The capsid anchor (Ca) is a single-pass transmembrane domain at the C terminus of the capsid protein (C) known to function as a signal for the translocation of PrM into the ER lumen. The objective of this study was to further examine the role of Ca in Zika virus life cycle, whether involved in the formation of nucleocapsid through association with C or in the formation of viral envelope. In this study, we show that Ca has a function beyond the one of translocation signal, controlling protein E stability and therefore its availability for assembly of infectious particles.
KEYWORDS: capsid anchor, flavivirus, morphogenesis, pseudovirus, viral assembly, Zika virus
ABSTRACT
The flavivirus capsid protein (C) is separated from the downstream premembrane (PrM) protein by a hydrophobic sequence named capsid anchor (Ca). During polyprotein processing, Ca is sequentially cleaved by the viral NS2B/NS3 protease on the cytosolic side and by signal peptidase on the luminal side of the endoplasmic reticulum (ER). To date, Ca is considered important mostly for directing translocation of PrM into the ER lumen. In this study, the role of Ca in the assembly and secretion of Zika virus was investigated using a pseudovirus-based approach. Our results show that, while Ca-mediated anchoring of C to the ER membrane is not needed for the production of infective particles, Ca expression in cis with respect to PrM is strictly required to allow proper assembly of infectious particles. Finally, we show that the presence of heterologous, but not homologous, Ca induces degradation of E through the autophagy/lysosomal pathway.
IMPORTANCE The capsid anchor (Ca) is a single-pass transmembrane domain at the C terminus of the capsid protein (C) known to function as a signal for the translocation of PrM into the ER lumen. The objective of this study was to further examine the role of Ca in Zika virus life cycle, whether involved in the formation of nucleocapsid through association with C or in the formation of viral envelope. In this study, we show that Ca has a function beyond the one of translocation signal, controlling protein E stability and therefore its availability for assembly of infectious particles.
INTRODUCTION
Similar to other members of the Flaviviridae family, which also includes significant human pathogens such as West Nile virus (WNV) and dengue virus (DENV), Zika virus (ZIKV) is an enveloped, single-stranded, positive-sense RNA virus (1). The surface structure of a typical flavivirus particle is formed by 180 copies of the envelope glycoprotein (E) and the membrane protein (M), which are anchored onto the host-derived lipid bilayer through their transmembrane domains (TMDs). The viral envelope encloses a nucleocapsid core composed of the single molecule of RNA genome and several copies of the capsid protein (C) (2–4).
In all flaviviruses, the viral genome encodes a single open reading frame which is translated into a polyprotein that is co- and posttranslationally cleaved into three structural (C, premembrane/membrane [PrM/M], and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). Cleavage into mature viral proteins is catalyzed by the virus-encoded NS2B-NS3 protease on the cytosolic side and by the host signal peptidase and furin at the luminal side of the endoplasmic reticulum (ER) and Golgi network, respectively (5). Processing of the polyprotein is crucial for virus replication and regulation of gene expression in the majority of positive-sense RNA viruses (6, 7). Replication and morphogenesis of flaviviruses occur in close association with intracellular membranes (8–11). After translation, virus assembly occurs by budding of the nucleocapsid into the ER, thus acquiring the membrane envelope together with proteins E and PrM. During traffic through the Golgi network, furin cleaves PrM to generate mature and infective virions. Following its cleavage, the Pr peptide remains associated with the viral particle and dissociates when the virus reaches neutral pH upon exit from the cell (10, 12–14).
During translation, the structural proteins PrM and E and the nonstructural proteins NS1 and NS4B are translocated into the ER lumen by TMDs present at the C terminus of the upstream proteins, which act concomitantly as signal peptides for ER translocation of the downstream protein and as a stop transfer signal for the upstream one. The capsid protein (C), the first one of the polypeptide, carries a single TMD, also termed a capsid anchor (Ca), which anchors C to the ER membrane before cleavage by NS2B/NS3 protease and is a translocation signal for PrM into the ER lumen. Instead, PrM and E proteins have double TMDs; the first acts as a stop transfer signal, while the second serves as a translocation signal for the following protein (E in the case of PrM and NS1 in the case of E) (15). Different studies performed to understand the role of PrM and E TMDs in viral assembly and release showed that mutations in these regions or interchange of them among flaviviruses severely impair virus assembly and release of viral particles (16–20). However, the role of Ca in the viral life cycle has not been extensively investigated.
The Ca region consists of three main segments and varies from 14 to 22 amino acids among the different flaviviruses. The amino-terminal region contains basic residues, which determine the orientation of the polyprotein insertion into the ER and provides the cleavage site for the viral protease, and is followed by a core formed by a series of uninterrupted hydrophobic amino acids and a carboxy-terminal cleavage region which is relatively hydrophobic but consistent with the (−3, −1) rule (21). The flavivirus Ca region has the requirement to be cleaved sequentially during processing of the polyprotein. The first cleavage takes place on the cytosolic side by the viral protease NS2B/NS3 releasing mature C in the cytosol, which is followed by cleavage by the host signal peptidase on the ER luminal side (22–25). This ordered processing, controlled by the Ca sequence, was shown to be required for efficient assembly of Murray Valley encephalitic virus (MVEV) and yellow fever virus (YFV) (23, 26).
To date, flavivirus Ca has been considered to function mainly as a signal sequence for PrM translocation. Here, we show that Ca plays an essential role in controlling stability of downstream structural polyprotein, thus participating in the fate of virus assembly.
RESULTS
Role of Ca on ZIKV packaging.
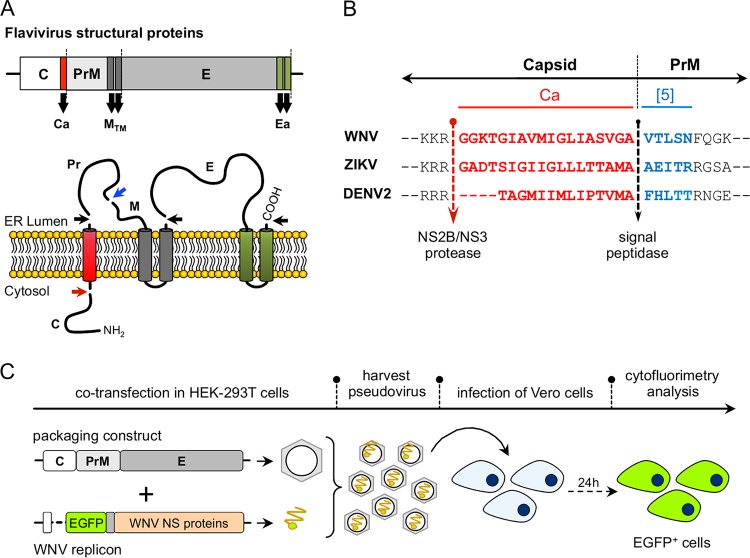
The role of the capsid anchor (Ca) on the production of ZIKV was investigated using a WNV replicon-packaging approach, which is based on the production of infectious pseudoviral particles by providing in trans the viral structural proteins expressed from appropriate constructs (27). A schematic representation of the structural proteins topology and the sequences at the C-Pr junction are shown in Fig. 1A and B, respectively. For the production of pseudoviruses, a WNV replicon that harbors an enhanced green fluorescent protein (EGFP) reporter (WNV-rep) (27) was cotransfected in HEK293T cells with packing constructs encoding C and PrME derived from WNV (positive control), ZIKV, or DENV2. The production of infective pseudoviral particles was analyzed by infecting Vero cells and determining the number of EGFP-positive cells by cytofluorimetry at 24 h postinfection (hpi) (Fig. 1C).
FIG 1.
