Abstract
Studies in yeasts have implicated the importance of Kin1 protein kinase, a member of the eukaryotic PAR1/MARK/MELK family, in polarized growth, cell division and septation through coordinated activity with the phosphatase, calcineurin. Kin1 is also required for virulence of the fungal pathogens Cryptococcus neoformans and Fusarium graminearum. Here we show that kin1 deletion in the human fungal pathogen Aspergillus fumigatus does not affect hyphal growth and septation but results in differential susceptibility to antifungals targeting the cell wall and cell membrane. The ∆kin1 strain remained virulent in a Galleria mellonella model of invasive aspergillosis. Expression of Kin1 tagged to GFP or RFP showed its stable localization at the septum. Co-localization experiments revealed calcineurin (CnaA) localization on either side of Kin1 at the septum suggesting possible interaction. Bimolecular fluorescence complementation assay confirmed the interaction of Kin1 with CnaA at the hyphal tips and septa in the presence of the antifungal caspofungin. Furthermore, phosphoproteomic analyses for the first time revealed Kin1 as a substrate of calcineurin providing novel insight into Kin1 regulation through calcineurin-mediated dephosphorylation mechanism.
Keywords: Aspergillus, Kin1, Calcineurin, Septum, Phosphorylation-dephosphorylation, Mass spectrometry
1. Introduction
Kin1 protein kinase is a member of the eukaryotic PAR-1/MARK/MELK family important for cell cycle, polarity and cytoskeletal dynamics in wide range of species from yeasts [1–3], fungi [4, 5], nematodes [6], to mammals [7]. In the budding yeast, Saccharomyces cerevisiae, Kin1 is not required for growth [8], but associates with plasma membrane and cell wall suggesting its role in sensing pathways [9]. In the fission yeast, Schizosaccharomyces pombe, kin1 mutation delayed cytokinesis, caused multi-septation [3], and differential susceptibility to α- and β-glucanases revealing cell wall alterations [2]. S. pombe Kin1 anchors to the plasma membrane [10], and its overexpression caused its accumulation at cell cortex and interfered with cell shape, actin and microtubule organization [11]. Majority of research on S. pombe Kin1 revealed its importance in cell division and septum synthesis [10, 12, 13]. In contrary to S. pombe, in the human pathogen, Cryptococcus neoformans, deletion of kin1 did not influence growth at 37°C, melanin or capsule synthesis but caused filamentation defects and importantly, decreased pathogenesis [5].
Filamentous fungi cause severe human diseases, but not much is known about Kin1 in these species. Kin1 has only been characterized in one filamentous plant pathogen, Fusarium graminearum, known to cause head blight, and is required for ascospore discharge, germination and infection [4]. Although F. graminearum Kin1 localizes at septal pores functional importance of localization remains unknown. In S. pombe, Kin1 and calcineurin co-regulate actin ring assembly and septum synthesis [13]. Based on these findings we were interested in understanding the role of Kin1 in the human fungal pathogen, Aspergillus fumigatus, known to cause invasive aspergillosis. We earlier showed that A. fumigatus calcineurin is required for virulence [14] and calcineurin localizes at hyphal septum to regulate proper septation [15], but the mechanism of how calcineurin regulates septation remains unknown.
Here we show that A. fumigatus Kin1 plays a very different role when compared to the yeasts and other organisms. A. fumigatus Kin1 is not required for polarized growth and septation but is important for cell wall stress response. A. fumigatus Kin1 localizes at septa, with calcineurin localizing on either side. Bimolecular complementation revealed association of Kin1 and calcineurin at the tips and septa under cell wall stress. Whole phosphoproteomics identified Kin1 as a phosphoprotein and as a substrate for calcineurin-mediated dephosphorylation. Our findings shed novel insight into Kin1 regulation in A. fumigatus.
2. Materials and methods
2.1. Strains and culture conditions
A. fumigatus strains akuBKU80 (wild-type; WT) and akuBKU80pyrG- auxotroph were used for growth and transformations. Strains were cultured on glucose minimal medium (GMM) at 37°C for 3–5 days. As required strains were cultured in presence or absence of anti-cell wall (caspofungin, nikkomycin Z, or congo red) and anti-cell membrane (voriconazole) antifungals in GMM agar or RPMI 1640 liquid medium. A. fumigatus was transformed as described [14].
2.2. A. fumigatus kin1 deletion and expression
A. fumigatus kin1 was deleted in akuBKU80pyrG- strain by replacing with A. parasiticus pyrG (Suppl. Fig. 1A). PCR-amplified kin1 promoter and kin1 terminator were cloned on either side of pyrG in pJW24 vector for kin1 deletion cassette, and the linearized construct was transformed into akuBKU80 pyrG-. Transformants were selected in the absence uracil/uridine [14]. Southern blotting confirmed homologous integration (Suppl. Fig. 1A).
