Influenza A virus is a continuously evolving respiratory pathogen. Endemic in swine, H1 and H3 subtype viruses sporadically cause human infections. As each zoonotic infection represents an opportunity for human adaptation, the emergence of a transmissible influenza virus to which there is little or no preexisting immunity is an ongoing threat to public health. Recently isolated variant H1 subtype viruses were shown to display extensive genetic diversity and in many instances were antigenically distinct from seasonal vaccine strains. In this study, we provide characterization of representative H1N1v and H1N2v viruses isolated since the 2009 pandemic. Our results show that although recent variant H1 viruses possess some adaptation markers of concern, these viruses have not fully adapted to humans and require further adaptation to present a pandemic threat. This investigation highlights the need for close monitoring of emerging variant influenza viruses for molecular changes that could facilitate efficient transmission among humans.
KEYWORDS: ferret, H1N1, H1N2, influenza, pathogenesis, risk assessment, variant virus
ABSTRACT
Influenza A virus pandemics are rare events caused by novel viruses which have the ability to spread in susceptible human populations. With respect to H1 subtype viruses, swine H1N1 and H1N2 viruses occasionally cross the species barrier to cause human infection. Recently isolated from humans (termed variants), swine viruses were shown to display great genetic and antigenic diversity, hence posing considerable public health risk. Here, we utilized in vitro and in vivo approaches to provide characterization of H1 subtype variant viruses isolated since the 2009 pandemic and discuss the findings in context with previously studied H1 subtype human isolates. The variant viruses were well adapted to replicate in the human respiratory cell line Calu-3 and the respiratory tracts of mice and ferrets. However, with respect to hemagglutinin (HA) activation pH, the variant viruses had fusion pH thresholds closer to that of most classical swine and triple-reassortant H1 isolates rather than viruses that had adapted to humans. Consistent with previous observations for swine isolates, the tested variant viruses were capable of efficient transmission between cohoused ferrets but could transmit via respiratory droplets to differing degrees. Overall, this investigation demonstrates that swine H1 viruses that infected humans possess adaptations required for robust replication and, in some cases, efficient respiratory droplet transmission in a mammalian model and therefore need to be closely monitored for additional molecular changes that could facilitate transmission among humans. This work highlights the need for risk assessments of emerging H1 viruses as they continue to evolve and cause human infections.
IMPORTANCE Influenza A virus is a continuously evolving respiratory pathogen. Endemic in swine, H1 and H3 subtype viruses sporadically cause human infections. As each zoonotic infection represents an opportunity for human adaptation, the emergence of a transmissible influenza virus to which there is little or no preexisting immunity is an ongoing threat to public health. Recently isolated variant H1 subtype viruses were shown to display extensive genetic diversity and in many instances were antigenically distinct from seasonal vaccine strains. In this study, we provide characterization of representative H1N1v and H1N2v viruses isolated since the 2009 pandemic. Our results show that although recent variant H1 viruses possess some adaptation markers of concern, these viruses have not fully adapted to humans and require further adaptation to present a pandemic threat. This investigation highlights the need for close monitoring of emerging variant influenza viruses for molecular changes that could facilitate efficient transmission among humans.
INTRODUCTION
Influenza A virus (IAV) is a serious respiratory pathogen that poses an ongoing public health problem, as it leads to annual epidemics and occasional pandemics. The latter are rare but recurrent events caused by novel viruses which have the ability to efficiently transmit in human populations with no or limited preexisting immunity. Unlike influenza epidemics, which take place annually during fall and winter months, pandemics are difficult to predict, as they do not occur at regular intervals. Several influenza pandemics have been documented in the past; the most recent was caused by a swine origin H1N1 virus that quickly spread worldwide, leading to approximately 200,000 deaths in 2009, mostly in persons <65 years of age (1).
Classical swine H1N1 influenza viruses, which emerged concurrent with the 1918 pandemic, were first isolated from pigs in the 1930s (2). Evolution of these viruses was furthered by reassortment due to the susceptibility of pigs to both human and avian IAVs (3). In the late 1990s, triple-reassortant swine H3N2 viruses containing genome segments from classical swine H1N1 viruses (nucleoprotein [NP], matrix protein [MP], and nonstructural protein [NS]), North American avian viruses (PB2 and PA), and seasonal human H3N2 viruses (hemagglutinin [HA], neuraminidase [NA], and PB1) were detected in North American pigs (4). These viruses subsequently reassorted with classical swine lineage H1 viruses and human lineage H1 viruses, which were introduced into the swine population, leading to the appearance of H1N1 and H1N2 viruses containing the triple-reassortant internal gene (TRIG) cassette and HA and NA genes originating from swine and/or human influenza viruses (5). Around 2009, quadruple-reassortant H1N1 viruses with a classical swine lineage HA and TRIG constellation containing NA and MP gene segments derived from Eurasian avian-like lineage viruses emerged and caused a pandemic (6).
Endemic in swine, H1 subtype viruses sporadically cause human infections, in most cases via direct exposure to infected pigs, and are termed H1 variant (H1v) viruses. The majority of variant influenza cases are associated with agricultural fairs and are reported during the summer and fall months (7). Prior to the 2009 pandemic, the largest cluster of classical swine H1N1 virus infections occurred in soldiers at Fort Dix, NJ, in 1976 (8, 9). The outbreak caused an estimated 230 cases, 13 hospitalizations, and 1 death. Limited human-to-human transmission was observed; however, the outbreak did not spread to other locations, likely due to poor transmissibility of this virus (10). Since 2005, dozens of H1N1v and H1N2v virus infections have been reported in the United States. As a result of continuous evolution and antigenic drift, ferret antisera produced against previously and currently circulating human seasonal H1 strains poorly cross-reacted with many of the variant viruses isolated in recent years (11, 12). In addition, poor reactivity of pooled human sera suggests that people may not have adequate humoral immunity to contemporary variant viruses. Although sustained human-to-human transmission of variant H1 viruses has not been documented since the 2009 pandemic (13), each human infection caused by a swine influenza virus provides the opportunity for novel influenza viruses to acquire the ability to cause disease and spread readily among humans. Furthermore, North American triple-reassortant H1 isolates have been shown to replicate and transmit among ferrets (13), which serve as the gold standard animal model for influenza virus pathogenesis and transmission risk assessment (14), underscoring the need to closely monitor variant viruses.
Because IAV pathogenicity and transmissibility represent polygenic traits and there is considerable heterogeneity among swine origin H1 subtype viruses, clear interpretation and contextualization of data available on variant virus strains present a challenge. In this study, we provide comparative in vitro and in vivo analysis of recent representative H1N1v (OH/09, IA/39) and H1N2v (MN/45, MN/19, and WI/71) viruses and assess their potential for sustained human-to-human transmission. We evaluated replication kinetics in a human respiratory tract cell line and pathogenesis and transmission in mammalian models, assessed HA activation pH and receptor binding preference, and analyzed molecular features. We found that the recent human infections with variant viruses were caused by strains possessing many mammalian adaptation markers in the HA and polymerase genes. We showed that all the tested variant viruses displayed a preference for alpha 2,6-linked sialic acid receptors (alpha-2,6 SA) but in some instances could also bind alpha-2,3 SA. Each of the viruses replicated efficiently in human airway epithelial cells and in the respiratory tracts of mice and ferrets. Similarities with swine H1 viruses were observed with respect to HA activation pH and transmission rates among cohoused ferrets. However, the ability of variant H1 viruses to transmit through the air among ferrets varied between virus strains but was not HA clade dependent. Together, these findings suggest that although some adaptation markers of concern have been noted, recent variant H1 viruses require further adaptations to present a pandemic threat to humans.
RESULTS
Replication of H1N1v and H1N2v influenza viruses in human airway cells.