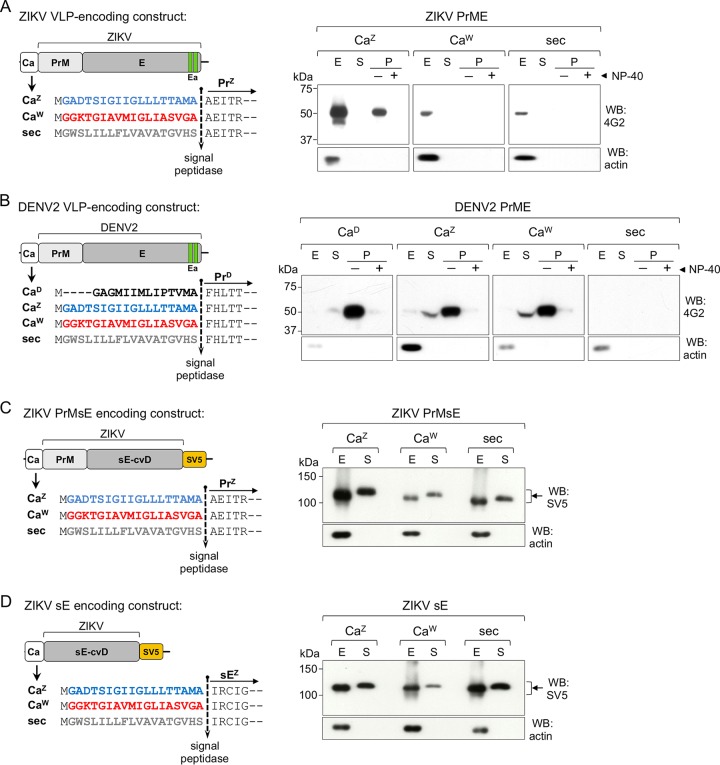
(A) Schematic representation of the genomic organization of flavivirus structural proteins (upper panel) and the membrane topology and cleavage sites for NS2B/NS3, furin and signal peptidase, indicated by red, blue, and black arrows, respectively (lower panel). (B) Sequence of capsid anchor region (Ca; in red) for WNV, ZIKV, and DENV2. The first 5 amino acids of the downstream PrM used in some of the constructs are indicated in blue. The NS2B/NS3 viral protease and signal peptidase cleavage sites are indicated by arrows. (C) Schematic representation of the protocol used for the production and detection of pseudoviruses.
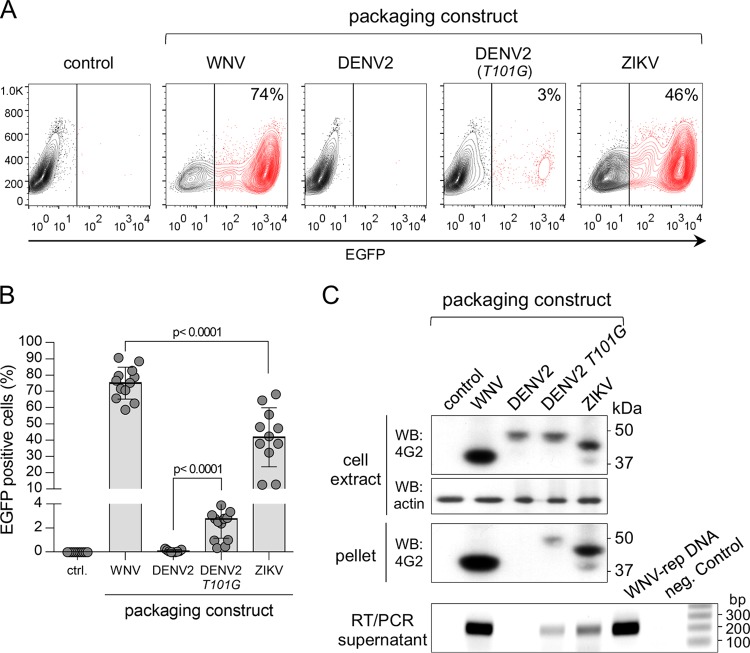
As shown in Fig. 2A and B, pseudoviruses were efficiently produced with both WNV and ZIKV packaging constructs, but not with DENV2, in agreement with previous studies showing context-dependent activity of the WNV NS2B/NS3 protease (28). A mutation in DENV2 (T101G) at the Ca cytosolic cleavage site improved proteolytic activity and partially restored pseudovirus production (Fig. 2A and B) (28). The production of pseudoviruses was further confirmed by Western blot analysis of virions (obtained from culture supernatants by ultracentrifugation) using anti-flavivirus E protein monoclonal antibody (mAb) 4G2 and by detection of replicon RNA in supernatants using reverse transcription-PCR (RT-PCR) (Fig. 2C). These results show that pseudovirus infectivity corresponds to the relative levels of secreted particles.
FIG 2.
Production of WNV, DENV2, and ZIKV pseudoviruses. (A) Cytofluorimetry profiles of Vero cells infected with pseudoviruses produced with the different packaging constructs shown in Fig. 1. The profiles are representative of several experiments. (B) Quantification of cytofluorimetry data shown in panel A (n = 12, 9, 15, and 11 for the WNV, DENV2, DENV2 T101G mutant, and ZIKV packaging constructs, respectively). (C) A nonreducing Western blot (upper panels) of cell extracts and pellets of ultracentrifuged supernatants of HEK293T cells cotransfected with the WNV-rep and the indicated packaging constructs is shown. Protein E was detected with the flavivirus-specific mAb 4G2. Actin was used as a loading control. The bottom panel shows the results of agarose gel analysis of RT-PCR products of the same supernatants. DNA stained with ethidium bromide.
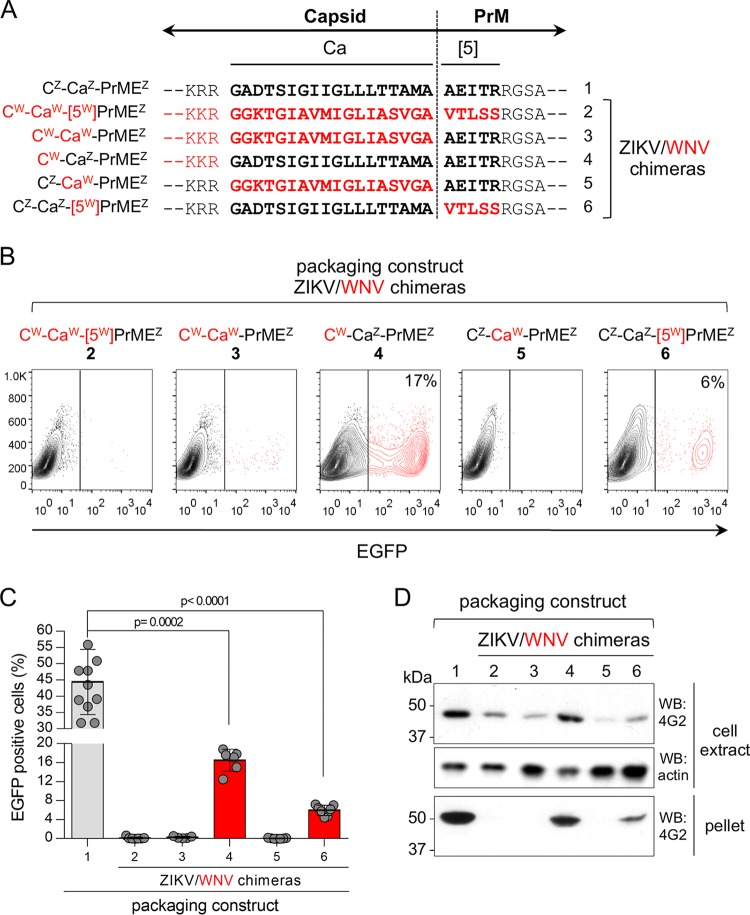
We then investigated the role of Ca in ZIKV life cycle. For this purpose, we generated different ZIKV chimeric packaging constructs in which the C and/or Ca region, and/or the first 5 amino acids of Pr ([5]), were substituted with the homologous regions from WNV (shown schematically in Fig. 3A). Cytofluorimetric analysis of cells infected with pseudoviruses obtained with the different packaging constructs showed that ZIKV chimeras carrying a heterologous Ca, i.e., CW-CaW-(5W)PrMEZ, CW-CaW-PrMEZ, and CZ-CaW-PrMEZ (Fig. 3A, constructs 2, 3, and 5, respectively), did not infect Vero cells, indicating that these replacements were lethal (Fig. 3B and C). In contrast, the chimera with homologous Ca, i.e., CW-CaZ-PrMEZ (construct 4), produced significant levels of infective particles, as well as the chimera with substitution of the five Pr N-terminal amino acids (CZ-CaZ-[5W]PrMEZ, construct 6), albeit in smaller amounts (Fig. 3B and C). In both cases, the number of infected cells were significantly lower than when using the wild-type ZIKV construct (construct 1). Pseudovirus production was further confirmed by Western blots. Consistent with the infection assay, secreted pseudoviruses were only detected in ZIKV constructs with the homologous Ca, suggesting a role for this region in the assembly and/or secretion of virion particles (Fig. 3D). Furthermore, the intracellular E levels were higher with constructs carrying ZIKV Ca (Fig. 3D). In agreement with the infection data, the chimera with the heterologous first 5 amino acids of Pr (CZ-CaZ-[5W]PrMEZ, construct 6) showed reduced level of intracellular and secreted E and was found to significantly compromise secretion of viral particles (Fig. 3D).