For expression and localization of A. fumigatus Kin1-GFP, kin1 genomic DNA was PCR-amplified and cloned into pJW24-GFP vector. Linearized Kin1-GFP construct was transformed into akuBKU80pyrG-, transformants were selected in the absence uracil/uridine and PCR screened for homologous integration (Suppl. Fig. 1B). For KinA-RFP and CnaA-GFP co-localization, similar procedure of kin1 cloning (into pJW24-RFP-NS vector) was followed. pUCGH-CnaApromo-CnaA-CnaAterm vector was previously obtained [15]. Linearized Kin1-RFP and CnaA-GFP constructs were co-transformed into akuBKU80pyrG- strain and transformants were selected in the absence of uracil/uridine and presence of hygromycin.
For bimolecular fluorescence complementation, kin1 and cnaA were cloned into vectors pJW24-GFP-C and pUCGH-GFP-N, respectively, each containing the split form of GFP (GFP-C or GFP-N). Cloning and constructs linearization was similar to description in the previous paragraph. Kin-GFP-C and CnaA-GFP-N constructs were co-transformed into akuBKU80pyrG-strain and transformants selected in the absence of uracil/uridine and presence of hygromycin, were PCR screened for homologous integration (Suppl. Fig. 2A and 2B). All strains and primers are listed in Suppl.Table 1 and Suppl.Table 2.
2.3. Sample preparation for protein expression and phosphopeptide analysis by LC-MS/MS
A. fumigatus WT and ∆cnaA strains (in biological replicates) cultured in GMM medium for 24 h (37°C; 200 rpm) were flash frozen in liquid nitrogen. Cultures were homogenized and whole proteins extracted as follows. Urea (12 g) was added to a concentration of 8 M and subjected to one round of tissue disruption and three rounds of sonication for 10 sec each (energy setting of 90%). After centrifugation (3,000 g; 4°C ; 5 min), supernatants protein concentrations were determined by Bradford assay. Concentrations ranged from 0.64–1.52 mg/ml with total protein ranging from 16–38 mg.
Each sample (10 mg) was normalized to 0.64 mg/ml with 50 mM ammonium bicarbonate containing 8 M urea. Samples were spiked with 300 pmol of casein for internal standard quality control. Each sample was split into twelve equal volumes in a 24-well (10 ml) plate for further processing. Samples were reduced for 45 min at 32°C with 10 mM dithiothreitol and alkylated for 30 min at room temperature with 25 mM iodoacetamide. Samples were diluted to 1.6 M urea with 50 mM ammonium bicarbonate and digested with Trypsin or GluC (18 h at 32°C; trypsin-1:25 enzyme/total protein, GluC-1:20 enzyme/total protein). Split samples were recombined, acidified with TFA, centrifuged (3,000 g; 4°C; 5 min), and subjected to C18 SPE cleanup (Sep-Pak, 500 mg bed). Following elution, samples were normalized to 10 ml. Fifty micrograms of each sample was kept for “open” analysis. Two hundred micrograms of each sample was TiO2 enriched and lyophilized.
For TiO2 enrichment samples were resuspended in 65 µl of 1 M glycolic acid in 80% acetonitrile/1% TFA (200 µg). Phosphopeptide enrichments were done using 10 µl GL Bioscience TiO2 spin tips as per standard protocol at: https://genome.duke.edu/cores-and-services/proteomics-and-metabolomics/protocols-reagents Eluted phosphopeptides were subjected to stage tip C18 cleanups, lyophilized, and resuspended in 12 µl of 10 mM citric acid in 1%TFA/2% acetonitr ile containing 10 fmol/µl ADH_YEAST. From each sample, 3 µl was used for QC pooling run periodically through the acquisition period.
2.4. Quantitative differential protein expression and phosphorylation analyses
Quantitative LC/MS/MS was performed on 2 µl sample, using nanoAcquity UPLC system (Waters Corp, USA) coupled to Thermo QExactive HF high resolution accurate mass tandem mass spectrometer (Thermo Scientific, USA) via nanoelectrospray ionization source. Sample was trapped on a Symmetry C18 20 mm × 180 µm column (5 µl/min at 99.9/0.1 v/v water/acetonitrile), and analytical separation performed on 1.7 µm Acquity BEH130 C18 75 µm × 250 mm column (Waters Corp, USA) with 90 min linear gradient of 5 to 30% acetonitrile with 0.1% formic acid (400 nl/min flow rate) at 55°C. Data collection on QExactive HF mass spectrometer was performed in data-dependent acquisition (DDA) mode with an r=120,000 (@ m/z 200) full MS scan from m/z 375–1600 with a target AGC value of 3e6 ions followed by 12 MS/MS scans at r=30,000 (@ m/z 200) at a target AGC value of 5e4 ions. A 20 sec dynamic exclusion was employed to increase depth of coverage. Analysis cycle time per sample injection was 2 h. Following 9 UPLC-MS/MS analyses for each open analysis set (excluding conditioning runs but including 3 replicate QC injections), data was imported into Rosetta Elucidator v 4.0 (Rosetta Biosoftware, USA), and analyses were aligned based on accurate mass and retention time of detected ions (“features”) using PeakTeller algorithm. Relative peptide abundance was calculated from area-under-the-curve (AUC) of selected ion chromatograms of aligned features across all runs. MS/MS data was searched against NCBI_Apergillus fumigatus database (May 2017) with proteins, including yeast ADH1, BSA, and an equal number of reversed-sequence “decoys” false discovery rate determination. Mascot Distiller and Mascot Server (v 2.5, Matrix Sciences, USA) were utilized for fragment ion spectra and database searches. Database searches included fixed modification on Cys (carbamidomethyl) and variable modifications on M (oxidation). Data was annotated at 1% peptide false discovery rate using Mascot Ion Score (Trypsin Open >25.91, GluC Open > 29.77). For scaling strategy protein intensities were normalized to a robust mean (excluding top and bottom 10% of the signals) and 3D PCA was run to see clustering of different groups.
