcbi.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luksza M, Lassig M. 2014. A predictive fitness model for influenza. Nature 507:57–61. doi: 10.1038/nature13087. [DOI] [PubMed] [Google Scholar]
- 28.Sun H, Yang J, Zhang T, Long LP, Jia K, Yang G, Webby RJ, Wan XF. 2013. Using sequence data to infer the antigenicity of influenza virus. mBio 4:e00230-. doi: 10.1128/mBio.00230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H, Yang Y, Wang S, Zhu R, Qiu T, Qiu J, Zhang Q, Jin L, He Y, Tang K, Cao Z. 2016. Predicting the mutating distribution at antigenic sites of the influenza virus. Sci Rep 6:20239. doi: 10.1038/srep20239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajao DS, Walia RR, Campbell B, Gauger PC, Janas-Martindale A, Killian ML, Vincent AL. 2017. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol 91:e01763-. doi: 10.1128/JVI.01763-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Z, Gomez J, Bowman AS, Ye J, Long LP, Nelson SW, Yang J, Martin B, Jia K, Nolting JM, Cunningham F, Cardona C, Zhang J, Yoon KJ, Slemons RD, Wan XF. 2013. Antigenic characterization of H3N2 influenza A viruses from Ohio agricultural fairs. J Virol 87:7655–7667. doi: 10.1128/JVI.00804-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pena L, Vincent AL, Ye J, Ciacci-Zanella JR, Angel M, Lorusso A, Gauger PC, Janke BH, Loving CL, Perez DR. 2011. Modifications in the polymerase genes of a swine-like triple-reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J Virol 85:456–469. doi: 10.1128/JVI.01503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitikoon P, Gauger PC, Anderson TK, Culhane MR, Swenson S, Loving CL, Perez DR, Vincent AL. 2013. Swine influenza virus vaccine serologic cross-reactivity to contemporary US swine H3N2 and efficacy in pigs infected with an H3N2 similar to 2011-2012 H3N2v. Influenza Other Respir Viruses 7(Suppl 4):S32–S41. doi: 10.1111/irv.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gauger PC, Vincent AL, Loving CL, Henningson JN, Lager KM, Janke BH, Kehrli ME Jr, Roth JA. 2012. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Pathol 49:900–912. doi: 10.1177/0300985812439724. [DOI] [PubMed] [Google Scholar]
- 35.Sandbulte MR, Platt R, Roth JA, Henningson JN, Gibson KA, Rajao DS, Loving CL, Vincent AL. 2014. Divergent immune responses and disease outcomes in piglets immunized with inactivated and attenuated H3N2 swine influenza vaccines in the presence of maternally-derived antibodies. Virology 464–465:45–54. doi: 10.1016/j.virol.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Souza CK, Rajao DS, Loving CL, Gauger PC, Perez DR, Vincent AL. 2016. Age at vaccination and timing of infection do not alter vaccine-associated enhanced respiratory disease in influenza A virus-infected pigs. Clin Vaccine Immunol 23:470–482. doi: 10.1128/CVI.00563-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vincent AL, Ma W, Lager KM, Richt JA, Janke BH, Sandbulte MR, Gauger PC, Loving CL, Webby RJ, Garcia-Sastre A. 2012. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J Virol 86:10597–10605. doi: 10.1128/JVI.01439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gauger PC, Loving CL, Khurana S, Lorusso A, Perez DR, Kehrli ME Jr, Roth JA, Golding H, Vincent AL. 2014. Live attenuated influenza A virus vaccine protects against A(H1N1)pdm09 heterologous challenge without vaccine associated enhanced respiratory disease. Virology 471–473:93–104. [DOI] [PubMed] [Google Scholar]
- 39.Richt JA, Lekcharoensuk P, Lager KM, Vincent AL, Loiacono CM, Janke BH, Wu WH, Yoon KJ, Webby RJ, Solorzano A, Garcia-Sastre A. 2006. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J Virol 80:11009–11018. doi: 10.1128/JVI.00787-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galliher-Beckley A, Pappan LK, Madera R, Burakova Y, Waters A, Nickles M, Li X, Nietfeld J, Schlup JR, Zhong Q, McVey S, Dritz SS, Shi J. 2015. Characterization of a novel oil-in-water emulsion adjuvant for swine influenza virus and Mycoplasma hyopneumoniae vaccines. Vaccine 33:2903–2908. doi: 10.1016/j.vaccine.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 41.Thomas M, Wang Z, Sreenivasan CC, Hause BM, Gourapura JR, Li F, Francis DH, Kaushik RS, Khatri M. 2015. Poly I:C adjuvanted inactivated swine influenza vaccine induces heterologous protective immunity in pigs. Vaccine 33:542–548. doi: 10.1016/j.vaccine.