FIG 3.
Generation and characterization of Zika pseudoviruses. (A) Scheme and sequence at the C-Pr junction of different C-PrME ZIKV/WNV chimeras used as packaging constructs and numbered 1 to 6. WNV-derived sequences are shown in red. The superscripts Z and W indicate sequences derived from ZIKV and WNV, respectively. (B) Representative cytofluorimetry profiles of Vero cells infected with the indicated ZIKV/WNV chimeric pseudoviruses. (C) Quantification of pseudovirus production determined by cytofluorimetry and expressed as a percentage of EGFP-expressing Vero cells at 24 hpi (n = 11, 7, 5, 6, 7, and 7, for packaging constructs 1 through 6, respectively). A multiple t test was used to analyze statistic differences between wild-type and chimeric pseudoviruses. (D) Western blot analysis of intracellular protein levels and secreted virions in cell lysates and pellets of ultracentrifuged supernatants of HEK293T cells transfected with the indicated packaging constructs. E protein was detected with the flavivirus-specific mAb 4G2. Actin was used as a loading control.
These results indicate a key role for Ca in the morphogenesis of ZIKV infectious particles, a function that goes beyond its signal peptide activity for PrM translocation into the ER lumen.
Cytosolic and ER luminal Ca processing.
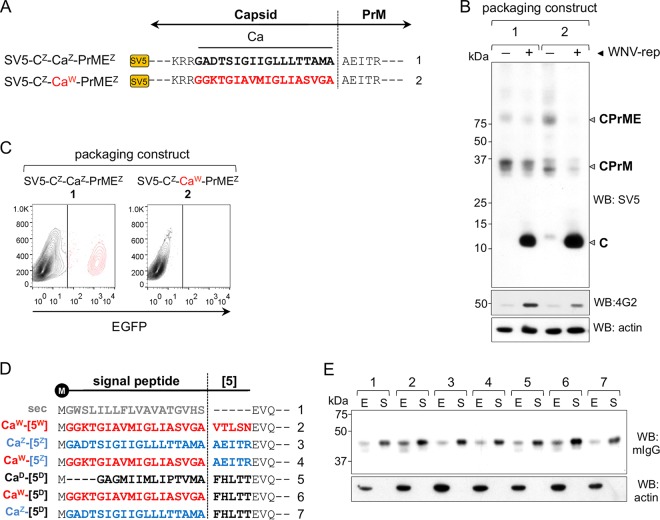
The processing of Ca is a coordinated step of cleavage by viral and host proteases. The chimeric constructs generated by replacements at the C-Pr junctions have modified cleavage site sequences, which could affect Ca processing and thus potentially affect the production of pseudoviruses. To examine the effect of these replacements on proteolytic cleavage, N-terminally SV5-tagged packaging constructs carrying the full ZIKV sequence and the Ca substitution were generated (SV5-CZ-CaZ-PrMEZ and SV5-CZ-CaW-PrMEZ, respectively) (Fig. 4A). These constructs were transfected into HEK293T cells in the presence or absence of the viral protease NS2B/NS3 provided by WNV-rep. Western blot analysis showed that in the absence of viral protease, C protein was not cleaved since only CPrM and CPrME unprocessed forms were detected at very low levels (Fig. 4B). Conversely, in the presence of NS2B/NS3, C was equally processed in both constructs, as represented by the high levels of SV5-tagged C (Fig. 4B). Similar results were obtained for E protein (detected with mAb 4G2), further confirming the downstream effect of Ca processing and the sequential requirement of cytosolic cleavage before signal peptidase luminal proteolysis (Fig. 4B). As shown in Fig. 4C, the ZIKV SV5-tagged packaging construct was able to produce infective particles although reduced in comparison to the untagged version while, as expected, the one with the CaW was not.
FIG 4.
Cleavage by NS2B/NS3 viral protease and signal peptidase. (A) Schematic representation of packaging construct carrying the SV5 tag at the C N terminus. Ca sequence derived from WNV (CaW) is shown in red. (B) Western blot analysis of cellular extracts of HEK293T cells transfected with the constructs shown in panel A in the absence (−) or presence (+) of the WNV-rep. The migration of C, CPrM, and CPrME are indicated. (C) Representative cytofluorimetry profile of cells infected with pseudoviruses produced with the packaging constructs shown in panel A. (D) Scheme of the Ca from ZIKV, WNV, and DENV2 (CaZ, blue; CaW, red; and CaD, black) used as signal peptides on an irrelevant secretory reporter protein. The nonviral signal peptide sec (gray) is also shown. The vertical line indicates signal peptidase cleavage site. All constructs contain the first 5 amino acids of viral Pr indicated as reported previously (5) using the same color code. (E) Western blot of cell extracts (E) and culture supernatants (S) of cells transfected with constructs shown in panel D. Reporter protein was developed with anti-mouse IgG. Actin was used as a loading control.
These results indicate that the lack of pseudovirus production with the heterologous CaW was not the consequence of impaired processing on the cytosolic side by the viral protease.
We next investigated processing by the signal peptidase at the Ca-Pr peptide junction of chimeric constructs and the performance of the different Ca as secretion signals. For this purpose, Ca from DENV2, ZIKV, and WNV with wild-type or chimeric Pr N-terminal amino acids were used as signal peptides on an irrelevant secretory soluble reporter protein (a VHH-Fcγ fusion protein). We utilized, as a positive control, an efficient immunoglobulin-derived signal peptide (sec) frequently used for different secretory proteins (29, 30) (Fig. 4D). Western blot analysis of intracellular and secreted protein levels showed that all three Cas allowed secretion of the reporter protein with an efficiency comparable to sec, irrespective of the 5 amino acids immediately downstream of the cleavage site (Fig. 4E). Thus, the comparable signal peptide activity of each Ca and their ability to be cleaved by the signal peptidase suggest that the differences observed in pseudovirus production are not a consequence of impaired translocation efficiency.
Effect of Ca on VLPs secretion.
We then analyzed the role of Ca in the production of virus-like particles (VLPs), which correspond to secretory vesicles harboring the PrME viral proteins without the nucleocapsid core. Although VLPs are not identical to viral particles, they represent structures that assemble into membranous vesicles, are secreted, and show antigenic properties similar to infectious viruses. For this purpose, we used constructs encoding ZIKV PrME with Ca from ZIKV or WNV as signal peptides (Fig. 5A, left panel). The nonviral sec signal peptide was used as a control. Western blot analysis of cell extracts and ultracentrifuged supernatants (to concentrate secreted VLPs) were in line with the production of pseudoviruses. VLPs were produced at higher levels and secreted only in the presence of its homologous CaZ (Fig. 5A, right panel). The vesicular nature of the secreted material was confirmed by treating supernatants with a nonionic detergent (NP-40, 0.5%) before ultracentrifugation. As expected, this treatment dissolved the membrane of the vesicles present in the supernatant, which did not sediment by ultracentrifugation. Further, we used DENV2 VLP constructs with different Cas to analyze whether these replacements had the same impact on a different flavivirus (Fig. 5B, left panel). The results showed that, in contrast to ZIKV, DENV2 VLPs were produced even when the homologous CaD was replaced with the ones from ZIKV and WNV but not with the control sec (Fig. 5B, right panel). This indicates that, while ZIKV strictly requires its homologous Ca, DENV is more permissive to other virus-derived sequences but remains refractory to the nonviral one.