Quantitative LC/MS/MS for phosphopeptides was similar to quantitative differential protein expression method, with a sample size of 4 µL. Following sample trapping, analytical separation was performed with 90 min linear gradient of 3 to 30% acetonitrile with 0.1% formic acid. After data collection and analysis, database search parameters included fixed modification on Cys (carbamidomethyl) and variable modifications on M (oxidation), STY (phosphorylation). Data was annotated at 1% peptide false discovery rate using Mascot Ion Score (Trypsin TiO2 > 18.24; GluC TiO2 > 30.50). To identify differences between the WT and ∆cnaA groups, two-tailed t-test was performed on log2 protein or phosphopeptide.
2.5. Galleria mellonella model for virulence assay
Galleria mellonella larvae infections were performed as described [14], and data plotted on a Kaplan-Meier curve with log rank pair-wise comparison and statistical analysis using Graph Pad Prism.
2.6. Fluorescence microscopy
Fluorescence microscopy was performed as described [15]. Effect of actin and microtubule disruptors was assessed on 18 h cultures treated with inhibitors for 2 h.
3. Results and Discussion
3.1. A. fumigatus contains a single ortholog of MARK/PAR-1 kinase family
Mammals contain 5 genes encoding Kin1/Par1/MARK family proteins. While S. cerevisiae has 2 genes, S. pombe, and C. neoformans, each have a single ortholog of Kin1 [1, 2, 5]. To date among filamentous fungi, Kin1 has only been characterized in the plant pathogen, F. graminearum [4]. To identify newer targets for treatment of invasive aspergillosis, we were interested in analyzing Kin1 function in growth and virulence of the human fungal pathogen A. fumigatus. A. fumigatus genome database search revealed the presence of a Kin1 ortholog (Afu1g11080), which contained typical domains characterized in mammals, with an N-terminal kinase domain, a variable middle domain and a C-terminal kinase A1 domain (KA1) (Fig. 1A). Akin to other fungi A. fumigatus Kin1 also contained the unique fungal kinase motif (FKM) in the middle domain. Within KA1 domain fungal sequences were highly homologous as opposed to human Kin1. Domains in Kin1 play distinct roles in their contribution to its function and localization in different species [4, 13]. Mutations in the kinase domain inactivated kinase activity [10, 12, 16]. While the mammalian C-terminal KA1 domain is essential for proper folding, the FKM motif and fungal specific extended region present in the KA1 domain are important for function and localization [13]. We therefore became interested in analyzing the function and localization of A. fumigatus Kin1.
FIG. 1.