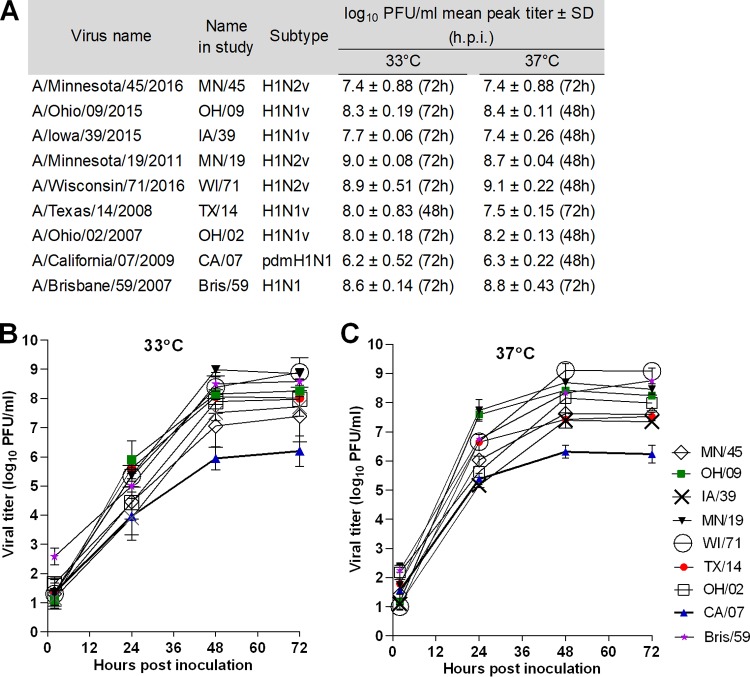
To test the capacity of swine H1v viruses isolated from human cases since the 2009 pandemic to replicate in human airway epithelium cells, the Calu-3 cell line was selected. These immortalized human bronchial epithelium cells, when grown on Transwell inserts, form tight, polarized monolayers that resemble the human airway epithelium (15). Because the ability to replicate efficiently at temperatures found in the upper (∼33°C) and lower (37°C) respiratory tracts of mammals is one of the human adaptation features of influenza viruses, replication kinetics were evaluated at these physiologically relevant temperatures. Five recent variant viruses were selected for comparison (for H1N1v, OH/09 and IA/39; for H1N2v, MN/45, MN/19, and WI/71). We also tested variant viruses isolated prior to the 2009 pandemic (for H1N1v, TX/14 and OH/02) and during the 2009 pandemic (H1N1pdm09 CA/07) and a representative human seasonal virus (H1N1 Bris/59). All viruses were capable of replication in Calu-3 cells. At the 24-h time point, each virus, except OH/02 and TX/14, had significantly higher titers in the cultures incubated at 37°C than in those incubated at 33°C (statistical analysis is included in Table S1 in the supplemental material); however, all of the viruses achieved a similar peak titer at both temperatures by 72 h (Fig. 1A). In comparison to other viruses, significantly lower mean peak titers were observed for the H1N1pdm09 virus, CA/07, which were 6.2 log10 PFU/ml (P < 0.5) at 33°C and 6.3 log10 PFU/ml (P < 0.001) at 37°C (Fig. 1B and C; see also Table S2 in the supplemental material). In contrast, the peak titers for all of the other viruses included in our study ranged between 7.4 and 9.1 log10 PFU/ml at both temperatures. MN/19, WI/71, OH/02, and OH/09 viruses replicated to titers similar to those of the human seasonal virus, Bris/59, while the titers of MN/45, IA/39, and TX/14 viruses were significantly lower at the 48-h and/or 72-h time point than the titer of Bris/59 virus. These data indicate that despite continuous genetic evolution of H1 viruses in swine, all of the variant viruses were ultimately capable of reaching higher titers in human airway cells than those viruses isolated during the 2009 pandemic and, in many instances, were able to replicate as efficiently as the human seasonal virus, Bris/59, warranting more extensive evaluation in additional cell sources and conditions.
FIG 1.
Replication kinetics of H1 influenza viruses in polarized human airway epithelial cells. Calu-3 cells grown on Transwell inserts were inoculated apically with virus at an MOI of 0.01. The cells were incubated at 33°C or 37°C, and culture supernatants were collected at 2, 24, 48, and 72 h postinoculation for viral titer determination by standard plaque assay. (A) Mean peak viral titers are shown as log10 PFU/milliliter ± standard deviation (SD). Hours postinoculation of peak titers are shown parenthetically. (B and C) Replication kinetics curves at 33°C (B) or 37°C (C) are expressed as mean titers from three time courses and are expressed as log10 PFU/milliliter ± SD. The limit of detection is 10 PFU. Statistical analysis is included in Tables S1 and S2 in the supplemental material.
Pathogenicity of H1N1v and H1N2v influenza viruses in mice.
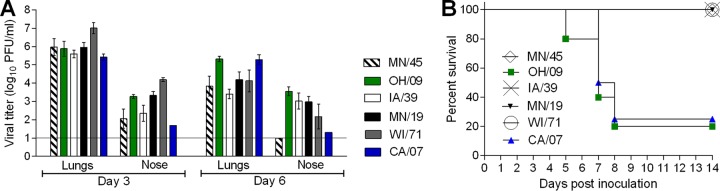
The emergence of new variant influenza strains with pandemic potential necessitates in vivo assessments of virulence. Mice offer numerous advantages as a model for influenza virus research and are routinely used to evaluate the pathogenicity of emerging influenza viruses (16). To determine the capacity of H1v viruses to cause disease in the mouse model, MN/45, OH/09, IA/39, MN/19, and WI/71 viruses were evaluated for their virulence in mice, and a representative H1N1pdm09 virus (CA/07) was included for comparison. High virus titers were detected in the lungs of mice on day 3 postinoculation (p.i.) (mean titer, 5.4 to 7.3 log10 PFU/ml). WI/71 virus displayed significantly higher titers in lungs and in most cases in nose tissues on day 3 p.i. than other tested viruses (see Table S3 in the supplemental material). Lung titers were reduced, but still detectable in all mice, at day 6 p.i. (mean titer, 3.4 to 5.3 log10 PFU/ml) for all of the viruses (Fig. 2A; Table 1). The influenza viruses that reached the highest titers in lung tissue on day 6 p.i. (OH/09 and CA/07) caused mortality rates of 80% and 75%, respectively, whereas all of the mice inoculated with MN/45, MN/19, or IA/39 virus did not display signs of severe disease and survived the challenge (Fig. 2B; Table 1). Virus titers in nose samples of mice averaged from 1.7 to 4.2 log10 PFU/ml on day 3 p.i. and persisted through day 6 p.i. (Fig. 2A) in all mice inoculated with MN/19, WI/71, OH/09, or IA/39 virus (2.2 to 3.6 log10 PFU/ml). CA/07 virus was detected in nose samples from only 1 of 3 mice on day 3 or 6 p.i., and no virus was detected in the MN/45 virus group at the later time point. Brain tissue was also collected from mice on day 3 p.i., but no virus was detected above the limit of detection for any of the viruses (data not shown).
FIG 2.
Pathogenicity of H1 influenza viruses in mice. (A) Mice were intranasally inoculated with 5.0 log10 PFU of virus, and then lung and nose tissues were collected on day 3 (n = 3) and day 6 (n = 3) p.i. for virus titer determination by plaque assay. Mean titers are expressed as log10 PFU/milliliter ± SD. The limit of detection is 10 PFU. (B) The remaining mice were monitored for signs of morbidity and mortality for 14 days p.i. (n = 4 for CA/07, n = 5 for all other viruses); percent survival is shown. Statistical analysis is included in Table S3 in the supplemental material.
TABLE 1.
Pathogenicity of H1N1 and H1N2 influenza viruses in mice
| Virus strain | Name in study | Subtype | % wt loss (day)a | Mortalityb | Mean viral titer log10 PFU/ml ± SD (no. testing positive/total no.)c |
|||
|---|---|---|---|---|---|---|---|---|
| Day 3 p.i. |
Day 6 p.i. |
|||||||
| Lung | Nose | Lung | Nose | |||||
| A/Minnesota/45/2016 | MN/45 | H1N2v | 13.0 (3) | 0 | 6.0 ± 0.5 (3/3) | 2.1 ± 0.5 (3/3) | 3.8 ± 0.5 (3/3) | NDd |
| A/Ohio/09/2015 | OH/09 | H1N1v | 23.0 (7) | 80 | 5.9 ± 0.4 (3/3) | 3.3 ± 0.1 (3/3) | 5.3 ± 0.1 (3/3) | 3.6 ± 0.2 (3/3) |
| A/Iowa/39/2015 | IA/39 | H1N1v | 11.1 (7) | 0 | 5.6 ± 0.2 (3/3) | 2.3 ± 0.4 (3/3) | 3.4 ± 0.3 (3/3) | 3.0 ± 0.4 (3/3) |
| A/Minnesota/19/2011 | MN/19 | H1N2v | 6.8 (5) | 0 | 6.0 ± 0.3 (3/3) | 3.3 ± 0.2 (3/3) | 4.2 ± 0.4 (3/3) | 3.0 ± 0.3 (3/3) |
| A/Wisconsin/71/2016 | WI/71 | H1N2v | 8.4 (6) | 0 | 7.3 ± 0.3 (3/3) | 4.2 ± 0.1 (3/3) | 4.1 ± 0.6 (3/3) | 2.2 ± 0.7 (3/3) |
| A/California/07/2009 | CA/07 | pdm09 H1N1 | 19.2 (7) | 75 | 5.4 ± 0.2 (3/3) | 1.7 (1/3) | 5.3 ± 0.3 (3/3) | 1.3 (1/3) |
Mean maximum percent weight loss (n = 4 for CA/07, n = 5 for all others) following inoculation with 5.0 log10 PFU.
Percent mortality (n = 4 for CA/07, n = 5 for all others) following inoculation with 5.0 log10 PFU.
From animals with detectable virus titer.
ND, not detected.
Pathogenicity and transmissibility of H1N2v influenza viruses in ferrets.
