Abstract
Background:
As a life threatening infectious disease, methicillin resistant Staphylococcus aureus (MRSA) infection has been turned into a global health concern recently. In a developing country like Bangladesh, the situation is vulnerable because of inadequate and inappropriate practices of control measures to prevent the spread of nosocomial infection.
Materials and Methods:
In this study, 100 clinical, nonclinical, and environmental samples were collected from different hospitals in Bangladesh and examined for the detection of methicillin resistant and multidrug resistant S. aureus by cultural methods. Antibiotic sensitivity pattern of isolates was also evaluated by Kirby-Bauer disc diffusion method.
Results:
Out of the total 100 samples, 66 isolates of S. aureus were determined. Among them, 53 isolates (80.30%) revealed a positive result for oxacillin test, but avoiding any false result, 53 oxacillin positive isolates were again examined by cefoxitin, whereas 43 isolates (65.15%) were resistant to cefoxitin and consequently confirmed as MRSA and rest of the 23 isolates (34.85%) were fixed as methicillin sensitive S. aureus. In antibiotic sensitivity test, S. aureus showed the highest (83%) resistance to gentamycin, oxacillin and cefotaxime than other antibiotics. Finally, the risk practices were assessed which are potential factors for spreading infections.
Conclusion:
Since, MRSA as well as other antibiotic resistant S. aureus are potent threat to human, therefore, we should consider it as a great concern to minimize or prevent the prevalence and adverse effects of MRSA.
Keywords: Azithromycin, cefoxitin, methicillin-resistant Staphylococcus aureus, oxacillin
INTRODUCTION
As the potent colonizers of skin of humans or animals, Staphylococcus aureus can accelerate nosocomial as well as community-acquired infections ranging from skin and soft-tissue infections, which are considered as mild condition, to sepsis, another severe and life-threatening condition.[1] In a hospital, methicillin-resistant S. aureus (MRSA) infections are normally spread out without introducing any surveillance program embedded with control procedures.[2] In the past couple of years, a significant number of MRSA cases have been noticed worldwide.[3] From clinical point of view, methicillin resistance is highly significant since a single genetic element can reveal resistance to antibiotics, called beta-lactam antibiotics including penicillins (oxacillin), carbapenems, and cephalosporins.[4] It has been reported that, the presence of resistance genes determine not only the resistance to antibiotics but also the expression of these genes which are regulated by environment, which can provoke the resistance to antibiotics. S. aureus, which has a high resistance pattern against a good number of antibiotics but sensitive to methicillin, is considered as methicillin-sensitive S. aureus (MSSA) as well as multidrug-resistant strains of S. aureus (MDRSA). In recent years, treatments against MDRSA have become burdensome and less successful.[5] It has been investigated that a large staphylococcal cassette chromosome, mec (SCCmec), mobile genetic element, is found in MRSA[6] which carries the mecA gene to code penicillin-binding protein, PBP2a, to all β-lactams.[7] Although some previous researches have been conducted on MRSA in Bangladesh using cultural methods, the result is not updated because the antibiotic resistance pattern of nosocomial microorganisms including MRSA and MDRSA has been changed in the last couple of years.[8]
The present study was conducted to investigate the recent prevalence of MRSA and MDR S. aureus from environmental and clinical samples in a hospital and aimed to evaluate their antibiotic resistance pattern.
MATERIALS AND METHODS
Bacterial isolates
A total of 100 swab samples were collected from various sources of different hospitals from Chittagong region in Bangladesh. Most of the samples were collected from skin-infected patients, hospital workers, utensils (such as forceps, sizer, trays, and several equipment used in operation theater), and drain water from hospital drainage line.