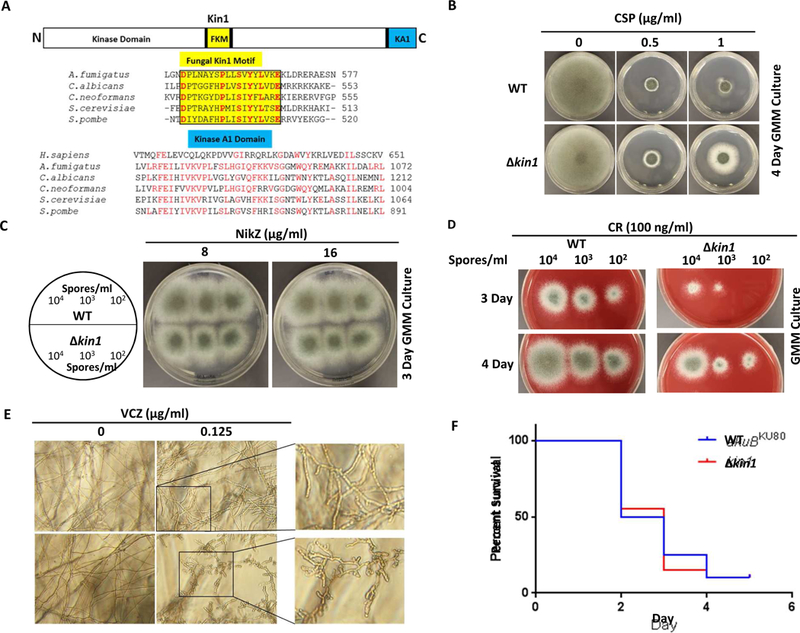
(A) A. fumigatus Kin1 domain organization. Fungal kinase motif (FKM) is shaded yellow. Conserved residues in FKM motif are colored red. Kinase A1 domain (KA1) across species is shaded blue. (B) Growth of wild-type (WT; akuBKU80) and kin1 strains after 4 days in the absence or presence of caspofungin (CSP). (C) Growth of WT and kin1 strains after 3 days in the absence or presence of nikkomycin-Z (NikZ). Circle indicates spores inoculated. (D) Growth of WT and kin1 strains after 3 and 4 days in the absence or presence of Congo red (CR). Spores inoculated are indicated. (E) Growth of WT and kin1 strains after 48 h RPMI liquid culture in the absence or presence of voriconazole (VCZ). Spores (104/ml) were inoculated. Marked regions are zoomed to show hyper branching of kin1 strain in presence of VCZ. (F) Galleria mellonella larvae infected with 5 µl of 2 × 10 5 spores (1 × 108 spores/ml) of WT and kin1 strains were incubated at 37°C with survival scored for 5 days.
3.2. A. fumigatus ∆kin1 mutant is differentially susceptible to antifungals and is avirulent
Deletion of kin1 in F. graminearum caused reduction in growth, conidiation and virulence [4]. In S. pombe, kin1 deletion caused cell wall defects resulting in aberrant hyphal morphology and polarity [2]. Conservation of A. fumigatus Kin1 with other fungal Kin1 orthologs and its role in growth, polarity, conidiation and pathogenesis prompted us to first examine the phenotypes of A. fumigatus kin1 deletion.
Surprisingly, ∆kin1 strain did not exhibit any growth or conidiation defects under normal growth conditions in comparison to the wild-type (WT; akuBKU80) (Fig. 1B). To verify if A. fumigatus ∆kin1 mutant has any inherent defects in cell wall synthesis/organization we probed the effect of anti-cell wall/anti-cell membrane antifungals (Caspofungin-CSP, Nikkomycin Z-NikZ and Congo red-CR, Voriconazole-VCZ). Intriguingly we noted differential growth effects for each of these agents on ∆kin1 mutant. While kin1 deletion partially rescued the CSP-mediated growth inhibition (1 µg/ml) in the WT (Fig. 1B), it did not alter sensitivity to NikZ (Fig. 1C; Suppl.Fig.3A), but showed clear sensitivity to CR (Fig. 1D). Treatment with sub-minimal inhibitory concentrations of VCZ (0.125 µg/ml) revealed higher sensitivity, hyper branching and polarity defects of kin1 mutant (Fig. 1E; Suppl.Fig.3B). While CSP targets β-glucan synthesis, NikZ and CR inhibit chitin synthesis, and VCZ targets membrane ergosterol biosynthesis.
To confirm any alterations in β-glucan and chitin distribution of cell walls, the WT and ∆kin1 strains were stained with aniline blue and calcofluor white, respectively. No significant changes were observed in β-glucan and chitin distribution at septa or cell walls of the ∆kin1 mutant (Suppl.Fig.3C and 3D), suggesting that loss of A. fumigatus Kin1 may not have any discernible effect on cell wall biosynthesis/organization. S. pombe kin1 mutant exhibited delayed septation, sensitivity to SDS and calcofluor white but not to CSP [10], indicating that Kin1 may function differently under different stress conditions.
Finally, to verify if A. fumigatus Kin1 is required for virulence, we utilized an invertebrate host aspergillosis infection model, Galleria mellonella. Infection of Galleria larvae with kin1 strain led to survival comparable to WT strain (p=0.92) (Fig. 1F). Similar melanization, an indication of immune response of the Galleria was noted, indicating that ∆kin1 strain is as virulent as the WT. Together these results suggest that although Kin1 is required for growth and pathogenesis in other fungi, such as C. neoformans and F. graminearum, it does not function similarly in A. fumigatus. In addition, A. fumigatus Kin1 is not a major determinant for polarized growth, cell wall synthesis and septation as shown in S. pombe and F. graminearum, but may be required for cell wall stress adaptation.
3.3. A. fumigatus Kin1 localizes to the septum and interacts with calcineurin
In S. cerevisiae Kin1 localizes to the plasma membrane [9]. In S. pombe Kin1 localizes to the tips during interphase and is recruited to cell division site during mitosis [12, 13]. Kin1 was also shown to dynamically associate with plasma membrane at sites of active cell surface remodeling in S. pombe [10]. In F. graminearum, Kin1 localized to the septal pore after completion of septation but was not observed at the hyphal tips [4].
