Influenza A virus assembly and particle release occur at the apical membrane of polarized epithelial cells. The integral membrane proteins encoded by the virus, HA, NA, and M2, are all targeted to the apical membrane and believed to recruit the other structural proteins to sites of virus assembly. By targeting M2 to the basolateral or endoplasmic reticulum membranes, influenza A virus replication was significantly reduced. Basolateral targeting of M2 reduced the infectious virus titers with minimal effects on virus particle release, while targeting to the endoplasmic reticulum resulted in reduced infectious and total virus particle release. Therefore, altering the expression and the intracellular targeting of M2 has major effects on virus replication.
KEYWORDS: apical, assembly, influenza, membrane transport, virus
ABSTRACT
The influenza A virus (IAV) M2 protein is a multifunctional protein with critical roles in virion entry, assembly, and budding. M2 is targeted to the apical plasma membrane of polarized epithelial cells, and the interaction of the viral proteins M2, M1, HA, and NA near glycolipid rafts in the apical plasma membrane is hypothesized to coordinate the assembly of infectious virus particles. To determine the role of M2 protein apical targeting in IAV replication, a panel of M2 proteins with basolateral plasma membrane (M2-Baso) or endoplasmic reticulum (M2-ER) targeting sequences was generated. MDCK II cells stably expressing M2-Baso, but not M2-ER, complemented the replication of M2-stop viruses. However, in primary human nasal epithelial cell (hNEC) cultures, viruses encoding M2-Baso and M2-ER replicated to negligible titers compared to those of wild-type virus. M2-Baso replication was negatively correlated with cell polarization. These results demonstrate that M2 apical targeting is essential for IAV replication: targeting M2 to the ER results in a strong, cell type-independent inhibition of virus replication, and targeting M2 to the basolateral membrane has greater effects in hNECs than in MDCK cells.
IMPORTANCE Influenza A virus assembly and particle release occur at the apical membrane of polarized epithelial cells. The integral membrane proteins encoded by the virus, HA, NA, and M2, are all targeted to the apical membrane and believed to recruit the other structural proteins to sites of virus assembly. By targeting M2 to the basolateral or endoplasmic reticulum membranes, influenza A virus replication was significantly reduced. Basolateral targeting of M2 reduced the infectious virus titers with minimal effects on virus particle release, while targeting to the endoplasmic reticulum resulted in reduced infectious and total virus particle release. Therefore, altering the expression and the intracellular targeting of M2 has major effects on virus replication.
INTRODUCTION
Virus infections frequently target the polarized epithelial cells lining the respiratory tract, digestive tract, or mucosal surfaces. Efficient production of progeny viruses requires directional coordination among virion components to traffic to either the apical or basolateral plasma membrane. Some viruses target their proteins to the lumen via the apical plasma membrane and can primarily spread only among epithelial cells. Other viruses target their proteins to the basolateral plasma membrane and are more prone to progress to systemic and disseminated infections (1, 2).
Influenza A virus (IAV) is in the family Orthomyxoviridae and has a genome consisting of eight negative-sense, single-stranded RNA segments encoding 10 to 14 proteins (3). All three integral membrane proteins, HA (4, 5), NA (6–8), and M2 (9), are targeted to the apical plasma membrane. M2 apical targeting is not dependent on its acylation or cholesterol binding residues (10). The viral matrix protein, M1, and the viral ribonucleoprotein (vRNP) complex traffic to the apical plasma membrane as well and must interact with the apically targeted viral surface proteins (11–14) for efficient virion assembly (15–17). M1 and vRNP traffic to the apical plasma membrane through interactions with the cytoskeleton (18), and NP has been shown to be intrinsically targeted to the apical plasma membrane (3, 19).
The influenza virus M2 protein is a 97-amino-acid integral membrane protein that forms disulfide-linked tetramers. M2 is predominantly associated with its well-characterized proton channel activity. During the virus entry process, this activity allows for the acidification of the virion interior, which permits vRNP release from M1 (3, 20–22). The C-terminal 54 amino acids of M2 form the highly conserved cytoplasmic tail, which is important for both the assembly and budding processes but has little effect on the M2 proton channel activity (23). The membrane-distal region of the cytoplasmic tail has been shown to be critical for the incorporation of vRNPs into budding particles (15–17, 24, 25). The membrane-proximal region of M2 can induce membrane curvature and has been implicated in ESCRT-independent membrane scission and budding of IAV particles (14, 26), although the extent to which this activity is needed appears to vary between virus strains and experimental systems (27–30).
To investigate the role M2 apical targeting plays in IAV replication, we generated M2 constructs targeted away from the apical plasma membrane, the site of virus budding and assembly. When M2 was targeted to the ER with a dilysine retrieval signal (31–33), virus particles were not released due to a defect in budding. When M2 was targeted to the basolateral plasma membrane, the effect on virus particle production was dependent on the polarization of the cell model being used. The data indicate the intracellular localization of M2 impacts infectious virus production.
RESULTS
Expression of mistargeted M2 constructs.
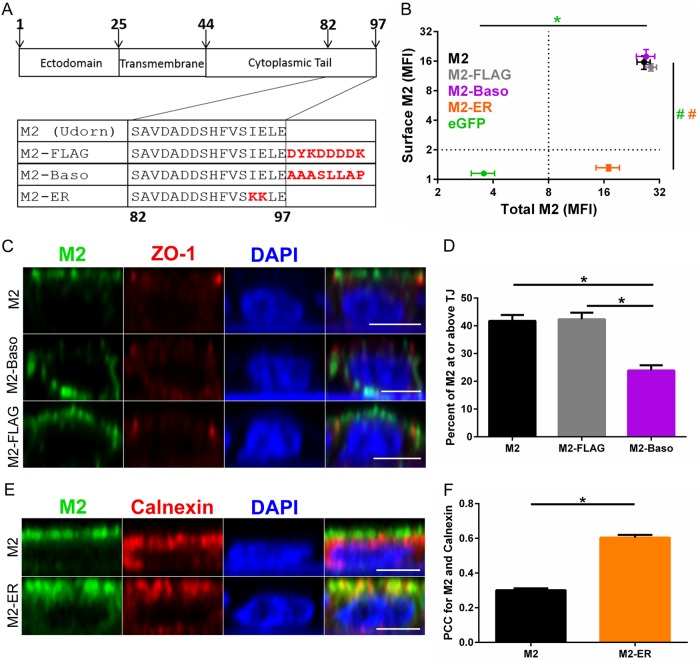
To investigate the role of M2 apical targeting on influenza virus replication, amino acid sequences were mutated (C-terminal KKXX motif) to introduce an endoplasmic reticulum (ER) retention signal (31–33) or added (C-terminal AAASLLAP) to create a basolateral plasma membrane-targeting motif (34) (Fig. 1A). As a control for the addition of amino acid sequences to the M2 C terminus, a FLAG-tagged M2 construct was created which contained the same number of added amino acids as the M2-Baso protein. Stable cell lines expressing the M2 cDNAs in MDCK II cells were generated, since this MDCK sublineage is often used for studies of polarized transport and targeting (35–37). The stable cell lines were characterized for surface and total M2 expression by flow cytometry using the anti-M2 monoclonal antibody 14C2 either before or after membrane permeabilization (Fig. 1B). Wild-type (WT) M2, M2-FLAG, M2-Baso, and M2-ER all express approximately the same amount of total M2. However, M2-ER is not present on the cell surface, while WT M2, M2-FLAG, and M2-Baso all express similar amounts of cell surface M2.
FIG 1.
Localization of M2 proteins. (A) Schematic of Udorn M2 proteins with mutations made to the cytoplasmic tail to alter intracellular membrane targeting. (B) Surface and total M2 staining of MDCK II cells stably expressing the indicated protein. Error bars indicate standard errors of the means (SEM). P values were <0.01 (*) (total) and <0.01 (#) (surface) for M2 compared to WT M2 (ANOVA with Bonferroni multiple-comparison posttest). The color of the significance indicator matches the image legend. n = 4 to 7 stably transfected cell lines. (C and E) Immunofluorescent confocal microscopy images of MDCK II cells stably expressing the indicated M2 construct and immunostained for the indicated proteins. Scale bar, 5 μm. (D) Quantification of apical targeting by percentage of M2 staining at or above the tight junctions (TJ). n = 40 total cells from 2 to 3 stably transfected cell lines. *, P < 0.01 (ANOVA with Bonferroni multiple comparison posttest). (F) Quantification of M2 and calnexin colocalization (Pearson's correlation coefficient). n = 100 total cells from 2 to 3 stably transfected cell lines. *, P < 0.01 (unpaired t test).
Confocal immunofluorescence microscopy was then used to assess the subcellular localization of these constructs. For determining the extent of basolateral targeting of M2-Baso, ZO-1 was used to stain the tight junctions and demarcate the border between the apical and basolateral plasma membranes (Fig. 1C). WT M2 and M2-FLAG primarily stain at or above the level of the tight junction, indicating apical plasma membrane localization. Alternatively, M2-Baso has increased staining below the tight junction and has significantly less apical staining than either WT M2 or M2-FLAG (Fig. 1D). To determine the localization of the M2-ER construct, the ER was stained with an anti-calnexin antibody (Fig. 1E), and then colocalization of M2 and calnexin was determined (Fig. 1F). M2-ER exhibits significantly increased ER colocalization compared to that of WT M2.