Bacteriological investigation
For initial screening, a loop of each sample was inoculated onto various selected media such as C.L.E.D. agar medium, mannitol salt agar medium, and Baird–Parker agar medium and incubated at 35°C ± 2°C for 24 h. Then, the positive isolates of S. aureus on selective medium were subjected to several biochemical tests such as catalase test according to Bergey's Manual of Bacteriology.[9]
Detection of MRSA by oxacillin and cefoxitin sensitivity assays
After the confirmation of S. aureus from cultural method, isolates were tested to identify MRSA by disc diffusion test according to NCCLS guidelines[10] using Mueller–Hinton agar and standard discs of methicillin (oxacillin) (1 μg) and cefoxitin (30 μg). ATCC culture of S. aureus (43300) was used as a positive control.
Antimicrobial susceptibility test to various drugs
Standard discs of azithromycin (15 μg), gentamicin (10 μg), tetracycline (30 μg), cefotaxime (30 μg), chloramphenicol (30 μg), and ciprofloxacin (5 μg) were applied to access the sensitivity of S. aureus for determining MDRSA. The procedure was performed followed by Kirby–Bauer disc diffusion method according to NCCLS guidelines.[10] ATCC culture of S. aureus (43300) was used as a positive control.
RESULTS
A total of 66 isolates of S. aureus were identified from the 100 samples through biochemical and cultural tests. MRSA was determined on the basis of measurement of zone of inhibition produced by oxacillin (<14 mm) and cefoxitin (<21 mm).[11] Fifty-three isolates (83.30%) gave a positive result for oxacillin test and, to avoid any false result, these 53 isolates were again tested by cefoxitin. Finally, 43 isolates (65.15%) gave a positive test for cefoxitin and confirmed as MRSA, whereas 23 isolates (34.85%) were confirmed as MSSA. The whole scenario is presented in Table 1.
Table 1.
Number of totally identified Staphylococcus aureus, methicillin-resistant Staphylococcus aureus, and methicillin-sensitive Staphylococcus aureus
| Source of samples | Number of isolated Staphylococcus aureus | MRSA | MSSA |
|---|---|---|---|
| Skin infection | 9 | 7 | 2 |
| Hospital workers | 7 | 3 | 4 |
| Utensils and equipment | 27 | 15 | 12 |
| Hospital drain water | 23 | 18 | 5 |
| Total | 66 | 43 | 23 |
MRSA: Methicillin-resistant Staphylococcus aureus, MSSA: Methicillin-sensitive Staphylococcus aureus
All the isolates of S. aureus were subjected to determine their antibiotic resistance pattern, whereas all MRSA showed a high antibiotic resistance and MSSA also revealed antibiotic resistance pattern as MDR S. aureus. Figure 1 shows the percentage of the antibiotic resistance pattern of total isolated S. aureus against different types of antibiotics.
Figure 1.
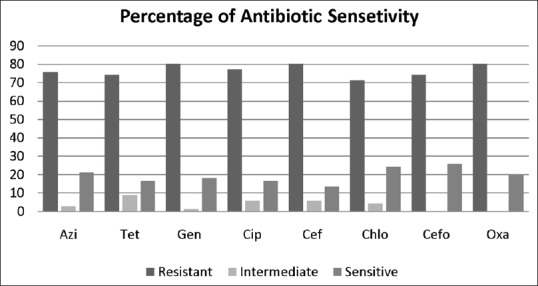
Antibiotic resistance pattern of isolated S. aureus against different antibiotics. Azi: Azithromycin, Tet: Tetracycline, Gen: Gentamicin, Cip: Ciprofloxacin, Cef: Cefotaxime, Chlo: Chloramphenicol, Cefo: Cefoxitin, and Oxa: Oxacillin
DISCUSSION
From this investigation, it was found that most of the isolates of S. aureus were resistant to cefoxitin, azithromycin, cefotaxime, tetracycline, gentamycin, chloramphenicol, and ciprofloxacin as both MRSA and MDR were highly potent for spreading nosocomial infection. This is consistent with the findings of Debnath et al.[12] The antibiotic activity of S. aureus isolates to beta-lactam antibiotic group members such as penicillin, cefoxitin, and tetracycline was also evident. A remarkable percentage of isolates were resistant to cefoxitin and tetracycline, which was almost similar to the finding of another study.[13]