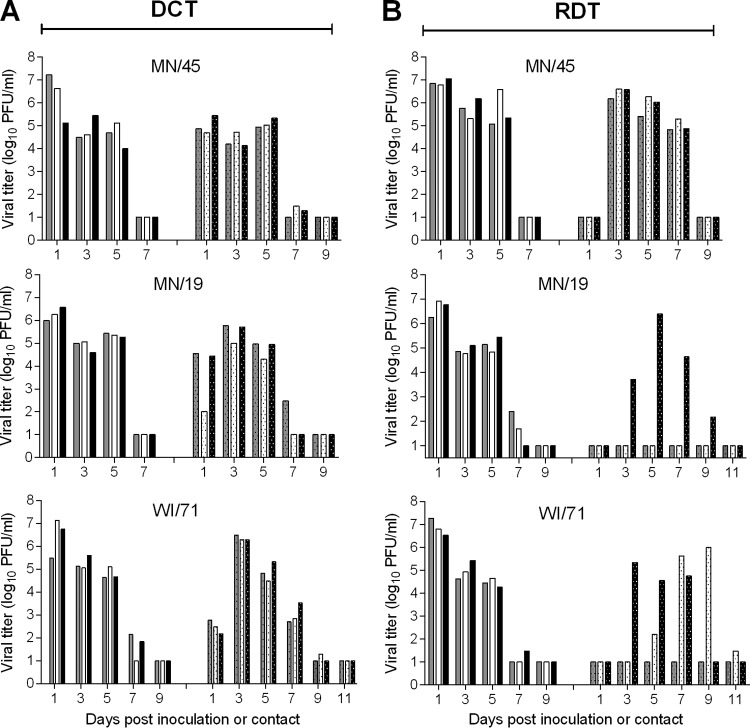
The ferret model has been used widely for the study of influenza virus pathogenicity and transmissibility because ferrets generally exhibit clinical signs and transmission patterns similar to those of humans after influenza virus infection (14). In addition, the ferret model has been indispensable in providing information necessary for evaluation of the pandemic potential of novel influenza viruses using the Influenza Risk Assessment Tool (IRAT) (17). Previous analysis of the pathogenicity and transmission of OH/09 and IA/39 viruses in ferrets showed that two H1N1v viruses replicated well in the ferret respiratory tract, transmitted efficiently among cohoused ferrets, but transmitted less efficiently than CA/07 virus in the respiratory droplet transmission model (11). Because none of the recently isolated variant H1N2v viruses has been tested in the ferret model, we selected 3 representative viruses (MN/45, MN/19, and WI/71) for risk assessment. Groups of 6 to 9 ferrets each were inoculated with 6.0 log10 PFU of H1N2v virus and were evaluated for their ability to transmit virus to naive ferrets in both direct contact and respiratory droplet settings. The direct contact model provides the opportunity for transmission by contact or inhalation, while the respiratory droplet model restricts transmission events to those occurring through the air. All animals inoculated with H1N2v influenza viruses exhibited mild lethargy and a mean increase in body temperature, 1.2 to 1.6°C above baseline (Table 2). Nasal discharge and/or sneezing was observed in some of the inoculated ferrets (5/6 MN/45, 5/9 MN/19, 3/6 WI/71), and ocular discharge was observed in one of the WI/71 virus-inoculated ferrets. MN/45 virus-infected animals had the greatest weight loss (mean maximum, 15.6%) among all the virus groups. All of the animals survived the full course of infection except for one ferret inoculated with WI/71 virus, which was euthanized due to excessive weight loss. Each of the viruses replicated well in inoculated animals; nasal wash titers peaked on day 1 p.i. (mean titer, 6.6 to 6.7 log10 PFU/ml) and were maintained at ≥4.0 log10 PFU/ml through day 5 p.i. (Fig. 3). In general, severity of disease and respiratory signs were comparable with those observed for other H1N1v and H1N1pdm09 viruses tested in this model (11, 18, 19). In the direct-contact transmission model, MN/45 virus was detected in all 3 contact animals at ≥4.7 log10 PFU/ml 1 day postcontact (p.c.), while MN/19 and WI/71 virus shedding was not as high in all of the contact animals until day 3 p.c. (Fig. 3A). In the respiratory droplet transmission model, MN/45 virus transmitted to all 3 contact animals by day 3 p.c. and peaked at 6.2 to 6.6 log10 PFU/ml. MN/19 and WI/71 influenza viruses did not transmit by respiratory droplets as readily; MN/19 virus was detected in a single contact animal, and WI/71 virus was detected in 2 of the contact ferrets (Fig. 3B). Consistently, seroconversion was observed only in those contact ferrets that had detectable virus in nasal wash samples.
TABLE 2.
Pathogenicity and transmissibility of H1N2 influenza viruses in ferrets
| Virus | NW titera | % wt lossb | Mean maximum temp (°C) changec | Lethargyd | Mortality (no. euthanized/total no.)e | No. of animals with detectable virus in NW or HI titer in serum/total no.f |
Mean titer ± SD (log10 PFU/ml or g)g |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DCT |
RDT |
|||||||||||
| Virus detection in NW | Seroconv | Virus detection | Seroconv | Nasal turbinates | Trachea | Lung | ||||||
| MN/45 | 6.7 ± 0.6 | 15.6 (6/6) | 1.2 (6/6) | 1.2 (6/6) | 0/6 | 3/3 | 3/3 | 3/3 | 3/3 | 5.3 ± 0.2 (3/3) | 4.5 ± 1.0 (3/3) | 3.8 ± 2.0 (3/3) |
| MN/19 | 6.6 ± 0.4 | 9.4 (5/9) | 1.6 (9/9) | 1.1 (9/9) | 0/9 | 3/3 | 3/3 | 1/3 | 1/3 | 4.9 ± 0.5 (3/3) | 4.6 ± 0.5 (3/3) | 4.1 ± 0.5 (3/3) |
| WI/71 | 6.7 ± 0.6 | 13.6 (5/6) | 1.6 (6/6) | 1.3 (6/6) | 1/6 | 3/3 | 3/3 | 2/3 | 2/3 | 4.2 ± 0.5 (3/3) | 4.1 ± 1.4 (3/3) | 3.7 ± 1.0 (3/3) |
Mean maximum nasal wash (NW) titer of inoculated ferrets (PFU/ml ± SD).
Mean percent maximum weight loss within 10 days postinoculation; in parentheses, no. of ferrets displaying weight loss/total no. of animals.
Mean maximum temperature change (°C) relative to baseline; in parentheses, no. of ferrets displaying temp change/total no. of animals.
As measured by the relative inactivity index (73); in parentheses, no. of ferrets displaying lethargy/total no. of animals.
Number of animals euthanized during the experiment due to excessive weight loss/total number of animals.
DCT, direct-contact transmission model; RDT, respiratory droplet transmission model; Seroconv, seroconversion (indicated by HI titers in serum).
Mean virus titers in tissues collected on day 3 postinoculation ± SD. Nasal turbinate viral titers are in log10 PFU/milliliter, and trachea and lung titers are in log10 PFU/gram of tissue; ; in parentheses, no. of ferrets with detectable titers/total no. of animals.
FIG 3.
Transmissibility of H1N2v influenza viruses in ferrets. Ferrets were inoculated with 6.0 log10 PFU of A/Minnesota/45/2016, A/Minnesota/19/2011 or A/Wisconsin/71/2016. After 24 h, the direct-contact transmission (DCT) model was used by adding a naive ferret to each cage housing an inoculated ferret (3 ferret pairs per virus) (A), and the respiratory droplet transmission (RDT) model was used by placing a naive ferret in an adjacent cage (3 ferret pairs per virus) (B). Virus titers in nasal wash samples collected from individual inoculated ferrets are shown on the left side of each panel, while those from individual contact ferrets are shown on the right side of each panel. The limit of detection is 10 PFU.
Most swine origin influenza viruses are capable of replication in both upper and lower respiratory tract tissues of ferrets (13). To evaluate the systemic spread of recently isolated variant H1 viruses, three groups of three ferrets each were inoculated with 6.0 log10 PFU of MN/45, MN/19, or WI/71 virus and were humanely euthanized on day 3 p.i. for assessment of virus titers in pulmonary and extrapulmonary tissues (Fig. 4). All of the H1N2v viruses were found throughout the respiratory tracts of all of the inoculated animals. Nasal turbinate and trachea mean virus titers were 4.1 to 5.3 log10 PFU/ml or g, and lung virus titers averaged 3.7 to 4.1 log10 PFU/g of tissue (Table 2). In most instances, virus was not detected in any of the extrapulmonary tissues examined, except for one ferret inoculated with MN/19 virus, which had detectable virus in the olfactory bulb (2.8 log10 PFU/g), and one ferret inoculated with WI/71 virus, which had detectable virus in the olfactory bulb and brain (2.6 and 2.0 log10 PFU/g, respectively) (Fig. 4). Overall, the H1N2v viruses replicated well in the ferret respiratory tract and displayed tissue distribution similar to those of the previously tested OH/09 and IA/39 H1N1v viruses; however, only the MN/45 virus transmitted as efficiently as the H1N1pdm09 virus, CA/07 (11).
FIG 4.