To define A. fumigatus Kin1 localization we expressed kin1 tagged to gfp and rfp under the control of kin1 native promoter. A. fumigatus Kin1-GFP and Kin1-RFP fusion proteins localized at septa (Fig. 2A). To examine importance of actin and microtubules for localization of Kin1, we used actin and microtubule disruptors, cytochalasin A (10 µg/ml) and nocodazole (80 µg/ml). However, none of these agents mislocalized Kin1 indicating that actin and microtubules are not required for Kin1 septal localization (Suppl.Fig. 4). Similarly in S. pombe the actin depolymerizing agent, latrunculin A, only affected Kin1 association with contractile actin ring during anaphase but not its localization at tips and septa [12].
FIG. 2.
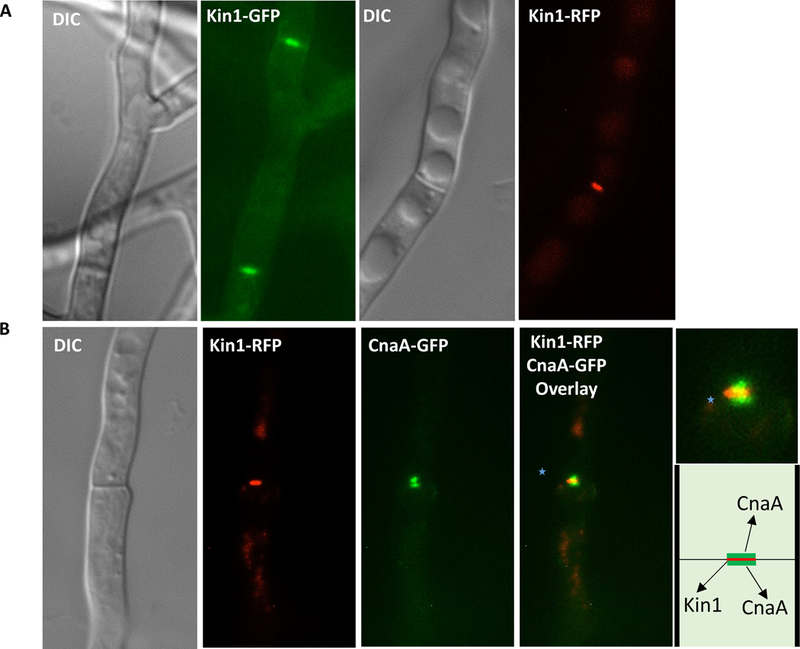
(A) Fluorescence microscopy showing Kin1-GFP or Kin1-RFP septal localization. Arrows indicate Kin1 at septa. (B) Co-localization showing Kin1-RFP and CnaA-GFP at the septum. Merged image shows localization of CnaA-GFP on either side of Kin1-RFP (inset zoomed; indicated by dotted arrows). Pictorial of Kin1 and CnaA at the septum.
Calcineurin and Kin1 coordinate actin ring assembly and septum synthesis in S. pombe, and S. pombe kin1 mutant exhibits polyseptation, and is synthetically lethal in calcineurin deletion background [13]. We previously showed that calcineurin inhibition in A. fumigatus causes aberrant hyperseptation and calcineurin localization at hyphal septa coordinated proper septation [15]. To investigate the link between A. fumigatus Kin1 and calcineurin, we co-expressed calcineurin catalytic subunit (CnaA), CnaA-GFP and Kin1-RFP, and found that calcineurin localized on either side of Kin1 at the septum (Fig. 2B). This suggests possible interaction between Kin1 and calcineurin to coordinate septation or for other functions at the septum. Because calcineurin deletion resulted in abnormal assembly of cell wall components [15], and also as kin1 strain showed differential susceptibility to anti-cell wall agents, we investigated if A. fumigatus calcineurin and Kin1 interacted by using bimolecular fluorescence complementation (BiFC) assay to detect protein-protein interaction in vivo.
For BiFC assay we co-expressed cnaA and kin1 fused to split forms of gfp (gfp-N and gfp-C). The cnaA gene was tagged to the N-terminal half of gfp (CnaA-GFP-N) and kin1 tagged to the C-terminal half of gfp (KinA-GFP-C) (Fig. 3A). While we did not find any GFP fluorescence indicative of interaction between CnaA and Kin1 under normal growth conditions (data not shown), treatment with the cell wall β-glucan synthase inhibitor, CSP (1 µg/ml), showed GFP fluorescence at hyphal tips (Fig. 3B). Hyphal lysis was evident at certain tips due to cell wall stress imposed by CSP (Fig. 3B and3C). In some lysed hyphae GFP fluorescence was also observed at center of the septum suggesting interaction of CnaA and Kin1 at the septum (Fig. 3C and3D). This suggests that calcineurin and Kin1 may interact in response to cell wall stress. We however cannot exclude the possibility of their interaction under normal growth conditions due to transient nature of certain enzymic interactions or limitations of BiFC assay. Differential susceptibility of kin1 mutant to cell wall inhibitors supports greater possibility that Kin1 may interact with calcineurin.