Complementation of viruses that do not express M2.
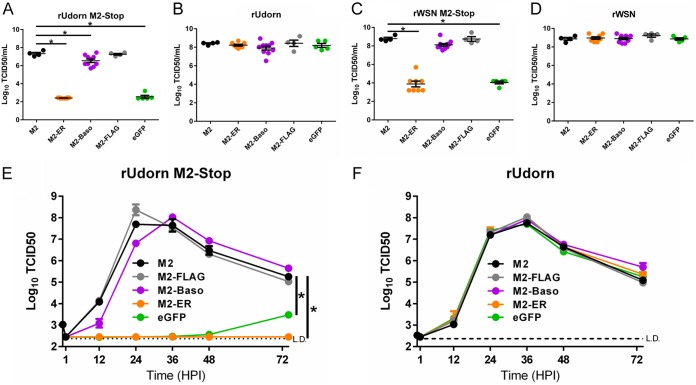
Infectious virus production requires minor amounts of M2, and M2 expressed in trans can complement the replication of recombinant viruses that do not encode the M2 protein (25). Accordingly, MDCK II cells stably transfected with WT M2 or M2-FLAG allowed for the replication of recombinant A/Udorn/72(H3N2) (rUdorn) M2-Stop virus (Fig. 2A). M2-Baso-expressing cells allowed for a significantly decreased amount of infectious virus particle production, while M2-ER- and enhanced green fluorescent protein (eGFP)-expressing cells are unable to support virus replication above the limit of detection (234 50% tissue culture infective doses [TCID50]/ml). Similarly, when recombinant A/WSN/33 (H1N1) (rWSN) M2-Stop virus was used, WT M2-, M2-FLAG-, and M2-Baso-expressing cells all supported similar levels of infectious virus particle production, while M2-ER- and eGFP-expressing cells detected significantly fewer virus particles. This indicates that the inhibitory effect of M2-Baso is not as prominent with the rWSN M2-Stop virus (Fig. 2C). In contrast, all cell lines supported similar levels of both rUdorn WT and rWSN WT infectious virus production, demonstrating that there are no nonspecific, detrimental effects associated with expressing M2 on the ER or basolateral plasma membrane and that the mistargeting M2 mutants do not have a dominant-negative phenotype (Fig. 2B and D). For a more sensitive measure of the ability of the M2 constructs to complement IAV infection, low-multiplicity-of-infection (MOI) growth curves were performed. MDCK II cells expressing WT M2, M2-FLAG, and M2-Baso all were able to replicate rUdorn M2-Stop to approximately 1 × 108 TCID50/ml by 24 to 36 h postinfection (Fig. 2E). The M2-Baso-expressing cells showed slightly slower kinetics of virus production and a later time of peak virus production, but these differences did not reach statistical significance. Neither M2-ER- nor eGFP-expressing cells were able to support rUdorn M2-Stop infectious virus production to appreciable levels. Additionally, WT M2-, M2-FLAG-, M2-Baso-, M2-ER-, and eGFP-expressing MDCK II cells all were able to replicate WT rUdorn to the same peak infectious virus titer with similar kinetics, again indicating that there are no nonspecific detrimental replication defects associated with expressing M2 on the ER or basolateral plasma membrane (Fig. 2F).
FIG 2.
Replication of IAV on MDCK II cells stably expressing M2 proteins. (A to D) rUdorn M2-Stop (A), rUdorn (B), rWSN M2-stop (C), and rWSN (D) endpoint dilution results on the indicated MDCK II cell lines. Stably transfected proteins are indicated on the x axis. Virus was incubated on cells for 5 days. *, P < 0.01 compared to WT M2 (one-way ANOVA). Data are pooled from two independent experiments. (E and F) rUdorn M2-Stop (E) and rUdorn (F) multistep growth curves performed on MDCK II cells stably expressing the indicated proteins. Infectious virus titers were determined on MDCK-M2 cells. Data are representative of two independent replicates. n = 3 wells. *, P < 0.01 compared to Udorn WT (two-way ANOVA).
Replication of recombinant viruses expressing mistargeted M2.
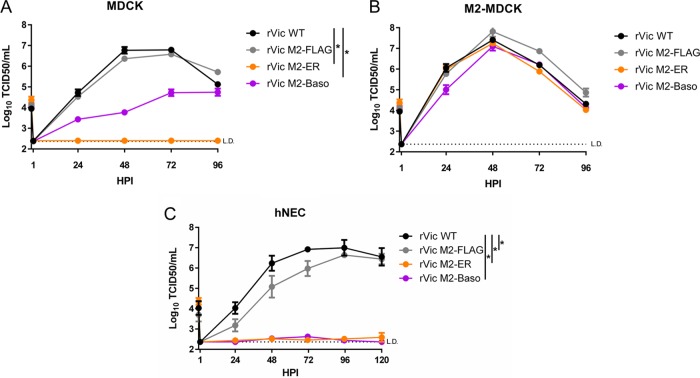
Recombinant IAV encoding the mistargeted M2 proteins were generated in the A/Victoria/361/2011(H3N2) (rVic) background to further study the effects of M2 localization on virus infection. The rVic M2-ER was unable to replicate to detectable levels in MDCK cells (Fig. 3A) but was able to replicate to levels similar to those of both rVic WT and rVic M2-FLAG in M2-MDCK cells (Fig. 3B), indicating that the rVic M2-ER mutation can be complemented by M2-WT protein. rVic M2-Baso replicated to lower peak titers and with slower kinetics in MDCK cells (Fig. 3A) but was indistinguishable from rVic WT and rVic M2-FLAG after infection of M2-MDCK cells (Fig. 3B), again indicating that the effect of targeting M2 to the basolateral membrane was not as detrimental to virus replication as targeting it to the ER.
FIG 3.
Replication of recombinant viruses expressing M2 constructs. (A to C) Multistep growth curves performed on MDCK cells (A), M2-MDCK cells (B), and hNECs (C). Data are pooled from two independent replicates for n = 6 to 7 wells. *, P < 0.01 compared to rVic WT (two-way ANOVA with Bonferroni multiple-comparison posttest).
To utilize a more relevant cell culture model for studying the rVic viruses, primary, differentiated human nasal epithelial cell (hNEC) cultures were used. In hNECs, both rVic M2-ER and rVic M2-Baso were unable to replicate to appreciable levels, while rVic M2-FLAG replicates with slightly decreased kinetics compared to rVic WT (Fig. 3C). Taken together, the data indicate that targeting M2 to the endoplasmic reticulum has a clear detrimental effect on influenza virus replication, while targeting it to the basolateral membrane results in a more variable, cell type-specific inhibition of virus replication.
High-MOI infection and protein analysis of rVic M2-Baso infection of hNECs.