FIG 5.
Secretion of VLPs and soluble E (sE). (A and B, left panels) Schemes of VLPs encoding constructs (PrME) of ZIKV (A) and DENV2 (B), with CaZ (blue), CaW (red), CaD (black) or the nonviral sec (gray), as signal peptides. Right panels show nonreducing Western blots of cellular extracts (E), culture supernatants (S), and ultracentrifuged pellet fraction (P; shown with [+] or without [−] 0.5% NP-40) of HEK293T cells transfected with the ZIKV (A) or DENV2 (B) VLP constructs. Protein E was detected with mAb 4G2. (C and D, left panels) Schemes of constructs encoding the covalent dimer (cvD) format of ZIKV PrMsE (C) or sE (D) with CaZ (blue), CaW (red), or the nonviral sec (gray), as signal peptides. Right panels show nonreducing Western blots of cellular extracts (E) and culture supernatants (S) of transfected HEK293T cells developed with anti-SV5. Arrows indicate the position of dimeric E.
In contrast to what we observed with VLP constructs, secretion of a soluble version of ZIKV dimeric E protein (sE-cvD, which lacks its own stem and C-terminal anchor sequences and carries a A264C mutation to induce covalent dimerization) (31) was efficient with both Ca and the control sec, independently of the presence or absence of PrM (Fig. 5C and D). We decided to use the cvD variant of ZIKV sE because, as we have previously shown (31), the dimeric protein is properly folded and secreted from mammalian cells and is closer to the settings in which vesicles are formed (dimerized E). Taken together, these results strongly suggest that Ca is not just a signal for PrM translocation but plays a crucial role in the assembly of VLP vesicles.
Ca determines E protein stability.
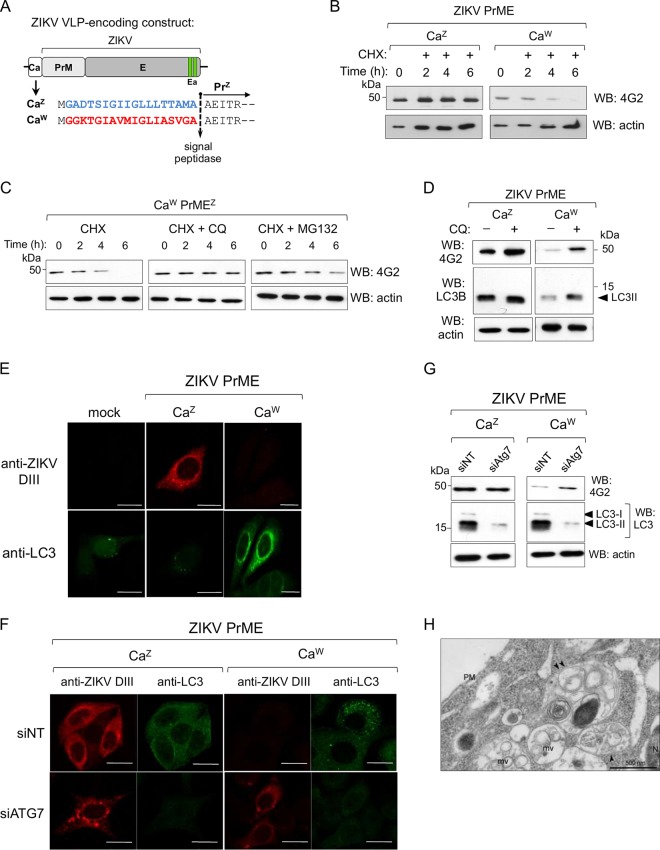
The largely reduced intracellular levels of the E protein expressed from the chimeric packaging and VLP constructs (Fig. 3B and 5A) suggested activation of a degradation pathway. This was confirmed in cells transfected with CaW-PrMEZ or CaZ-PrMEZ after cycloheximide (CHX) treatment (Fig. 6A). While E expressed from the chimeric construct with the heterologous CaW was actively degraded, the one produced from the construct with the homologous CaZ was not (Fig. 6B). To analyze the degradation pathway, transfected cells were treated with a lysosomal inhibitor (chloroquine [CQ]) or a proteasome inhibitor (MG132) along with CHX. CQ treatment completely rescued expression, while treatment with MG132 did it only partially (Fig. 6C). Further, Western blot analysis of cells extracts from VLP-transfected cells after CQ treatment showed accumulation of protein E and LC3-II (microtubule-associated protein 1 light chain 3), a marker of autophagy (32) (Fig. 6D), indicating that degradation occurs mainly through the autophagy-lysosomal pathway. LC3-II is known to be partially degraded with the target protein (33). Subsequently, activation of autophagy was studied by confocal microscopy with an anti-LC3 antibody recognizing both nonlipidated (LC3-I) and lipidated (LC3-II) forms, and an anti-E serum specific for ZIKV domain III (DIII) produced by immunizing BALB/c mice. As shown in Fig. 6E robust expression of E and a low number of LC3 puncta was observed in cells transfected with the CaZ-PrMEZ construct. In contrast, but in agreement with the activation of autophagy, in cells transfected with the chimeric CaW-PrMEZ, E was undetectable but a large increase in LC3 puncta was observed. Further evidence was obtained by small interfering RNA (siRNA) silencing of Atg7. LC3 is produced as a precursor protein, which is first cleaved by Atg4 to generate LC3-I and then by Atg3 and Atg7 to generate the lipidated form LC3-II that is associated with autophagic vesicles. Confocal microscopy showed that, whereas Atg7 silencing had no effect on E expressed from the wild-type construct, it did induce E accumulation when expressed from the chimeric CaW-PrMEZ compared to cells treated with an irrelevant siRNA (Fig. 6F). These results were confirmed by Western blot analysis of E and endogenous LC3 protein levels as well (Fig. 6G). Furthermore, electron microscopy studies have shown that the cytoplasm of cells expressing CaW-PrMEZ was enriched in late endosome/multivesicular bodies and organelles of the degradative compartment/autolysosomes (Fig. 6H). Overall, these results indicate that the single replacement of the homologous Ca with a closely related one was sufficient to induce activation of the autophagy pathway resulting in degradation of protein E and therefore impaired pseudovirus assembly.
FIG 6.
Degradation of ZIKV E protein. (A) Schematic representation of ZIKV VLP encoding constructs. (B) Western blot analysis at different time points of cellular extracts after CHX (+, 100 μg/ml) treatment at 16 h posttransfection. (C) CHX treatment of CaW-PrMEZ-transfected cells in the presence of chloroquine (CQ; 100 μM) or MG132 (20 μM). (D) Western blots of CaZ-PrMEZ- and CaW-PrMEZ-transfected cells treated (+) or not treated (−) with CQ (50 μM). An arrowhead indicates LC3-II. (E and F) Confocal immunofluorescence of HeLa cells transfected with ZIKV VLP (wt and chimeric) using anti-ZIKV DIII sera and anti-LC3. In panel F, HeLa cells were transfected with the indicated siRNAs 48 h before transfection with ZIKV VLP constructs. siNT is a control nontargeting siRNA. Scale bar, 20 μm. (G) Western blot analysis of proteins E and LC3 in HEK293T cellular extracts treated with the indicated siRNAs and transfected with the wt and chimeric ZIKV VLP constructs. (H) Representative electron microscopy image of a cell cotransfected with WNV-rep and the CZ-CaW-PrMEZ packaging construct. Arrowheads indicate autolysosomes/degradative compartments. PM, plasma membrane; N, nucleus; mv, multivesicular bodies. Scale bar, 500 nm.