FIG. 3.
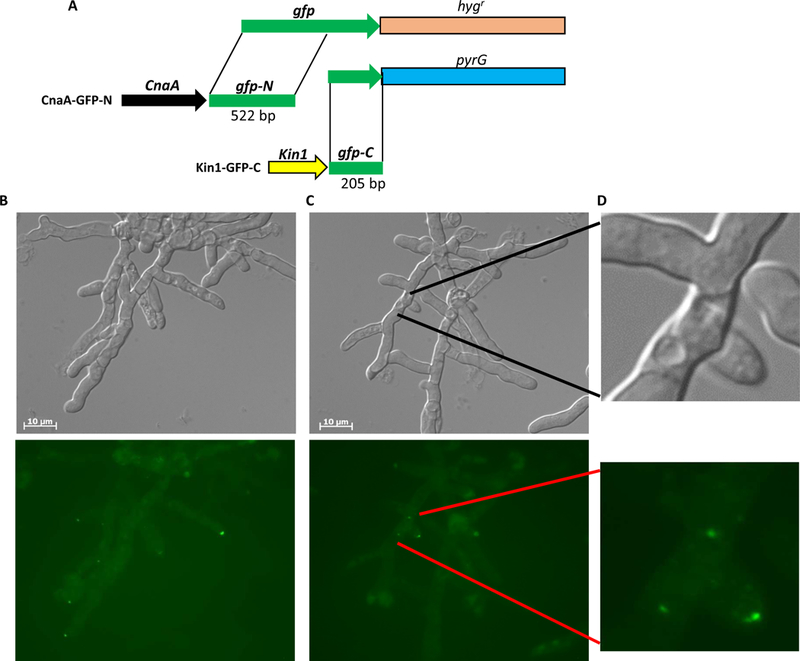
(A) Strategy for bimolecular fluorescence complementation. cnaA and kin1 are tagged to respective split form of gfp (gfp-N and gfp-C) with hygromycin resistance (hygr) and pyrG marker genes for selection. Both constructs were co-transformed into akuBKU80pyrG strain for expression and visualization of interaction. (B-D) BiFC microscopy showing interaction of Kin1-GFP-C and CnaA-GFP-N at tips and septa (indicated by arrowheads and arrows in lower panels) following caspofungin induced cell wall stress. Tip lysis seen in DIC images (upper panels; indicated by dotted arrows).
3.4. A. fumigatus Kin1 is a phosphorylated protein and is dephosphorylated by calcineurin
We hypothesized that interaction of calcineurin with Kin1 may involve dephosphorylation of Kin1 by phosphatase activity of calcineurin. Kin1 orthologs are phosphorylated in different species including yeasts [8, 16], and vertebrates [17–19]. However, nothing is known on Kin1 phosphorylation in filamentous fungi. We therefore performed whole phosphoproteomic analyses to identify phosphorylated proteins in A. fumigatus WT versus the ∆cnaA background to identify proteins that are specifically dephosphorylated by calcineurin and identified Kin1 as a substrate of calcineurin.
Following TiO2 phosphopeptide enrichment and LC-MS/MS, 4 phosphorylated sites were detected in A. fumigatus Kin1 at Ser395, Ser783, Ser955 and Ser995 (Fig. 4). Quantitative comparison of Kin1 phosphosites between WT and ∆cnaA strains revealed that phosphorylation at Ser395 and Ser955 increased by 6.8-fold and 8.2-fold, respectively, in ∆cnaA (Fig. 4A). In contrast, phosphorylation at Ser783 and Ser995 decreased by 2.7-fold and 23.3-fold in ∆cnaA (Fig. 4B). Together these suggested that A. fumigatus Kin1 is a phosphoprotein and a substrate for calcineurin-mediated dephosphorylation because the phosphorylation at Ser395 and Ser955 was increased in ∆cnaA background. In support of these bioinformatic analysis of A. fumigatus Kin1 using Eukaryotic Linear Motif resource (http://elm.eu.org/) revealed 4 putative LxVP motifs on Kin1. LxVP is a secondary docking motif that facilitates substrate interaction with calcineurin complex (catalytic and regulatory subunit) [20]. The primary docking motif (PxIxIT) enables the interaction of substrate with only calcineurin catalytic subunit. Although F. graminearum Kin1 contains a concensus PxIxIT motif within the FKM motif, this sequence is not conserved in the other fungi (Fig. 1A).
FIG. 4.