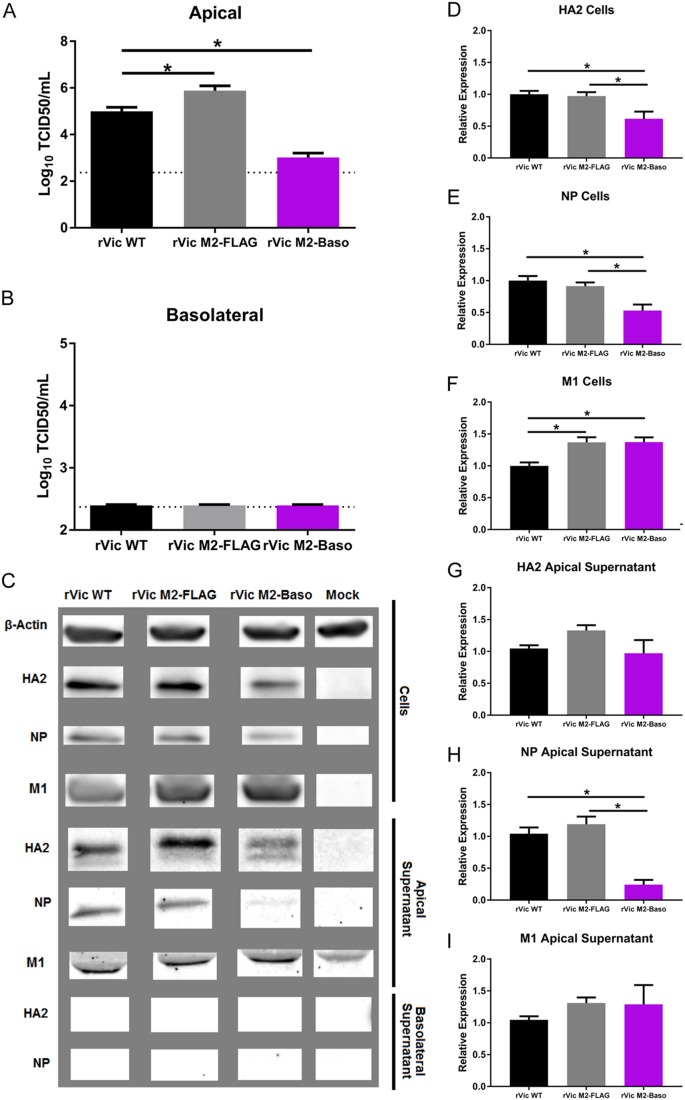
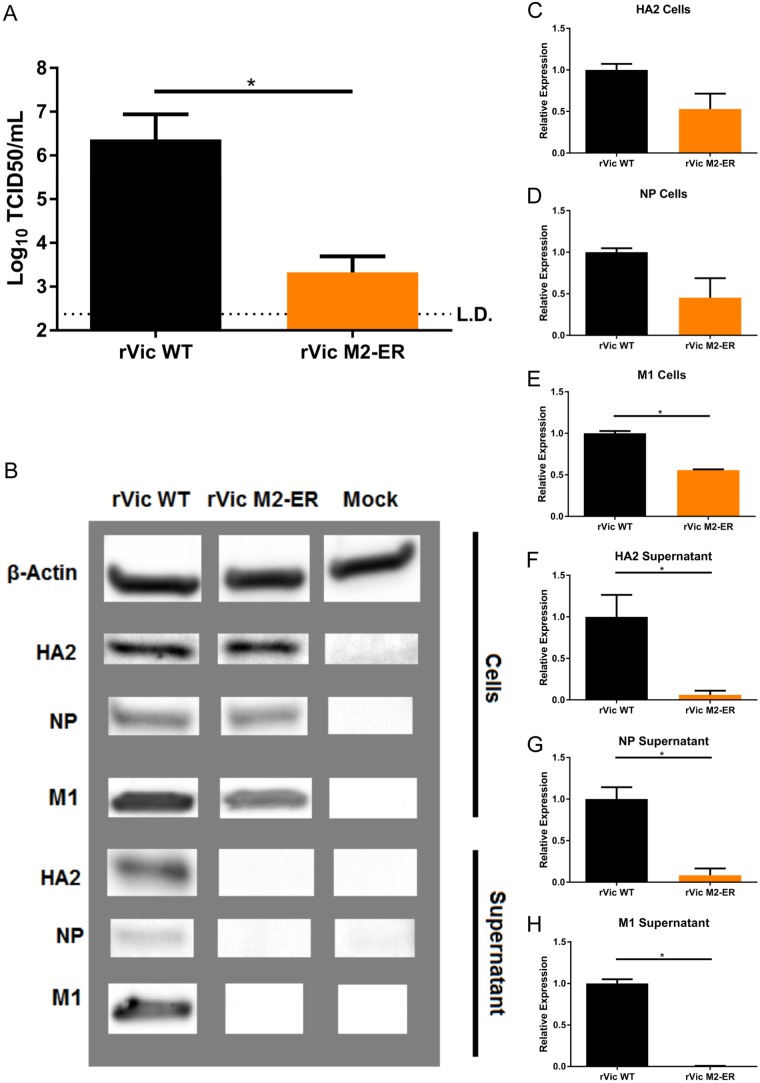
Following high-MOI infection, rVic M2-Baso replicates to significantly lower titers than rVic WT on hNECs (Fig. 4A), consistent with the low-MOI growth curve results. In contrast to the low-MOI data, rVic M2-FLAG replicates to a higher titer than rVic WT during high-MOI infections on hNECs (Fig. 4A). Infectious particles and viral proteins were never detected in the basolateral chamber of hNECs infected with rVic WT, rVic M2-FLAG, or rVic M2-Baso (Fig. 4B and C), indicating there was no aberrant virus budding occurring at the basolateral membranes. Cells infected with rVic M2-Baso expressed slightly smaller amounts of HA2 and NP protein than rVic WT- or rVic M2 FLAG-infected cells (Fig. 4C, D, and E). The levels of cell-associated M1 protein were equal to or higher than those in rVic M2 Baso-infected cells compared to either rVic WT or rVic M2-FLAG (Fig. 4C and F). In the apical supernatant, all three viruses had equivalent levels of HA2 and M1 (Fig. 4C, G, and I), but the amount of NP secreted from rVic M2-Baso-infected cells was significantly reduced, suggesting virus particle release was not significantly reduced but that the proportion of viral proteins in released particles was altered in rVic M2-Baso-infected hNEC cultures. Taken together, the data indicate that the reduced amount of infectious virus particles in the rVic M2-Baso-infected hNECs is due not to reduced virus budding but a decrease in the infectivity of the released virus particles related to the reduced packaging of viral NP.
FIG 4.
Replication and protein expression of rVic M2-Baso and rVic M2-FLAG on hNECs. High-MOI infections were performed with the indicated viruses, and 24 hpi virus titers in the apical (A) and basolateral (B) chambers were determined. Data are pooled from three independent replicates. n = 3 wells in each replicate. *, P < 0.01 (one-way ANOVA). (C) Infected cell supernatants and cells were lysed and analyzed by Western blotting with antibodies against HA2, NP, and M1. (D and G) HA2 quantification for cells (D) and apical supernatant (G). (E and H) NP quantification for cells (E) and apical supernatant (H). (F and I) M1 quantification for cells (F) and apical supernatant (I). Data are pooled from two or three independent replicates. n = 3 wells in each replicate. *, P < 0.01 (one-way ANOVA).
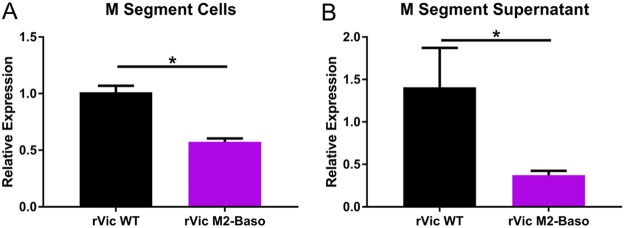
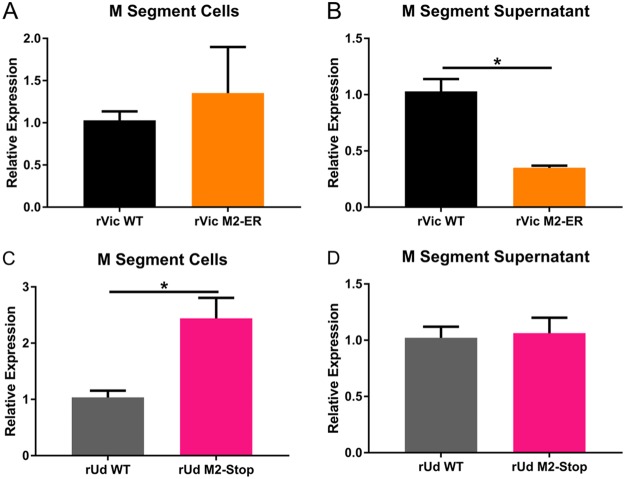
The phenotype of reduced NP incorporation and reduced overall virus infectivity is consistent with mutations in the membrane-distal portion of the M2 cytoplasmic tail (15, 17, 25). This phenotype is often associated with reduced packaging of viral RNA, so we used real-time reverse transcription-quantitative PCR (RT-qPCR) to assess the secretion of M segment RNA in hNEC cultures infected with rVic WT or rVic M2-Baso (Fig. 5A and B). While the amount of M segment RNA was reduced in cells infected with rVic M2-Baso, even smaller amounts were present in the supernatant of rVic M2-Baso-infected cells (Fig. 5A), suggesting reduced viral RNA packaging into particles secreted from rVic M2-Baso-infected cells (Fig. 5B).
FIG 5.
Relative M gene segment expression following infection of rVic M2-Baso and rVic M2-FLAG. High-MOI infections were performed with the indicated viruses, and cells (A) and apical supernatant (B) were assessed by RT-qPCR for relative expression of the virus M gene segment. Two or three independent experiments were performed in triplicate.
rVic M2-Baso replication and cell polarization.
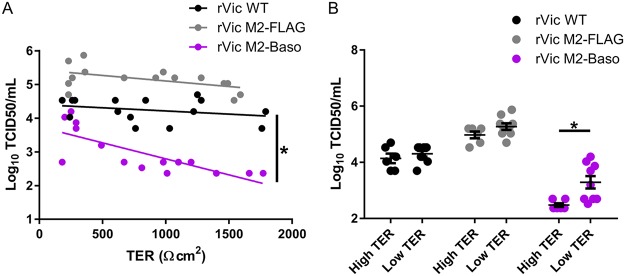
High-MOI infections on hNEC cultures with different degrees of polarization, as determined by trans-epithelial cell electrical resistance (TER), were performed to assess the degree to which the reduced replication of rVic M2-Baso was dependent upon the polarization state of the hNEC cultures. Both rVic WT and rVic M2-FLAG replicated to equivalent levels in hNEC cultures with low or high TER levels, as indicated by linear regression slopes that do not significantly deviate from 0 (Fig. 6A), and similar amounts of infectious virus were detected in cells with a TER reading greater than 650 Ω · cm2 or cells with a TER reading less than 650 Ω · cm2 (Fig. 6B). This indicated that the polarization of the cultures did not affect infectious virus production with these two virus strains. However, rVic M2-Baso replicates less efficiently in hNEC cultures with high TER readings than in cultures with low TER readings, as evidenced by the negative correlation between virus replication and hNEC polarity (Fig. 6A) and a slope that is statistically different from both 0 and the slope of the rVic WT and rVic M2-FLAG lines. Consistent with these results, rVic M2-Baso replicates to a higher titer in cells with a TER reading of less than 650 Ω · cm2 than in cells with a TER reading of 650 Ω · cm2 or greater (Fig. 6B). These data indicate that rVic M2-Baso replicates more efficiently in cells that are not highly polarized, perhaps because the localization of the M2-Baso protein is not as restricted under conditions of reduced cell polarization.