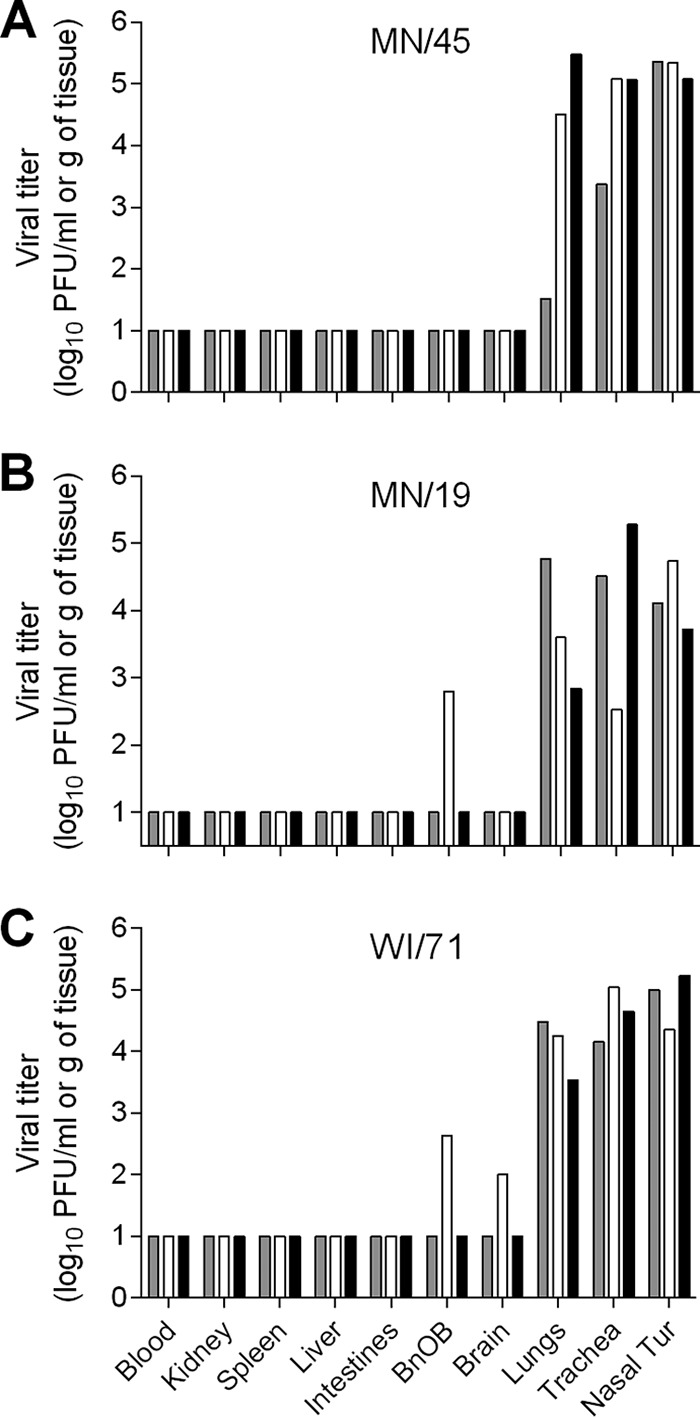
Detection of H1N2v influenza viruses in tissues of ferrets. Viral titers in tissues from individual ferrets on day 3 following inoculation with 6.0 log10 PFU of A/Minnesota/45/2016 (A), A/Minnesota/19/2011 (B), and A/Wisconsin/71/2016 (C). Blood and nasal turbinate (Nasal Tur) viral titers are presented as log10 PFU/milliliter, and kidney, spleen, liver, intestines (pooled duodenum, jejuno-ileal loop, and descending colon), olfactory bulb (BnOB), brain (pooled anterior and posterior brain), lungs, and trachea are presented as log10 PFU/gram of tissue. The limit of detection is 10 PFU.
H1 subtype influenza virus acid stability.
Stability in the acidic environment has been associated with host adaptation of influenza viruses. In general, the HA activation pH for influenza viruses ranges from about 5.0 to 6.0, but the activation pH is higher for avian influenza viruses (∼5.6 to 6.0) than for human influenza viruses (∼5.0 to 5.5). As HA stabilization prevents virus inactivation in the mildly acidic mammalian upper respiratory tract (20), adaptation of an H5 virus to the upper respiratory tract of ferrets and enhanced transmissibility were associated with mutations that led to a lower HA activation pH (21, 22). Similarly, evolution leading to lower HA activation pH was observed between precursor swine H1N1 viruses (pH 5.5 to 6.0), early human isolates (∼5.5), and the later human isolates of H1N1pdm09 viruses (5.2 to 5.4). A pH of ≤5.5 was shown to be necessary for human-adapted viruses (23). Here, the measured HA activation pH was ≤5.5 for the human seasonal virus Bris/59 and H1N1pdm09 viruses TX/15 and CA/07 (Fig. 5). In agreement with previous observations for classical swine and triple-reassortant H1 viruses (23), NJ/08, OH/02, TX/14 and all the variant viruses tested had HA activation pHs of 5.5 to 5.7. The lowest HA activation pH (5.5 to 5.6) was observed for the most transmissible virus in the respiratory droplet contact model, MN/45 virus, which suggests that although the variant viruses have not fully adapted to humans, lower activation pH may be associated with increased respiratory droplet transmissibility.
FIG 5.
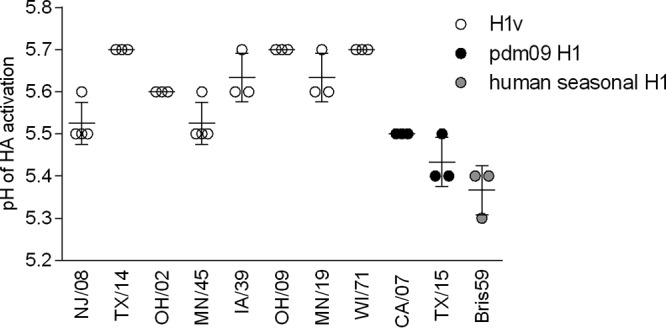
HA activation pH of selected H1 viruses. HA activation was determined by syncytium formation in infected Vero cells exposed to pH 4.8 to 7.4 fusion buffers increasing by 0.1 increments. The means and ranges from 3 or 4 independent experiments are shown. Variant H1 viruses: A/New Jersey/08/1976, A/Ohio/02/2007, A/Texas/14/2008, A/Minnesota/45/2016, A/Ohio/09/2015, A/Iowa/39/2015, A/Minnesota/19/2011, A/Wisconsin/71/2016. H1N1pdm09 viruses: A/California/07/2009, A/Texas/15/2009. Human seasonal H1N1 virus: A/Brisbane/59/2007.
Receptor binding specificity and molecular marker analysis.
Zoonotic viruses require host adaptation to replicate and transmit efficiently among humans. Receptor binding specificity has been identified as a critical determinant of efficient virus transmissibility. The combination of amino acid residues in H1 subtype HA at positions 187 and 222 (H1 numbering) is important for the prediction of receptor binding specificity of emerging strains. However, additional residues located in the HA receptor binding site may contribute to the receptor binding specificity. In this comparison, the combination of amino acids in positions 187 and 222 of all the variant viruses (D187/D222 [IA/39 and WI/71], D187/G222 [MN/45 and OH/09], and N187/D222 [MN/19]) indicated a preference for alpha-2,6 SA binding specificity or dual alpha-2,6 SA and alpha-2,3 SA binding specificity (Table 3). All of the tested variant viruses contained Q223, a residue previously shown to be critical for the H1N1pdm09 virus to bind to human-type receptors and transmit among mammals (11, 12). In order to evaluate SA binding specificity of H1v viruses isolated after the 2009 pandemic (MN/45, OH/09, IA/39, MN/19, WI/71), a resialylated red blood cell assay was used. Following the removal of SA receptors from turkey red blood cells (TRBCs) using Vibrio cholerae neuraminidase, specific sialyltransferases were used to restore either alpha-2,3 SA or alpha-2,6 SA. Avian influenza viruses preferentially bind alpha-2,3 SA receptors, while human influenza viruses preferentially bind alpha-2,6 SA receptors (24). As expected, the control avian H7N7 (ml/NL/12) virus showed binding to TRBCs bearing alpha-2,3 SA but not alpha-2,6 SA (Table 3). Swine origin influenza viruses predominantly have alpha-2,6 SA and, less frequently, dual (alpha-2,6 and alpha 2,3 SA) receptor specificity (24, 25), which is in accord with the reported similarity in SA receptor distribution in the respiratory tracts of swine and humans (26). Consistent with previous data (25), control H1N1v viruses displayed binding to TRBCs containing only alpha-2,6 SA receptors (OH/02 virus) or bound to both types of receptors (TX/14 virus). The H1v viruses MN/45 and IA/39 displayed binding to alpha-2,6 SA TRBCs, while OH/09, MN/19, and WI/71 viruses displayed higher HA titers with alpha-2,6 SA TRBCs but could also bind alpha-2,3 SA TRBCs. Each of the tested variant viruses, whether possessing alpha-2,6 SA or dual receptor binding specificity, was capable of transmission in the respiratory droplet model. However, the frequency of transmission differed among strains and could not be predicted by the receptor binding preference alone.
TABLE 3.