Effect of Ca on the assembly of infective particles.
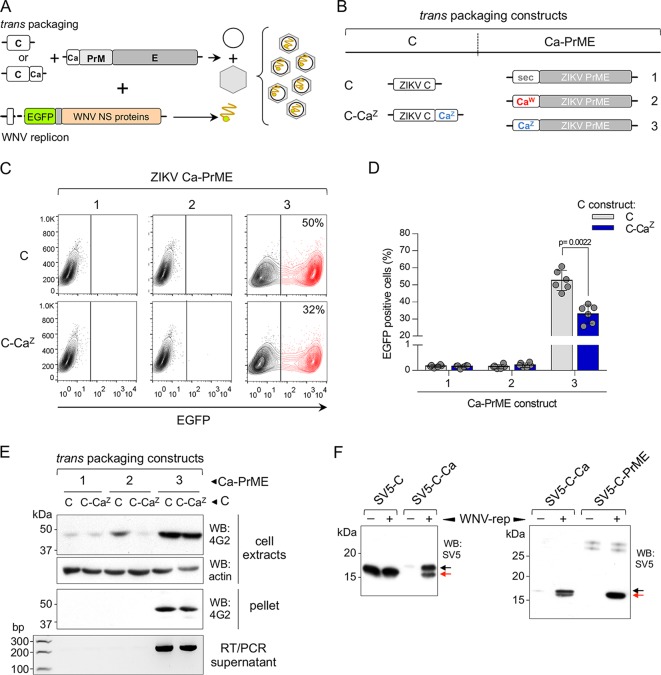
We then investigated whether, in addition to the Ca role in determining stability of the packaging structural proteins, Ca-mediated anchoring of C to the ER membrane is also required for virus morphogenesis. To achieve this, we analyzed the production of pseudoviruses using a tripartite system: C and PrME were expressed in trans from two different constructs instead of the classical cis C-PrME and cotransfected with the WNV replicon (Fig. 7A). We used ZIKV C in two different versions: one with its own CaZ and one without any anchor. The ZIKV PrME constructs instead had different Cas or a sec (as a nonviral control sequence) as a secretion signal (Fig. 7B). Pseudoviruses were produced with PrME constructs containing CaZ, but not with CaW or sec, regardless of whether C was anchored to the ER membrane (C-CaZ) or not (C) (Fig. 7C and D). Secretion of ZIKV pseudoviral particles was confirmed by Western blotting and RT-PCR (Fig. 7E). The higher efficiency of pseudovirus production when C was expressed without any anchor at its carboxy terminus was most likely due to the lower levels of soluble C produced from the C-CaZ construct in the presence of the nonstructural viral proteins (WNV-rep) because of incomplete processing (Fig. 7F, left panel). Interestingly, we observed that cleavage efficiency by the viral protease was largely improved (almost complete) in the presence of PrM (Fig. 7F, right panel).
FIG 7.
Tripartite production of pseudoviruses. (A) Schematic representation of the tripartite system of transfection for the production of pseudoviruses. (B) Packaging constructs used. ZIKV C was expressed alone (C) or fused to its own anchor CaZ (blue, C-CaZ), while ZIKV PrME was expressed with the indicated Ca (or sec), as signal peptides. (C) Representative cytofluorimetry profiles of Vero cells infected with ZIKV pseudoviruses produced with the indicated set of trans packaging constructs. (D) Quantification of ZIKV pseudoviruses production determined by cytofluorimetry of infected Vero cells at 24 hpi (n = 6 in all cases). (E) Nonreducing Western blot (upper panels) of cell extracts and pellets of ultracentrifuged supernatants of HEK293T cells, transfected with the indicated packaging constructs and developed with mAb 4G2. The bottom panel shows an agarose gel of RT-PCR products of the same supernatants stained with ethidium bromide. (F) Western blot analysis of cellular extracts of HEK293T cells cotransfected with ZIKV SV5-tagged constructs C and C-Ca (left panel) or C-Ca and C-PrME (right panel) and without (−) or with (+) WNV-rep. Cleaved and uncleaved C are indicated by red and black arrows, respectively.
Taken together, our results demonstrate that (i) the viral Ca is strictly required at the N-terminal side of PrM for proper assembly of infective particles, since PrM constructs with other signal peptides that allow efficient translocation into the lumen were unable to assemble even when coexpressed with C-Ca, and (ii) C anchorage to the ER membrane is not essential for the formation of nucleocapsid and virus assembly, which is consistent with the requirement of sequential cleavage on the cytosolic side before cleavage by the signal peptidase.
DISCUSSION
The topological distribution of structural and nonstructural proteins is a conserved characteristic feature of flaviviruses, which have a replication and assembly strategy centered on attachment to the ER membranes. With the exception of NS1, most of the nonstructural proteins are mainly facing the cytosolic side and remain attached to the ER membrane either by their own TMDs or through interactions with other NS proteins. The two structural proteins M and E associated with the virus envelope have both double-spanning transmembrane domains that serve as anchors to expose the proteins to the lumen of the ER. The role of their TMDs in the assembly and release of infective particles has been extensively studied in the context of other flaviviruses like tick borne encephalitic virus and Yellow fever virus (16, 20). Conversely, little is known of the Ca transmembrane domain, which has long been considered as a signal peptide to allow translocation of PrM into the ER lumen. Here, we show that Ca plays a significant role in ZIKV morphogenesis by controlling E protein stability, thus making it an essential determinant for assembly of infective particles.
In other viruses signal sequences have also been shown to play additional roles. In hepatitis C virus (HCV), a member of the Flaviviridae family (Hepacivirus genus), the signal peptide at the C terminus of the core protein (equivalent to Ca in ZIKV) allows secretion of nonenveloped nucleocapsids a phenomenon that contributes to persistent infection and escape from immune surveillance (34, 35). It has also been shown that this transmembrane signal peptide is responsible for lipid accumulation in infected cells causing steatosis (36). In alphaviruses, such as Semliki Forest virus and Sindbis virus, the signal peptide of p62 is found in mature viral particles associated with the complex formed by E1-E2 glycoproteins (37), whereas in the lymphocytic choriomeningitis virus the long extended N-terminal sequence (58 amino acids) of glycoprotein C remains anchored to the viral membrane after cleavage by the signal peptidase and plays a role in glycoprotein maturation and virus infectivity (38).
In this study, we used a pseudovirus-based system that allows production of infective particles. Infectivity was determined as a function of the expression of the EGFP reporter encoded in the WNV replicon. This approach has been previously used to study different aspects of flavivirus life cycle (39–44).
To understand the role of Ca in viral life cycle, different chimeras were generated by replacements of C and/or Ca on the ZIKV packaging constructs. We took advantage of the strict requirement shown by ZIKV, which only permits its own Ca to produce pseudoviruses and VLPs. This was surprising since the Cas of other viruses, like that of WNV, have a closely related sequence. Previous studies in other flaviviruses have shown that replacement of TBEV Ca with that of JEV, a distantly related flavivirus, did not impair virion formation or infectivity of viral particles (20), highlighting a permissiveness not present in ZIKV. In contrast, replacement of PrME from DENV2 with that of ZIKV without the exchange of Ca failed to produce infectious chimeric viruses (45). This was also true when pseudoviruses were produced in other cell lines such as HeLa and Vero cells (data not shown). In addition, a study on a ZIKV VLP-based vaccine showed that replacement of the signal sequence of PrM (i.e., Ca) and the TMDs of E with those of JEV improved the secretion of VLPs (46). These contrasting data indicate that different flaviviruses show markedly different permissiveness following the replacement of structural components.