FIG 6.
M2-Baso replication defect is dependent on the polarization of the hNEC culture. High-MOI infections performed with the indicated viruses for 24 h. TER was measured before infection. (A) The correlation between hNEC polarization (Ω · cm2) and virus titer at 24 hpi (MOI of 1) was quantified using a linear regression model. n = 15 per virus pooled from two independent experiments. *, P < 0.05. (B) Virus replication at 24 h on hNEC stratified by TER level (low TER < 650 Ω · cm2 < high TER). *, P < 0.01 Wilcoxon rank sum test. n = 15 per virus pooled from two independent experiments.
High-MOI infection and protein analysis of rVic M2-ER in MDCK cells.
Consistent with the low-MOI growth curve results, rVic M2-ER replicates to a significantly decreased titer compared to that of rVic WT during high-MOI infections of MDCK cells (Fig. 7A). Cells infected with either rVic WT or rVic M2-ER express similar levels of both HA2 and NP (Fig. 7B, C, and D) but a slightly reduced amount of M1 (Fig. 7B and E). However, while there is detectable HA2, NP, and M1 in the supernatant of rVic WT-infected cells, there are virtually no detectable viral proteins in the supernatant of rVic M2-ER-infected cells (Fig. 7B, E, F, and G). This indicates that the defect in rVic M2-ER replication occurs at the level of virus particle budding, in contrast to the defect seen in rVic M2-Baso-infected cells, which appears to be at the level of infectious virus production and not overall particle budding.
FIG 7.
Replication and protein expression of rVic M2-ER on MDCK cells. High-MOI infections were performed with the indicated viruses for 15 to 24 h. (A) Virus titer at endpoint. Data are representative of three independent experiments. n = 3 wells per experiment. *, P < 0.01 (unpaired t test). (B) Infected cell supernatants and cells were lysed and analyzed by Western blotting with antibodies against HA2, NP, and M1. (C to H) HA2 quantification for cells (C) and supernatant (F), NP quantification for cells (D) and supernatant (G), and M1 quantification for cells (E) and supernatant (H). Two or three independent replicates were performed in triplicate, and average data from each replicate were used. *, P < 0.01 (unpaired t test).
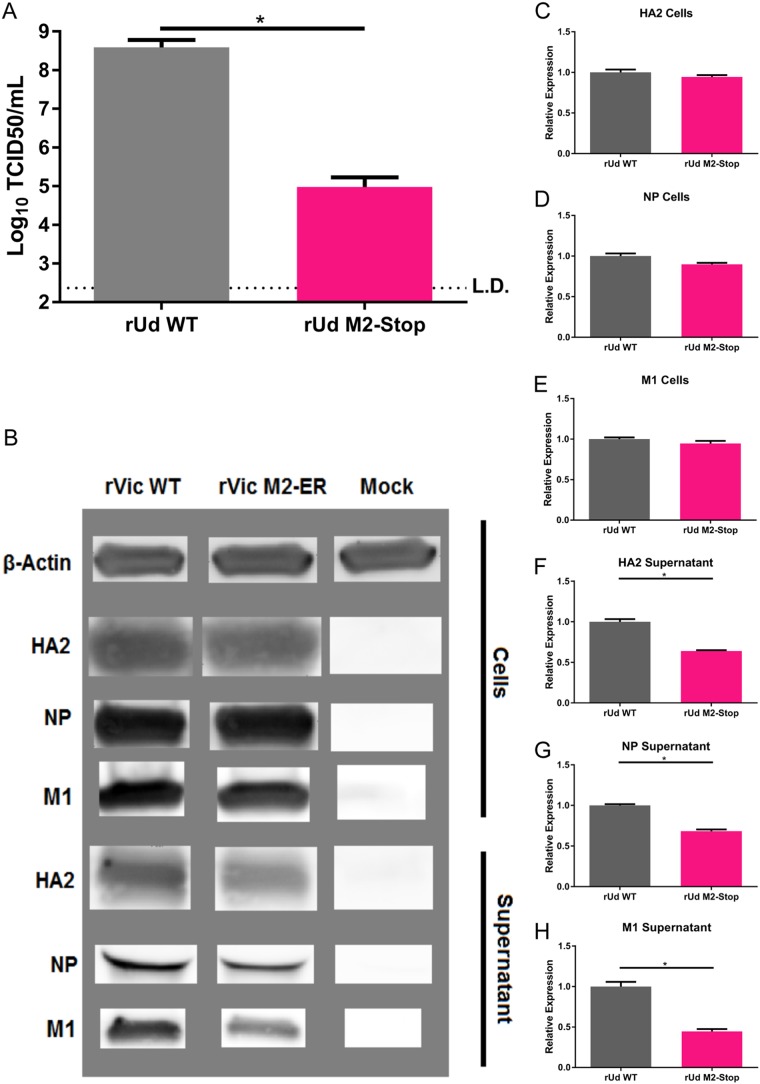
Like rVic M2-ER, rUdorn M2-Stop replicates to a significantly lower titer than rUdorn WT virus in MDCK cells (Fig. 8A). While cells infected with rUdorn M2-Stop express quantities of viral proteins HA2, NP, and M1 similar to those of cells infected with rUdorn WT (Fig. 8B, C, D, and E), rUdorn M2-Stop-infected cell supernatants have detectable amounts of HA2, NP, and M1, although they are reduced compared to rUdorn WT (Fig. 8B, F, G, and H), indicating that viral budding is taking place in rUdorn M2-stop-infected cells, in contrast to what is seen with rVic M2-ER. Not surprisingly, there was very little viral M segment RNA detected in the supernatant of rVic M2-ER-infected MDCK cells (Fig. 9A and B). Compared to the amount of viral M segment RNA present in infected cells, there was also a significant reduction in rUdorn M2-Stop packaging of viral M RNA (Fig. 9C and D).
FIG 8.
Replication and protein expression of rUdorn M2-Stop on MDCK cells. High-MOI infections were performed with the indicated viruses for 24 h. (A) Virus titer at endpoint. Data are representative of three independent experiments. n = 3 wells per experiment. *, P < 0.01 (unpaired t test). (B) Infected cell supernatants and cells were lysed and analyzed by Western blotting with antibodies against HA2, NP, and M1. (C to H) HA2 quantification for cells (C) and supernatant (F), NP quantification for cells (D) and supernatant (G), and M1 quantification for cells (E) and supernatant (H). Data are pooled from two or three independent experiments (n = 3). *, P < 0.01 (unpaired t test).
FIG 9.
Relative M gene segment expression following infection with rVic M2-ER and rUdorn M2-Stop. High-MOI infections were performed with either rVic M2-ER (A and B) or rVic M2-Stop (C and D), and infected cells (A and C) and supernatant (B and D) were assessed by RT-qPCR for relative expression of the virus M gene segment. Two independent experiments were performed in triplicate.
Together, the data indicate that rVic M2-ER has significantly lower release of viral proteins than rUdorn M2-stop, suggesting mislocalizing M2 to the endoplasmic reticulum has a stronger effect on virus particle release than deleting the protein completely.
DISCUSSION
IAV infects and buds primarily from the apical plasma membrane during infection of polarized epithelial cells. All three viral integral membrane proteins are targeted to the apical plasma membrane (4–9). Because of this, it would be expected that critical egress processes like scission and genome packaging are disrupted if viral proteins critical for these roles were targeted to other locations in the cell. When a basolateral localization signal is added to the HA cytoplasmic tail, HA loses its strict apical targeting, but virus budding still occurs at the apical plasma membrane without a drastic decrease in virus replication (38–40). This has been seen with other viral glycoproteins as well. When vesicular stomatitis virus glycoprotein loses its strict targeting to the basolateral plasma membrane, virus was still predominantly released at the basolateral plasma membrane (41). During measles virus infection of polarized epithelial cells, the glycoproteins H (apical and basolateral) and F (basolateral) are not strictly targeted to the apical plasma membrane, but virus is preferentially released apically (42). However, when basolateral targeting signals in these proteins were disrupted, virus was unable to spread by syncytium formation and had decreased lateral spread in vivo, indicating a role for basolateral glycoprotein targeting in measles virus cell-to-cell spread (43). Furthermore, measles virus matrix protein interacts with the cytoplasmic tails of the glycoproteins, permitting M-glycoprotein cotargeting to the apical plasma membrane and allowing for virus airway shedding (44). Similarly, the data described here indicate that M2 apical targeting is required for IAV replication.