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Artiaga BL, Yang G, Hackmann TJ, Liu Q, Richt JA, Salek-Ardakani S, Castleman WL, Lednicky JA, Driver JP. 2016. alpha-Galactosylceramide protects swine against influenza infection when administered as a vaccine adjuvant. Sci Rep 6:23593. doi: 10.1038/srep23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Reeth K, Gracia JCM, Trus I, Sys L, Claes G, Versnaeyen H, Cox E, Krammer F, Qiu Y. 2017. Heterologous prime-boost vaccination with H3N2 influenza viruses of swine favors cross-clade antibody responses and protection. NPJ Vaccines 2:11. doi: 10.1038/s41541-017-0012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plant EP, Fredell LJ, Hatcher BA, Li X, Chiang MJ, Kosikova M, Xie H, Zoueva O, Cost AA, Ye Z, Cooper MJ. 2017. Different repeat annual influenza vaccinations improve the antibody response to drifted influenza strains. Sci Rep 7:5258. doi: 10.1038/s41598-017-05579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephenson I, Nicholson KG, Hoschler K, Zambon MC, Hancock K, DeVos J, Katz JM, Praus M, Banzhoff A. 2008. Antigenically distinct MF59-adjuvanted vaccine to boost immunity to H5N1. N Engl J Med 359:1631–1633. doi: 10.1056/NEJMc0805274. [DOI] [PubMed] [Google Scholar]
- 46.Galli G, Hancock K, Hoschler K, DeVos J, Praus M, Bardelli M, Malzone C, Castellino F, Gentile C, McNally T, Del Giudice G, Banzhoff A, Brauer V, Montomoli E, Zambon M, Katz J, Nicholson K, Stephenson I. 2009. Fast rise of broadly cross-reactive antibodies after boosting long-lived human memory B cells primed by an MF59 adjuvanted prepandemic vaccine. Proc Natl Acad Sci U S A 106:7962–7967. doi: 10.1073/pnas.0903181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen DL, Karasin A, Olsen CW. 2001. Immunization of pigs against influenza virus infection by DNA vaccine priming followed by killed-virus vaccine boosting. Vaccine 19:2842–2853. doi: 10.1016/S0264-410X(01)00014-7. [DOI] [PubMed] [Google Scholar]
- 48.Van Reeth K, Labarque G, Pensaert M. 2006. Serological profiles after consecutive experimental infections of pigs with European H1N1, H3N2, and H1N2 swine influenza viruses. Viral Immunol 19:373–382. doi: 10.1089/vim.2006.19.373. [DOI] [PubMed] [Google Scholar]
- 49.Wesley RD, Lager KM. 2006. Overcoming maternal antibody interference by vaccination with human adenovirus 5 recombinant viruses expressing the hemagglutinin and the nucleoprotein of swine influenza virus. Vet Microbiol 118:67–75. doi: 10.1016/j.vetmic.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 50.Paules CI, Sullivan SG, Subbarao K, Fauci AS. 2018. Chasing seasonal influenza: the need for a universal influenza vaccine. N Engl J Med 378:7–9. doi: 10.1056/NEJMp1714916. [DOI] [PubMed] [Google Scholar]
- 51.Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, Wilson PC, Compans RW, Skountzou I, Monto AS. 2018. NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio 9:e02332-. doi: 10.1128/mBio.02332-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, Henry C, Palm AE, Stamper CT, Lan LY, Topham DJ, Treanor J, Wrammert J, Ahmed R, Eichelberger MC, Georgiou G, Krammer F, Wilson PC. 2018. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173:417–429.e410. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, Davey RT Jr, Taubenberger JK. 2016. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. mBio 7:e00417-. doi: 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DiPiazza A, Richards KA, Knowlden ZA, Nayak JL, Sant AJ. 2016. The role of CD4 T cell memory in generating protective immunity to novel and potentially pandemic strains of influenza. Front Immunol 7:10. doi: 10.3389/fimmu.2016.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Souquette A, Thomas PG. 2018. Past life and future effects: how heterologous infections alter immunity to influenza viruses. Front Immunol 9:1071. doi: 10.3389/fimmu.2018.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nussing S, Sant S, Koutsakos M, Subbarao K, Nguyen THO, Kedzierska K. 2018. Innate and adaptive T cells in influenza disease. Front Med 12:34–47. doi: 10.1007/s11684-017-0606-8. [DOI] [PubMed] [Google Scholar]