Receptor binding specificity and molecular marker analysis
| Virus | Subtype | H1 HA clade | GSAIDe ID | Passage | HA titera |
Amino acid sequence |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HAb |
PB2 |
PA |
PB1-F2 |
||||||||||||||||||
| Untreated TRBCs | No SA TRBCs | α-2,6 SA TRBCs | α-2,3 SA TRBCs | 187 | 222 | 223 | 158 | 271 | 588 | 590 | 591 | 627 | 701 | 35 | 97 | 66 | |||||
| ml/NL/12 | H7N7 | NAc | NA | NA | 256 | <1 | <1 | 256 | NA | NA | NA | NA | |||||||||
| OH/02 | H1N1v | Gamma | 30054 | C4 | 16 | <1 | 64 | <1 | D | D | Q | E | A | T | S | R | E | D | F | T | N |
| TX/14 | H1N1v | Beta | 83280 | C1/E2 | 256 | <1 | 256 | 32 | D | N | Q | E | A | T | S | R | E | D | F | T | N |
| MN/45 | H1N2v | Alpha | 221825 | C2 | 512 | <1 | 1,024 | <1 | D | G | Q | E | A | T | S | R | E | D | F | T | —d |
| OH/09 | H1N1v | Gamma | 178264 | E2 | 256 | <1 | 512 | 64 | D | G | Q | E | A | I | S | R | E | D | F | T | S |
| IA/39 | H1N1v | Gamma | 194942 | E2 | 128 | <1 | 512 | <1 | D | D | Q | E | A | T | S | R | E | D | F | T | N |
| MN/19 | H1N2v | Delta | 99810 | C3 | 1,024 | <1 | 1,024 | 128 | N | D | Q | E | A | T | S | R | E | D | F | T | N |
| WI/71 | H1N2v | Delta | 230639 | C2 | 512 | <1 | 512 | 64 | D | D | Q | E | S | T | S | R | E | D | F | T | N |
HA titers were measured with untreated turkey red blood cells (TRBCs), Vibrio cholerae-treated TRBCs, or α-2,6 or α-2,3 resialylated TRBCs. BPL-inactivated A/mallard/Netherlands/12/2000 (H7N7)-IBCDC-1 (ml/NL/12) virus (70), OH/02, and TX/14 viruses were used as controls (25). The experiment was repeated 3 times; the results from one representative experiment are shown.
H1 numbering.
NA, not applicable.
Truncated protein containing 8 amino acids (MN/45). IA/39 contains 79 amino acids. The full-length protein contains 90 amino acids.
GISAID, Global Initiative on Sharing All Influenza Data.
A second critical determinant of host adaptation is the composition of the influenza polymerase complex. The amino acid substitutions T271A, T588I, G590S/Q591R, E627K, and D701N in PB2 have been shown to play a role in overcoming species barriers by increasing replication efficiency in mammalian cells (27–32). E158G in PB2 (33), F35L (34) and T97I (35) in PA, and S66 in PB1-F2 (36, 37) were also shown to increase pathogenicity in the mouse model. All tested variant viruses were capable of efficient replication in a human bronchial epithelial cell line at both 33 and 37°C and in mouse lungs and ferret respiratory tracts. Consistently, most of the amino acids encoded by key locations in the polymerase genes are similar in the variant viruses and in earlier isolates containing polymerase gene segments derived from the TRIG cassette (TX/14, OH/02, CA/07, TX/15). Interestingly, two markers previously associated with increased virulence in mice (588I in PB2 and S66 in PB1-F2) are observed in the OH/09 virus. This virus was isolated from a fatal human case (11), and in addition, it caused mortality in 1/9 inoculated ferrets and 80% of inoculated mice, suggesting that mutations at these positions may be associated with increased virulence for this virus.
DISCUSSION
The pandemic potential of swine influenza viruses was made clear in 2009 when a novel H1N1 virus containing Eurasian avian-like lineage NA and MP gene segments jumped from pigs to humans and spread worldwide within months of its emergence. Although human infections with swine H1 viruses are rare, each such event is an opportunity for a novel virus to adapt to humans and possibly cause a pandemic. Antigenic analyses have revealed that some of the recent H1v viruses isolated from humans, including MN/45, OH/09, MN/19, and WI/71 viruses, are antigenically distinct from previously and currently circulating human seasonal viruses (11, 12) and thus may not be adequately covered by vaccination. Here, we provide side-by-side analysis of molecular determinants of virulence and adaptation of variant H1 viruses isolated between 2011 and 2016 and discuss the phenotypic traits associated with airborne transmission to better understand the potential for sustained human-to human transmission of these viruses and to aid in pandemic risk assessments.
Previous phylogenetic analyses revealed that recently isolated H1v viruses possess highly diverse gene segment constellations (11, 38). The five variant viruses described in this study cluster into three distinct H1 HA clades: alpha (MN/45), gamma (OH/09 and IA/39), and delta (MN/19 and WI/71). Viruses in the alpha and gamma clades contain a classical swine lineage HA, while viruses in the delta clade contain a human seasonal lineage HA. There are also differences in the subtype and origin of NA among these viruses; OH/09 and IA/39 viruses possess classical swine lineage N1 NA, whereas MN/45, MN/19, and WI/71 viruses possess human seasonal lineage N2 NA. Each of the recent variant viruses possesses a TRIG constellation, characteristic of pre-2009 pandemic triple-reassortant viruses (11, 38). Following the introduction of Eurasian avian-like lineage NA and MP gene segments during the 2009 pandemic (6), a plethora of H1v isolates containing various combinations of H1N1pdm09-derived virus gene segments have been detected (39). Each of the tested variant viruses, except for MN/19 virus, contained a Eurasian avian-like lineage MP, among which IA/39 and MN/45 also contained H1N1pdm09-derived PA and NP gene segments. The Eurasian avian-like lineage MP gene was the most frequently observed gene among swine isolates, suggesting that acquisition of this gene confers a fitness advantage in guinea pig and pig models (40, 41). Multiple studies demonstrated that an optimal gene segment constellation is required for respiratory droplet transmission to occur. Eurasian avian-like lineage virus gene segments MP and NA were shown to be important for respiratory droplet transmission of H1N1pdm09 viruses (40, 42, 43). Barman et al. assessed the transmissibility of triple-reassortant viruses isolated from pigs prior to the pandemic and found an association between transmissibility in the ferret model and the origin of HA and NA genes. Efficient transmission via respiratory droplets was observed for viruses containing human-like HA and NA, moderate transmission efficiency was observed for viruses containing mixed-origin HA and NA, and poor respiratory droplet transmission was observed for viruses containing swine origin HA and NA (13). In agreement, previously studied variant H1 viruses containing swine origin HA and NA (TX/14 [beta clade] and OH/02 [gamma clade]), did not have the ability to transmit efficiently via respiratory droplets (19). However, this consensus did not hold true for H1N1pdm09 viruses or H1v viruses that emerged as a result of reassortment with H1N1pdm09 viruses. In our study, transmissibility did not correlate with gene origin or H1 clade. Overall, all of the H1v viruses included in this and previous studies transmitted efficiently between cohoused ferrets (11, 18, 19). Of the five variant viruses isolated between 2001 and 2016 (MN/45, OH/09, IA/39, MN/19, WI/71), the most transmissible via respiratory droplets variant virus was MN/45; it transmitted with efficacy similar to that of human seasonal and H1N1pdm09 viruses (11, 18).
Despite great heterogeneity between H1 viruses, the recently isolated H1v viruses share many known markers of mammalian adaptation for virulence and transmission in the HA and polymerase genes with H1N1pdm09 viruses. The importance of alpha-2,6 SA binding for transmission has been demonstrated using mammalian models (44–46). The respiratory tracts of humans, pigs, and ferrets share similarities with respect to the distribution of sialic acids, with upper respiratory tract epithelia enriched for alpha-2,6 SA while the lower respiratory tract is enriched with alpha-2,3 SA (26, 47, 48). Consistently, swine and human viruses have been shown to have preference for alpha-2,6 SA receptors (49). A role for the soft palate in the selection of alpha-2,6 SA binding mutants in ferrets has been recently identified (50). In the current study, all the variant viruses displayed binding to TRBCs presenting alpha-2,6 SA receptors, while OH/09, MN/19, and WI/71 were also capable of binding to alpha-2,3 SA. Although airborne transmission requires a gain of long-chain alpha-2,6 SA binding, an accompanying loss of alpha-2,3 SA binding is not necessary for transmission (50). Viruses with HA proteins containing E187/G222, E187/D222, D187/G222, and N187/D222 have been previously shown to bind alpha-2,3 SA in addition to binding alpha-2,6 SA receptors (25, 51, 52). The D222G substitution and mixed-receptor binding specificity were detected in the 1918 (53) and 2009 (54, 55) pandemic viruses. H1N1pdm09 viruses that possess D222 were associated with a number of severe human infection outcomes, presumably because of their ability to infect epithelial cells presenting alpha-2,3 SA receptors that line the lower respiratory tract, bronchi, bronchioles, and type II pneumocytes (56–58). Two of the tested viruses, MN/45 and OH/09, contained G222. Interestingly, the OH/09 virus was associated with increased virulence in humans and mammalian models (11); however, it is not clear whether the G222 substitution contributed to the severity of disease. Unlike the other H1v viruses tested in the ferret respiratory droplet transmission model, MN/45 was the only virus that was capable of efficient transmission between all ferret pairs. Although the presence of D187/G222 in the HA of both MN/45 and OH/09 suggested dual binding specificity, MN/45 did not bind alpha-2,3 SA-containing TRBCs, which may have contributed to the highly transmissible phenotype of this virus.