In contrast to HA (38, 39), M2 targeted to the basolateral plasma membrane (Fig. 1A, C, and D) or the ER (Fig. 1A, E, and F) resulted in substantial replication defects. This was demonstrated for rVic M2-ER virus in MDCK cells (Fig. 3A) and hNECs (Fig. 3C) and with rUdorn M2-ER in the transcomplementation system (Fig. 2E). For M2-Baso, the replication defect was apparent on MDCK cells infected with rVic M2-Baso (Fig. 3A) and hNECs (Fig. 3C) but not in the transcomplementation system (Fig. 2E). The phenotype penetrates more strongly when M2-Baso is expressed by the recombinant virus (Fig. 3A) rather than the cells being infected (Fig. 2E). Since M2 expression is driven by a strong, constitutively active promoter in the cell lines, more M2 protein is present than for a virus infection, and this may result in some of the tagged protein being expressed on the apical membrane if protein levels exceed the capacity of the basolateral transport system. Only a small amount of M2 is required to complement infection of an M2-stop virus (25), so even a small amount of spillover to the apical compartment could rescue replication to nearly WT levels. When rUdorn expressing WT M2 infects MDCK II cells stably transfected with M2-ER, M2-Baso, or WT M2, it replicates to similar levels with similar kinetics (Fig. 2F), indicating that expressing M2 at these membranes does not induce a dominant-negative phenotype and does not disrupt the ability of the cell to support replication of the WT virus. This is consistent with a prior report that M2-ER does not inhibit the transport of HA to the cell surface (31). Furthermore, MDCK cells stably transfected with WT M2 support the replication of rVic WT, rVic M2-ER, and rVic M2-Baso to the same levels (Fig. 3B), demonstrating that there are no detrimental effects associated with encoding the mutations in the virus genome.
Both rVic M2-Baso and rVic M2-ER are unable to replicate in hNEC cultures. However, rVic M2-ER replication is barely detectable on MDCK cells, while rVic M2-Baso does produce some infectious virus particles. MDCK cells and hNECs differ in numerous characteristics, but critically for these results, hNECs, as ex vivo nasal epithelial cells, are more highly polarized than MDCK cells, an immortalized cell line derived from canine kidney cells. Accordingly, rVic M2-Baso replicates to significantly lower titers on highly polarized hNEC cultures than poorly polarized hNEC cultures (Fig. 6A and B), indicating that the M2-Baso replication phenotype is cell polarization dependent.
M2 plays several roles during IAV replication that could account for the emergence of a replication defect when targeting M2 away from the apical plasma membrane. The C-terminal end of the M2 cytoplasmic tail has been demonstrated to be essential for incorporation of vRNPs into budding particles (15, 17, 25). The membrane-proximal region of the M2 cytoplasmic tail has been demonstrated to contribute to virus membrane scission and budding in an ESCRT-independent manner in some studies (14, 26, 45). However, other studies have shown that many of the individual amino acids in this region of M2 are either dispensable or have a modest phenotype, suggesting the virus strain or the cell culture system utilized plays an important role in phenotype penetration (27–30). During infection of hNECs, rVic M2-Baso and rVic WT secrete similar amounts of virus protein into the apical supernatant (Fig. 4B) despite rVic M2-Baso having significantly reduced levels of infectious virus (Fig. 4A). In vivo epithelial cells and hNECs produce mucus that can interact with IAV particles. After removing the inoculum during high-MOI infections of hNECs, the apical surface was washed with an HA-inactivating acidic PBS wash (PBS containing calcium and magnesium [PBS+] titrated to a pH of 5.2 with citric acid [46]) to prevent infectious virus left over from the inoculum being detected at subsequent time points. However, this inactivated virus may still be present at these time points and detected as either cell associated or secreted into the supernatant, confounding the hNEC protein results.
High-MOI infection with rVic M2-ER on MDCK cells resulted in a significant decrease in virus replication (Fig. 7A) associated with a near-complete absence in virus particle release (Fig. 7B). Similarly, when M2 was not expressed (M2-stop virus), virus replication was reduced (Fig. 8A) but there were detectable amounts of viral proteins present in the apical supernatant (Fig. 8B). Taken together, the data indicate that targeting M2 to the endoplasmic reticulum results in a greater reduction of virus particle budding than deleting the protein completely. This indicates that M2-ER is taking other viral or cellular factors needed for virus budding away from the apical membrane, thereby sequestering them away from the sites of virus budding. M2 has a well-characterized interaction with M1 (15–17), and targeting M2 away from the apical plasma membrane may result in the relocalization of M1 and/or the vRNP. The putative cellular ubiquitin ligase UBR4 has been shown to associate with M2 and promote plasma membrane localization of not only M2 but also HA and NA (47). Furthermore, M2 has been demonstrated to relocalize the essential autophagy protein LC3 to the plasma membrane, resulting in viral subversion of autophagy (48). Annexin A6 has been shown to interact with the M2 protein and alter virus budding (49). Finally, M2 and both cellular Rab11 and the β subunit of the F1Fo-ATPase have been demonstrated to be important for IAV filament budding (50, 51). Therefore, targeting M2 away from the apical plasma membrane may disrupt any number of virus and cellular protein interactions critical for influenza virus assembly, budding, and replication.
Membrane targeting by viral proteins is a critical determinant of tropism and pathogenesis (2). Sendai virus is a pneumotropic mouse virus and buds predominantly from the apical plasma membrane. However, a mutant that buds nonspecifically from the apical and basolateral plasma membranes was isolated and determined to expand tropism and alter pathogenesis (52–55). Similarly, for coronaviruses, apical release is associated with local spread and transmission, while basolateral release is associated with systemic infection (56, 57). In human airway epithelial cells, IAV is released from the apical surface of infected cells (58, 59). IAV is principally an acute respiratory illness. However, systemic infections and complications are often associated with poor prognosis. The mechanisms of IAV systemic dissemination are poorly understood but may involve the depolarization of epithelial cells during infection and subsequent infection of endothelial cells (59), in addition to the well-characterized addition of an HA multibasic cleavage site (60–64). The data described here indicate that targeting M2 to the ER causes a defect in budding and replication, but relocalizing it to the basolateral membrane has more modest effects on the apical budding of IAV. The contribution of M2 mislocalization and the mislocalization of viral/cellular factors associated with M2 on the altered virus budding and infectious virus production needs to be determined.
MATERIALS AND METHODS
Plasmids.
The plasmid pCAGGS (pC) (65), expressing the cDNA for the M2 protein from A/Udorn/72(H3N2) (rUdorn), was described previously (25, 66) and was used as the starting material for all mutagenesis and subsequent establishment of cell lines stably expressing M2 proteins. The pHH21 plasmids expressing full-length influenza virus gene segments were used to generate recombinant viruses (25) as described below. pHH21 plasmids encoding the A/Victoria/361/2011(H3N2) (rVic) virus RNA segments were generated using sequences available in GenBank and the NIAID Influenza Research Database (67–69). The plasmid pBABE, which expresses puromycin N-acetyltransferase, was used for puromycin resistance in stably transfected cells. All mutations were introduced using the QuikChange Lightning site-directed mutagenesis kit (Agilent). All inserts and mutations were confirmed by sequencing. Primer sequences are available upon request.
Cells.
Madin-Darby canine kidney (MDCK) cells and human embryonal kidney cells (293T) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM GlutaMAX (Gibco). M2-MDCK cells stably transfected with the pCAGGS plasmid expressing the M2 protein from A/Udorn/72 (H3N2) were cultured as described for MDCK cells, except the medium was supplemented with 5 μM amantadine hydrochloride and 7.5 μg/ml puromycin dihydrochloride (25). MDCK type II cells (MDCK II) (a gift from Ann Hubbard) were cultured in minimum essential medium (MEM) supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM GlutaMAX. This MDCK subclone is routinely utilized for studies of polarized transport of proteins, so it was deemed the most appropriate for studies of M2 subcellular localization. Human nasal epithelial cell (hNEC) cultures were derived from epithelial cells isolated from nondiseased tissue after endoscopic sinus surgery for non-infection-related conditions and grown at the air-liquid interface (ALI) as previously described (70–73). hNEC differentiation medium (DM), infection medium (IM), and culture conditions have previously been described in detail (73). hNEC polarization was monitored by measuring transepithelial resistance with a Millicell-ERS after adding 250 μl of warm IM without N-acetyl trypsin (NAT) to the apical chamber.
Antibodies.
M2 was detected with an anti-M2 rabbit polyclonal antibody (pAb) (GeneTex GTX125951) or the anti-M2 monoclonal antibody (MAb) 14C2, which recognizes the extracellular domain of M2. HA was detected with pAb anti-H3 (goat) serum derived from A/Aichi/2/1968 (BEI V314-591-157). The antibody that recognizes calnexin is a rabbit pAb (GenScript A01240-40), and the ZO-1A12 MAb (33-9100; ThermoFisher) recognizes ZO-1. β-Actin was detected by a mouse MAb (ab6276; Abcam). The antibody that recognizes NP is a rabbit pAb (GTX125989; GeneTex). All secondary antibodies used are ThermoFisher Scientific Alexa Fluor (AF) fluorescent antibodies conjugated to AF488, AF555, or AF647.
Stable cell line production and culture.