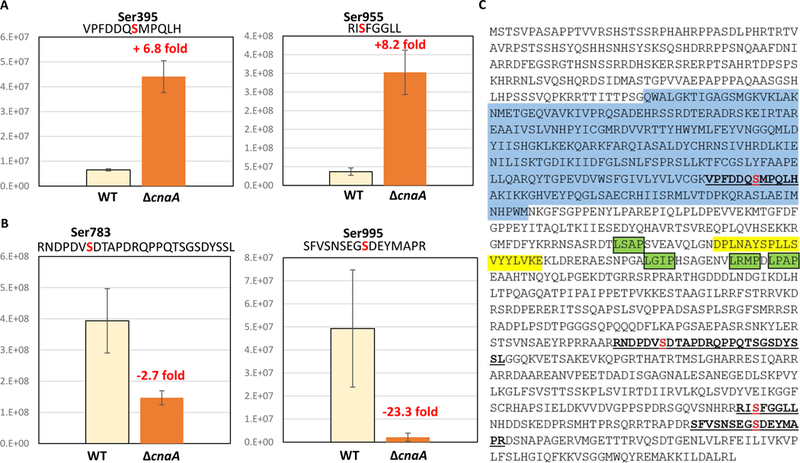
Phosphorylation of A. fumigatus Kin1 in WT and cnaA mutant. TiO2 phosphopeptide enrichment and LC-MS/MS indicated four phosphosites (p value < 0.05, fold change >2.0). (A) Ser395 (+6.8 fold, p 0.0003), Ser955 (+8.2 fold, p 0.0005); (B) Ser783 (−2.7 fold, p 0.0075), and Ser995 (−23.3 fold, p 0.0060). Expression values measured are accurate mass retention time alignment and area-under-the-curve quantitation. P-values were two-tailed heteroscedastic t-tests from log2 expression values. (C) A. fumigatus Kin1 showing phosphopeptides (underlined) with phosphorylated residues colored in red and putative LxVP calcineurin-binding motifs highlighted green. Kin1 activation loop residues are highlighted blue and FKM motif residues are highlighted yellow.
Phosphorylation of S.pombe Kin1 by Ssp1, the Ca2+/calmodulin-dependent (CaMMK)-like protein kinase, was also noted at T299 residue in the activation loop leading to the activation of Kin1 and S. pombe Kin1 was autophosphorylated at 19 sites [16]. Although T299 is conserved in A.fumigatus Kin1 we did not detect its phosphorylation in the WT or ∆cnaA background. However, we detected the phosphorylation of S395 in the activation loop of the catalytic domain which was dependent on calcineurin for dephosphorylation (Fig. 4C). Interestingly, while the calcineurin-dependent dephosphorylated residues, Ser395 and Ser955, are not conserved in S. pombe the two other phosphosites, Ser783 and Ser995 are conserved in S. pombe and present in the non-catalytic domain and close to the KA1 domain. Though Kin1 is conserved in A. fumigatus it does not function in a similar manner as characterized in yeasts, S. cerevisiae, S. pombe, and pathogenic fungi C. neoformans and F. graminearum. BiFC and whole phosphoproteomic approaches revealed Kin1 interaction with calcineurin under cell wall stress, is phosphorylated in vivo, and is dephosphorylated by calcineurin. Further studies should address Kin1 phosphorylation-dephosphorylation mechanisms in regulating other stress responses in this important human pathogen.
Supplementary Material
Highlights.