To understand how the replacement of Ca in ZIKV structural polyprotein affected production of viral and subviral particles, we first ruled out impairment of the cytosolic and luminal cleavages of Ca using different packaging constructs. The signal peptide activity was tested on both viral sE and an irrelevant secretory protein, and the results showed that this function was not dependent on the downstream-encoded protein. However, different Cas clearly showed distinct activities when we assessed their ability to produce VLPs, which are the result of coexpression of PrME and require incorporation of the two membrane proteins into a lipid vesicle. In this context, only the homologous Ca was permissive for assembly of VLPs. Interestingly, the nonviral leader peptide (sec) was completely incompetent in allowing production of pseudoviruses or VLPs, even with the DENV2 construct, which is highly permissive to heterologous Ca sequences. This suggests that the virus-derived Ca sequences have been selected for a function that goes beyond their capacity to allow PrM translocation to the ER lumen.
It was clear that the replacement of Ca abolished the production of pseudoviral particles by strongly reducing the intracellular levels of protein E. The experiments performed in the presence of inhibitors indicated that enhanced E degradation took place preferentially through activation of the autophagy/lysosomal pathway. These results were further confirmed by electron microscopy and immunofluorescence analysis using anti-E and anti-LC3 antibodies and Atg7 silencing. Other studies have shown that the accumulation of large protein aggregates in the ER triggers the autophagy-dependent degradation pathway (47). One possibility is that Ca actively participates in the folding of PrM and/or E. However, since proper folding of the extracellular domains of E (sE) can be efficiently achieved even in the absence of Ca and PrM (31), this putative role for Ca might be dependent on anchorage of the proteins to the ER membrane through their TM domains. In this scenario, Ca might be required to allow proper interactions and assembly of PrME complexes, which are otherwise actively targeted to degradation. Since Ca is a short hydrophobic sequence embedded in the ER membrane, the different tolerances of exogenous Ca sequences by different flaviviruses suggest complex interactions taking place between Ca and the TM domains of M and/or E. Surprisingly, the lack of permissive interactions leads to activation of a degradation pathway. In agreement with the existence of these interactions, the secretion of E with exogenous Ca in the absence of PrM and of its own TMs was also well tolerated by ZIKV.
Interestingly, the experiments with the tripartite system for the production of pseudoviruses showed a strict requirement for CaZ to be expressed in cis with respect to PrME. In fact, providing CaZ in trans (as when expressed fused to C in C-CaZ construct) was not sufficient to allow assembly into vesicles. In agreement with our results, transcomplementation studies on MVEV, YFV, and Kunjin viruses have also shown reduced pseudovirus assembly when C-Ca and PrME proteins were translated from separate coding units (23, 41, 48, 49).
It was not surprising to find that protein C does not require anchorage to the ER membrane for the formation of the nucleocapsid and assembly into infectious particles, since this is consistent with the sequential cleavage requirement on the cytosolic side before processing by the signal peptidase. Moreover, soluble C was more efficient in packaging WNV-rep than the membrane-anchored C, most likely because in the absence of the downstream Pr peptide, Ca processing efficiency on the cytosolic side was affected. These results are consistent with a previous report showing that in YFV efficient cleavage was dependent on the Pr peptide sequence downstream (49).
In conclusion, our study shows that ZIKV assembly and maturation rearrangements involve not only the transmembrane domains of the lipid-embedded M and E proteins, but also Ca, which determines E stability controlling activation of the autophagy/lysosomal pathway.
MATERIALS AND METHODS
Cell lines, monoclonal antibodies, and chemicals.
HEK293T (CRL-11268; ATCC, Rockville, MD) and Vero (ATCC CCL-81) cells were cultured in Dulbecco modified Eagle medium (DMEM; Life Technologies, Paisley, UK) supplemented with 10% heat-inactivated fetal calf serum (FCS; Life Technologies), 50 μg/ml gentamicin, and 2 mM l-glutamine. Unless indicated otherwise, all cells were incubated at 37°C in the presence of 5% CO2. For the expression of covalent E dimers, transfected cells were incubated at 28°C instead of 37°C. Flavivirus E protein-specific mAb 4G2 was expressed as a scFv (50; NCBI accession codes KJ438784 and KJ438785) fused to avidin in a format called avibody (A. Predonzani and O. R. Burrone, unpublished data) and used as cultured supernatant from HEK293T-transfected cells. mAb SV5 was obtained as previously described (30). The ZIKV-specific anti-DIII serum was produced by immunizing BALB/c mice with a DNA construct encoding the DIII-CH3 as reported previously (30).
For protein degradation analysis, HEK293T cells were treated with the translation inhibitor cycloheximide (100 μg/ml; Sigma), the proteasomal inhibitor MG132 (20 μM; Calbiochem), or the autophagy inhibitor chloroquine (50 μM; Sigma) at 16 h posttransfection, and the cells were collected at different time intervals after treatment to analyze the protein levels.
Plasmid DNA constructs.
The gene fragments encoding the structural proteins (CPrME) from ZIKV MR766 strain (GenBank accession number AEN75266.1) and DENV2 NGC strain (GenBank accession number AAA42941) were obtained as a synthetic, mammalian-codon optimized genes (GenScript, Piscataway, NJ) and cloned into pVAX1 expression vectors (Life Technologies). The WNV packaging construct and the DNA-launched WNV subgenomic replicon expressing EGFP (here named WNV-rep) were kindly provided by Theodore Pierson (National Institute of Allergies and Infectious Diseases, Bethesda, MD) (27). The chimeric packaging constructs were obtained either by cloning or site-directed mutagenesis (QuikChange XL site-directed mutagenesis kit; Agilent Technologies, La Jolla, CA) according to the manufacturer's instructions.
Capsid, PrME, and PrMsE encoding constructs were obtained by cloning the sequence of interest into pVAX1 expression vector after a PCR amplification of the fragment from the packaging constructs. PrMsE constructs were fused to a carboxy-terminal SV5 tag (GKPIPNPLLGLD) for the detection of the soluble protein (PrMsE-SV5) (51). sE-SV5 constructs were obtained after directed deletion of PrM from PrMsE-SV5 plasmids.
Production of pseudoviral particles and VLPs.
Pseudoviral particles were produced as previously described (52). Briefly, the WNV-rep and plasmids expressing the wild-type (WT) or chimeric packaging constructs of WNV, ZIKV, and DENV2 (1:3 WNV-rep/packaging construct ratio) were cotransfected into HEK293T cells using linear polyethylenimine (molecular weight, 25,000; Polysciences, Warrington, PA) in a 1:3 DNA/polymer ratio. When the structural genes were provided in trans, the WNV-rep, PrME, and C gene constructs were used at a 1:1:4 ratio, respectively. At 16 h posttransfection, the culture medium was replaced by DMEM with 7% FCS, and pseudoviruses were harvested after 24 h of incubation at 37°C. Supernatants were clarified by centrifugation and stored at −20°C until use. Using the same protocol, VLPs were obtained by transfecting the PrME constructs into HEK293T cells. When needed, the culture supernatant was ultracentrifuged with a Beckman Coulter Airfuge centrifuge using a A100/18 rotor at 149,000 × g for 2 h at 4°C, and the pellet containing either pseudoviruses or VLPs was resuspended in nonreducing sample buffer for Western blot analysis.
Infection of Vero cells.
A total of 4 × 104 Vero cells were seeded in 24-multiwell plates 24 h before infection. The culture medium was removed, and the cells were infected with 200 μl of pseudoviral preparations for 3 h at 37°C. Then, 0.5 ml of DMEM with 2% FBS was added, and the cells were cultured for 24 h. Cell infection was determined by measuring the percentage of EGFP-positive cells in cytofluorimetric analysis in a FACSCalibur (BD Biosciences, San Jose, CA).
Expression of secretory proteins.
Standard calcium phosphate transfections were performed on HEK293T cells in 6-multiwell plates as previously described (53). After overnight incubation at 37°C, the culture medium was replaced with serum-free DMEM, and the cells were incubated for another 24 h at 37 or 28°C, depending on the construct. The culture supernatants were then cleared by centrifugation, and cellular extracts were prepared in 100 μl of TNN lysis buffer (100 mM Tris-HCl [pH 8], 250 mM NaCl, 0.5% NP-40) supplemented with protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Samples were stored at −20°C until use.
siRNA transfection.