Transmission of influenza viruses containing avian-origin polymerase genes may be limited by inefficient replication at temperatures found in the mammalian upper respiratory tract (59). Unlike human influenza viruses, which have adapted to replicate in temperatures ranging from 32°C in the upper respiratory tract to 37°C in the lower respiratory tract (60, 61), avian influenza viruses have evolved to replicate at the higher temperatures, 40 to 41°C, of the avian enteric tract (62). The E627K substitution in PB2 has been identified as a marker associated with adaptation to enhanced replication in the temperatures of the human respiratory tract. In some virus subtypes, the absence of E627K could be compensated with a D701N substitution (28). In this study, only the human seasonal virus, Bris/59, and the classical swine virus NJ/08, which was associated with the largest cluster of human infections prior to the 2009 pandemic, contained K627 in the PB2 gene. The H1v viruses with a PB2 gene of avian origin contained 627E, 701D, and compensatory substitutions, S590/R591, critical for viral replication and virulence of swine influenza viruses (31, 32). The lack of K627 and the presence of the S590/R591 polymorphism were commonly observed among triple-reassortant and H1N1pdm09 viruses (13). Each of the H1v viruses was able to replicate efficiently in a human airway epithelial cell line at both 33°C and 37°C, as well as in the upper and lower respiratory tracts of mice and ferrets, consistent with observations for previously tested swine origin viruses (13, 18, 19, 63). Except for WI/71 virus (S271), all variant viruses contained A271, which was shown to play a role in increased polymerase activity in mammalian cells (30). Several substitutions in the polymerase genes have been previously associated with increased virulence in mammalian models. In our study, OH/09 virus caused mortality in both mouse and ferret models and possessed markers in the polymerase genes for increased virulence in mammal models. These include S66 in PB1-F2, which was shown to contribute to pathogenesis in mice (36, 37), and I588 in PB2, which was shown to enhance H1N1pdm09 virus virulence by increasing replication and inhibition of interferon signaling (29).
Multiple studies have suggested a correlation between HA activation pH and transmissibility between ferrets, underscoring the importance of monitoring fusion pH to aid in the identification of viruses with pandemic potential. Mutations that led to acid stabilization of HA were associated with adaptation of H5 viruses to the upper respiratory tract of ferrets and gain of transmissibility (21, 22). Adaptation of H1N1pdm09 viruses from swine to humans also coincided with stepwise acid stabilization of HA pH from 5.5 to 6.0 observed for precursor H1N1pdm09 viruses to approximately 5.5 for early 2009 pandemic isolates, and 5.2 to 5.4 for isolates that had adapted to humans. A study by Russier et al. (23) suggested that an activation pH of ≤5.5 was necessary for human adaptation. Further analysis of swine H1N1 viruses isolated after the 2009 pandemic revealed a shift in HA activation pH. Multiple post-2009 pandemic isolates had activation pH values that overlapped those of viruses that were adapted to humans, suggesting pigs as a bridging host for HA stabilization (64). In the current study, the lowest activation pH was observed for H1 viruses that were capable of sustained transmission in humans: Bris/59 (pH 5.3 to 5.4) and H1N1pdm09 viruses CA/07 and TX/15 (pH 5.4 to 5.5). Variant viruses examined here had activation pH of >5.5, which was similar to that of most classical swine and North American triple-reassortant isolates (23). The lowest activation pH was observed for NJ/08 and MN/45 viruses (pH 5.5 to 5.6). Interestingly, NJ/08 virus was associated with an outbreak in 1976, during which limited human-to-human transmission was observed (10). No human-to-human transmission was observed in the case of H1N2v MN/45 virus; however, this virus transmitted efficiently via the respiratory droplets between all ferret pairs. Other tested variant viruses had activation pH values of ≥5.6 and transmitted less efficiently between ferrets, but it is not clear whether poor transmissibility of these viruses was attributed to the higher HA activation pH and/or additional viral traits.
Growing evidence indicates that swine can facilitate adaptations of zoonotic influenza viruses to humans, including increased binding to alpha-2,6 SA receptors, increased replication at temperatures found in mammalian respiratory tracts, increased pathogenicity and transmissibility, and decreased HA activation pH (64, 65). As virulence and transmissibility are multifactorial traits, additional determinants such as virus morphology, HA-NA balance, and the ability to evade innate immune responses may also be important (66). Continuous emergence of genetically and antigenically distinct variant influenza viruses that possess humanizing adaptations necessitates in vitro and in vivo evaluations of these viruses. Contextualization of current and previous observations is essential to recognize trends and similarities among viruses, which will ultimately improve our risk assessment of emerging strains of influenza so that we are better prepared for the next pandemic.
MATERIALS AND METHODS
Viruses.
Stocks of A/Minnesota/45/2016 (MN/45) H1N2v, A/Wisconsin/71/2016 (WI/71) H1N2v, A/Minnesota/19/2011 (MN/19) H1N2v, A/Ohio/02/2007 (OH/02) H1N1v, and A/Texas/15/2009 (TX/15) H1N1pdm09 viruses were propagated in MDCK cells at 37°C for 48 h. Virus stocks of A/Ohio/09/2015 (OH/09) H1N1v, A/Iowa/39/2015 (IA/39) H1N1v, A/Texas/14/2008 (TX/14) H1N1v, A/New Jersey/08/1976 (NJ/08) H1N1v, A/California/07/2009 (CA/07) H1N1pdm09, and A/Brisbane/59/2007 (Bris/59) H1N1 were propagated in the allantoic cavity of 10-day-old embryonated hens' eggs at 35°C for 48 h (67). Pooled cell supernatants or allantoic fluid were clarified by centrifugation and frozen in aliquots at −80°C. Virus titers of each stock were determined by standard plaque assay in MDCK cells. The stock viruses were exclusivity tested by real-time reverse transcription (RT)-PCR to rule out the presence of other subtypes of influenza virus. All the stocks have been sequenced to ensure preservation of the consensus sequence of the original isolate. All work with variant influenza viruses was conducted in a biosafety level 2 (BSL2)-enhanced or higher laboratory.
Replication kinetics in Calu-3 cells.
The human bronchial epithelial cell line, Calu-3 (ATCC), was cultured on Transwell membrane inserts in a 12-well plate (Corning, Inc.) for 1 week under liquid-liquid interface conditions as previously described (15). Virus was applied to cells apically at a multiplicity of infection (MOI) of 0.01 and incubated for 1 h, after which the cells were washed with medium and cultured at either 37°C or 33°C. Culture supernatants were harvested at 2, 24, 48, and 72 h p.i. and frozen at −80°C until titration for determination of infectious virus titers in MDCK cells by standard plaque assay.
HA activation pH.
A previously described virus-induced syncytium formation assay was used to determine the pH of HA activation (68). Briefly, Vero cells (ATCC, Manassas, VA) were inoculated with influenza viruses at an MOI of 1 to 20, standardized to achieve a minimum of 50% infectivity (based on NP staining). Sixteen hours p.i., infected cells were treated with 5 μg/ml of N-p-tosyl-l-phenylalanine chloromethyl ketone (TPCK)-treated trypsin for 15 min at 37°C to induce fusion and then incubated for 5 min at 37°C in prewarmed fusion buffers (20 mM HEPES, 2 mM CaCl2, 150 mM NaCl, 20 mM citric acid monohydrate/sodium citrate tribasic dehydrate) at pH values ranging between 4.8 and 7.4, increasing by 0.1 increments. Next, the cells were incubated with Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) for 3 h, followed by fixation with 4% paraformaldehyde. Fixed monolayers were stained with mouse anti-NP primary antibody (clone A1, A3 blend; Millipore) and goat anti-mouse IgG (H+L) Alexa Fluor Plus 488 secondary antibody (Thermo Fisher Scientific) and viewed by immunofluorescence microscopy. The HA activation pH was defined as the highest pH value at which ≥50% syncytia were observed.
HA binding specificity.
Hemagglutination assays using resialylated TRBCs were performed as previously described with minor modifications (69). Briefly, TRBCs were desialylated using Vibrio cholerae neuraminidase (Sigma-Aldrich) followed by resialylation using CMP-sialic acid (Sigma) and either alpha-2-6-(N)-sialyltransferase or alpha-2-3-(N)-sialyltransferase (Prozyme). Betapropiolactone (BPL)-inactivated A/mallard/Netherlands/12/2000 (H7N7)-IBCDC-1 (70), OH/02, and TX/14 viruses were used as controls (25).
Mouse experiments.
All animal experiments were performed under the guidance of the Centers for Disease Control and Prevention's Institutional Animal Care and Use Committee and were conducted in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited animal facility. Three groups of 6- to 8-week-old female BALB/c mice (Charles River Laboratories, Wilmington, MA) were anesthetized intraperitoneally with 0.2 ml of 2,2,2-tribromoethanol in tert-amyl alcohol (Avertin; Acros Organics) and inoculated intranasally (i.n.) with 50 μl of 5.0 log10 PFU of either MN/45, CA/07, OH/09, IA/39, WI/71, or MN/19 virus diluted in phosphate-buffered saline (PBS). Five mice per group were monitored for 14 days p.i. for clinical signs of infection and loss of body weight; any mouse that lost >25% of preinoculation body weight was humanely euthanized. On days 3 and 6 p.i., 3 mice from each group were euthanized for determination of viral titers in lungs and noses by plaque assay (71).