Kin1 deletion in A. fumigatus does not affect septation, growth and virulence
Fluorescence microscopy revealed Kin1 localization at the center of the septum
Bimolecular fluorescence assay confirmed interaction of Kin1 and calcineurin
Phosphoproteomics identified Kin1 as a substrate of calcineurin
Kin1 is differentially phosphorylated in vivo in the absence of calcineurin
Acknowledgements
This work was supported by an NIH/NIAID R21 award AI127551 to PRJ and WJS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Levin DE, Hammond CI, Ralston RO, Bishop JM, Two yeast genes that encode unusual protein kinases, Proceedings of the National Academy of Sciences, 84 (1987) 6035–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Levin DE, Bishop JM, A putative protein kinase gene (kin1+) is important for growth polarity in Schizosaccharomyces pombe, Proceedings of the National Academy of Sciences, 87 (1990) 8272–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Drewes G, Nurse P, The protein kinase kin1, the fission yeast orthologue of mammalian MARK/PAR-1, localises to new cell ends after mitosis and is important for bipolar growth, FEBS Letters, 554 (2003) 45–49. [DOI] [PubMed] [Google Scholar]
- [4].Luo Y, Zhang H, Qi L, Zhang S, Zhou X, Zhang Y, Xu J-R, FgKin1 kinase localizes to the septal pore and plays a role in hyphal growth, ascospore germination, pathogenesis, and localization of Tub1 beta-tubulins in Fusarium graminearum, New Phytologist, 204 (2014) 943–954. [DOI] [PubMed] [Google Scholar]
- [5].Mylonakis E, Idnurm A, Moreno R, El Khoury J, Rottman JB, Ausubel FM, Heitman J, Calderwood SB, Cryptococcus neoformans Kin1 protein kinase homologue, identified through a Caenorhabditis elegans screen, promotes virulence in mammals, Molecular Microbiology, 54 (2004) 407–419. [DOI] [PubMed] [Google Scholar]
- [6].Guo S, Kemphues KJ, par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed, Cell, 81 (1995) 611–620. [DOI] [PubMed] [Google Scholar]
- [7].Tassan J-P, Goff X, An overview of the KIN1/PAR-1/MARK kinase family, Biology of the Cell, 96 (2004) 193–199. [DOI] [PubMed] [Google Scholar]
- [8].Lamb A, Tibbetts M, Hammond CI, The product of the KIN1 locus in Saccharomyces cerevisiae is a serine/threonine-specific protein kinase, Yeast, 7 (1991) 219–228. [DOI] [PubMed] [Google Scholar]
- [9].Tibbetts M, Donovan M, Roe S, Stiltner AM, Hammond CI, KIN1 and KIN2 Protein Kinases Localize to the Cytoplasmic Face of the Yeast Plasma Membrane, Experimental Cell Research, 213 (1994) 93–99. [DOI] [PubMed] [Google Scholar]
- [10].Cadou A, Couturier A, Le Goff C, Soto T, Miklos I, Sipiczki M, Xie L, Paulson JR, Cansado J, Le Goff X, Kin1 is a plasma membrane-associated kinase that regulates the cell surface in fission yeast, Molecular Microbiology, 77 (2010) 1186–1202. [DOI] [PubMed] [Google Scholar]
- [11].Carbona S, Allix C, Philippe M, Goff X, The protein kinase kin1 is required for cellular symmetry in fission yeast, Biology of the Cell, 96 (2004) 169–179. [DOI] [PubMed] [Google Scholar]
- [12].Cadou A, La Carbona S, Couturier A, Le Goff C, Le Goff X, Role of the protein kinase Kin1 and nuclear centering in actomyosin ring formation in fission yeast, Cell Cycle, 8 (2009) 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cadou A, Couturier A, Le Goff C, Xie L, Paulson JR, Le Goff X, The Kin1 kinase and the calcineurin phosphatase cooperate to link actin ring assembly and septum synthesis in fission yeast, Biology of the Cell, 105 (2013) 129–148. [DOI] [PubMed] [Google Scholar]
- [14].Steinbach WJ, Cramer RA, Perfect BZ, Asfaw YG, Sauer TC, Najvar LK, Kirkpatrick WR, Patterson TF, Benjamin DK, Heitman J, Perfect JR, Calcineurin Controls Growth, Morphology, and Pathogenicity in Aspergillus fumigatus, Eukaryotic Cell, 5 (2006) 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Juvvadi PR, Fortwendel JR, Rogg LE, Burns KA, Randell SH, Steinbach WJ, Localization and activity of the calcineurin catalytic and regulatory subunit complex at the septum is essential for hyphal elongation and proper septation in Aspergillus fumigatus, Molecular Microbiology, 82 (2011) 1235–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lee ME, Rusin SF, Jenkins N, Kettenbach AN, Moseley JB, Mechanisms Connecting the Conserved Protein Kinases Ssp1, Kin1, and Pom1 in Fission Yeast Cell Polarity and Division, Current Biology, 28 (2018) 84–92.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Blot J, Chartrain I, Roghi C, Philippe M, Tassan J-P, Cell Cycle Regulation of pEg3, a New Xenopus Protein Kinase of the KIN1/PAR-1/MARK Family, Developmental Biology, 241 (2002) 327–338. [DOI] [PubMed] [Google Scholar]
- [18].Patel H, Zich J, Serrels B, Rickman C, Hardwick KG, Frame MC, Brunton VG, Kindlin-1 regulates mitotic spindle formation by interacting with integrins and Plk-1, Nature Communications, 4 (2013) 2056. [DOI] [PubMed] [Google Scholar]
- [19].Davezac N, Baldin V, Blot J, Ducommun B, Tassan J-P, Human pEg3 kinase associates with and phosphorylates CDC25B phosphatase: a potential role for pEg3 in cell cycle regulation, Oncogene, 21 (2002) 7630. [DOI] [PubMed] [Google Scholar]
- [20].Rodríguez A, Roy J, Martínez-Martínez S, López-Maderuelo MD, Niño-Moreno P, Ortí L, Pantoja-Uceda D, Pineda-Lucena A, Cyert MS, Redondo JM, A Conserved Docking Surface on Calcineurin Mediates Interaction with Substrates and Immunosuppressants, Molecular Cell, 33 (2009) 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