Stable MDCK II cell lines expressing M2, M2-ER, M2-Baso, M2-FLAG, and eGFP were established by cotransfecting pBABE (0.5 μg/well) with pC plasmids encoding the cDNA of interest (1.0 μg/well) and Lipofectamine 2000 (9 μl/well). Transfections were performed in suspension. At 20 h posttransfection the transfection mixture was removed and replaced with culture medium supplemented with 5 μM amantadine hydrochloride (Sigma). At 48 h posttransfection cells were detached with trypsin and plated at low densities onto 96-well plates with culture medium supplemented with amantadine hydrochloride and 3.75 μg/ml puromycin dihydrochloride (Sigma). The puromycin-resistant cells were screened visually for clonality (wells containing a single colony). M2 expression was detected using a plate reader or fluorescence-activated cell sorting (FACS). Live cells were stained on ice with 14C2 (1:500 dilution) followed by AF488-conjugated goat anti-mouse antibody (1:500 dilution). Fluorescence was measured with a FilterMax F5 multimode microplate reader (Molecular Devices). Since M2-ER and M2-Baso are not expressed on the apical surface, 18 to 24 clonal wells were expanded and analyzed by FACS as described below. Stably transfected cells were then grown in medium supplemented with 5 μM amantadine hydrochloride and 3.75 μg/ml puromycin dihydrochloride for MDCK II cells and 7.5 μg/ml puromycin dihydrochloride for MDCK cells.
Flow cytometry.
Cells were detached with trypsin, fixed for 15 min at room temperature with 2% paraformaldehyde (PFA) in PBS, permeabilized with 0.2% Triton X-100 (Sigma) in PBS, blocked for 1 h in PBS with 3% donkey serum and 0.5% bovine serum albumin (BSA; Sigma), and stained for 1 h with 14C2 (diluted 1:500 in blocking solution), followed by staining for 1 h with AF-labeled donkey anti-mouse antibodies (diluted 1:500 in blocking solution). Cells were washed between each step with PBS. Surface M2 staining was performed as described above, except without permeabilization. The cells were analyzed by flow cytometry on a BD FACSCalibur, and the data were processed and analyzed using FlowJo software.
Viruses.
rUdorn and recombinant A/WSN/33 (H1N1) (rWSN) have been described previously (17, 25, 74–76). rVic virus was rescued using the 12-plasmid reverse genetics system (74–77). Briefly, 293T cells were transfected with pHH21 plasmids encoding all 8 influenza A virus gene segments along with protein expression plasmids encoding the NP, PA, PB1, and PB2 cDNAs. Transfected cells were then cocultured with M2-MDCK cells and sampled every 24 h for the release of virus particles. Viruses were then plaque purified and confirmed by sequencing. Stocks of virus were generated by infecting M2-MDCK cells at a low multiplicity of infection (MOI) as described below.
Plaque assays.
Plaque assays were performed in 6-well plates of confluent M2-MDCK cells. Serial 10-fold dilutions of the virus inoculum were generated in IM (DMEM supplemented with 0.3% BSA [Sigma], 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM GlutaMAX [Gibco], and 4 μg/ml N-acetyl trypsin [NAT; Sigma]). Cells were washed twice with PBS containing calcium and magnesium (PBS+), and 250 μl of inoculum was added. The cells were incubated for 1 h at room temperature with rocking. The inoculum was removed and replaced with IM containing 1% agarose. Cells were then incubated at 37°C for 2 to 5 days, fixed in 2% formaldehyde, and stained in a naphthol blue-black solution. For recombinant viruses, plaque picks were collected before fixing by removing the agar above individual plaques with a sterile pipette, aspirating the agar plug into 1 ml of IM, and storing the suspension at −80°C.
TCID50 assay.
MDCK or M2-MDCK cells were plated in 96-well plates, grown to confluence, and washed twice with PBS+. Tenfold serial dilutions of the virus inoculum were made, and 200 μl of dilution was added to each of 6 wells in the plate, followed by incubation for 5 days at 37°C. Cells were then fixed by adding 100 μl of 4% formaldehyde in PBS to each well, followed by staining with a naphthol blue-black solution.
Low-MOI infection.
Before all low-MOI infections, cells were first washed twice with PBS+. Seed stocks (SS) were generated by incubating 250 μl of the plaque pick solution on fully confluent MDCK or M2-MDCK cells for 1 h at room temperature. Inoculum was removed, cells were washed twice with PBS+, and then 1 ml of IM was added before returning the cells to the incubator at 37°C. Virus supernatant was harvested when 50% of the cells displayed cytopathic effect upon inspection with light microscopy (usually 24 to 48 h postinfection [hpi]). The infectious virus titer of the SS was then determined by TCID50 assay and used to generate working stocks (WS). The WS was generated as described above for seed stocks, except infections were performed in 75- or 150-cm2 flasks and the inoculum used was SS diluted to an MOI of 0.01 in IM.
Multistep virus growth curves were performed at an MOI of 0.001 in MDCK cells and 0.1 in hNECs. For MDCK cell infections, the inoculum was diluted in IM, added to cells, and allowed to incubate at room temperature with rocking for 1 h. Cells were then washed 3 times with PBS+ and incubated with fresh IM at 37°C. At the indicated time postinfection, all medium was removed and replaced with fresh IM. The amount of infectious virus in each sample was determined by TCID50 assay on M2-MDCK cells. For hNECs, the apical surface was washed with IM without NAT, and the basolateral DM was replaced immediately before infection. Inoculum diluted in IM without NAT was then added and allowed to incubate on cells at 32°C for 1 to 2 h. The inoculum was then removed, and the cells were washed three times with PBS+, left at the ALI, and returned to 32°C. At the indicated times postinfection, IM without NAT was added to the apical chamber of the wells and allowed to incubate at 32°C for 5 min and removed, and the cells were returned to the incubator at ALI. Basolateral DM was removed and replaced every 48 h for the duration of the growth curves. All samples were stored at −80°C.
High-MOI infection.
High-MOI (1 or 2 TCID50 per cell) infections were performed in the same way as low-MOI infections, except that at the indicated time postinfection, in addition to collecting virus supernatant, cells were lysed in 1% SDS in PBS, collected, and stored at −20°C. Infection supernatant was cleared of cell debris by centrifugation at 1,400 × g for 10 min, and infectious virus titers were determined by TCID50. For hNECs, prior to the first time point, the apical surface was washed with an acidic PBS wash (PBS+ adjusted to a pH of 5.2 with citric acid) (46).
Microscopy.
MDCK II cells stably transfected with the indicated constructs were grown for 6 days on collagen-coated polystyrene Transwell inserts (Costar). Cells were then fixed with 2% PFA at room temperature for 15 min or ice-cold methanol for 10 min at −20°C. Cells were then washed and stored in PBS at 4°C. PFA-fixed cells (but not methanol-fixed cells) were permeabilized with 0.2% Triton X-100 (Sigma) in PBS for 15 min at room temperature before blocking. PFA-fixed cells were then blocked in PBS with 2% donkey serum and 0.5% BSA (blocking solution) and then stained with polyclonal rabbit anti-M2 antibody (1:400 in blocking solution) and ZO-1A12 (1:50 in blocking solution), followed by secondary AF488-linked donkey anti-rabbit antibody and AF555-linked donkey anti-mouse antibody (both 1:500 in blocking solution). Methanol-fixed cells were stained the same way, except 14C2 and rabbit anti-calnexin antibody were used as primary antibodies and AF488-linked donkey anti-rabbit and AF55-linked donkey anti-mouse were used as secondary antibodies (all 1:500 in blocking solution). Cells were washed after each step by dipping membranes in PBS with 0.2% Tween 20 (PBST) 12 times. Cells were washed one final time in MilliQ water before mounting in Prolong gold antifade with 4′,6-diamidino-2-phenylindole (DAPI) (ThermoFisher). Cells were imaged using a Zeiss LSM 700 microscope with Zen software for image acquisition. Images were taken with a 63× objective in Z-stack mode with 0.25- or 0.5-μm spacing between stacks. Images were analyzed and processed with Volocity and FIJI.
SDS-PAGE and Western blotting.
Virus supernatant from high-MOI infections was concentrated through a 35% sucrose cushion at 118,000 × g for 1 h in an Optima Max-TL Beckman Ultracentrifuge. Alternatively, concentrating viral supernatants with Amicon Ultra 0.5-ml centrifugal units (Ultracel-100K; Millipore) or using nonconcentrated infection supernatant yielded comparable results. All samples were mixed with 4× SDS-PAGE sample buffer containing dithiothreitol (DTT; ThermoFisher) before heating for 5 min at 100°C. Samples were loaded alongside Precision Plus Protein all blue protein ladder standards (Bio-Rad) into a 4 to 15% Mini-PROTEAN TGX gel (Bio-Rad) and run at 100 V. Proteins were transferred onto an Immobilon FL polyvinyl difluoride membrane at 100 V for 1 h. Afterwards, membranes were blocked in PBS containing 0.2% Tween 20 and 5% blotting-grade blocker (Bio-Rad). The membrane was then stained with primary antibodies (1:1,000 goat anti-Aichi, 1:1,000 rabbit anti-NP, 1:500 rabbit anti-M1, or 1:10,000 mouse anti-β-actin) and then stained with the appropriate AF-conjugated secondary antibodies (1:1,000). Between each step, the membrane was washed 3 times with PBS containing 0.2% Tween 20. Protein was detected using a FluorChem Q system (Proteinsimple).
Viral RNA quantification by RT-qPCR.