Ferret experiments.
Five- to 7-month-old male Fitch ferrets (Triple F Farms, Sayre, PA), serologically negative for currently circulating influenza viruses, were housed in Duo-Flo Bioclean mobile units (Lab Products Incorporated, Seaford, DE) during experimentation. Ferrets (n = 9) were inoculated i.n. with 1 ml of 6.0 log10 PFU of MN/19, MN/45, or WI/71 virus diluted in PBS. The following day, a serologically naive ferret was placed in the same cage as each of three inoculated ferrets (direct-contact transmission model) or in a cage with a perforated side wall adjacent to each of three additional inoculated ferrets (respiratory droplet transmission model) (72). Clinical signs of infection, including weight loss, fever, respiratory tract signs, and food and water intake, were monitored daily for 14 days p.i. Lethargy was assessed using the Reuman scale (73). Nasal wash samples were collected every 2 days for virus titer determination. The 3 remaining inoculated ferrets in each virus group were euthanized on day 3 p.i. for the assessment of virus spread to pulmonary and extrapulmonary tissues as previously described (71). Convalescent-phase sera collected from all the ferrets 3 weeks p.i. or p.c. were tested by hemagglutination inhibition assay using homologous virus and 0.5% TRBCs to determine seroconversion.
Genetic analysis.
Alignments of full-length coding sequences were done using BioEdit (v7.1.3.0) (74) and Muscle (75) software. Protein identity data were obtained using MegAlign version 14.1.0 software.
Statistical analysis.
Experimental results were analyzed by two-way analysis of variance (ANOVA) followed by Tukey's posttest. Analyses were performed using GraphPad Prism 6.0 software.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Comparative Medicine Branch for excellent care of the animals used in this study.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry. Nicole Brock is a contractor with Chickasaw Nation Industries.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01444-18.
REFERENCES
- 1.Simonsen L, Spreeuwenberg P, Lustig R, Taylor RJ, Fleming DM, Kroneman M, Van Kerkhove MD, Mounts AW, Paget WJ, GLaMOR Collaborating Teams. 2013. Global mortality estimates for the 2009 influenza pandemic from the GLaMOR project: a modeling study. PLoS Med 10:e1001558. doi: 10.1371/journal.pmed.1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shope RE. 1931. Swine influenza. III. Filtration experiments and etiology. J Exp Med 54:373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao S, Anderson TK, Walia RR, Dorman KS, Janas-Martindale A, Vincent AL. 2017. The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016. J Gen Virol 98:2001–2010. doi: 10.1099/jgv.0.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen CW. 2002. The emergence of novel swine influenza viruses in North America. Virus Res 85:199–210. doi: 10.1016/S0168-1702(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 5.Vincent AL, Lager KM, Ma W, Lekcharoensuk P, Gramer MR, Loiacono C, Richt JA. 2006. Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States. Vet Microbiol 118:212–222. doi: 10.1016/j.vetmic.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 6.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC Information on swine influenza/variant influenza virus. CDC, Atlanta, GA: https://www.cdc.gov/flu/swineflu/index.htm Accessed 10 August 2017. [Google Scholar]
- 8.Gaydos JC, Top FH Jr, Hodder RA, Russell PK. 2006. Swine influenza A outbreak, Fort Dix, New Jersey, 1976. Emerg Infect Dis 12:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodder RA, Gaydos JC, Allen RG, Top FH Jr, Nowosiwsky T, Russell PK. 1977. Swine influenza A at Fort Dix, New Jersey (January-February 1976). III. Extent of spread and duration of the outbreak. J Infect Dis 136(Suppl):S369–S375. [DOI] [PubMed] [Google Scholar]
- 10.Lessler J, Cummings DA, Fishman S, Vora A, Burke DS. 2007. Transmissibility of swine flu at Fort Dix, 1976. J R Soc Interface 4:755–762. doi: 10.1098/rsif.2007.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulit-Penaloza JA, Jones J, Sun X, Jang Y, Thor S, Belser JA, Zanders N, Creager HM, Ridenour C, Wang L, Stark TJ, Garten R, Chen LM, Barnes J, Tumpey TM, Wentworth DE, Maines TR, Davis CT. 2018. Antigenically diverse swine-origin H1N1 variant influenza viruses exhibit differential ferret pathogenesis and transmission phenotypes. J Virol 92:e00095-18. doi: 10.1128/JVI.00095-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization September 2017 report. http://www.who.int/influenza/vaccines/virus/characteristics_virus_vaccines/en/ World Health Organization, Geneva, Switzerland: Accessed August 2018. [Google Scholar]
- 13.Barman S, Krylov PS, Fabrizio TP, Franks J, Turner JC, Seiler P, Wang D, Rehg JE, Erickson GA, Gramer M, Webster RG, Webby RJ. 2012. Pathogenicity and transmissibility of North American triple reassortant swine influenza A viruses in ferrets. PLoS Pathog 8:e1002791. doi: 10.1371/journal.ppat.1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belser JA, Eckert AM, Tumpey TM, Maines TR. 2016. Complexities in ferret influenza virus pathogenesis and transmission models. Microbiol Mol Biol Rev 80:733–744. doi: 10.1128/MMBR.00022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng H, Goldsmith C, Thawatsupha P, Chittaganpitch M, Waicharoen S, Zaki S, Tumpey TM, Katz JM. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J Virol 81:12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thangavel RR, Bouvier NM. 2014. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods 410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trock SC, Burke SA, Cox NJ. 2015. Development of framework for assessing influenza virus pandemic risk. Emerg Infect Dis 21:1372–1378. doi: 10.3201/eid2108.141086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belser JA, Gustin KM, Maines TR, Blau DM, Zaki SR, Katz JM, Tumpey TM. 2011. Pathogenesis and transmission of triple-reassortant swine H1N1 influenza viruses isolated before the 2009 H1N1 pandemic. J Virol 85:1563–1572. doi: 10.1128/JVI.02231-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer H, Widdicombe JH. 2006. Mechanisms of acid and base secretion by the airway epithelium. J Membr Biol 211:139–150. doi: 10.1007/s00232-006-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linster M, van Boheemen S, de Graaf M, Schrauwen EJA, Lexmond P, Manz B, Bestebroer TM, Baumann J, van Riel D, Rimmelzwaan GF, Osterhaus A, Matrosovich M, Fouchier RAM, Herfst S. 2014. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell 157:329–339. doi: 10.1016/j.cell.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russier M, Yang G, Rehg JE, Wong SS, Mostafa HH, Fabrizio TP, Barman S, Krauss S, Webster RG, Webby RJ, Russell CJ. 2016. Molecular requirements for a pandemic influenza virus: an acid-stable hemagglutinin protein. Proc Natl Acad Sci U S A 113:1636–1641. doi: 10.1073/pnas.1524384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers GN, D'Souza BL. 1989. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 25.Chen LM, Rivailler P, Hossain J, Carney P, Balish A, Perry I, Davis CT, Garten R, Shu B, Xu X, Klimov A, Paulson JC, Cox NJ, Swenson S, Stevens J, Vincent A, Gramer M, Donis RO. 2011. Receptor specificity of subtype H1 influenza A viruses isolated from swine and humans in the United States. Virology 412:401–410. doi: 10.1016/j.virol.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelli RK, Kuchipudi SV, White GA, Perez BB, Dunham SP, Chang KC. 2010. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet Res 6:4. doi: 10.1186/1746-6148-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol 67:1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Z, Yi C, Zhao L, Wang S, Zhou L, Hu Y, Zou W, Chen H, Jin M. 2014. PB2-588I enhances 2009 H1N1 pandemic influenza virus virulence by increasing viral replication and exacerbating PB2 inhibition of beta interferon expression. J Virol 88:2260–2267. doi: 10.1128/JVI.03024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T. 2010. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J Virol 84:4395–4406. doi: 10.1128/JVI.02642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Q, Qiao C, Marjuki H, Bawa B, Ma J, Guillossou S, Webby RJ, Richt JA, Ma W. 2012. Combination of PB2 271A and SR polymorphism at positions 590/591 is critical for viral replication and virulence of swine influenza virus in cultured cells and in vivo. J Virol 86:1233–1237. doi: 10.1128/JVI.05699-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehle A, Doudna JA. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A 106:21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou B, Li Y, Halpin R, Hine E, Spiro DJ, Wentworth DE. 2011. PB2 residue 158 is a pathogenic determinant of pandemic H1N1 and H5 influenza A viruses in mice. J Virol 85:357–365. doi: 10.1128/JVI.01694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seyer R, Hrincius ER, Ritzel D, Abt M, Mellmann A, Marjuki H, Kuhn J, Wolff T, Ludwig S, Ehrhardt C. 2012. Synergistic adaptive mutations in the hemagglutinin and polymerase acidic protein lead to increased virulence of pandemic 2009 H1N1 influenza A virus in mice. J Infect Dis 205:262–271. doi: 10.1093/infdis/jir716. [DOI] [PubMed] [Google Scholar]