For siRNA experiments, 5 × 104 HeLa cells/well and 1 × 105 HEK293T cells were seeded in 12-multiwell plates and transfected with 0.1 nmol of annealed duplex siRNA (Sigma) using 5 μl of RNAiMAX Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol. The following siRNAs were transfected: siAtg7 (5′-GGUCAAAGGAUGAAGAUAA-3′) and a control nontargeted siRNA, siNT (5′-UCGUCUUCUACAACGUCAATT-3′). At 48 h posttransfection, the medium was changed, and the cells were allowed to grow for 24 h, followed by transfection with VLP constructs. The next day, HeLa cells were used for immunofluorescence, and HEK293T cells were used for Western analysis.
Western blotting.
Western blot analyses of untagged and SV5-tagged proteins were done as previously reported (30). Briefly, samples were separated by nonreducing SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore, Temecula, CA) and blocked with 5% milk solution in phosphate-buffered saline (PBS-milk). Afterward, the membranes were incubated for 1 h with the 4G2 avibody or anti-SV5 mAb and washed. For SV5-tagged proteins, membranes were incubated for 1 h with anti-SV5 mAb (1 μg/ml), washed, and further probed with HRP-linked anti-mouse IgG goat antibodies (catalog no. 074-1809, 1:10,000; KPL, Gaithersburg, MA) for 1 h. 4G2 avibody was previously treated with biotinylated-HRP (ThermoFisher-Pierce, Rockford, IL, USA), incubated for 1 h, and washed. Mouse HRP-conjugated anti-actin (clone AC-15, 1:30,000; Sigma-Aldrich) was used as loading control. Signals were developed by ECL (Thermo Fisher-Pierce, Rockford, IL).
RT-PCR.
WNV replicon-derived RNA was isolated from 100 μl of pseudoviral preparations using the RNAzol-BEE solution (Tel-Test, Friendswood, TX) and then treated with DNase I (Promega, Madison, WI) according to the manufacturer's instructions. Reverse transcription was performed using random hexamers (Sigma) and Moloney murine leukemia virus reverse transcriptase (Life Technologies) according to the manufacturer's protocol. cDNAs were then amplified by PCR using WNV-3′ untranslated region-specific primers (forward, 5′-CAGTGTCAGACCACACTTTAATGT-3′; reverse, 5′-GCTTACAGCTTCAGCCAAG-3′).
Immunofluorescence microscopy.
For immunofluorescence, 5 × 104 HeLa cells/well were seeded in 12-multiwell plate and transfected with VLP constructs using calcium phosphate. The cells were fixed with 4% paraformaldehyde (PFA) at 24 h posttransfection, and immunofluorescence experiments were performed as described previously (54) using the following antibody dilutions: mouse ZIKV anti-DIII sera (1:100), anti-LC3B rabbit antibody (1:200; Sigma), Alexa Fluor 647 conjugated anti-mouse (1:500; Life Technologies), Alexa Fluor 488-conjugated anti-rabbit (1:500; Life Technologies). Samples were analyzed by an LSM510 confocal microscope (Zeiss, Gottingen, Germany) equipped with a 100× NA 1.3 objective.
Electron microscopy.
HEK293T cells were fixed at 20 h posttransfection as a monolayer in 2% glutaraldehyde–0.1 M cacodylate buffer for 30 min at room temperature, scraped, collected as a pellet, and supplemented with fresh fixative for 24 h at room temperature. Cell pellets were further processed and embedded in epoxy resin. Thin sections were observed with a Philips CM10 transmission electron microscope, and images were acquired at a final magnification of 25,000× to 34,000× using a Morada CDD camera (Olympus, Munster, Germany).
Statistical analysis.
Data were obtained from at least six independent experiments performed in duplicate or triplicate. Unless indicated otherwise, arithmetic means ± standard deviations were calculated. A nonparametric Mann-Whitney test for median comparison was used when required (Prism 6.0; GraphPad Software, Inc., La Jolla, CA); in all cases P values of <0.005 were considered significant. The sample size was not statistically assessed, and no randomization was done. The researchers involved were not blind for data collection or analysis.
ACKNOWLEDGMENTS
We are very grateful to Theodore Pierson (National Institute of Allergy and Infectious Diseases, Bethesda, MD) for kindly providing the WNV packaging construct and the DNA-launched WNV subgenomic replicon expressing EGFP.
J.R. and J.L.S.C. were supported by Arturo Falaschi ICGEB postdoctoral fellowships.
REFERENCES
- 1.Pierson TC, Diamond MS. 2013. Flaviviruses, p 747–794. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. 2003. Structures of immature flavivirus particles. EMBO J 22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostyuchenko VA, Zhang Q, Tan JL, Ng TS, Lok SM. 2013. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J Virol 87:7700–7707. doi: 10.1128/JVI.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apte-Sengupta S, Sirohi D, Kuhn RJ. 2014. Coupling of replication and assembly in flaviviruses. Curr Opin Virol 9:134–142. doi: 10.1016/j.coviro.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindenbach BD, Rice CM. 2001. Flaviviridae: the viruses and their replication, p 991–1042. In Knipe DM, Howley PM (ed), Fields virology, 4th ed Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Hellen CUT, Kräusslich HG, Wimmer E. 1989. Proteolytic processing of polyproteins in the replication of RNA viruses. Biochem 28:9881–9890. doi: 10.1021/bi00452a001. [DOI] [PubMed] [Google Scholar]
- 7.Yamshchikov VF, Compans RW. 1993. Regulation of the late events in flavivirus protein processing and maturation. Virol 192:38–51. doi: 10.1006/viro.1993.1006. [DOI] [PubMed] [Google Scholar]
- 8.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol 84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol 71:6650–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junjhon J, Pennington JG, Edwards TJ, Perera R, Lanman J, Kuhn RJ. 2014. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J Virol 88:4687–4697. doi: 10.1128/JVI.00118-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stadler K, Allison SL, Schalich J, Heinz FX. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol 71:8475–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers TJ, Weir RC, Grakoui A, McCourt DW, Bazan JF, Fletterick RJ, Rice CM. 1990. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A 87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanyi-Nagy R, Darlix JL. 2010. Intrinsic disorder in the core proteins of flaviviruses. Protein Peptide Lett 17:1019–1025. doi: 10.2174/092986610791498911. [DOI] [PubMed] [Google Scholar]
- 15.Lindenbach BD, Murray CL, Thiel HJ, Rice CM. 2013. Flaviviridae, p 713–746. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 16.Op De Beeck A, Molenkamp R, Caron M, Ben Younes A, Bredenbeek P, Dubuisson J. 2003. Role of the transmembrane domains of prM and E proteins in the formation of yellow fever virus envelope. J Virol 77:813–820. doi: 10.1128/JVI.77.2.813-820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orlinger KK, Hoenninger VM, Kofler RM, Mandl CW. 2006. Construction and mutagenesis of an artificial bicistronic tick-borne encephalitis virus genome reveals an essential function of the second transmembrane region of protein E in flavivirus assembly. J Virol 80:12197–12208. doi: 10.1128/JVI.01540-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh SC, Tsai WY, Wang WK. 2010. The length of and nonhydrophobic residues in the transmembrane domain of dengue virus envelope protein are critical for its retention and assembly in the endoplasmic reticulum. J Virol 84:4782–4797. doi: 10.1128/JVI.01963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz R, Blazevic J, Taucher C, Pangerl K, Heinz FX, Stiasny K. 2011. The unique transmembrane hairpin of flavivirus fusion protein E is essential for membrane fusion. J Virol 85:4377–4385. doi: 10.1128/JVI.02458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blazevic J, Rouha H, Bradt V, Heinz FX, Stiasny K. 2016. Membrane anchors of the structural flavivirus proteins and their role in virus assembly. J Virol 90:6365–6378. doi: 10.1128/JVI.00447-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Heijne G. 1990. The signal peptide. J Membr Biol 115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 22.Amberg SM, Nestorowicz A, McCourt DW, Rice CM. 1994. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J Virol 68:3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobigs M, Lee E, Ng ML, Pavy M, Lobigs P. 2010. A flavivirus signal peptide balances the catalytic activity of two proteases and thereby facilitates virus morphogenesis. Virol 401:80–89. doi: 10.1016/j.virol.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Lobigs M. 1993. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc Natl Acad Sci U S A 90:6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stocks CE, Lobigs M. 1998. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J Virol 72:2141–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E, Stocks CE, Amberg SM, Rice CE, Lobigs M. 2000. Mutagenesis of the signal sequence of yellow fever virus prM protein: enhancement of signalase cleavage In vitro is lethal for virus production. J Virol 74:24–32. doi: 10.1128/JVI.74.1.24-32.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierson TC, Sanchez MD, Puffer BA, Ahmed AA, Geiss BJ, Valentine LE, Altamura LA, Diamond MS, Doms RW. 2006. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virol 346:53–65. doi: 10.1016/j.virol.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 28.VanBlargan LA, Davis KA, Dowd KA, Akey DL, Smith JL, Pierson TC. 2015. Context-Dependent cleavage of the capsid protein by the West Nile virus protease modulates the efficiency of virus assembly. J Virol 89:8632–8642. doi: 10.1128/JVI.01253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casini A, Olivieri M, Vecchi L, Burrone OR, Cereseto A. 2015. Reduction of HIV-1 infectivity through endoplasmic reticulum associated degradation-mediated Env depletion. J Virol 89:2966–2971. doi: 10.1128/JVI.02634-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poggianella M, Slon Campos JL, Chan KR, Tan HC, Bestagno M, Ooi EE, Burrone OR. 2015. Dengue E protein domain III-based DNA immunisation induces strong antibody responses to all four viral serotypes. PLoS Neg Trop Dis 9:e0003947. doi: 10.1371/journal.pntd.0003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slon Campos JL, Marchese S, Rana J, Mossenta M, Poggianella M, Bestagno M, Burrone OR. 2017. Temperature-dependent folding allows stable dimerization of secretory and virus-associated E proteins of dengue and Zika viruses in mammalian cells. Sci Rep 7:966. doi: 10.1038/s41598-017-01097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabeya YY, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. 2005. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 34.Maillard P, Krawczynski K, Nitkiewicz J, Bronnert C, Sidorkiewicz M, Gounon P, Dubuisson J, Faure G, Crainic R, Budkowska A. 2001. Nonenveloped nucleocapsids of hepatitis C virus in the sera of infected patients. J Virol 75:8240–8250. doi: 10.1128/JVI.75.17.8240-8250.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi SH, Park KJ, Kim SY, Choi DH, Park JM, Hwang SB. 2005. C-terminal domain of hepatitis C virus core protein is essential for secretion. World J Gastroenterol 11:3887–3892. doi: 10.3748/wjg.v11.i25.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jhaveri R, Qiang G, Diehl AM. 2009. Domain 3 of Hepatitis C virus core protein is sufficient for intracellular lipid accumulation. J Infect Dis 200:1781–1788. doi: 10.1086/648094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lobigs M, Zhao HX, Garoff H. 1990. Function of Semliki Forest virus E3 peptide in virus assembly: replacement of E3 with an artificial signal peptide abolishes spike heterodimerization and surface expression of E1. J Virol 64:4346–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Froeschke M, Basler M, Groettrup M, Dobberstein B. 2003. Long-lived signal peptide of lymphocytic choriomeningitis virus glycoprotein pGP-C. J Biol Chem 278:41914–41920. doi: 10.1074/jbc.M302343200. [DOI] [PubMed] [Google Scholar]
- 39.Davis CW, Mattei LM, Nguyen HY, Ansarah-Sobrinho C, Doms RW, Pierson TC. 2006. The location of asparagine-linked glycans on West Nile virions controls their interactions with CD209 (dendritic cell-specific ICAM-3 grabbing nonintegrin). J Biol Chem 281:37183–37194. doi: 10.1074/jbc.M605429200. [DOI] [PubMed] [Google Scholar]
- 40.Goto A, Yoshii K, Obara M, Ueki T, Mizutani T, Kariwa H, Takashima I. 2005. Role of the N-linked glycans of the prM and E envelope proteins in tick-borne encephalitis virus particle secretion. Vaccine 23:3043–3052. doi: 10.1016/j.vaccine.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 41.Khromykh AA, Westaway EG. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J Virol 71:1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholle F, Girard YA, Zhao Q, Higgs S, Mason PW. 2004. trans-Packaged West Nile virus-like particles: infectious properties in vitro and in infected mosquito vectors. J Virol 78:11605–11614. doi: 10.1128/JVI.78.21.11605-11614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitby K, Pierson TC, Geiss B, Lane K, Engle M, Zhou Y, Doms RW, Diamond MS. 2005. Castanospermine, a potent inhibitor of dengue virus infection in vitro and in vivo. J Virol 79:8698–8706. doi: 10.1128/JVI.79.14.8698-8706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshii K, Goto A, Kawakami K, Kariwa H, Takashima I. 2008. Construction and application of chimeric virus-like particles of tick-borne encephalitis virus and mosquito-borne Japanese encephalitis virus. J Gen Virol 89:200–211. doi: 10.1099/vir.0.82824-0. [DOI] [PubMed] [Google Scholar]
- 45.Xie X, Yang Y, Muruato AE, Zou J, Shan C, Nunes BTD, Medeiros DBA, Vasconcelos PFC, Weaver SC, Rossi SL, Shi PY. 2017. Understanding Zika virus stability and developing a chimeric vaccine through functional analysis. mBio 8:e02134-16. doi: 10.1128/mBio.02134-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dowd KA, Morabito K, Yang ES, Pelc RS, DeMaso CR, Castilho LR, Abbink P, Boyd M, Nityanandam R, et al. 2016. Rapid development of a DNA vaccine for Zika virus. Science 354:237–240. doi: 10.1126/science.aai9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. 2006. Endoplasmic reticulum stress triggers autophagy. J Biol Chem 281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harvey TJ, Liu WJ, Wang XJ, Linedale R, Jacobs M, Davidson A, Le TT, Anraku I, Suhrbier A, Shi PY, Khromykh AA. 2004. Tetracycline inducible packaging cell line for production of flavivirus replicon particles. J Virol 78:531–538. doi: 10.1128/JVI.78.1.531-538.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mason PW, Shustov AV, Frolov I. 2006. Production and characterization of vaccines based on flaviviruses defective in replication. Virol 351:432–443. doi: 10.1016/j.virol.2006.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett KM, Gorham RD Jr, Gusti V, Trinh L, Morikis D, Lo DD. 2015. Hybrid flagellin as a T cell independent vaccine scaffold. BMC Biotechnol 15:71. doi: 10.1186/s12896-015-0194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanke T, Szawlowski P, Randall RE. 1992. Construction of solid matrix-antibody-antigen complexes containing simian immunodeficiency virus p27 using tag-specific monoclonal antibody and tag-linked antigen. J Gen Virol 73:653–660. doi: 10.1099/0022-1317-73-3-653. [DOI] [PubMed] [Google Scholar]
- 52.Mossenta M, Marchese S, Poggianella M, Slon Campos JL, Burrone OR. 2017. Role of N-glycosylation on Zika virus E protein secretion, viral assembly and infectivity. Biochem Biophys Res Commun 492:579–586. doi: 10.1016/j.bbrc.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 54.Eichwald C, Rodriguez JF, Burrone OR. 2004. Characterisation of rotavirus NSP2/NSP5 interaction and dynamics of viroplasms formation. J Gen Virol 85:625–634. doi: 10.1099/vir.0.19611-0. [DOI] [PubMed] [Google Scholar]