RNA from virus particles secreted into the supernatant was isolated and reverse transcribed into cDNA as described previously (78). RNA from infected cells was isolated using TRIzol and the PureLink RNA minikit according to the manufacturer's protocol (Thermo/Ambion). cDNA from infected cells was generated using a high-capacity cDNA reverse transcription kit with RNase inhibitor (Thermo) according to the manufacturer's protocol, except 20-μl reaction mixtures were used. The viral cDNA was quantified from both secreted virus particles (10 μl cDNA per reaction mix) and infected cells (2 μl cDNA per reaction) by qPCR of the M gene using TaqMan qPCR assays (Applied Biosystems). The forward primers (rVic, 5′-AAGACCAATTCTGTCACCTCTGA-3′; rUdorn, 5′-AAGACCAATCCTGTCACCTCTGA-3′), revere primers (rVic and rUdorn, 5′-CAAAGCGTCTACGCTGCAGTCC3′), and probes (rVic, 5′-/56-FAM/TTTGTTTTC/ZEN/ACGCTCACCGT/3IABkFQ/-3′; rUdorn, 5′-/56-FAM/TTTGTATTC/ZEN/ACGCTCACCGT/3IABkFQ/-3′) were purchased from Integrated DNA Technologies (79). The qPCR assay was performed as described previously (78), except the sample volumes and primers described above were used. Each sample was assayed in triplicate. The number of relative M gene copies in each sample was assessed by normalizing the threshold cycle of each sample to the threshold cycle of WT infection.
Statistical analyses.
Multistep growth curves were analyzed by two-way analysis of variance (ANOVA). FACS, M2-Baso mislocalization, endpoint dilution assays, high-MOI-infection virus replication, protein expression, RNA levels, and TER dependence on titer were analyzed by ANOVA. M2-ER localization and M2-ER and M2-Stop high-MOI infections were analyzed with unpaired t tests. All statistical analyses were performed in GraphPad Prism 7.0.
ACKNOWLEDGMENTS
We thank the members of the Pekosz laboratory, Sabra Klein, members of the Klein laboratory, Kimberly Davis, and members of the Davis laboratory for useful and critical discussions of the data. We also thank and acknowledge Cailin Deal, Jean-Marc Guedon, Melanie Simpson, Theodore Oliphant, Matthew McCown, Hsuan Liu, and Farah El Najjar for technical assistance.
The work was supported by the Shikani/El Hibri Prize for Discovery and Innovation (A.P.), R01 AI097417 (A.P.), HHSN272201400007C (A.P.), R01 AI072502 (A.P.L.), T32 AI007417 (N.W.), and S10OD016374 (Zeiss LSM 700 confocal laser scanning microscope; Scot C. Kuo, principal investigator).
REFERENCES
- 1.Rodriguez Boulan E, Sabatini DD. 1978. Asymmetric budding of viruses in epithelial monlayers: a model system for study of epithelial polarity. Proc Natl Acad Sci U S A 75:5071–5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Compans RW. 1995. Virus entry and release in polarized epithelial cells. Curr Top Microbiol Immunol 202:209–219. [DOI] [PubMed] [Google Scholar]
- 3.Shaw MLP. 2013. Orthomyxoviridae, p 1151–1185. In Knipe DM, Howley, Peter M (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Roth MG, Compans RW, Giusti L, Davis AR, Nayak DP, Gething MJ, Sambrook J. 1983. Influenza virus hemagglutinin expression is polarized in cells infected with recombinant SV40 viruses carrying cloned hemagglutinin DNA. Cell 33:435–443. doi: 10.1016/0092-8674(83)90425-7. [DOI] [PubMed] [Google Scholar]
- 5.Lin S, Naim HY, Rodriguez AC, Roth MG. 1998. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J Cell Biol 142:51–57. doi: 10.1083/jcb.142.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones LV, Compans RW, Davis AR, Bos TJ, Nayak DP. 1985. Surface expression of influenza virus neuraminidase, an amino-terminally anchored viral membrane glycoprotein, in polarized epithelial cells. Mol Cell Biol 5:2181–2189. doi: 10.1128/MCB.5.9.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barman S, Nayak DP. 2000. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J Virol 74:6538–6545. doi: 10.1128/JVI.74.14.6538-6545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kundu A, Avalos RT, Sanderson CM, Nayak DP. 1996. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J Virol 70:6508–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughey PG, Compans RW, Zebedee SL, Lamb RA. 1992. Expression of the influenza A virus M2 protein is restricted to apical surfaces of polarized epithelial cells. J Virol 66:5542–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thaa B, Siche S, Herrmann A, Veit M. 2014. Acylation and cholesterol binding are not required for targeting of influenza A virus M2 protein to the hemagglutinin-defined budozone. FEBS Lett 588:1031–1036. doi: 10.1016/j.febslet.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Ali A, Avalos RT, Ponimaskin E, Nayak DP. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J Virol 74:8709–8719. doi: 10.1128/JVI.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enami M, Enami K. 1996. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J Virol 70:6653–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin H, Leser GP, Zhang J, Lamb RA. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J 16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grantham ML, Stewart SM, Lalime EN, Pekosz A. 2010. Tyrosines in the influenza A virus M2 protein cytoplasmic tail are critical for production of infectious virus particles. J Virol 84:8765–8776. doi: 10.1128/JVI.00853-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen BJ, Leser GP, Jackson D, Lamb RA. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol 82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCown MF, Pekosz A. 2006. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol 80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avalos RT, Yu Z, Nayak DP. 1997. Association of influenza virus NP and M1 proteins with cellular cytoskeletal elements in influenza virus-infected cells. J Virol 71:2947–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrasco M, Amorim MJ, Digard P. 2004. Lipid raft-dependent targeting of the influenza A virus nucleoprotein to the apical plasma membrane. Traffic 5:979–992. doi: 10.1111/j.1600-0854.2004.00237.x. [DOI] [PubMed] [Google Scholar]
- 20.Helenius A. 1992. Unpacking the incoming influenza virus. Cell 69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 21.Martin K, Helenius A. 1991. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol 65:232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinto LH, Holsinger LJ, Lamb RA. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517–528. doi: 10.1016/0092-8674(92)90452-I. [DOI] [PubMed] [Google Scholar]
- 23.Tobler K, Kelly ML, Pinto LH, Lamb RA. 1999. Effect of cytoplasmic tail truncations on the activity of the M(2) ion channel of influenza A virus. J Virol 73:9695–9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwatsuki-Horimoto K, Horimoto T, Noda T, Kiso M, Maeda J, Watanabe S, Muramoto Y, Fujii K, Kawaoka Y. 2006. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J Virol 80:5233–5240. doi: 10.1128/JVI.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCown MF, Pekosz A. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J Virol 79:3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart SM, Wu W-H, Lalime EN, Pekosz A. 2010. The cholesterol recognition/interaction amino acid consensus motif of the influenza A virus M2 protein is not required for virus replication but contributes to virulence. Virology 405:530–538. doi: 10.1016/j.virol.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart SM, Pekosz A. 2011. Mutations in the membrane-proximal region of the influenza A virus M2 protein cytoplasmic tail have modest effects on virus replication. J Virol 85:12179–12187. doi: 10.1128/JVI.05970-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thaa B, Tielesch C, Moller L, Schmitt AO, Wolff T, Bannert N, Herrmann A, Veit M. 2012. Growth of influenza A virus is not impeded by simultaneous removal of the cholesterol-binding and acylation sites in the M2 protein. J Gen Virol 93:282–292. doi: 10.1099/vir.0.038554-0. [DOI] [PubMed] [Google Scholar]
- 30.Roberts KL, Leser GP, Ma C, Lamb RA. 2013. The amphipathic helix of influenza A virus M2 protein is required for filamentous bud formation and scission of filamentous and spherical particles. J Virol 87:9973–9982. doi: 10.1128/JVI.01363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakaguchi T, Leser GP, Lamb RA. 1996. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J Cell Biol 133:733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson MR, Nilsson T, Peterson PA. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J 9:3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson MR, Nilsson T, Peterson PA. 1993. Retrieval of transmembrane proteins to the endoplasmic reticulum. J Cell Biol 121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bello V, Goding JW, Greengrass V, Sali A, Dubljevic V, Lenoir C, Trugnan G, Maurice M. 2001. Characterization of a di-leucine-based signal in the cytoplasmic tail of the nucleotide-pyrophosphatase NPP1 that mediates basolateral targeting but not endocytosis. Mol Biol Cell 12:3004–3015. doi: 10.1091/mbc.12.10.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dukes JD, Whitley P, Chalmers AD. 2011. The MDCK variety pack: choosing the right strain. BMC Cell Biol 12:43. doi: 10.1186/1471-2121-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barker G, Simmons NL. 1981. Identification of two strains of cultured canine renal epithelial cells (MDCK cells) which display entirely different physiological properties. Q J Exp Physiol 66:61–72. doi: 10.1113/expphysiol.1981.sp002529. [DOI] [PubMed] [Google Scholar]
- 37.Richardson JC, Scalera V, Simmons NL. 1981. Identification of two strains of MDCK cells which resemble separate nephron tubule segments. Biochim Biophys Acta 673:26–36. doi: 10.1016/0304-4165(81)90307-X. [DOI] [PubMed] [Google Scholar]