- 57.Kappes MA, Sandbulte MR, Platt R, Wang C, Lager KM, Henningson JN, Lorusso A, Vincent AL, Loving CL, Roth JA, Kehrli ME Jr. 2012. Vaccination with NS1-truncated H3N2 swine influenza virus primes T cells and confers cross-protection against an H1N1 heterosubtypic challenge in pigs. Vaccine 30:280–288. doi: 10.1016/j.vaccine.2011.10.098. [DOI] [PubMed] [Google Scholar]
- 58.Loving CL, Vincent AL, Pena L, Perez DR. 2012. Heightened adaptive immune responses following vaccination with a temperature-sensitive, live-attenuated influenza virus compared to adjuvanted, whole-inactivated virus in pigs. Vaccine 30:5830–5838. doi: 10.1016/j.vaccine.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masic A, Lu X, Li J, Mutwiri GK, Babiuk LA, Brown EG, Zhou Y. 2010. Immunogenicity and protective efficacy of an elastase-dependent live attenuated swine influenza virus vaccine administered intranasally in pigs. Vaccine 28:7098–7108. doi: 10.1016/j.vaccine.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 60.Jegaskanda S, Luke C, Hickman HD, Sangster MY, Wieland-Alter WF, McBride JM, Yewdell JW, Wright PF, Treanor J, Rosenberger CM, Subbarao K. 2016. Generation and protective ability of influenza virus-specific antibody-dependent cellular cytotoxicity in humans elicited by vaccination, natural infection, and experimental challenge. J Infect Dis 214:945–952. doi: 10.1093/infdis/jiw262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ. 2013. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol 87:5512–5522. doi: 10.1128/JVI.03030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan GS, Leon PE, Albrecht RA, Margine I, Hirsh A, Bahl J, Krammer F. 2016. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog 12:e1005578. doi: 10.1371/journal.ppat.1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanderven HA, Jegaskanda S, Wheatley AK, Kent SJ. 2017. Antibody-dependent cellular cytotoxicity and influenza virus. Curr Opin Virol 22:89–96. doi: 10.1016/j.coviro.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 64.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 66.DiLillo DJ, Palese P, Wilson PC, Ravetch JV. 2016. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest 126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, Winnall WR, Stratov I, Brooks AG, Reading PC, Kent SJ. 2013. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol 190:1837–1848. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- 68.Jegaskanda S, Vandenberg K, Laurie KL, Loh L, Kramski M, Winnall WR, Kedzierska K, Rockman S, Kent SJ. 2014. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity in intravenous immunoglobulin as a potential therapeutic against emerging influenza viruses. J Infect Dis 210:1811–1822. doi: 10.1093/infdis/jiu334. [DOI] [PubMed] [Google Scholar]
- 69.Tang Y, Lee CW, Zhang Y, Senne DA, Dearth R, Byrum B, Perez DR, Suarez DL, Saif YM. 2005. Isolation and characterization of H3N2 influenza A virus from turkeys. Avian Dis 49:207–213. doi: 10.1637/7288-101304R. [DOI] [PubMed] [Google Scholar]
- 70.World Health Organization. 2002. WHO manual on animal influenza diagnosis and surveillance. WHO, Geneva, Switzerland: WHO/CDS/CSR/NCS/2002.5 Rev. 1 http://www.who.int/csr/resources/publications/influenza/en/whocdscsrncs20025rev.pdf. [Google Scholar]
- 71.Couzens L, Gao J, Westgeest K, Sandbulte M, Lugovtsev V, Fouchier R, Eichelberger M. 2014. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J Virol Methods 210:7–14. doi: 10.1016/j.jviromet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 72.Gauger PC, Vincent AL, Loving CL, Lager KM, Janke BH, Kehrli ME Jr, Roth JA. 2011. Enhanced pneumonia and disease in pigs vaccinated with an inactivated human-like (delta-cluster) H1N2 vaccine and challenged with pandemic 2009 H1N1 influenza virus. Vaccine 29:2712–2719. doi: 10.1016/j.vaccine.2011.01.082. [DOI] [PubMed] [Google Scholar]
- 73.Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. 1995. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol 32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- 74.Gauger PC, Vincent AL. 2014. Serum virus neutralization assay for detection and quantitation of serum-neutralizing antibodies to influenza A virus in swine. Methods Mol Biol 1161:313–324. doi: 10.1007/978-1-4939-0758-8_26. [DOI] [PubMed] [Google Scholar]
- 75.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]