- 35.Song MS, Pascua PN, Lee JH, Baek YH, Lee OJ, Kim CJ, Kim H, Webby RJ, Webster RG, Choi YK. 2009. The polymerase acidic protein gene of influenza A virus contributes to pathogenicity in a mouse model. J Virol 83:12325–12335. doi: 10.1128/JVI.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamarin D, Ortigoza MB, Palese P. 2006. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol 80:7976–7983. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu B, Garten R, Emery S, Balish A, Cooper L, Sessions W, Deyde V, Smith C, Berman L, Klimov A, Lindstrom S, Xu X. 2012. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990-2010. Virology 422:151–160. doi: 10.1016/j.virol.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 39.Kong W, Wang F, Dong B, Ou C, Meng D, Liu J, Fan ZC. 2015. Novel reassortant influenza viruses between pandemic (H1N1) 2009 and other influenza viruses pose a risk to public health. Microb Pathog 89:62–72. doi: 10.1016/j.micpath.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, Garcia-Sastre A, Palese P, Hai R. 2011. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol 85:11235–11241. doi: 10.1128/JVI.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma J, Shen H, Liu Q, Bawa B, Qi W, Duff M, Lang Y, Lee J, Yu H, Bai J, Tong G, Hesse RA, Richt JA, Ma W. 2015. Pathogenicity and transmissibility of novel reassortant H3N2 influenza viruses with 2009 pandemic H1N1 genes in pigs. J Virol 89:2831–2841. doi: 10.1128/JVI.03355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lakdawala SS, Lamirande EW, Suguitan AL Jr, Wang W, Santos CP, Vogel L, Matsuoka Y, Lindsley WG, Jin H, Subbarao K. 2011. Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS Pathog 7:e1002443. doi: 10.1371/journal.ppat.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen HL, Liang CH, Wu CY, Forrest HL, Ferguson A, Choy KT, Jones J, Wong DD, Cheung PP, Hsu CH, Li OT, Yuen KM, Chan RW, Poon LL, Chan MC, Nicholls JM, Krauss S, Wong CH, Guan Y, Webster RG, Webby RJ, Peiris M. 2011. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A 108:14264–14269. doi: 10.1073/pnas.1111000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, Garcia-Sastre A. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 45.Pappas C, Viswanathan K, Chandrasekaran A, Raman R, Katz JM, Sasisekharan R, Tumpey TM. 2010. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PLoS One 5:e11158. doi: 10.1371/journal.pone.0011158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Zhang Q, Gao Y, He X, Kong H, Jiang Y, Guan Y, Xia X, Shu Y, Kawaoka Y, Bu Z, Chen H. 2012. Key molecular factors in hemagglutinin and PB2 contribute to efficient transmission of the 2009 H1N1 pandemic influenza virus. J Virol 86:9666–9674. doi: 10.1128/JVI.00958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trebbien R, Larsen LE, Viuff BM. 2011. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol J 8:434. doi: 10.1186/1743-422X-8-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol 171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gambaryan AS, Karasin AI, Tuzikov AB, Chinarev AA, Pazynina GV, Bovin NV, Matrosovich MN, Olsen CW, Klimov AI. 2005. Receptor-binding properties of swine influenza viruses isolated and propagated in MDCK cells. Virus Res 114:15–22. doi: 10.1016/j.virusres.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Lakdawala SS, Jayaraman A, Halpin RA, Lamirande EW, Shih AR, Stockwell TB, Lin X, Simenauer A, Hanson CT, Vogel L, Paskel M, Minai M, Moore I, Orandle M, Das SR, Wentworth DE, Sasisekharan R, Subbarao K. 2015. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 526:122–125. doi: 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Y, Wu Y, Zhang W, Qi J, Gao GF. 2014. Enabling the ‘host jump’: structural determinants of receptor-binding specificity in influenza A viruses. Nat Rev Microbiol 12:822–831. doi: 10.1038/nrmicro3362. [DOI] [PubMed] [Google Scholar]
- 52.Fei Y, Sun YS, Li Y, Yu H, Lau K, Landry JP, Luo Z, Baumgarth N, Chen X, Zhu X. 2015. Characterization of receptor binding profiles of influenza A viruses using an ellipsometry-based label-free glycan microarray assay platform. Biomolecules 5:1480–1498. doi: 10.3390/biom5031480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. 2006. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol 355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, Childs RA, Matrosovich T, Wharton S, Palma AS, Chai W, Daniels R, Gregory V, Uhlendorff J, Kiso M, Klenk HD, Hay A, Feizi T, Matrosovich M. 2010. Altered receptor specificity and cell tropism of D222G hemagglutinin mutants isolated from fatal cases of pandemic A(H1N1) 2009 influenza virus. J Virol 84:12069–12074. doi: 10.1128/JVI.01639-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, Carney P, Stevens J. 2010. Structure and receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr 2:RRN1152. doi: 10.1371/currents.RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen H, Wen X, To KK, Wang P, Tse H, Chan JF, Tsoi HW, Fung KS, Tse CW, Lee RA, Chan KH, Yuen KY. 2010. Quasispecies of the D225G substitution in the hemagglutinin of pandemic influenza A(H1N1) 2009 virus from patients with severe disease in Hong Kong, China. J Infect Dis 201:1517–1521. doi: 10.1086/652661. [DOI] [PubMed] [Google Scholar]
- 57.Kilander A, Rykkvin R, Dudman SG, Hungnes O. 2010. Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009-2010. Euro Surveill 15:19498. doi: 10.2807/ese.15.09.19498-en. [DOI] [PubMed] [Google Scholar]
- 58.Potdar VA, Chadha MS, Jadhav SM, Mullick J, Cherian SS, Mishra AC. 2010. Genetic characterization of the influenza A pandemic (H1N1) 2009 virus isolates from India. PLoS One 5:e9693. doi: 10.1371/journal.pone.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, Garcia-Sastre A, Sasisekharan R, Katz JM, Tumpey TM. 2009. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A 106:3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McFadden ER Jr, Pichurko BM, Bowman HF, Ingenito E, Burns S, Dowling N, Solway J. 1985. Thermal mapping of the airways in humans. J Appl Physiol 58:564–570. [DOI] [PubMed] [Google Scholar]
- 61.Lindemann J, Leiacker R, Rettinger G, Keck T. 2002. Nasal mucosal temperature during respiration. Clin Otolaryngol Allied Sci 27:135–139. doi: 10.1046/j.1365-2273.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- 62.Scull MA, Gillim-Ross L, Santos C, Roberts KL, Bordonali E, Subbarao K, Barclay WS, Pickles RJ. 2009. Avian influenza virus glycoproteins restrict virus replication and spread through human airway epithelium at temperatures of the proximal airways. PLoS Pathog 5:e1000424. doi: 10.1371/journal.ppat.1000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Belser JA, Wadford DA, Pappas C, Gustin KM, Maines TR, Pearce MB, Zeng H, Swayne DE, Pantin-Jackwood M, Katz JM, Tumpey TM. 2010. Pathogenesis of pandemic influenza A (H1N1) and triple-reassortant swine influenza A (H1) viruses in mice. J Virol 84:4194–4203. doi: 10.1128/JVI.02742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russier M, Yang G, Marinova-Petkova A, Vogel P, Kaplan BS, Webby RJ, Russell CJ. 2017. H1N1 influenza viruses varying widely in hemagglutinin stability transmit efficiently from swine to swine and to ferrets. PLoS Pathog 13:e1006276. doi: 10.1371/journal.ppat.1006276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otte A, Marriott AC, Dreier C, Dove B, Mooren K, Klingen TR, Sauter M, Thompson KA, Bennett A, Klingel K, van Riel D, McHardy AC, Carroll MW, Gabriel G. 2016. Evolution of 2009 H1N1 influenza viruses during the pandemic correlates with increased viral pathogenicity and transmissibility in the ferret model. Sci Rep 6:28583. doi: 10.1038/srep28583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schrauwen EJ, Fouchier RA. 2014. Host adaptation and transmission of influenza A viruses in mammals. Emerg Microbes Infect 3:e9. doi: 10.1038/emi.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balish AL, Katz JM, Klimov AI. 2013. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol Chapter 15:Unit 15G1. doi: 10.1002/9780471729259.mc15g01s29. [DOI] [PubMed] [Google Scholar]
- 68.Belser JA, Pulit-Penaloza JA, Sun X, Brock N, Pappas C, Creager HM, Zeng H, Tumpey TM, Maines TR. 2017. A novel A(H7N2) influenza virus isolated from a veterinarian caring for cats in a New York City animal shelter causes mild disease and transmits poorly in the ferret model. J Virol 91: e00672-17. doi: 10.1128/JVI.00672-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. 2005. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol 79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramos I, Krammer F, Hai R, Aguilera D, Bernal-Rubio D, Steel J, Garcia-Sastre A, Fernandez-Sesma A. 2013. H7N9 influenza viruses interact preferentially with alpha2,3-linked sialic acids and bind weakly to alpha2,6-linked sialic acids. J Gen Virol 94:2417–2423. doi: 10.1099/vir.0.056184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HH, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol 79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai le Q, Sedyaningsih ER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM. 2006. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A 103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reuman PD, Keely S, Schiff GM. 1989. Assessment of signs of influenza illness in the ferret model. J Virol Methods 24:27–34. doi: 10.1016/0166-0934(89)90004-9. [DOI] [PubMed] [Google Scholar]
- 74.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 75.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.