- 38.Mora R, Rodriguez-Boulan E, Palese P, Garcia-Sastre A. 2002. Apical budding of a recombinant influenza A virus expressing a hemagglutinin protein with a basolateral localization signal. J Virol 76:3544–3553. doi: 10.1128/JVI.76.7.3544-3553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barman S, Adhikary L, Kawaoka Y, Nayak DP. 2003. Influenza A virus hemagglutinin containing basolateral localization signal does not alter the apical budding of a recombinant influenza A virus in polarized MDCK cells. Virology 305:138–152. doi: 10.1006/viro.2002.1731. [DOI] [PubMed] [Google Scholar]
- 40.Brewer CB, Roth MG. 1991. A single amino acid change in the cytoplasmic domain alters the polarized delivery of influenza virus hemagglutinin. J Cell Biol 114:413–421. doi: 10.1083/jcb.114.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmer G, Zimmer K-P, Trotz I, Herrler G. 2002. Vesicular stomatitis virus glycoprotein does not determine the site of virus release in polarized epithelial cells. J Virol 76:4103–4107. doi: 10.1128/JVI.76.8.4103-4107.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maisner A, Klenk H-D, Herrler G. 1998. Polarized budding of measles virus is not determined by viral surface glycoproteins. J Virol 72:5276–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moll M, Pfeuffer J, Klenk HD, Niewiesk S, Maisner A. 2004. Polarized glycoprotein targeting affects the spread of measles virus in vitro and in vivo. J Gen Virol 85:1019–1027. doi: 10.1099/vir.0.19663-0. [DOI] [PubMed] [Google Scholar]
- 44.Naim HY, Ehler E, Billeter MA. 2000. Measles virus matrix protein specifies apical virus release and glycoprotein sorting in epithelial cells. EMBO J 19:3576–3585. doi: 10.1093/emboj/19.14.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. 2010. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J Virol 84:5078–5088. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russier M, Yang G, Rehg JE, Wong SS, Mostafa HH, Fabrizio TP, Barman S, Krauss S, Webster RG, Webby RJ, Russell CJ. 2016. Molecular requirements for a pandemic influenza virus: an acid-stable hemagglutinin protein. Proc Natl Acad Sci U S A 113:1636–1641. doi: 10.1073/pnas.1524384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, Yanguez E, Andenmatten D, Pache L, Manicassamy B, Albrecht RA, Gonzalez MG, Nguyen Q, Brass A, Elledge S, White M, Shapira S, Hacohen N, Karlas A, Meyer TF, Shales M, Gatorano A, Johnson JR, Jang G, Johnson T, Verschueren E, Sanders D, Krogan N, Shaw M, Konig R, Stertz S, Garcia-Sastre A, Chanda SK. 2015. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. 2014. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe 15:239–247. doi: 10.1016/j.chom.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma H, Kien F, Maniere M, Zhang Y, Lagarde N, Tse KS, Poon LL, Nal B. 2012. Human annexin A6 interacts with influenza a virus protein M2 and negatively modulates infection. J Virol 86:1789–1801. doi: 10.1128/JVI.06003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruce EA, Digard P, Stuart AD. 2010. The Rab11 pathway is required for influenza A virus budding and filament formation. J Virol 84:5848–5859. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorai T, Goto H, Noda T, Watanabe T, Kozuka-Hata H, Oyama M, Takano R, Neumann G, Watanabe S, Kawaoka Y. 2012. F1Fo-ATPase, F-type proton-translocating ATPase, at the plasma membrane is critical for efficient influenza virus budding. Proc Natl Acad Sci U S A 109:4615–4620. doi: 10.1073/pnas.1114728109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tashiro M, Pritzer E, Khoshnan MA, Yamakawa M, Kuroda K, Klenk HD, Rott R, Seto JT. 1988. Characterization of a pantropic variant of Sendai virus derived from a host range mutant. Virology 165:577–583. doi: 10.1016/0042-6822(88)90601-0. [DOI] [PubMed] [Google Scholar]
- 53.Tashiro M, Yamakawa M, Tobita K, Seto JT, Klenk HD, Rott R. 1990. Altered budding site of a pantropic mutant of Sendai virus, F1-R, in polarized epithelial cells. J Virol 64:4672–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tashiro M, James I, Karri S, Wahn K, Tobita JK, Klenk HD, Rott R, Seto JT. 1991. Pneumotropic revenants derived from a pantropic mutant, F1-R, of Sendai virus. Virology 184:227–234. doi: 10.1016/0042-6822(91)90839-4. [DOI] [PubMed] [Google Scholar]
- 55.Tashiro M, Seto JT, Choosakul S, Yamakawa M, Klenk HD, Rott R. 1992. Budding site of Sendai virus in polarized epithelial cells is one of the determinants for tropism and pathogenicity in mice. Virology 187:413–422. doi: 10.1016/0042-6822(92)90443-S. [DOI] [PubMed] [Google Scholar]
- 56.Rossen JW, Horzinek MC, Rottier PJ. 1995. Coronavirus infection of polarized epithelial cells. Trends Microbiol 3:486–490. doi: 10.1016/S0966-842X(00)89018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cong Y, Ren X. 2014. Coronavirus entry and release in polarized epithelial cells: a review. Rev Med Virol 24:308–315. doi: 10.1002/rmv.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson CI, Barclay WS, Zambon MC, Pickles RJ. 2006. Infection of human airway epithelium by human and avian strains of influenza a virus. J Virol 80:8060–8068. doi: 10.1128/JVI.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan MC, Chan RW, Yu WC, Ho CC, Chui WH, Lo CK, Yuen KM, Guan YI, Nicholls JM, Peiris JS. 2009. Influenza H5N1 virus infection of polarized human alveolar epithelial cells and lung microvascular endothelial cells. Respir Res 10:102. doi: 10.1186/1465-9921-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webster RG, Rott R. 1987. Influenza virus A pathogenicity: the pivotal role of hemagglutinin. Cell 50:665–666. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- 61.Klenk HD, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol 2:39–43. doi: 10.1016/0966-842X(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 62.Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 63.Schrauwen EJ, Bestebroer TM, Munster VJ, de Wit E, Herfst S, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2011. Insertion of a multibasic cleavage site in the haemagglutinin of human influenza H3N2 virus does not increase pathogenicity in ferrets. J Gen Virol 92:1410–1415. doi: 10.1099/vir.0.030379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schrauwen EJ, Herfst S, Leijten LM, van Run P, Bestebroer TM, Linster M, Bodewes R, Kreijtz JH, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Kuiken T, van Riel D. 2012. The multibasic cleavage site in H5N1 virus is critical for systemic spread along the olfactory and hematogenous routes in ferrets. J Virol 86:3975–3984. doi: 10.1128/JVI.06828-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 66.McCown M, Diamond MS, Pekosz A. 2003. The utility of siRNA transcripts produced by RNA polymerase i in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses. Virology 313:514–524. doi: 10.1016/S0042-6822(03)00341-6. [DOI] [PubMed] [Google Scholar]
- 67.Cox NJ, Kitame F, Kendal AP, Maassab HF, Naeve C. 1988. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2). Virology 167:554–567. [PubMed] [Google Scholar]
- 68.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Res 44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Squires RB, Noronha J, Hunt V, García-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, Scheuermann RH. 2012. Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respir Viruses 6:404–416. doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lane AP, Saatian B, Yu XY, Spannhake EW. 2004. mRNA for genes associated with antigen presentation are expressed by human middle meatal epithelial cells in culture. Laryngoscope 114:1827–1832. doi: 10.1097/00005537-200410000-00028. [DOI] [PubMed] [Google Scholar]
- 71.Ramanathan M Jr, Lane AP. 2007. Innate immunity of the sinonasal cavity and its role in chronic rhinosinusitis. Otolaryngol Head Neck Surg 136:348–356. doi: 10.1016/j.otohns.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 72.Ramanathan M Jr, Lee WK, Dubin MG, Lin S, Spannhake EW, Lane AP. 2007. Sinonasal epithelial cell expression of toll-like receptor 9 is decreased in chronic rhinosinusitis with polyps. Am J Rhinol 21:110–116. doi: 10.2500/ajr.2007.21.2997. [DOI] [PubMed] [Google Scholar]
- 73.Fischer WA II, King LS, Lane AP, Pekosz A. 2015. Restricted replication of the live attenuated influenza A virus vaccine during infection of primary differentiated human nasal epithelial cells. Vaccine 33:4495–4504. doi: 10.1016/j.vaccine.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96:9345–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. 2002. Influenza a virus M2 ion channel activity is essential for efficient replication in tissue culture. J Virol 76:1391–1399. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grantham ML, Wu WH, Lalime EN, Lorenzo ME, Klein SL, Pekosz A. 2009. Palmitoylation of the influenza A virus M2 protein is not required for virus replication in vitro but contributes to virus virulence. J Virol 83:8655–8661. doi: 10.1128/JVI.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neumann G, Ozawa M, Kawaoka Y. 2012. Reverse genetics of influenza viruses. Methods Mol Biol 865:193–206. doi: 10.1007/978-1-61779-621-0_12. [DOI] [PubMed] [Google Scholar]
- 78.Liu H, Grantham ML, Pekosz A. 2018. Mutations in the influenza A virus M1 protein enhance virus budding to complement lethal mutations in the M2 cytoplasmic tail. J Virol 92:e00858-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ward CL, Dempsey MH, Ring CJ, Kempson RE, Zhang L, Gor D, Snowden BW, Tisdale M. 2004. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol 29:179–188. doi: 10.1016/S1386-6532(03)00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]