In this study, it was noticed that 80.30% and 65.15% of isolates were resistant to oxacillin and cefoxitin, respectively. The feasible reason behind this variation could be the false resistance to oxacillin because of the elevated production of β-lactamase by bacteria.[14] From molecular point of view, MRSA isolates which have no mecA could be phenotypically resistant to methicillin.[15] It has been reported that S. aureus, which does not carry any mecA gene, is capable of producing huge amounts of β-lactamase which can make the isolates resistant to oxacillin.[16] On the contrary, it has been also observed that some mecA-positive S. aureus were susceptible to both methicillin and oxacillin.[17] The study by van Griethuysen et al. mentioned that the isolates might lost the mecA gene during the storage of MRSA isolates.[18] Previous other studies also suggested that, besides mecA, there are other genes which are also accountable for converting S. aureus into MRSA.[19] Hence, to block the resistance to antibiotics, addition of β-lactamase inhibitors including clavulanic acid or sulbactam could be an effective strategy.[20]
Several risk practices as well as factors were also noticed which were highly considerable for spreading nosocomial pathogens. Various practices including the use of contaminated needle or forceps, improper disposal of antibiotics in the environment, contaminated dustbins positioned besides to the patient's bed, hospital discharges’ accumulation into drainage system in hospitals, lack of public awareness, use of unsterilized equipment, and faulty drainage system in hospitals were noticed. These practices are extremely correlated as the independent risk factors for spreading MRSA at higher rate in hospitals. Although the mode of transmission of MRSA is quite similar to other strains of S. aureus, their host colonization efficiency is different.[21] It has been reported that MRSA can be transmitted from animals to humans, humans to animals, and humans to humans by direct and indirect contacts.[22] As the potential source of contamination, the surface of equipment, which are used for food handling or processing, is also recognized as a potent harbor for microbial growth.[23] Indiscriminate use of antibiotics is also responsible to develop resistance.[24] Therefore, we suggested that effective control measures should be implemented to prevent the outbreak of S. aureus in hospitals and the surrounding environments.
CONCLUSION
Presence of methicillin and other antibiotics resistant to S. aureus indicates risks to public health. Although very few preventive measures are commonly practiced in hospitals, therefore, to minimize the spread of MRSA, the study recommends taking proper measures such as practice of hand hygiene, use of personal protective equipment, disinfection, sterilization, and maintenance of waste disposal system to prevent infectious diseases. It is also highly recommended to ensure auditing on a regular basis to make sure of compliance to infection control measures.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors are grateful to Noakhali Science and Technology University, University of Chittagong, and Chittagong Veterinary Animal Science University for supporting us to conduct this research.
REFERENCES
- 1.Ki V, Rotstein C. Bacterial skin and soft tissue infections in adults: A review of their epidemiology, pathogenesis, diagnosis, treatment and site of care. Can J Infect Dis Med Microbiol. 2008;19:173–84. doi: 10.1155/2008/846453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurlenda J, Grinholc M, Jasek K, Wegrzyn G. RAPD typing of methicillin-resistant Staphylococcus aureus: A 7-year experience in a Polish hospital. Med Sci Monit. 2007;13:MT13–8. [PubMed] [Google Scholar]
- 3.Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, et al. Methicillin-resistant Staphylococcus aureus (MRSA): Global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents. 2012;39:273–82. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 4.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–85. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 5.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–41. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–55. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fishovitz J, Hermoso JA, Chang M, Mobashery S. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life. 2014;66:572–7. doi: 10.1002/iub.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samanipour A, Dashti-Khavidaki S, Abbasi MR, Abdollahi A. Antibiotic resistance patterns of microorganisms isolated from nephrology and kidney transplant wards of a referral academic hospital. J Res Pharm Pract. 2016;5:43–51. doi: 10.4103/2279-042X.176559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchanan RE, Gibbon NE. 9th Edition. Baltimore, USA: Williams and Wilkins Co; 1974. Bergey's Manual of Determinative Bacteriology. [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. Performance Standards of Antimicrobial Susceptibility Testing. Eleven Informational Supplement. 2011;21 [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Sixteen Informational Supplement 2007, CLSI document M100-S17. 2007 [Google Scholar]
- 12.Debnath T, Bhowmik S, Islam T, Chowdhury MM. Presence of multidrug resistant bacteria on mobile phones of healthcare workers accelerates the spread of nosocomial infections and regarded as a threat to public health in Bangladesh. J Microsc Ultrastruct. 2017 doi: 10.4103/JMAU.JMAU_30_18. [In press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derese Y, Hailu E, Assefa T, Bekele Y, Mihret A, Aseffa A, et al. Comparison of PCR with standard culture of fine needle aspiration samples in the diagnosis of tuberculosis lymphadenitis. J Infect Dev Ctries. 2012;6:53–7. doi: 10.3855/jidc.2050. [DOI] [PubMed] [Google Scholar]
- 14.Felten A, Grandry B, Lagrange PH, Casin I. Evaluation of three techniques for detection of low-level methicillin-resistant Staphylococcus aureus (MRSA): A disk diffusion method with cefoxitin and moxalactam, the Vitek 2 system, and the MRSA-screen latex agglutination test. J Clin Microbiol. 2002;40:2766–71. doi: 10.1128/JCM.40.8.2766-2771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, Jeong JM, Park YH, Choi SS, Kim YH, Chae JS, et al. Evaluation of the methicillin-resistant Staphylococcus aureus (MRSA)-screen latex agglutination test for detection of MRSA of animal origin. J Clin Microbiol. 2004;42:2780–2. doi: 10.1128/JCM.42.6.2780-2782.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stapleton PD, Taylor PW. Methicillin resistance in Staphylococcus aureus: Mechanisms and modulation. Sci Prog. 2002;85:57–72. doi: 10.3184/003685002783238870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olonitola OS, Olayinka BO, Onaolapo JA. Absence of mecA in MRSA isolated from non-hospital source in Zaria, Nigeria. Int J Nat Appl Sci. 2007;13:160–4. [Google Scholar]
- 18.van Griethuysen A, van Loo I, van Belkum A, Vandenbroucke-Grauls C, Wannet W, van Keulen P, et al. Loss of the mecA gene during storage of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2005;43:1361–5. doi: 10.1128/JCM.43.3.1361-1365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonas D, Speck M, Daschner FD, Grundmann H. Rapid PCR-based identification of methicillin-resistant Staphylococcus aureus from screening swabs. J Clin Microbiol. 2002;40:1821–3. doi: 10.1128/JCM.40.5.1821-1823.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papp-Wallace KM, Bonomo RA. New β-lactamase inhibitors in the clinic. Infect Dis Clin North Am. 2016;30:441–64. doi: 10.1016/j.idc.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawada M, Okuzumi K, Hitomi S, Sugishita C. Transmission of Staphylococcus aureus between healthy, lactating mothers and their infants by breastfeeding. J Hum Lact. 2003;19:411–7. doi: 10.1177/0890334403257799. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira JP, Anderson KL, Correa MT, Lyman R, Ruffin F, Reller LB, et al. Transmission of MRSA between companion animals and infected human patients presenting to outpatient medical care facilities. PLoS One. 2011;6:e26978. doi: 10.1371/journal.pone.0026978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somers EB, Johnson ME, Wong AC. Biofilm formation and contamination of cheese by nonstarter lactic acid bacteria in the dairy environment. J Dairy Sci. 2001;84:1926–36. doi: 10.3168/jds.S0022-0302(01)74634-6. [DOI] [PubMed] [Google Scholar]
- 24.Chowdhury MM, Kubra K, Islam MT, Rahman MM, Mehedy ME. Indiscriminate uses of antibiotics as a threat to public health demand implementation of effective drug practices and enhancement of public awareness in Bangladesh. Eur J Sci Res. 2015;133:187–95. [Google Scholar]


